Abstract
According to controversial theories and results of studies, foods with animal origins play an important role in the transmission of H. pylori to human. The aim of this study was to determine the distribution of vacA genotypes of H. pylori, isolated from milk and meat samples of cow, sheep, goat, camel, and buffalo. Eight hundred and twenty raw milk and meat samples were collected from various parts of Iran. Samples were cultured and those found positive for H. pylori were analyzed for the presence of various genotypes of vacA gene. Out of 420 milk and 400 meat samples, 92 (21.90%) and 105 (26.25%) were positive for H. pylori, respectively. The most commonly detected genotypes in the vacA gene were s1a (86.80%), m1a (79.18%), s1b (69.54%), and m1b (63.45%) and detected combined genotypes were mostly m1as1a (68.52%), m1as1b (60.40%), m1bs1b (55.83%), and m1bs1a (53.29%). High presence of bacteria in the milk and meat samples of sheep represents that sheep may be the natural host of H. pylori. High presence of H. pylori strains in milk and meat samples similar to vacA genotypes in human being suggests that milk and meat samples could be the sources of bacteria for human.
1. Introduction
Although Helicobacter pylori (H. pylori) has been accepted as a major cause of gastrointestinal disorders and especially gastric adenocarcinoma, type B gastritis, mucosa associated lymphoid tissue lymphoma, and peptic ulcer disease, its route of transmission, sources, and also the role of foods are still unknown. H. pylori is a Gram negative, coccoid flagellated bacterium with 2 to 4 μm in length and 0.5 to 1 μm in width, which is the first formally recognized bacterial carcinogen and one of the most successful human pathogens [1–5]. High prevalence of H. pylori in the stomach of domestic animals, milk, meat, and gastric biopsies suggests that food with animal origin, and also domestic animals, may be its reservoirs [1–5]. Higher prevalence of H. pylori in meat eaters than vegetarians which was achieved in the previous investigation supports the significant role of foods with animal origins in the transmission of bacteria to humans [6]. Appropriate condition of meat and milk including acidic PH, nutritional values, salt concentration, and also high amount of activated water (AW) facilitate the growth and survival of H. pylori and provide adequate setting for transmission of H. pylori to human [7]. High prevalence of antibodies against H. pylori in the serum samples taken from veterinarians, butchers, and staffs of the slaughterhouses and milking rooms can support the zoonoses aspects of this bacterium [1, 8].
To evaluate the pathogenicity of H. pylori, apprising virulence factors is requisite. The most commonly identified virulence factor among H. pylori strains is vacuolating cytotoxin (vacA) [9, 10]. VacA belongs to the group of genes with mutable genotypes associated with damage to gastric epithelial cells. This gene exists in practically all strains of H. pylori. This gene is polymorphic and comprises variable signal regions (type s1 or type s2) and midregions (type m1 or type m2) [9, 10]. The s-region is classified into s1 and s2 and the m-region is categorized as m1 and m2. The s1 type is further subtyped into s1a, s1b, and s1c and the subcategories of m1 are m1a and m1b, respectively. Higher cytotoxicity and acuity have been done by this mosaic pattern [11, 12]. Genotyping using vacA alleles is considered as one of the best methods to study the associations of H. pylori strains in various samples.
Industrialized information designated the fact that closely 50% of the world population and also 60–90% of Iranian people are infected with virulent strains of H. pylori [4, 13]. According to the high prevalence of H. pylori in Iran and other parts of the world, and also with respect to the indistinct situation of H. pylori in foods with animal origins, the present investigation was carried out in order to study the vacA genotype status of H. pylori isolated from Iranian raw milk and meat samples.
2. Materials and Methods
2.1. Sample Collection
In all, 420 raw milk samples were collected: cow (n = 120), sheep (n = 120), goat (n = 80), buffalo (n = 50), and camel (n = 50) raw milk samples were collected from farm bulk tanks and milk collection centers from several geographic regions of Iran, from March 2013 to March 2014. Cow and buffalo milk samples were collected throughout this time period. Because the lactating periods of ewes and goats in Iran are seasonal (from March through May and September to November of the subsequent year), goat and sheep milk samples were only available through these months within the fore-mentioned time frame. At each site, sampling of milk was performed according to the International Dairy Federation guidelines (IDF 1995). Samples (100 mL, in sterile glass containers) were transported to the laboratory at ca. 4°C within a maximum of 6–12 h after sampling. For raw meat samples, 100 cow, 100 sheep, 100 goat, 50 buffalo, and 50 camel meat samples were purchased from butchers of various parts of Iran. All samples were kept under refrigeration in plastic bags; information about dates of production and assigned shelf lives was not presented. Meat samples were collected over a period of eight months from August 2013 to February 2014, and they were analyzed on the day of acquisition. Samples were transported under refrigeration (4–6°C) in thermal boxes containing ice packs and were tested immediately after collection.
2.2. Isolation of Helicobacter pylori
Twenty-five mL of each homogenized sample was added to 225 mL of Wilkins Chalgren anaerobe broth (Oxoid, UK) supplemented with 5% of horse serum (Sigma, St. Louis, MO, USA) and colistin methanesulfonate (30 mg/L), cycloheximide (100 mg/L), nalidixic acid (30 mg/L), trimethoprim (30 mg/L), and vancomycin (10 mg/L) (Sigma, St. Louis, MO, USA) and colistin methanesulfonate (30 mg/L), cycloheximide (100 mg/L), nalidixic acid (30 mg/L), trimethoprim (30 mg/L), and vancomycin (10 mg/L) (Sigma, St. Louis, MO, USA) and incubated for 7 days at 37°C with shaking under microaerophilic condition. Then, 0.1 mL of the enrichment selective broth was plated onto Wilkins Chalgren anaerobe agar (Oxoid, UK) supplemented with 5% of defibrinated horse blood and 30 mg/L colistin methanesulfonate, 100 mg/L cycloheximide, 30 mg/L nalidixic acid, 30 mg/L trimethoprim, and 10 mg/L vancomycin (Sigma, St. Louis, MO, USA) and incubated for 7 days at 37°C under microaerophilic condition. For comparison, a reference strain of H. pylori (ATCC 43504) was employed.
2.3. DNA Extraction and Helicobacter pylori 16S rRNA Gene Amplification
Based on the PCR technique, suspected colonies were identified as H. pylori. Genomic DNA was extracted from the colonies with typical characters of H. pylori using a DNA extraction kit for cells and tissues (Roche Applied Science, Germany, 11814770001) according to the manufacturer's instructions and its density which was assessed by optic densitometry. Extracted DNA was amplified for the 16S rRNA gene (primers: HP-F: 5′-CTGGAGAGACTAAGCCCTCC-3′ and HP-R: 5′-ATTACTGACGCTGATTGTGC-3′) [14]. PCR reactions were performed in a final volume of 50 µL containing 5 µL 10x buffer + MgCl2, 2 mM dNTP, 2 unit Taq DNA polymerase, 100 ng genomic DNA as a template, and 25 picomole of each primer. PCR was performed using a thermal cycler (Eppendorf Co., Germany) under the following condition: an initial denaturation for 2 minutes at 94°C; 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 8 min.
2.4. Genotyping of vacA Gene of Helicobacter pylori
Presence of the genotypes of vacA alleles (s1a, s1b, s1c, m1a, m1b, and m2) was determined by PCR. The primer sequences are shown in Table 1 [15].
Table 1.
Oligonucleotide primers used for genotyping of Helicobacter pylori isolated from foods with animal origin in Iran [15].
| vacA alleles | Primer sequence (5′-3′) | Size of product (bp) |
|---|---|---|
| s1a | F: CTCTCGCTTTAGTAGGAGC R: CTGCTTGAATGCGCCAAAC |
213 |
|
| ||
| s1b | F: AGCGCCATACCGCAAGAG R: CTGCTTGAATGCGCCAAAC |
187 |
|
| ||
| s1c | F: CTCTCGCTTTAGTGGGGYT R: CTGCTTGAATGCGCCAAAC |
213 |
|
| ||
| s2 | F: GCTAACACGCCAAATGATCC R: CTGCTTGAATGCGCCAAAC |
199 |
|
| ||
| m1a | F: GGTCAAAATGCGGTCATGG R: CCATTGGTACCTGTAGAAAC |
290 |
|
| ||
| m1b | F: GGCCCCAATGCAGTCATGGA R: GCTGTTAGTGCCTAAAGAAGCAT |
291 |
|
| ||
| m2 | F: GGAGCCCCAGGAAACATTG R: CATAACTAGCGCCTTGCA |
352 |
The PCR was performed in a total volume of 50 μL containing 1 μM of each primer, 1 μL of genomic DNA (approximately 200 ng), 1 mM of dNTPs mix (invitrogen), 2 mM of Mgcl2, and 0.05 U/μL Taq DNA polymerase (invitrogen). PCR amplifications were performed in an automated thermal cycler (Biometra Co., Germany). The following cycle conditions were used for PCR amplification: 32 cycles of 45 s at 95°C, 50 s at 64°C, and 70 s at 72°C. All runs included one negative DNA control consisting of PCR grade water and two or more positive controls (H. pylori 26695, H. pylori J99, H. pylori SS1, H. pylori Tx30, H. pylori 88-23, and H. pylori 84-183).
2.5. Gel Electrophoresis
The PCR amplification products (10 μL) were subject to electrophoresis in a 1% agarose gel in 1x TBE buffer at 80 V for 30 min and stained with ethidium bromide, and images were obtained in UVIdoc gel documentation systems (UK). The PCR products were identified by 100 bp DNA size marker (Fermentas, Germany).
2.6. Statistical Analysis
Using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA), Chi-square test and Fisher's exact two-tailed test analysis were performed, and differences were considered significant at values of P < 0.05. Distributions of genotypes of H. pylori isolated from food stuff were statistically analyzed.
3. Results and Discussion
All of the milk and meat samples were examined using the culture and PCR techniques. Table 2 shows the distribution of H. pylori in the milk and meat samples. Of 820 meat and milk samples, 197 (24.02%) were positive for H. pylori. Of 420 milk and 400 meat samples, 92 (21.90%) and 105 (26.25%) were positive for H. pylori, respectively (Table 2).
Table 2.
Distribution of Helicobacter pylori in various types of raw milk and meat samples.
| Types of samples | Number of samples collected | Positive results for H. pylori (%) |
|---|---|---|
| Cow milk | 120 | 25 (20.83) |
| Sheep milk | 120 | 35 (29.16) |
| Goat milk | 80 | 15 (18.75) |
| Buffalo milk | 50 | 12 (24) |
| Camel milk | 50 | 5 (10) |
| Total raw milk | 420 | 92 (21.90) |
| Cow meat | 100 | 25 (25) |
| Sheep meat | 100 | 37 (37) |
| Goat meat | 100 | 22 (22) |
| Buffalo meat | 50 | 14 (28) |
| Camel meat | 50 | 7 (14) |
| Total raw meat | 400 | 105 (26.25) |
| Total | 820 | 197 (24.02) |
The most commonly contaminated milk and meat samples were raw sheep milk (29.16%) and raw sheep meat (37%). There were no statistically significant differences among the incidence of bacteria in milk and meat samples. There were statistically significant differences in the incidence of H. pylori between sheep and camel milk (P = 0.033) and between sheep and camel meat (P = 0.048). Distribution of vacA genotypes of the H. pylori strains of meat and milk samples is shown in Table 3.
Table 3.
Distribution of vacA genotypes in Helicobacter pylori strains of meat and milk samples.
| Types of samples (number of positive results) | Distribution of vacA genotypes (%) | ||||||
|---|---|---|---|---|---|---|---|
| s1a | s1b | s1c | s2 | m1a | m1b | m2 | |
| Cow milk (25) | 20 | 18 | 14 | 9 | 19 | 17 | 8 |
| Sheep milk (35) | 32 | 23 | 16 | 12 | 27 | 20 | 10 |
| Goat milk (15) | 12 | 10 | 8 | 5 | 10 | 9 | 4 |
| Buffalo milk (12) | 10 | 8 | 5 | 7 | 8 | 8 | 2 |
| Camel milk (5) | 5 | 4 | — | 2 | 3 | 3 | — |
| Total raw milk (92) | 79 (85.86) | 63 (68.47) | 43 (46.73) | 35 (38.04) | 67 (72.82) | 57 (61.95) | 24 (26.08) |
| Cow meat (25) | 22 | 19 | 14 | 10 | 20 | 19 | 9 |
| Sheep meat (37) | 34 | 26 | 20 | 13 | 29 | 21 | 13 |
| Goat meat (22) | 19 | 15 | 11 | 8 | 17 | 16 | 7 |
| Buffalo meat (14) | 11 | 9 | 6 | 9 | 10 | 9 | 5 |
| Camel meat (7) | 6 | 5 | 1 | 3 | 3 | 3 | 1 |
| Total raw meat (105) | 92 (87.61) | 74 (70.47) | 52 (49.52) | 43 (40.95) | 89 (84.76) | 68 (64.76) | 35 (33.33) |
| Total (197) | 171 (86.80) | 137 (69.54) | 95 (48.22) | 78 (39.59) | 156 (79.18) | 125 (63.45) | 59 (29.94) |
Results of the gel electrophoresis of PCR products for amplification of various genotypes of vacA gene are shown in Figures 1 –3.
Figure 1.
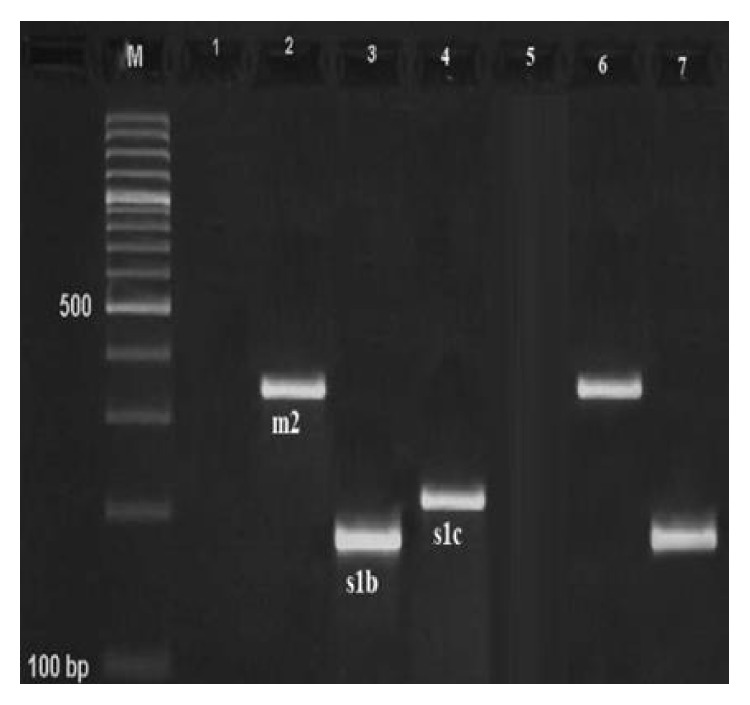
Results of the gel electrophoresis for identification of m2, s1b, and s1c genotypes of the H. pylori strains of milk and meat samples. Line 5: negative control, line 7: positive controls, 1: negative sample, M: 100 bp DNA ladder (Fermentas, Germany), and numbers 2–4: positive samples for m2 (352 bp), s1b (187 bp), and s1c (213 bp) alleles.
Figure 2.
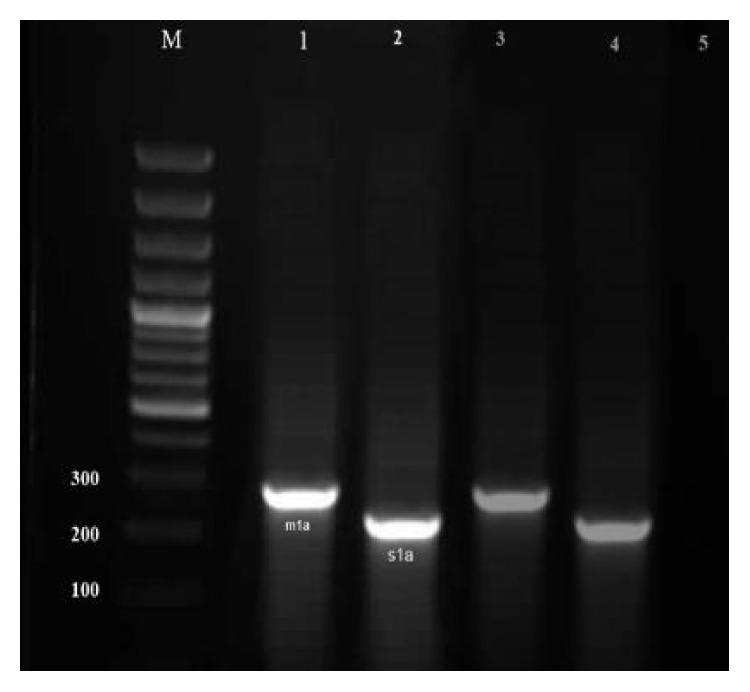
Results of the gel electrophoresis for identification of m1a and s1a genotypes of the H. pylori strains of milk and meat samples. Lanes 3 and 4: positive controls, 5: negative control, M: 100 bp DNA ladder (Fermentas, Germany), and numbers 1-2: positive samples for m1a (290 bp) and s1a (213 bp) alleles, respectively.
Figure 3.

Results of the gel electrophoresis for identification of m1b and s2 genotypes of the H. pylori strains of milk and meat samples. Lanes 3 and 4: positive controls, 5: negative control, M: 100 bp DNA ladder (Fermentas, Germany), and numbers 1-2: positive samples for s2 (199 bp) and m1b (291 bp) alleles, respectively.
Significant difference was found between the type of samples and prevalence of genotypes (P < 0.05). Fourteen different genotypic combinations are shown in Table 4. The most commonly detected combined genotypes were m1as1a (68.52%), m1as1b (60.40%), m1bs1b (55.83%), and m1bs1a (53.29%).
Table 4.
Distribution of combined genotypes of Helicobacter pylori isolated from Iranian raw milk and meat samples.
| Genotypes | Prevalence (%)∗ |
|---|---|
| M1as1a | 135 (68.52) |
| M1as1b | 119 (60.40) |
| M1bs1a | 105 (53.29) |
| M1bs1b | 110 (55.83) |
| M1as1c | 70 (35.53) |
| M1bs1c | 58 (29.44) |
| M2s1a | 40 (20.30) |
| M2s1b | 31 (15.73) |
| M2s1c | 26 (13.19) |
| M2s2 | 19 (9.64) |
| M1as2 | 56 (28.42) |
| M1bs2 | 44 (22.33) |
∗Percent of positive genes from total of 197 positive samples.
Results of current study showed that raw milk and meat samples were reservoir for H. pylori. Total prevalences of H. pylori in raw cow, sheep, goat, buffalo, and camel milk samples of our survey were 20.83%, 29.16%, 18.75%, 24%, and 10%, respectively. Rahimi and Kheirabadi [3] reported that the incidence of H. pylori in raw cow, sheep, goat, buffalo, and camel milk samples of Iranian herds was 1.41%, 12.20%, 8.70%, 23.4%, and 3.6%, respectively, which was lower than our results. In a study carried out in Italy, H. pylori was detected in 50%, 33%, and 25.6% of raw cow, sheep, and goat milk, respectively, which was higher than our results [16]. In a study conducted in Japan, H. pylori was detected in 72.2% of raw cow milk samples [17]. Total distribution of H. pylori in the milk samples of Greek [18] and American [4] herds was 20% and 60%, respectively. Recent clinical investigation among Iranian cows showed that 16% of milk and 40% of feces samples of seropositive herds were infected with H. pylori [2].
Total prevalence of H. pylori in cow, sheep, goat, buffalo, and camel meat samples of our survey was 25%, 37%, 22%, 28%, and 14%, respectively, which was entirely contrary to the results of Stevenson et al. [19]. They suggested that transmission of H. pylori from beef and beef products is not a primary factor in the high prevalence of this bacterium in humans. Moreover, Mhaskar et al. [20] reported that the prevalence of peptic ulcer and H. pylori infection were entirely higher in those patients who have used meat and meat products (Odds Ratio (OR): 2.35, 95% and Confidence Interval (CI): 1.30–4.23) and restaurant foods (OR: 3.77, 95% CI: 1.39–10.23) in their main meals.
The possibility that H. pylori may be a zoonosis first rose the publication of two epidemiological studies that exhibited that the prevalence of H. pylori infection in abattoir and meat workers was significantly increased in comparison with the subjects that were not involved in handling animals or meat [21, 22]. This hypothesis is further reinforced by the demonstration of H. pylori in the gastric mucosa of calves, pigs, and horses and its isolation from sheep's gastric tissue and milk [4], suggesting that these animal species may act as reservoirs and spreaders of H. pylori. Findings of Momtaz et al. [1] have conclusively proved the zoonotic aspects of H. pylori. They showed that the vacA s1a/m1a was prominent H. pylori genotype in all cow, sheep, and human beings clinical samples. They showed 3.4–8.4% variability and 92.9–98.5% homology between sheep and human samples [1].
High prevalence of H. pylori in the milk (29.16%) and meat (37%) samples of sheep of our study suggested that sheep may be the natural host of H. pylori. Dore et al. [4] reported that H. pylori DNA was demonstrated in 60% (38/63) of milk samples and in 30% (6/20) of sheep tissue samples. They showed that the vacA gene was amplified in five of 38 milk samples, and in two of six sheep tissue samples, respectively. Sequence analysis of 16S rRNA PCR products from H. pylori strains [4] investigation demonstrated 99% identity with H. pylori.
Genotyping using vacA virulence marker gene is considered as one of the best approaches for studying of the correlations between H. pylori isolates from different samples [1]. H. pylori strains of milk and meat samples of our study showed similar status in the distribution of vacA genotypes. Totally, the most commonly detected genotypes in milk and meat samples were s1a (86.80%), m1a (79.18%), s1b (69.54%), and m1b (63.45%). Various genotypes of vacA strains were the most commonly detected genotypes in the studies of Linpisarn et al. (Thailand) [23], López-Vidal et al. (Mexico) [24], and Rudi et al. (Germany) [25]. The high presence of m1as1a and m1as1b genotypes has been reported previously from Iran [13] and Germany [25] but far different results have been reported from Thailand [23] and Mexico [24].
According to the high prevalence of pathogenic strains of H. pylori in milk and meat samples especially in those collected from sheep and also based on the considerable consumption of milk and meat in their raw forms in some areas of the world [26–28], consumption of these food products in their raw forms should be stopped. On the other hand, thorough cooking of meat and pasteurization of milk can prevent the presence and also transmission of pathogenic bacteria like H. pylori.
4. Conclusions
H. pylori which is harbored from milk and meat samples are similar in genotype of the vacA allele with isolates recovered from human. Also, since there was a high similarity in the genotyping pattern of H. pylori DNA among milk and meat samples and human specimens of other investigations, it is suggested that raw milk and meat samples are the sources of the bacteria and that they entered the human population in period of time. On the other hand, diversity of H. pylori genotypes between milk and meat samples with the clinical isolation of other studies suggested that consumption of contaminated milk and meat with H. pylori strains may be a threat to human health.
Acknowledgments
The authors would like to thank Dr. F. Safarpoor Dehkordi at the Department of Food Hygiene and Quality Control, University of Tehran, Tehran, Iran, for his important technical and clinical support.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Momtaz H., Dabiri H., Souod N., Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterology. 2014;14, article 61 doi: 10.1186/1471-230x-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yahaghi E., Khamesipour F., Mashayekhi F., et al. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. BioMed Research International. 2014;2014:11. doi: 10.1155/2014/757941.757941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahimi E., Kheirabadi E. K. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathogens and Disease. 2012;9(5):453–456. doi: 10.1089/fpd.2011.1060. [DOI] [PubMed] [Google Scholar]
- 4.Dore M. P., Sepulveda A. R., El-Zimaity H., et al. Isolation of helicobacter pylori from sheep—implications for transmission to humans. The American Journal of Gastroenterology. 2001;96(5):1396–1401. doi: 10.1016/s0002-9270(01)02335-8. [DOI] [PubMed] [Google Scholar]
- 5.Mousavi S., Dehkordi F., Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. Journal of Venomous Animals and Toxins including Tropical Diseases. 2014;20, article 51 doi: 10.1186/1678-9199-20-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webberley M. J., Webberley J. M., Newell D. G., Lowe P., Melikian V. Seroepidemiology of Helicobacter pylori infection in vegans and meat-eaters. Epidemiology and Infection. 1992;108(3):457–462. doi: 10.1017/s0950268800049967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X.-G., Chua A., Li T.-G., Zeng Q.-S. Survival of Helicobacter pylori in milk and tap water. Journal of Gastroenterology and Hepatology. 1998;13(11):1096–1098. doi: 10.1111/j.1440-1746.1998.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 8.Husson M.-O., Vincent P., Grabiaud M.-H., Furon D., Leclerc H. Anti-Helicobacter pylori IgG levels in abattoir workers. Gastroentérologie Clinique et Biologique. 1991;15(10):723–726. [PubMed] [Google Scholar]
- 9.Jafari F., Shokrzadeh L., Dabiri H., et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Japanese Journal of Infectious Diseases. 2008;61(4):290–293. [PMC free article] [PubMed] [Google Scholar]
- 10.Atherton J. C., Cao P., Peek R. M., Jr., Tummuru M. K. R., Blaser M. J., Cover T. L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. The Journal of Biological Chemistry. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 11.Momtaz H., Souod N., Dabiri H. Comparison of the virulence factors of Helicobacter pylori isolated in stomach and saliva in Iran. The American Journal of the Medical Sciences. 2010;340(5):345–349. doi: 10.1097/maj.0b013e3181d94fbc. [DOI] [PubMed] [Google Scholar]
- 12.Torres L. E., Melián K., Moreno A., et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World Journal of Gastroenterology. 2009;15(2):204–210. doi: 10.3748/wjg.15.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momtaz H., Souod N., Dabiri H., Sarshar M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World Journal of Gastroenterology. 2012;18(17):2105–2111. doi: 10.3748/wjg.v18.i17.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho S.-A., Hoyle J. A., Lewis F. A., et al. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. Journal of Clinical Microbiology. 1991;29(11):2543–2549. doi: 10.1128/jcm.29.11.2543-2549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki S., Yamakawa A., Okuda T., et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. Journal of Clinical Microbiology. 2005;43(8):3906–3916. doi: 10.1128/jcm.43.8.3906-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quaglia N. C., Dambrosio A., Normanno G., et al. High occurrence of Helicobacter pylori in raw goat, sheep and cow milk inferred by glmM gene: a risk of food-borne infection? International Journal of Food Microbiology. 2008;124(1):43–47. doi: 10.1016/j.ijfoodmicro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura S., Kawamura T., Kato S., Tateno H., Watanabe A. Detection of Helicobacter pylori in cow's milk. Letters in Applied Microbiology. 2002;35(6):504–507. doi: 10.1046/j.1472-765x.2002.01229.x. [DOI] [PubMed] [Google Scholar]
- 18.Angelidis A. S., Tirodimos I., Bobos M., Kalamaki M. S., Papageorgiou D. K., Arvanitidou M. Detection of Helicobacter pylori in raw bovine milk by fluorescence in situ hybridization (FISH) International Journal of Food Microbiology. 2011;151(2):252–256. doi: 10.1016/j.ijfoodmicro.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Stevenson T. H., Bauer N., Lucia L. M., Acuff G. R. Attempts to isolate Helicobacter from cattle and survival of Helicobacter pylori in beef products. Journal of Food Protection. 2000;63(2):174–178. doi: 10.4315/0362-028x-63.2.174. [DOI] [PubMed] [Google Scholar]
- 20.Mhaskar R. S., Ricardo I., Azliyati A., et al. Assessment of risk factors of Helicobacter pylori infection and peptic ulcer disease. Journal of Global Infectious Diseases. 2013;5(2):60–67. doi: 10.4103/0974-777X.112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris A., Nicholson G., Lloyd G., Haines D., Rogers A., Taylor D. Seroepidemiology of Campylobacter pyloridis . The New Zealand Medical Journal. 1986;99(809):657–659. [PubMed] [Google Scholar]
- 22.Vaira D., Holton J., Londei M., et al. Campylobacter pylori in abattoir workers: is it a zoonosis? The Lancet. 1988;332(8613):725–726. doi: 10.1016/s0140-6736(88)90196-1. [DOI] [PubMed] [Google Scholar]
- 23.Linpisarn S., Suwan W., Lertprasertsuk N., et al. Helicobacter pylori cagA, vacA and iceA genotypes in northern Thai patients with gastric disease. Southeast Asian Journal of Tropical Medicine and Public Health. 2007;38(2):356–362. [PubMed] [Google Scholar]
- 24.López-Vidal Y., Ponce-de-León S., Castillo-Rojas G., Barreto-Zúñiga R., Torre-Delgadillo A. High diversity of vacA and cagA Helicobacter pylori genotypes in patients with and without gastric cancer. PLoS ONE. 2008;3(12) doi: 10.1371/journal.pone.0003849.e3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudi J., Kolb C., Maiwald M., et al. Diversity of Helicobacter pylori vaca and caga genes and relationship to vaca and caga protein expression, cytotoxin production, and associated diseases. Journal of Clinical Microbiology. 1998;36(4):944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver S. P., Boor K. J., Murphy S. C., Murinda S. E. Food safety hazards associated with consumption of raw milk. Foodborne Pathogens and Disease. 2009;6(7):793–806. doi: 10.1089/fpd.2009.0302. [DOI] [PubMed] [Google Scholar]
- 27.Robesyn E., De Schrijver K., Wollants E., Top G., Verbeeck J., Van Ranst M. An outbreak of hepatitis A associated with the consumption of raw beef. Journal of Clinical Virology. 2009;44(3):207–210. doi: 10.1016/j.jcv.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Bremer V., Bocter N., Rehmet S., Klein G., Breuer T., Ammon A. Consumption, knowledge, and handling of raw meat: a representative cross-sectional survey in Germany, March 2001. Journal of Food Protection. 2005;68(4):785–789. doi: 10.4315/0362-028x-68.4.785. [DOI] [PubMed] [Google Scholar]


