Abstract
Increasing empirical evidence suggests that many genetic variants influence multiple distinct phenotypes. When cross-phenotype effects exist, multivariate association methods that consider pleiotropy are often more powerful than univariate methods that model each phenotype separately. Although several statistical approaches exist for testing cross-phenotype effects for common variants, there is a lack of similar tests for gene-based analysis of rare variants. In order to fill this important gap, we introduce a statistical method for cross-phenotype analysis of rare variants using a nonparametric distance-covariance approach that compares similarity in multivariate phenotypes to similarity in rare-variant genotypes across a gene. The approach can accommodate both binary and continuous phenotypes and further can adjust for covariates. Our approach yields a closed-form test whose significance can be evaluated analytically, thereby improving computational efficiency and permitting application on a genome-wide scale. We use simulated data to demonstrate that our method, which we refer to as the Gene Association with Multiple Traits (GAMuT) test, provides increased power over competing approaches. We also illustrate our approach using exome-chip data from the Genetic Epidemiology Network of Arteriopathy.
Keywords: pleiotropy, rare variant, gene mapping, complex human traits
Introduction
The 1980s were an era of debate in the theoretical quantitative genetics community between two competing schools of thought.1 The question of interest was “What is the nature of genetic variation contributing to complex traits?” On one hand there was the infinitesimal school,2 which argued that complex traits were the result of mutation/selection balance under stabilizing selection. The variants that contributed to traits were a combination of very rare alleles of potentially large effect combined with many common alleles of exceedingly small effect. The opposing camp, sometimes called Neo-Darwinian,3 argued that a substantial fraction of genetic variation was contributed by high-frequency alleles of large effect, whose frequency was maintained through balancing selection.4 The neo-Darwinian’s school leveled two interrelated and potentially fatal criticisms at the infinitesimal camp: believing in the infinitesimal model requires one to simultaneously accept that (1) much of the standing genetic variation is due to extremely rare alleles of large effect and (2) a large fraction of the genome of an organism is contributing to nearly every phenotype.3 That means that nearly every rare, large-effect allele must simultaneously be contributing to a large number of different traits. The neo-Darwinian school argued that the only alternative to believing in this worldview was to suppose that a substantial fraction of the variation in complex traits was contributed to by common alleles of large effect.
Perhaps without explicitly acknowledging it,5, 6, 7 the genome-wide association study (GWAS) era was fundamentally testing the predictions of the neo-Darwinian school. We now know that, by and large, common alleles of large effect do not exist. When considered collectively, common variants can explain a sizable proportion of the heritability for many complex traits like height, body-mass index, and cardiovascular disease.8, 9, 10 However, common trait-influencing variants identified and replicated by GWASs tend to have very modest effect sizes. Much of the genetic contributors to complex traits still remain undiscovered and are presumably due to very rare variation. Thus, although it might be time to reject the neo-Darwinian worldview in favor of the infinitesimal model, we can not logically do so without simultaneously embracing the central Neo-Darwinian critique of the infinitesimal school: most traits should be affected by a large fraction of the genome, and rare alleles of large effect should be generally highly pleiotropic for seemingly unrelated phenotypes. Moreover, if we adopt this worldview whole-heartedly, it suggests a paradigm shift in how we should approach genetic association studies.
If rare alleles of large effect are both ubiquitous and generally highly pleiotropic, we can leverage this to discover genes involved in complex traits. When pleiotropy exists, an analysis that models multiple phenotypes simultaneously in a multivariate or “cross-phenotype” framework will provide greater statistical power than a standard univariate method that considers each phenotype separately.11, 12 Because underlying genetic pleiotropy will induce phenotypic correlation, a genetic association that exists with multiple traits will be more readily detectable through cross-phenotype analyses due to the extra information provided by cross-phenotype correlation. This information is ignored in univariate analyses. Additionally, when pleiotropy is suspected, allowing for cross-phenotype associations might yield a more biologically plausible statistical model and potentially help to explain shared pathogenesis.11, 13
Cross-phenotype association tests for common variants using SNPs have demonstrated considerable success.14, 15 For example, common-variant cross-phenotype association has been reported among Crohn disease and ulcerative colitis,16 different facial morphology measures,17 and among bipolar disorder, autism spectrum disorder, ADHD, major depressive disorder, and schizophrenia.18 However, although there are several excellent statistical methods appropriate for cross-phenotype analysis of common genetic variants,19, 20, 21, 22, 23, 24 theory tells us that rare alleles cannot be ignored and that pleiotropy due to rare alleles should be more pronounced. Unfortunately, there is a shortage of analogous statistical approaches to assess cross-phenotype associations of rare genetic variants.
Currently, most cross-phenotype association methods are designed to assess the effect of a single polymorphism at a time; however, in rare variant analysis, a test typically requires aggregation of information from multiple rare variants within a gene simultaneously. One possible rare-variant cross-phenotype test is a modification of the common-variant method of Maity et al.23 Although the Maity approach was developed to study the relationship between multiple SNPs in a gene and multiple correlated phenotypes using mixed models, it could be adapted to consider rare variants rather than common SNPs. Additionally, Wang et al. proposed an alternative gene-level test of pleiotropy that uses multivariate functional linear models (MFLM).25 However, we note that the approaches of Maity and Wang allow only for continuous phenotypes and thus cannot be applied to important categorical phenotypes like presence or absence of a disease. Ideally, a cross-phenotype test of rare variation should be able to handle both continuous and categorical phenotypes and be able to scale efficiently to handle an arbitrary number of phenotypes. Here, we present a method that meets both these criteria.
We propose a method called Gene Association with Multiple Traits (GAMuT) for association testing of high-dimensional phenotype data with high-dimensional genotype data. GAMuT relies on a machine-learning framework called kernel distance-covariance (KDC)26, 27, 28, 29, 30 to provide a nonparametric test of independence between a set of phenotypes and a set of genetic variants. The KDC framework used by GAMuT assesses whether pairwise phenotypic similarity in a sample is independent of pairwise rare-variant genotypic similarity in a gene or region of interest. The framework allows for an arbitrary number of phenotypes that can be both continuous and/or categorical in nature and similarly allows for an arbitrary number of genotypes, thereby permitting gene-based testing of rare variants. GAMuT can correct for important covariates, such as measures of ancestry to account for population stratification. Furthermore, GAMuT is a closed form test that yields analytic p values, thus scaling easily to genome-wide analysis.
This manuscript is organized as follows. First, we develop GAMuT using the KDC framework and show how we derive analytic p values for this test. We also describe how we can adjust for covariates in GAMuT. Additionally, we describe an efficient resampling strategy that can be used if one wishes to construct a GAMuT test multiple times using different similarity measures for phenotypes and/or genotypes. This resampling strategy appropriately corrects for multiple testing but is far less computationally intensive than standard permutations. Next, we present simulation work comparing GAMuT to MFLM and univariate SKAT31 analysis of rare variants under various trait-influencing models and demonstrate that our analytic strategy can be considerably more powerful than these competing approaches, both when pleiotropy truly exists and also when variants influence only one of the phenotypes under consideration. Finally, we apply GAMuT to perform exome-chip analysis of multivariate phenotypic measures of cardiovascular health using data from the Genetic Epidemiology Network of Arteriopathy (GENOA).32
Material and Methods
Assumptions and Notation
We assume a sample of N subjects who have been measured for multiple phenotypes of interest and possess sequencing or exome-chip data in a target gene or region. For subject j (j = 1,…,N), we define Pj = (Pj,1, Pj,2, …, Pj,L) as the L phenotypes of the subject and allow such phenotypes to be continuous and/or categorical in nature. We then define a matrix of phenotypes for the entire sample , which is of dimension N × L. Similarly, we define Gj = (Gj,1, Gj,2, …, Gj,V) to be the genotypes of subject j at V rare-variant sites in the gene of interest, where Gj,v is coded as the number of copies of the minor allele that the subject possesses at variant v. We then construct the matrix of rare-variant genotypes for the sample as which is of dimension N × V.
GAMuT Test of Cross-Phenotype Associations
We create GAMuT to examine the relationship between phenotypes P and rare-variant genotypes G. GAMuT is based on a KDC machine-learning technique,26, 27, 28, 29, 30 which allows nonparametric tests of independence between two distinct sets of multivariate variables. For each set of multivariate variables, KDC constructs an N × N matrix with individual elements of the matrix corresponding to similarity (or dissimilarity) in the variables among different pairs of subjects. KDC then evaluates whether the pairwise elements in the similarity matrix of one set of multivariate variables is independent of the pairwise elements in the similarity matrix for the other set of multivariate variables.
Leveraging the KDC framework, we create a rare-variant test of pleiotropy to test for independence between P (N × L matrix of multivariate phenotypes) and G (N × V matrix of multivariate rare-variant genotypes). To do this, we first develop an N × N phenotypic-similarity matrix Y (based on P) and an N × N genotypic-similarity matrix X (based on G). The choice of how to model pairwise similarity or dissimilarity for a set of multivariate outcomes is quite flexible. For example, for phenotypes P, we can model the matrix Y using a projection matrix,33, 34 such that Y = P(PTP)−1PT. We can also construct the model Y using user-selected kernel functions.31, 35, 36, 37 Denote the kernel function y(Pi, Pj) as the measure of similarity between subjects i and j across the L phenotypes. We can model y(Pi, Pj) using kernel similarity functions like the linear kernel, ; a quadratic kernel, ; or a Gaussian kernel, , where δ is a tuning parameter.
For genotypes G, we model the corresponding matrix X using kernel functions x(Gi, Gj) that can take the same form (e.g., linear, quadratic, or Gaussian) used to construct y(Pi, Pj). A few genetic-specific kernel functions also exist, like the identity-by-state (IBS) kernel, , where IBS(Gi,v Gj,v) denotes the number of alleles (0, 1, or 2) shared IBS by subjects i and j at variant v. Also, we might wish to further augment x(Gi, Gj) to preferentially upweight the contributions of particular rare variants in G over others in the gene. For example, we may wish to give more weight to variants that are more rare in the population or to variants that are predicted to be deleterious in nature.38, 39, 40 We can do this by creating a diagonal weight matrix W = diag(w1, w2, …, wV), where wv reflects the relative weight for the vth variant in the gene. Using W, we can then create a weighted linear kernel function as X = GWGT. Derivation of other weighted kernel functions is straightforward.
Once we construct the similarity matrixes Y and X, we derive our GAMuT approach as a test of independence between the elements of these two matrices. We first center each matrix as Yc = HYH and Xc = HXH. Here, is a centering matrix with property HH = H, I is an identity matrix of dimension N, and 1N is an N × 1 vector with each element equal to 1. Using Yc and Xc, we construct our GAMuT test of independence of the two matrices as
| (Equation 1) |
Under the null hypothesis where the two matrices are independent, TGAMuT follows the same asymptotic distribution as
| (Equation 2) |
where λX,i is the ith ordered non-zero eigenvalue of Xc, λY,j is the jth ordered non-zero eigenvalue of Yc, and are independent and identically distributed variables.30 Given L phenotypes and V rare-variant sites, and further assuming sample size N is larger than both L and V, the maximum number of possible elements in the summation will be L∗V.
Based on the KDC literature, we could derive the p value of the GAMuT test approximately using a gamma distribution26 or instead via permutation techniques.28, 30 In our experience, the gamma approximation is accurate for p values as small as 0.01 but becomes less accurate in the more extreme tails of the distribution (results not shown). Given that large-scale genetic studies require p values much smaller than 0.01 to declare significance in the presence of multiple testing, the gamma approximation is not suitable in this setting. The derivation of p values using permutations is a valid alternative, but computationally demanding and difficult to scale to genome-wide analyses. Consequently, we instead derive p values for GAMuT using Davies’ exact method,41 which is a computationally efficient method to provide accurate p values in the extreme tails of tests that follow mixtures of chi-square variables.31 An implementation of Davies’ method is available in the R package CompQuadForm.42
Relationship of GAMuT to Other Multivariate Association Tests
Although the form of the GAMuT test is quite general, we note that specific choices of Y and X can lead to test statistics that have similar forms to other multivariate association tests previously published in the literature. If we assume a projection matrix Y for the phenotypes (with each phenotype mean centered prior to analysis) and assume X is the Gower distance (or some other measure of genetic dissimilarity as opposed to similarity), the GAMuT test has a form similar to the numerator of existing multivariate distance matrix regression (MDMR) tests.33, 34, 43 We note, however, that MDMR procedures typically require permutations for inference whereas we can derive analytic p values of GAMuT directly via Davies’ method. MDMR tests’ reliance on permutations limits application of these techniques to smaller-scale studies such as candidate-gene investigations. On the other hand, GAMuT’s efficient derivation of analytic p values enables the approach to be applied efficiently to whole-exome and whole-genome sequencing projects.
In addition to MDMR, we also note that applying GAMuT using a linear kernel to model the phenotype similarity matrix Y and to further model the genotype similarity matrix X results in a test that becomes a rare-variant version of the multivariate kernel-machine test of Maity et al.23, 27 created for the analysis of common variants. The approach of Maity, however, required perturbations to calculate p values of individual tests where again GAMuT can derive p values analytically via Davies’ method.
GAMuT Testing Assuming Multiple Candidate Matrices
The GAMuT test in the previous section requires a priori selection of the functions used to construct the phenotypic similarity matrix Y and genotypic similarity matrix X. In practice, though, it is often unclear what the optimal choices for Y and X should be. For example, an investigator might want to model phenotypes P in the matrix Y using both the projection matrix and the linear kernel function. Also, an investigator might want to construct the genotype-similarity matrix X under different kernel functions (e.g., linear and IBS) and assuming different weight functions (e.g., minor allele frequency [MAF] weights, functionality weights). If we construct GAMuT tests under multiple different phenotypic and genotypic similarity matrices, we then need to adjust for the additional tests that were performed. To adjust for additional tests, one could use a Bonferroni correction or apply permutations. However, a Bonferroni correction probably will lead to conservative inference because these tests are correlated, whereas permutations are computationally demanding and unappealing on a genome-wide level.
Rather than use Bonferroni or permutations, we follow the ideas of Zhang et al.30 and Wu et al.44 to develop a perturbation (resampling) approach to correct for testing of multiple candidate matrices in GAMuT that is more computationally efficient than standard permutations. Assume we test M different combinations of Y and X. For combination m (m = 1,…,M), we let p(m) denote the uncorrected GAMuT p value and further let and denote the vectors of all non-zero eigenvalues for Yc and Xc, respectively, for that combination.
We wish to determine whether the minimum observed p value across the M tested combinations is significant after adjustment for the M correlated tests. To do this, we use perturbations to create an empirical distribution of minimum p values across the same M combinations under the null hypothesis of no association. We then calculate our corrected p value by comparing our minimum observed p value to the empirical minimum p values generated under the null hypothesis induced by the perturbation process. In particular, we implement the following:
-
(1)
Calculate the minimum observed p value across the M different combinations as .
-
(2)
For perturbation k (k = 1,…K), generate a set of independent variables z∗k of length equal to K∗V.
-
(3)
For each combination m, calculate the test and obtain a new p value via Davies’ method.
-
(4)
Evaluate the minimum p value across all M combinations for perturbation k as .
-
(5)
Repeat steps 2–4 a total of K times and obtain the empirical distribution of uncorrected minimum p values .
-
(6)
Derive the final p value as
Adjusting for Covariates
Pleiotropic tests must adjust for important covariates, such as principal components of ancestry, to avoid potential confounding of results. We can control for confounders before applying GAMuT by regressing each phenotype separately on covariates of interest and then using the residuals to form the phenotypic similarity matrix Y. Although residualizing binary phenotypes is not standard, studies have suggested that this procedure does not affect the validity of genetic association tests in case-control studies.45, 46 As we describe in the Results, the residualizing procedure provides an effective correction for confounders in the analysis of binary outcomes within our simulated datasets.
Simulations
We conducted simulations to verify that GAMuT properly preserves type I error and to assess power of GAMuT relative to competing approaches for genetic analysis of multiple phenotypes. To create genetic data for these simulations, we generated 20,000 haplotypes of 30 kb in size using COSI, a coalescent model that mimics LD pattern, local recombination rate, and population history for individuals of European descent.47 To create multivariate phenotype data, we assume either six or ten phenotypes for each subject generated from a multivariate normal distribution with mean vector 0 and L × L residual correlation matrix Σ. To model the residual correlation matrix, we considered scenarios of low residual correlation among phenotypes (pairwise correlation among phenotypes selected from a uniform (0, 0.3) distribution), moderate residual correlation (pairwise correlation selected from a uniform (0.3, 0.5) distribution), and high residual correlation (pairwise correlation selected from a uniform (0.5, 0.7) distribution). To generate binary traits, we defined phenotype measurements for the top quartile as affected (Pi,l = 1) and defined 1st–3rd quartile measurements as controls (Pi,l = 0). We considered sample size N of either 1,000 or 2,500 subjects.
To investigate the performance of GAMuT under confounding and to assess whether the approach can successfully adjust for relevant covariates in this setting, we also simulated phenotypes under a confounding model where phenotypes were independent of genotypes, but both phenotypes and genotypes are associated with a normally distributed covariate Z. We simulated phenotypes correlated with the covariate Z under the model , where Z denotes the N × 1 sample vector of covariates. To simulate correlation between rare-variant genotypes and covariate, we let 5% of the rare variants in our haplotypes be causal. We set effect size, βZ,r, of each causal genetic variant r on Z, as , where MAFr is the minor allele frequency of causal variant r. Evaluating type I error under this model allows us to verify that our approach to controlling for confounders is valid.
We also performed type I error calculations to examine the validity of our resampling approach to adjust for multiple similarity matrices when applying GAMuT. For a given null dataset, we applied GAMuT using three combinations of phenotype similarity matrices Y and genotype similarity matrices X:
-
(1)
Model phenotypes using a projection matrix, model genotypes using a weighted linear kernel.
-
(2)
Model phenotypes using a linear kernel, model genotypes using a weighted linear kernel.
-
(3)
Model phenotypes using a projection matrix, model genotypes using an unweighted linear kernel.
We then implement the perturbation procedure described above to obtain a p value accounting for testing the three combinations of similarity matrices. For both continuous and binary null simulations, we applied GAMuT to 10,000 simulated datasets.
For power models, we considered simulation designs similar to those proposed in the original SKAT paper.31 We simulated datasets in which 5% of the rare variants in our haplotypes were modeled as causal. We set effect size of each causal variant, r, for phenotype l, βr,l, as . This formulation sets mean effect size of causal variant r as inversely proportional to its MAF, such that very rare variants have on average a larger effect size than less rare variants. The mean effect size is based on the simulations performed for Wu et al.’s original evaluation of SKAT.31 Allowing βr,l to vary around a normal distribution maintains the relationship between MAF and effect size while allowing the variant to have a slightly different effect size for each phenotype.
We performed power simulations both in situations where there was no pleiotropy (i.e., only one of the modeled phenotypes was associated with the rare causal variants) and also when there was pleiotropy. Under pleiotropy, we varied the number of phenotypes associated with the rare variants, such that not all of the tested phenotypes will be dependent on the gene of interest. Under models assessing ten phenotypes, we consider situations where one, two, four, six, or eight phenotypes are actually associated with the gene. Under models assessing six phenotypes, we consider situations where only one, three, or five phenotypes are associated. We control correlation among phenotypes through consideration of the relative variance of phenotype explained by the R causal variants. We define this relative variance for phenotype l as . As in Galesloot et al.,11 we define the overall correlation between phenotypes l and l’ as where Σl,l’ is (l,l’) element of the L × L residual phenotypic correlation matrix. This allows the residual correlation structure among phenotypes to stay at the defined values.
For demonstration purposes, we also estimated power for limited simulations where we considered multiple combinations of phenotypic/genotypic similarity matrices for analyses. For such simulations, we considered a weighted linear kernel to form X and either the projection matrix or linear kernel to form Y. We then implement the perturbation procedure described above to obtain a p value accounting for the testing of the two similarity matrices.
For all simulations and analyses reported here, unless specified otherwise, we implement a weighting scheme based on the MAF of each variant that weights very rare variants more heavily than less rare variants. We selected the weighting scheme recommended by Wu et al.,31 setting wv = Beta(MAFv, 1, 25)/Beta(0, 1,25).
We evaluate GAMuT using the simulated data and compare our approach to competing strategies. For the analysis of continuous phenotypes, we compared GAMuT to the MFLM approach of Wang et al.25 Our implementation of MFLM used the B-spline basis based on Pillai-Bartlett trace, selecting the default parameters suggested by the authors for data analysis. Additionally, we compared GAMuT to a standard rare-variant association approach that ignored pleiotropy. Here, we consider the standard approach to be application of the popular SKAT31 test, a powerful, kernel-based univariate test for sequencing data. We applied SKAT to each of the simulated phenotypes and then based inference on the minimum SKAT p value across phenotypes analyzed. Because we perform SKAT testing on each of our L phenotypes, we must correct for multiple hypothesis testing. Although a permutation-based procedure is the gold standard for multiple test correction, it is computationally intensive and unlikely to scale to genome-wide analysis. Instead, we perform multiple testing correction using two approaches. First, we implement a simple Bonferroni correction of αBONFERRONI = αe/L, where αe is the experimental-wise error rate. Unfortunately, this approach can be conservative, especially for tightly correlated phenotypes. We therefore also consider a more liberal threshold by estimating the effective number of independent tests, Leff, where Leff is the number of principal components necessary to explain either 98% or 90% of phenotypic variance in L phenotypes.48 We can then calculate a more liberal correction of αEFFECTIVE = αe/Leff. Although thresholds of 90%–98% of phenotypic variance are more liberal than 99.5% threshold recommended by Gao et al.,48 we wanted to estimate the upper bounds of power to detect an effect using SKAT. Correction using the permutation approach should therefore fall somewhere between the conservative Bonferroni approach and the liberal principal component approaches.
For the analysis of binary phenotypes, we are unaware of existing methods for testing cross-phenotype effects of rare variants. Hence, we compared GAMuT to univariate SKAT testing only as described in the previous paragraph.
Analysis of GENOA Study
High body mass index (BMI), low high-density lipoprotein (HDL), and high blood pressure are interrelated conditions that increase risk of developing cardiovascular disease, stroke, kidney disease, and type 2 diabetes. These conditions are moderately heritable. The heritability of BMI has been estimated to be between 17%9 and 34%49 depending on methods used for the estimation. Similarly, heritability of HDL is estimated at 40%–48%,49, 50 and the estimates of heritability of blood pressure range from 30%49 to 48%–67%.51 Understanding genetic factors underlying these conditions is of considerable clinical importance. Several GWASs, including pleiotropic analyses of common variants, have been performed on one or more of the conditions.52, 53, 54, 55, 56, 57, 58 These studies have been tremendously successful in identification of common genetic variants; however, much of the genetic underpinnings of the conditions remains unexplained.59
The GENOA study32, 60 seeks to identify genetic variants that influence risk for hypertension and arteriosclerotic complications of hypertension. The GENOA resources include a cohort of African American sibships from Jackson, Mississippi. In the initial phase of the GENOA study, all members of sibships containing ≥2 individuals diagnosed with hypertension prior to age 60 were invited to participate, including both hypertensive and normotensive siblings. GENOA investigators collected extensive phenotypic information on each participant, including BMI, HDL, systolic blood pressure (SBP), and diastolic blood pressure (DBP). We selected these continuous measures for analysis. Additionally, GENOA investigators genotyped 1,429 subjects on the Illumina HumanExome Beadchip. We used the HumanExome-12 support files provided by Illumina to identify 48,712 non-singleton, rare or less-common autosomal genetic variants (MAF < 3%; hereafter referred to as “rare-variant”) that fell within known genes. We further excluded genes with fewer than 5 rare-variant sites within the GENOA dataset, leaving 3,277 genes in our analysis. Although GENOA collects data on sibs, GAMuT assumes study subjects are unrelated. Therefore, we randomly selected one sibling from each family for inclusion in our analysis.
We performed standard data cleaning, removed subjects who did not fast for at least 10 hr prior to phenotype collection, and removed related subjects that were either identified as relatives via pedigree information or identified as first-degree cryptic relatives identified with the program RELPAIR.61 The final sample for analysis consisted of 539 unrelated subjects with measures of all four phenotypes. For each of the study participants, we also obtained gender, age, smoking status (ever smoked at least 100 cigarettes), and use of anti-hypertension or lipid-lowering medication, and we calculated the top ten genetic principal components using ancestry informative markers included on the Illumina array. We applied GAMuT using both a projection matrix and a linear kernel to measure pairwise phenotypic similarity. We also ran univariate SKAT on each of the four phenotypes and adjusted for multiple testing. For all GAMuT and SKAT tests, we used a weighted linear kernel (selecting the weighting scheme recommended by Wu et al.,31 described above, as we used in our simulation work) to measure pairwise genotypic similarity. We also applied MFLM to the GENOA dataset as a comparison. The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and proper informed consent was obtained.
Results
Type I Error Simulations
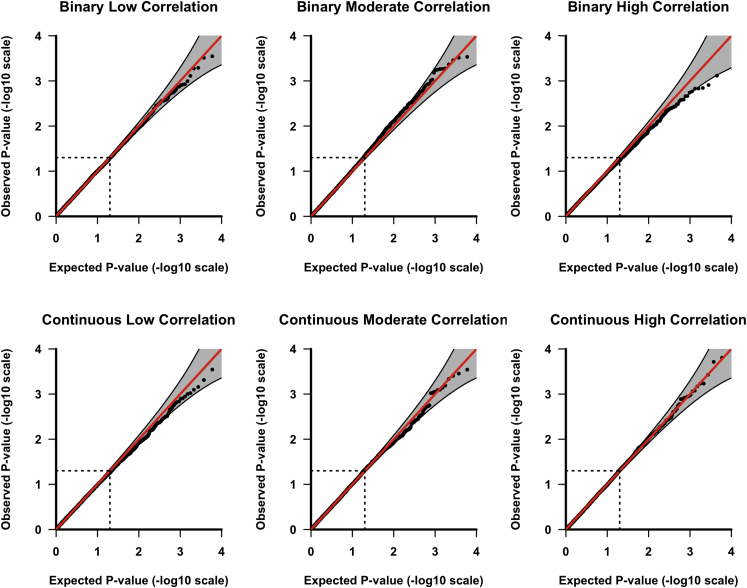
Figure 1 shows the quantile-quantile (QQ) plots based on application of GAMuT to null datasets consisting of 1,000 subjects assayed for ten phenotypes. We present QQ plots both for binary and continuous phenotypes assuming low, moderate, or high residual phenotypic correlation. We provide additional QQ plots of the GAMuT test for other combinations of phenotypes considered and sample size in Figures S1–S3. For all models tested, GAMuT properly controls for type I error, even at the extreme tails of the test. We further investigated the type I error of GAMuT in the presence of confounding due to a continuous covariate (see Material and Methods section) where we adjusted for confounding by residualizing the phenotypes on the covariate prior to analysis. Our QQ plots in Figure S4 show that this residualization effectively controls for the confounding for both binary and continuous phenotypes that, unadjusted, would yield inflated results.
Figure 1.
GAMuT QQ Plots
The QQ plots applying GAMuT to 10,000 simulated null datasets assuming a sample size of 1,000. In each simulation, 10 phenotypes are tested. Top row assumes binary phenotypes; bottom row assumes continuous phenotypes. Left column shows low residual phenotypic correlation (correlation 0–0.3), middle column shows moderate residual correlation (correlation 0.3–0.5), and right column shows high residual correlation (correlation 0.5–0.7).
Table 1 shows type I error at α ≥ 0.001 of GAMuT, MFLM, and univariate SKAT analyses of ten phenotypes for N = 1,000 and N = 2,500, and Table S1 shows similar results when analyzing six phenotypes. As expected based on the QQ plots in Figures 1 and S1–S3, the GAMuT approach maintains appropriate type I error across a range of assumptions and significance thresholds. Meanwhile, we observed appropriate type I error rates of the MFLM as well as SKAT tests after multiple-testing correction. The difference in type I error between the three SKAT approaches was minor, particularly at smaller significance thresholds. This finding is consistent with previous publications,48, 62 particularly given the small number of tests performed (either six or ten phenotypes).
Table 1.
Empirical Type I Error Rates Assuming Ten Phenotypes
| Sample Size | Type of Phenotypes | Phenotypic Correlation |
α = 0.05 |
α = 0.01 |
α = 0.001 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAMuT | MFLM |
SKAT |
GAMuT | MFLM |
SKAT |
GAMuT | MFLM |
SKAT |
|||||||||
| Bonf. | PC: 98% | PC: 90% | Bonf. | PC: 98% | PC: 90% | Bonf. | PC: 98% | PC: 90% | |||||||||
| 1,000 | continuous | low | .0453 | .0503 | .0455 | .0455 | .0545 | .0076 | .0099 | .0096 | .0096 | .0112 | .0007 | .0009 | .0010 | .0010 | .0011 |
| moderate | .0504 | .0481 | .0423 | .0423 | .0503 | .0085 | .0095 | .0097 | .0097 | .0115 | .0013 | .0007 | .0012 | .0012 | .0013 | ||
| high | .0517 | .0484 | .0462 | .0498 | .0509 | .0104 | .0138 | .0100 | .0102 | .0129 | .0009 | .0013 | .0010 | .0011 | .0011 | ||
| binary | low | .0488 | – | .0447 | .0447 | .0481 | .0093 | – | .0134 | .0134 | .0140 | .0006 | – | .0023 | .0023 | .0023 | |
| moderate | .0537 | – | .0429 | .0429 | .0461 | .0128 | – | .0105 | .0105 | .0115 | .0013 | – | .0028 | .0028 | .0029 | ||
| high | .0439 | – | .0474 | .0487 | .0509 | .0076 | – | .0082 | .0088 | .0096 | .0003 | – | .0013 | .0013 | .0014 | ||
| 2,500 | continuous | low | .0512 | .0493 | .0447 | .0474 | .0567 | .0090 | .0099 | .0077 | .0101 | .0127 | .0014 | .0012 | .0007 | .0007 | .0007 |
| moderate | .0538 | .0506 | .0402 | .0416 | .0547 | .0107 | .0114 | .0080 | .0090 | .0113 | .0012 | .0008 | .0010 | .0010 | .0012 | ||
| high | .0457 | .0496 | .0496 | .0502 | .0510 | .0091 | .0115 | .0090 | .0093 | .0101 | .0009 | .0018 | .0012 | .0012 | .0012 | ||
| binary | low | .0491 | – | .0360 | .0480 | .0529 | .0107 | – | .0092 | .0107 | .0116 | .0015 | – | .0017 | .0017 | .0017 | |
| moderate | .0524 | – | .0384 | .0450 | .0491 | .0121 | – | .0098 | .0102 | .0113 | .0018 | – | .0015 | .0015 | .0015 | ||
| high | .0450 | – | .0455 | .0457 | .0503 | .0081 | – | .0110 | .0117 | .0120 | .001 | – | .0012 | .0014 | .0014 | ||
Empirical size for GAMuT, MFLM, and SKAT analyses at significance thresholds of 0.05, 0.01, and 0.001. Empirical size calculated from 10,000 null simulations. Simulations assume analysis of 10 phenotypes. Sample size was set at either 1,000 or 2,500. Phenotypes were either continuous or dichotomous. Phenotypic correlation was low (correlation < 0.3), moderate (correlation 0.3–0.5), or high (correlation 0.5–0.7).
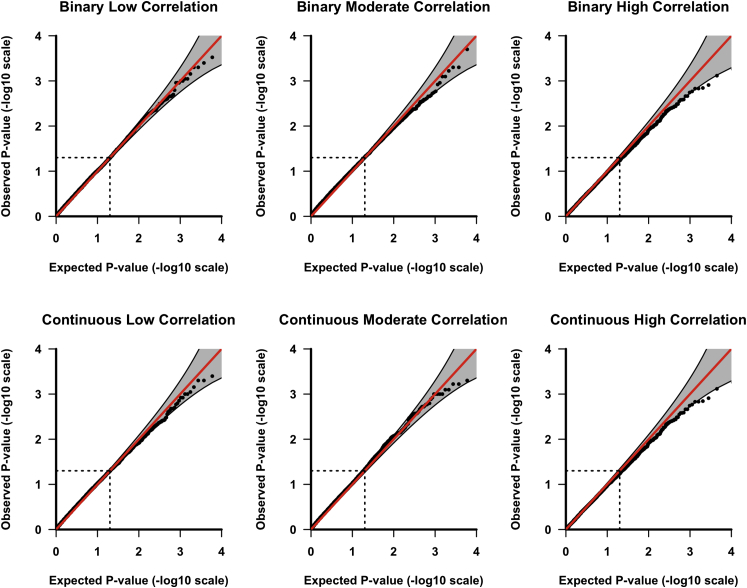
Figure 2 shows GAMuT QQ plots for binary and continuous phenotypes where we adjusted for multiple candidate matrices (see Material and Methods section). The perturbation procedure properly accounts for testing three combinations of Y and X and properly controls for false positive rate for a range of assumptions. By contrast, as we show in Figures S5 (binary outcomes) and S6 (continuous outcomes), using the minimum p value of GAMuT across matrices tested (i.e., without multiple-testing correction) yields inflated results, whereas the Bonferroni correction yields deflated results.
Figure 2.
QQ Plots for GAMuT Assuming Multiple Matrices Tested
The QQ plots applying GAMuT to 10,000 simulated null datasets assuming a sample size of 1,000. p values using three candidate matrices combinations were obtained for each simulation. We then implement a perturbation procedure to obtain a p value accounting for testing the three combinations of similarity matrices. In each simulation, ten phenotypes are tested. Top row assumes binary phenotypes; bottom row assumes continuous phenotypes. Left column shows low residual phenotypic correlation (correlation 0–0.3), middle column shows moderate residual correlation (correlation 0.3–0.5), and right column shows high residual correlation (correlation 0.5–0.7).
Power Simulations
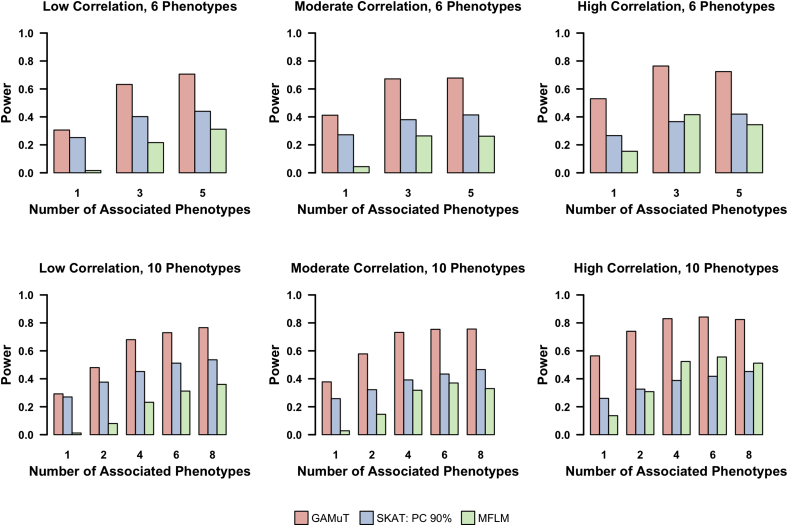
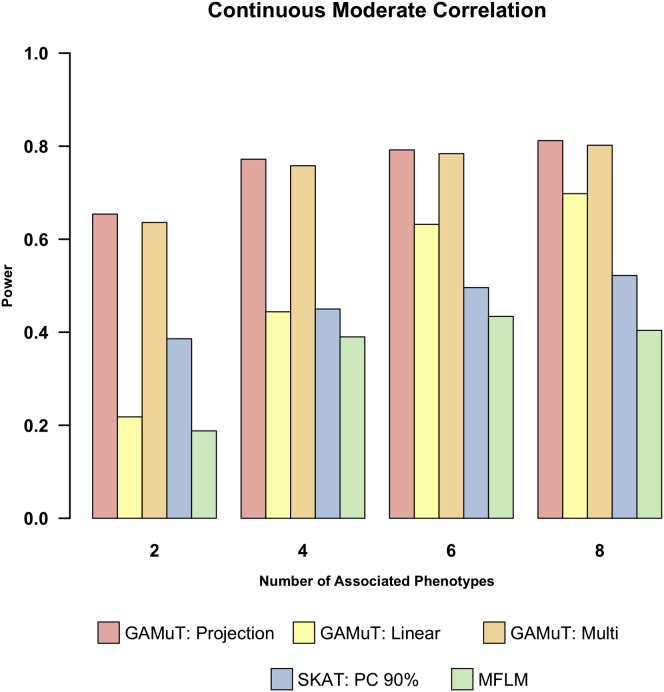
Next we compared the power of GAMuT with MFLM for continuous traits and univariate SKAT analysis (using three different multiple-testing corrections) for both continuous traits and binary traits. For these power simulations, we set sample size to 1,000. Power was estimated as the proportion of p values < 2.5 × 10−6 (reflecting a genome-wide correction for 20,000 genes) and was evaluated based on 500 replicates of the data per model. Figure 3 shows the power results when we analyze continuous phenotypes. We plot power as a function of the number of phenotypes that are truly associated with the causal variants. The figure clearly shows that GAMuT outperforms both MFLM and the standard univariate SKAT approach for all models considered. The difference in power between the three SKAT approaches was negligible; therefore, we show only 90% cutoff to determine the effective number of independent tests, because it is the most anti-conservative correction method. As expected, GAMuT performs particularly well against SKAT and MFLM as the ratio of associated to unassociated phenotypes increases (i.e., as the gene is increasingly pleiotropic). In addition, under models of no pleiotropy where rare causal variants were associated with only one of the phenotypes under consideration, we observed the power of GAMuT to be approximately equal or better than SKAT.
Figure 3.
Power to Detect Cross-Phenotype Effects: Continuous Phenotypes
Power for GAMuT (red), univariate SKAT using a 90% cutoff to determine effective number of independent tests (blue), and MFLM (green) is plotted as a function of number of continuous phenotypes associated with the gene of interest. Top row assumes six continuous phenotypes are tested in each simulation, and bottom row assumes ten continuous phenotypes are tested. Left column shows low residual phenotypic correlation (correlation 0–0.3), middle column shows moderate residual correlation (correlation 0.3–0.5), and right column shows high residual correlation (correlation 0.5–0.7).
MFLM performs poorly in all of our assumptions. We therefore simulated data that mimics the assumptions presented in the top row of Wang et al.’s Figure 4.25 The differing assumptions are detailed in Figure S7; in brief, the differences in our assumptions compared with the Wang et al. manuscript are that the latter work assumes smaller number of phenotypes, smaller genes, larger effect sizes, a more lenient significance threshold, and a larger percentage of causal variants. When we implement the simulation strategy of Wang et al., we observe increases in power for MLFM versus SKAT that are similar to those in their paper. GAMuT performance is approximately equivalent to MLFM under the simulation assumptions of Wang et al.
Figure 4 shows similar results when binary phenotypes are modeled. Because MFLM is valid only for continuous outcomes, we compare GAMuT only to univariate SKAT for binary outcomes. We observed similar improvements of power for GAMuT compared to SKAT in our binary simulations as we did for our continuous simulations. Under pleiotropic models, the improvement in power of GAMuT over SKAT grows more noticeable as the number of phenotypes associated with the gene increases. At the same time, even under power models where there is no pleiotropy (only one phenotype associated with the rare variants), our results indicate GAMuT is at least as powerful compared with the univariate SKAT approaches under models assuming low correlation, and in fact is more powerful than the univariate approach under moderate and high correlation structure.
Figure 4.
Power to Detect Cross-Phenotype Effects: Binary Phenotypes
Power for GAMuT (red) and univariate SKAT using a 90% cutoff to determine effective number of independent tests (blue) is plotted as a function of number of binary phenotypes associated with the gene of interest. Top row assumes six binary phenotypes are tested in each simulation, and bottom row assumes ten binary phenotypes are tested. Left column shows low residual phenotypic correlation (correlation 0–0.3), middle column shows moderate residual correlation (correlation 0.3–0.5), and right column shows high residual correlation (correlation 0.5–0.7).
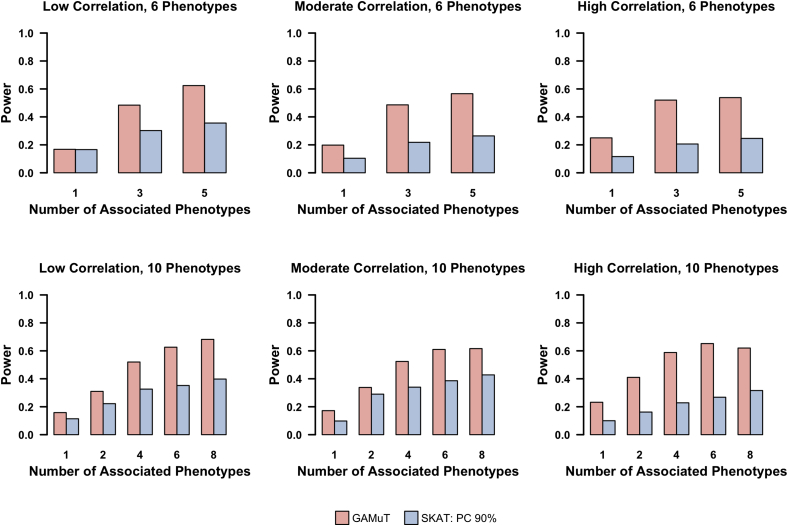
We also implemented the perturbation approach to model phenotypic similarity using both the projection matrix and the linear kernel. For both cases, we used the weighted linear kernel to model genotypic similarity. In Figure 5 we compare power of GAMuT using the projection matrix against power when two candidate matrices are considered (projection and linear kernel), implementing the perturbation procedure to account for testing two combinations of Y. Power in Figure 5 is defined as the proportion of p values less than 1.5 × 10−5, to reflect the study-wide significance threshold we will use for the GENOA data. We also show power using the linear kernel to model phenotypic similarity. Although the linear kernel was not as powerful as the projection matrix on our simulated data, simulations indicate that the perturbation procedure retains much of the power of the optimal kernel approach.
Figure 5.
Power to Detect Pleiotropic Effect using Multiple Similarity Matrices
Power for GAMuT assuming a projection matrix (red), GAMuT assuming a linear kernel (yellow), GAMuT assuming testing of both projection matrix and linear kernel (orange), univariate SKAT using a 90% cutoff to determine effective number of independent tests (blue), and MFLM (green). In each simulation, ten continuous phenotypes with moderate residual correlation (correlation 0.3–0.5) are tested.
Application to GENOA Dataset
We use the GENOA dataset to test for associations between BMI, HDL, SBP, DBP, and rare variants in 3,277 genes. Prior to analysis by GAMuT, we controlled for gender, age, smoking status, use of anti-hypertension medication, use of lipid-lowering medication, and ancestry on the 539 unrelated subjects. After adjusting for covariates, correlation of the four phenotypes was low to moderate with the largest pairwise correlation (0.67, Pearson’s product-moment correlation p value < 2.2 × 10−16) between SBP and DBP (see Table 2). We applied GAMuT using both a projection matrix and a linear kernel to measure pairwise phenotypic similarity. For comparison, we ran MFLM as well as univariate SKAT on each of the four phenotypes and adjusted for multiple testing. For all GAMuT and SKAT tests, we used a weighted linear kernel to measure pairwise genotypic similarity. We set a stringent study-wise significance threshold of 1.5 × 10−5, which corresponds to a Bonferroni correction based on the number of genes tested (3,277): αBONFERRONI = 0.05/3,277. We considered p values less than p < 1 × 10−3 as suggestive.
Table 2.
Correlation of GENOA Phenotypes
| BMI | HDL | SBP | DBP | |
|---|---|---|---|---|
| BMI | 1 | -0.17∗ | 0.09∗ | 0.02 |
| HDL | – | 1 | −0.01 | −0.03 |
| SBP | – | – | 1 | 0.67∗ |
| DBP | – | – | – | 1 |
Correlation among the four GENOA phenotypes: body mass index (BMI), high-density lipoprotein (HDL), systolic blood pressure (SBP), and diastolic blood pressure (DBP). Asterisk indicates correlations are nominally significant (Pearson’s product-moment correlation p value < 0.05).
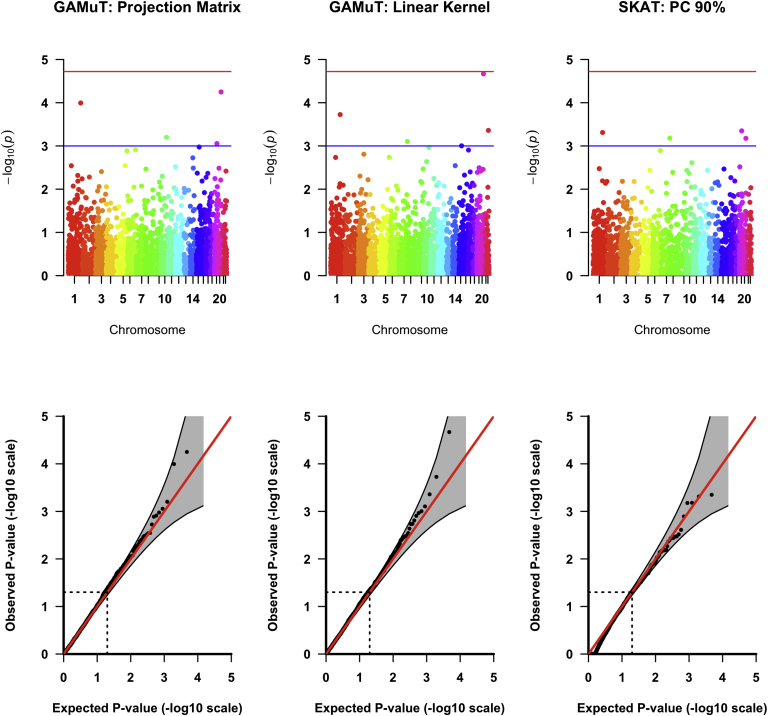
Figure 6 provides genome-wide results using GAMuT and univariate SKAT analyses with top findings highlighted in Table 3. None of the methods identified any genes associated at the study-wide significance threshold. Using the linear kernel, GAMuT identified five genes of suggestive significance. Of note, SELP, which was identified as suggestive significance by GAMuT (p = 1.9 × 10−4), has previously been associated with traits related to the four GENOA phenotypes. Haplotypes or common polymorphisms in SELP have been associated with myocardial infarction63, 64 and thromboembolic stroke.65 Levels of P-selectin, the protein encoded by SELP, is increased in hypercholesterolemic individuals66 and individuals with unstable angina.67 P-selectin levels were significantly associated with carotid artery stiffness and wall thickness among Japanese individuals with type II diabetes, hypertension, or hyperlipidemia.68 The same study found that percentage of P-selectin-positive platelets was positively associated with BMI, SBP, and DBP and inversely associated with HDL.
Figure 6.
Results of GENOA Analyses
Left column shows Manhattan and QQ plots for GAMuT using a projection matrix for phenotypes. Middle column shows Manhattan and QQ plots for GAMuT using a linear kernel for phenotypes. Right column shows Manhattan and QQ plots for SKAT, using a 90% cutoff to determine the effective number of independent tests. Horizontal blue line indicates suggestive significance threshold. Horizontal red line indicates study-wide significance.
Table 3.
Top GENOA Results
| Gene Symbol | MIM Number | Chromosome | Number Rare Variants |
GAMuT |
SKAT: 90% PC | ||
|---|---|---|---|---|---|---|---|
| Projection Matrix | Linear Kernel | Combined (Perturbation) | |||||
| SELP | 173610 | 1 | 8 | 4.8 × 10−3 | 1.9 × 10−4 | 2.8 × 10−4 | 4.9 × 10−4 |
| DISP1 | 607501 | 1 | 8 | 1.0 × 10−4 | 8.1 × 10−3 | 1.4 × 10−4 | 7.3 × 10−3 |
| ARHGEF10 | 608136 | 8 | 14 | 2.8 × 10−2 | 7.9 × 10−4 | 1.0 × 10−3 | 6.6 × 10−4 |
| COL17A1 | 113811 | 10 | 11 | 6.3 × 10−4 | 1.1 × 10−3 | 9.2 × 10−4 | 9.0 × 10−3 |
| STRA6 | 610745 | 15 | 7 | 1.1 × 10−3 | 9.9 × 10−4 | 1.5 × 10−3 | 3.4 × 10−3 |
| ZNF222 | NA | 19 | 5 | 8.8 × 10−4 | 3.6 × 10−3 | 1.4 × 10−3 | 4.5 × 10−4 |
| COL9A3 | 120270 | 20 | 5 | 5.6 × 10−5 | 2.2 × 10−5 | 2.3 × 10−5 | 6.7 × 10−4 |
| FAM83F | NA | 22 | 5 | 3.8 × 10−3 | 4.4 × 10−4 | 6.6 × 10−4 | 9.3 × 10−3 |
We identified eight genes in the GENOA dataset with p values of at least suggestive significance (p < 1 × 10−3) using either GAMuT or SKAT, using a 90% cutoff to determine the effective number of independent tests. For the eight genes we provide gene name, chromosomal location of gene, number of rare variants (MAF < 3%) found in each gene in the GENOA dataset, and p values for the four approaches.
The projection matrix form of GAMuT identified four genes of suggestive significance. p values from the two forms of GAMuT were strongly correlated (Pearson correlation = 0.90). After accounting for confounders, GAMuT did not demonstrate any systematic inflation across the genome (see QQ plots in Figure 6).
In order to correct for using two phenotypic similarity matrices for GAMuT, we performed the perturbation approach described in the Material and Methods on the eight genes with p values of less 1 × 10−3 for either GAMuT or SKAT. The p values obtained through combined perturbation method are also shown in Table 3. Of the eight genes identified as suggestive by either or both of the GAMuT approaches, five remained suggestive after correcting for use of two GAMuT similarity matrices (including SELP).
We investigated whether our top genetic associations for the modeled phenotypes (SBP/DBP/HDL/BMI) in Table 3 were possibly spurious due to the fact that the phenotypes analyzed are secondary phenotypes collected in a study ascertained on a correlated primary phenotype (hypertension). To verify that a confounding association between rare-variant genotypes at our top genes and hypertension was not driving our results, we performed univariate SKAT testing of our top genes in Table 3 on the primary hypertension variable. We observed none of our top genes to be significantly associated with hypertension.
The SKAT p values using the three multiple testing correction methods were identical across all genes tested. SKAT did not identify any genes at genome-wide significance. It identified four genes at the suggestive significance threshold, all of which were identified by one or both of the GAMuT tests. When we applied MFLM to the GENOA data, we observed sizeable inflation of the p values. The p value inflation was not resolved by inverse-normal transforming the phenotypes, as performed in Wang et al.25 See Figure S8 for QQ plots of the untransformed and transformed analyses.
Running the GAMuT analyses on a single-threaded R script on an Intel i7-2720QM CPU took 22.3 min using either the linear kernel or the projection matrix to model phenotypic similarity. Implementing the perturbation approach (1 × 106 replicates per gene) required approximately 44.5 min of computing time per gene analyzed.
Discussion
Some patterns in the genetic basis of complex traits have emerged in prior studies. First, common variants of relatively small individual effect located throughout the genome collectively explain a large fraction of the total genetic variance.9, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78 Second, for some disorders such as autism,79, 80 more than a thousand genes appear capable of harboring exceedingly rare, large-effect mutations. Although it is still unclear whether these two patterns are ubiquitous, they are central predictions of the infinitesimal model of allele effects. Moreover, we know from detailed theoretical analysis3 that if the infinitesimal model is true for most phenotypes, then most rare large-effect mutations should be highly pleiotropic.
We have presented GAMuT, a framework for cross-phenotype analysis of rare variants using a nonparametric distance-covariance approach.26, 27, 30 This approach can accommodate both binary and continuous phenotypes and can adjust for covariates. The GAMuT test derives analytic p values based on Davies’ exact method, thereby improving computational efficiency and permitting application on a genome-wide scale. Like the popular SKAT framework for univariate rare variant analysis, our approach allows for inclusion of prior information, such as biological plausibility of the variants under study, and further remains powerful when a gene harbors a mixture of rare causal variants that act in different directions on phenotype. Our approach demonstrates greater power than SKAT and MFLM when pleiotropy exists. Further, simulations indicate that even if only one phenotype is associated with the gene of interest (i.e., no pleiotropy is occurring), GAMuT is at least as powerful as univariate SKAT analyses after multiple-testing adjustment. These results hold for both continuous and binary outcomes.
We provide R software implementing GAMuT on our website (see Web Resources), which can be run through software packages like PLINK, PLINK-SEQ, or EPACTS if desired. GAMuT analysis of simulated datasets comprised of 1,000 subjects and 10 phenotypes takes 0.52 s per gene for either continuous or binary phenotypes using a R script running single-threaded on an a 1.7 GHz Intel Core i7 CPU processor. Increasing the number of phenotypes or rare variants tested does not substantially increase GAMuT’s run-time. However, increasing sample size does increase run time. For sample sizes of N = 2,500, 5,000, 10,000, 20,000, and 30,000 subjects, we found that GAMuT takes approximately 4.1 s, 13.2 s, 68.6 s, 580 s, and 3,600 s per gene, respectively, for either continuous or binary phenotypes. Based on these estimates, we feel genome-wide analysis using GAMuT is feasible even with enormous sample sizes with the aid of parallel computing.
GAMuT’s perturbation approach to adjust for multiple combinations of phenotype/genotype similarity matrices when testing a gene is computationally far more efficient than permutations but still remains intensive. For a sample size of 10K, the total computation time required to run K = 106 perturbations for a single gene is ∼5–6 hr for M = 2 combinations and ∼10–12 hr for M = 4 combinations. Computation timescales linearly with number of perturbations performed and number of combinations assessed. Sample size has only a minor effect on perturbation run time; for example, increasing sample size by a factor of 10 increases computation time only by a factor of approximately 2. Although perturbations are computationally demanding, we note that we can circumvent this computational issue in a couple of ways. First, we can elect to apply the perturbation approach to just the small set of genes with a minimum unadjusted p value (across the M combinations considered) smaller than the unadjusted genome-wide significance threshold; genes that fail to meet this criteria will be of little or no interest for follow up. This strategy is a variation of the strategy we applied in our GENOA analyses. Alternatively, if one wanted to apply the perturbation procedure to each of 20,000 genes, then one could consider an adaptive perturbation strategy similar in logic to the adaptive permutation procedure in PLINK81 to adjust for multiple testing in GWASs. We will explore this idea in future work.
We applied GAMuT to exome-chip data from the GENOA study to identify genes harboring rare variants with cross-phenotype effects on four phenotypes: BMI, HDL levels, SBP, and DBP. Using the linear kernel to model phenotypic similarity and the weighted linear kernel to model genotypic similarity, we detected eight genes that were suggestively associated with our phenotypes. Of note, common variants and gene product levels of one such gene, Selectin P (SELP [MIM: 173610]), have previously been associated with BMI, SBP, DBP, and HDL.66, 68
GAMuT’s KDC framework is amenable to several promising extensions that we will explore in future work. Because GAMuT is an omnibus test, an association of the gene with just one of the tested phenotypes (i.e., no pleiotropy) could result in a significant finding. Although the result is valid, researchers will often wish to identify which underlying phenotype(s) of those modeled are directly associated with the gene of interest. Additionally, if we identify a cross-phenotype association, a follow-up analysis could be to assess whether the cross-phenotype effect is due to biological pleiotropy (a causal locus directly affecting more than one trait) or mediation pleiotropy (a causal locus affecting only one trait, which in turn affects another trait). Existing mediation analyses are not intended to handle high-dimensional traits; we propose the creation of KDC procedures to identify whether an observed cross-phenotype association is mediated by a different set of phenotypes. Additionally, we could also perform post hoc GAMuT of different subgroupings of the phenotypes to identify the true phenotypes associated with the gene and adjust for multiple testing using perturbations. We will pursue these ideas in future work.
GAMuT currently assumes unrelated subjects; however, it should be reasonably straightforward to extend GAMuT to allow for case-parent trio studies. The work by Jiang et al.82 provides a framework for transforming genotypic data for trios into data that is amenable to a kernel-based framework. Specifically, the Jiang method uses the quantitative transmission disequilibrium test introduced by Abecasis et al.83 to decompose observed genotypes into between-family and within-family components, and then integrates within-family genetic components into a kernel-machine regression framework. Although the Jiang method uses a KMR approach and is therefore appropriate only for univariate phenotype analyses, an analogous approach, using GAMuT, should allow for high-dimensional phenotype data. Finally, one might be interested in combining cross-phenotype association results from multiple studies through a meta-analysis. GAMuT is designed to test for rare variant cross-phenotype associations in a single dataset. However, the meta-analysis approach in Lee et al.,84 which is designed to combine results of multiple KMR-based studies, should be readily extendible to KDC results, such as those obtained via GAMuT.
That pleiotropy might be ubiquitous should come as no surprise. The central organismal level result of pleiotropy will be the frequent occurrence of comorbid diagnoses. Neuropsychiatric disorders, for instance, are particularly laden with comorbid diagnoses. The National Institute of Mental Health (NIMH) estimates that as many as 45% of individuals diagnosed with a mental disorder meet criteria for two or more disorders.85 Likewise, nearly 75% of adults with diabetes also have hypertension,86 and individuals with rheumatoid arthritis are about twice as likely to suffer from myocardial infarction as individuals without arthritis.87 Although some of these overlapping phenotypes are ultimately due to environmental risk factors, other comorbidities are almost certainly explained by common genetic pathways. Ignoring comorbidity, or worse, setting inclusion criteria that exclude individuals suffering a comorbid diagnosis, will limit biological understanding of complex traits and might limit our ability to detect missing heritability.
Acknowledgments
This work was supported by NIH grants HG007508, HL086694, HL119443, MH071537, GM117946, and AR060893. For purposes of disclosing duality of interest, M.P.E. is a consultant for Amnion Laboratories.
Published: March 3, 2016
Footnotes
Supplemental Data include eight figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.01.017.
Web Resources
The URLs for data presented herein are as follows:
Epstein software, http://www.genetics.emory.edu/labs/epstein/software
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Barton N.H., Turelli M. Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 1989;23:337–370. doi: 10.1146/annurev.ge.23.120189.002005. [DOI] [PubMed] [Google Scholar]
- 2.Lande R. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 2007;89:373–387. doi: 10.1017/S0016672308009555. [DOI] [PubMed] [Google Scholar]
- 3.Turelli M. Heritable genetic variation via mutation-selection balance: Lerch’s zeta meets the abdominal bristle. Theor. Popul. Biol. 1984;25:138–193. doi: 10.1016/0040-5809(84)90017-0. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie J.H. Oxford University Press; 1994. The Causes of Molecular Evolution. [Google Scholar]
- 5.Lander E.S. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 6.Collins F.S., Guyer M.S., Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti A. Population genetics--making sense out of sequence. Nat. Genet. 1999;21(1, Suppl):56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 8.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Manolio T.A., Pasquale L.R., Boerwinkle E., Caporaso N., Cunningham J.M., de Andrade M., Feenstra B., Feingold E., Hayes M.G. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonson M.A., Wills A.G., Keller M.C., McQueen M.B. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med. Genet. 2011;12:146. doi: 10.1186/1471-2350-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galesloot T.E., van Steen K., Kiemeney L.A., Janss L.L., Vermeulen S.H. A comparison of multivariate genome-wide association methods. PLoS ONE. 2014;9:e95923. doi: 10.1371/journal.pone.0095923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison D.B., Thiel B., St Jean P., Elston R.C., Infante M.C., Schork N.J. Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am. J. Hum. Genet. 1998;63:1190–1201. doi: 10.1086/302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavali S., Barrenas F., Kanduri K., Benson M. Network properties of human disease genes with pleiotropic effects. BMC Syst. Biol. 2010;4:78. doi: 10.1186/1752-0509-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivakumaran S., Agakov F., Theodoratou E., Prendergast J.G., Zgaga L., Manolio T., Rudan I., McKeigue P., Wilson J.F., Campbell H. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees C.W., Barrett J.C., Parkes M., Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 17.Liu F., van der Lijn F., Schurmann C., Zhu G., Chakravarty M.M., Hysi P.G., Wollstein A., Lao O., de Bruijne M., Ikram M.A. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8:e1002932. doi: 10.1371/journal.pgen.1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira M.A., Purcell S.M. A multivariate test of association. Bioinformatics. 2009;25:132–133. doi: 10.1093/bioinformatics/btn563. [DOI] [PubMed] [Google Scholar]
- 20.Huang J., Johnson A.D., O’Donnell C.J. PRIMe: a method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27:1201–1206. doi: 10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Reilly P.F., Hoggart C.J., Pomyen Y., Calboli F.C., Elliott P., Jarvelin M.R., Coin L.J. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE. 2012;7:e34861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ried J.S., Döring A., Oexle K., Meisinger C., Winkelmann J., Klopp N., Meitinger T., Peters A., Suhre K., Wichmann H.E., Gieger C. PSEA: Phenotype Set Enrichment Analysis--a new method for analysis of multiple phenotypes. Genet. Epidemiol. 2012;36:244–252. doi: 10.1002/gepi.21617. [DOI] [PubMed] [Google Scholar]
- 23.Maity A., Sullivan P.F., Tzeng J.Y. Multivariate phenotype association analysis by marker-set kernel machine regression. Genet. Epidemiol. 2012;36:686–695. doi: 10.1002/gepi.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu A., Mills J.L., Boehnke M., Wilson A.F., Bailey-Wilson J.E., Xiong M., Wu C.O., Fan R. Pleiotropy analysis of quantitative traits at gene level by multivariate functional linear models. Genet. Epidemiol. 2015;39:259–275. doi: 10.1002/gepi.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gretton A., Fukumizu K., Teo C.H., Song L., Schölkopf B., Smola A.J. A kernel statistical test of independence. Adv. Neural Inf. Process. Syst. 2008:585–592. [Google Scholar]
- 27.Hua W.Y., Ghosh D. Equivalence of kernel machine regression and kernel distance covariance for multidimensional phenotype association studies. Biometrics. 2015;71:812–820. doi: 10.1111/biom.12314. [DOI] [PubMed] [Google Scholar]
- 28.Kosorok M.R., Rizzo M.L. On Brownian distance covariance and high dimensional data. Ann. Appl. Stat. 2009;3:1266–1269. doi: 10.1214/09-AOAS312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szekely G.J., Rizzo M.L., Bakirov N.K. Measuring and testing dependence by correlation of distances. Ann. Stat. 2007;35:2769–2794. [Google Scholar]
- 30.Zhang, K., Peters, J., Janzing, D., and Schölkopf, B. (2012). Kernel-based conditional independence test and application in causal discovery. arXiv, arXiv:12023775.
- 31.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels P.R., Kardia S.L., Hanis C.L., Brown C.A., Hutchinson R., Boerwinkle E., Turner S.T., Genetic Epidemiology Network of Arteriopathy study Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am. J. Med. 2004;116:676–681. doi: 10.1016/j.amjmed.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 33.Zapala M.A., Schork N.J. Statistical properties of multivariate distance matrix regression for high-dimensional data analysis. Front. Genet. 2012;3:190. doi: 10.3389/fgene.2012.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessel J., Schork N.J. Generalized genomic distance-based regression methodology for multilocus association analysis. Am. J. Hum. Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwee L.C., Liu D., Lin X., Ghosh D., Epstein M.P. A powerful and flexible multilocus association test for quantitative traits. Am. J. Hum. Genet. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaid D.J. Genomic similarity and kernel methods II: methods for genomic information. Hum. Hered. 2010;70:132–140. doi: 10.1159/000312643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M.C., Kraft P., Epstein M.P., Taylor D.M., Chanock S.J., Hunter D.J., Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 40.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies R.B. Algorithm AS 155: the distribution of a linear combination of 2 random variables. J. R. Stat. Soc. Ser. C Appl. Stat. 1980;29:323–333. [Google Scholar]
- 42.Duchesne P., Lafaye De Micheauz P. Computing the distribution of quadratic forms: Further comparisons between the Liu-Tang-Zhang approximation and exact methods. Comput. Stat. Data Anal. 2010;54:858–862. [Google Scholar]
- 43.Schork N.J., Wessel J., Malo N. DNA sequence-based phenotypic association analysis. Adv. Genet. 2008;60:195–217. doi: 10.1016/S0065-2660(07)00409-9. [DOI] [PubMed] [Google Scholar]
- 44.Wu M.C., Maity A., Lee S., Simmons E.M., Harmon Q.E., Lin X., Engel S.M., Molldrem J.J., Armistead P.M. Kernel machine SNP-set testing under multiple candidate kernels. Genet. Epidemiol. 2013;37:267–275. doi: 10.1002/gepi.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X., Becker L.C., Becker D.M., Starmer J.D., Province M.A. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet. Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vattikuti S., Guo J., Chow C.C. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarkesh M., Daneshpour M.S., Faam B., Fallah M.S., Hosseinzadeh N., Guity K., Hosseinpanah F., Momenan A.A., Azizi F. Heritability of the metabolic syndrome and its components in the Tehran Lipid and Glucose Study (TLGS) Genet. Res. 2012;94:331–337. doi: 10.1017/S001667231200050X. [DOI] [PubMed] [Google Scholar]
- 51.Hottenga J.J., Boomsma D.I., Kupper N., Posthuma D., Snieder H., Willemsen G., de Geus E.J. Heritability and stability of resting blood pressure. Twin Res. Hum. Genet. 2005;8:499–508. doi: 10.1375/183242705774310123. [DOI] [PubMed] [Google Scholar]
- 52.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., International Consortium for Blood Pressure Genome-Wide Association Studies. CARDIoGRAM consortium. CKDGen Consortium. KidneyGen Consortium. EchoGen consortium. CHARGE-HF consortium Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zabaneh D., Balding D.J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE. 2010;5:e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kraja A.T., Vaidya D., Pankow J.S., Goodarzi M.O., Assimes T.L., Kullo I.J., Sovio U., Mathias R.A., Sun Y.V., Franceschini N. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avery C.L., He Q., North K.E., Ambite J.L., Boerwinkle E., Fornage M., Hindorff L.A., Kooperberg C., Meigs J.B., Pankow J.S. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet. 2011;7:e1002322. doi: 10.1371/journal.pgen.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., Wellcome Trust Case Control Consortium. Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herbert A., Gerry N.P., McQueen M.B., Heid I.M., Pfeufer A., Illig T., Wichmann H.E., Meitinger T., Hunter D., Hu F.B. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 59.Fall T., Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol. Cell. Endocrinol. 2014;382:740–757. doi: 10.1016/j.mce.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 60.Lange L.A., Lange E.M., Bielak L.F., Langefeld C.D., Kardia S.L., Royston P., Turner S.T., Sheedy P.F., 2nd, Boerwinkle E., Peyser P.A. Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler. Thromb. Vasc. Biol. 2002;22:418–423. doi: 10.1161/hq0302.105721. [DOI] [PubMed] [Google Scholar]
- 61.Epstein M.P., Duren W.L., Boehnke M. Improved inference of relationship for pairs of individuals. Am. J. Hum. Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moskvina V., Schmidt K.M. On multiple-testing correction in genome-wide association studies. Genet. Epidemiol. 2008;32:567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- 63.Tregouet D.A., Barbaux S., Escolano S., Tahri N., Golmard J.L., Tiret L., Cambien F. Specific haplotypes of the P-selectin gene are associated with myocardial infarction. Hum. Mol. Genet. 2002;11:2015–2023. doi: 10.1093/hmg/11.17.2015. [DOI] [PubMed] [Google Scholar]
- 64.Herrmann S.M., Ricard S., Nicaud V., Mallet C., Evans A., Ruidavets J.B., Arveiler D., Luc G., Cambien F. The P-selectin gene is highly polymorphic: reduced frequency of the Pro715 allele carriers in patients with myocardial infarction. Hum. Mol. Genet. 1998;7:1277–1284. doi: 10.1093/hmg/7.8.1277. [DOI] [PubMed] [Google Scholar]
- 65.Zee R.Y., Cook N.R., Cheng S., Reynolds R., Erlich H.A., Lindpaintner K., Ridker P.M. Polymorphism in the P-selectin and interleukin-4 genes as determinants of stroke: a population-based, prospective genetic analysis. Hum. Mol. Genet. 2004;13:389–396. doi: 10.1093/hmg/ddh039. [DOI] [PubMed] [Google Scholar]
- 66.Davì G., Romano M., Mezzetti A., Procopio A., Iacobelli S., Antidormi T., Bucciarelli T., Alessandrini P., Cuccurullo F., Bittolo Bon G. Increased levels of soluble P-selectin in hypercholesterolemic patients. Circulation. 1998;97:953–957. doi: 10.1161/01.cir.97.10.953. [DOI] [PubMed] [Google Scholar]
- 67.Ikeda H., Takajo Y., Ichiki K., Ueno T., Maki S., Noda T., Sugi K., Imaizumi T. Increased soluble form of P-selectin in patients with unstable angina. Circulation. 1995;92:1693–1696. doi: 10.1161/01.cir.92.7.1693. [DOI] [PubMed] [Google Scholar]
- 68.Koyama H., Maeno T., Fukumoto S., Shoji T., Yamane T., Yokoyama H., Emoto M., Shoji T., Tahara H., Inaba M. Platelet P-selectin expression is associated with atherosclerotic wall thickness in carotid artery in humans. Circulation. 2003;108:524–529. doi: 10.1161/01.CIR.0000081765.88440.51. [DOI] [PubMed] [Google Scholar]
- 69.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S.H., DeCandia T.R., Ripke S., Yang J., Sullivan P.F., Goddard M.E., Keller M.C., Visscher P.M., Wray N.R., Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ) International Schizophrenia Consortium (ISC) Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C., Sullivan P.F., Sklar P., International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Mägi R., MAGIC. Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Visscher P.M., Yang J., Goddard M.E. A commentary on ‘common SNPs explain a large proportion of the heritability for human height’ by Yang et al. (2010) Twin Res. Hum. Genet. 2010;13:517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- 75.Davies G., Tenesa A., Payton A., Yang J., Harris S.E., Liewald D., Ke X., Le Hellard S., Christoforou A., Luciano M. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S.H., Harold D., Nyholt D.R., Goddard M.E., Zondervan K.T., Williams J., Montgomery G.W., Wray N.R., Visscher P.M., ANZGene Consortium. International Endogene Consortium. Genetic and Environmental Risk for Alzheimer’s disease Consortium Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum. Mol. Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S.H., Ripke S., Neale B.M., Faraone S.V., Purcell S.M., Perlis R.H., Mowry B.J., Thapar A., Goddard M.E., Witte J.S., Cross-Disorder Group of the Psychiatric Genomics Consortium. International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plomin R., Haworth C.M., Meaburn E.L., Price T.S., Davis O.S., Wellcome Trust Case Control Consortium 2 Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychol. Sci. 2013;24:562–568. doi: 10.1177/0956797612457952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desachy G., Croen L.A., Torres A.R., Kharrazi M., Delorenze G.N., Windham G.C., Yoshida C.K., Weiss L.A. Increased female autosomal burden of rare copy number variants in human populations and in autism families. Mol. Psychiatry. 2015;20:170–175. doi: 10.1038/mp.2014.179. [DOI] [PubMed] [Google Scholar]
- 80.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang Y., Conneely K.N., Epstein M.P. Flexible and robust methods for rare-variant testing of quantitative traits in trios and nuclear families. Genet. Epidemiol. 2014;38:542–551. doi: 10.1002/gepi.21839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee S., Teslovich T.M., Boehnke M., Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am. J. Hum. Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kessler R.C., Chiu W.T., Demler O., Merikangas K.R., Walters E.E. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long A.N., Dagogo-Jack S. Comorbidities of diabetes and hypertension: mechanisms and approach to target organ protection. J. Clin. Hypertens. (Greenwich) 2011;13:244–251. doi: 10.1111/j.1751-7176.2011.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Solomon D.H., Goodson N.J., Katz J.N., Weinblatt M.E., Avorn J., Setoguchi S., Canning C., Schneeweiss S. Patterns of cardiovascular risk in rheumatoid arthritis. Ann. Rheum. Dis. 2006;65:1608–1612. doi: 10.1136/ard.2005.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.