Abstract
Ampelopsis brevipedunculata is an economically important plant that belongs to the Vitaceae family of angiosperms. The phylogenetic placement of Vitaceae is still unresolved. Recent phylogenetic studies suggested that it should be placed in various alternative families including Caryophyllaceae, asteraceae, Saxifragaceae, Dilleniaceae, or with the rest of the rosid families. However, these analyses provided weak supportive results because they were based on only one of several genes. Accordingly, complete chloroplast genome sequences are required to resolve the phylogenetic relationships among angiosperms. Recent phylogenetic analyses based on the complete chloroplast genome sequence suggested strong support for the position of Vitaceae as the earliest diverging lineage of rosids and placed it as a sister to the remaining rosids. These studies also revealed relationships among several major lineages of angiosperms; however, they highlighted the significance of taxon sampling for obtaining accurate phylogenies. In the present study, we sequenced the complete chloroplast genome of A. brevipedunculata and used these data to assess the relationships among 32 angiosperms, including 18 taxa of rosids. The Ampelopsis chloroplast genome is 161,090 bp in length, and includes a pair of inverted repeats of 26,394 bp that are separated by small and large single copy regions of 19,036 bp and 89,266 bp, respectively. The gene content and order of Ampelopsis is identical to many other unrearranged angiosperm chloroplast genomes, including Vitis and tobacco. A phylogenetic tree constructed based on 70 protein-coding genes of 33 angiosperms showed that both Saxifragales and Vitaceae diverged from the rosid clade and formed two clades with 100% bootstrap value. The position of the Vitaceae is sister to Saxifragales, and both are the basal and earliest diverging lineages. Moreover, Saxifragales forms a sister clade to Vitaceae of rosids. Overall, the results of this study will contribute to better support of the evolution, molecular biology and genetic improvement of the plant Ampelopsis.
Keywords: Porcelain berry, Ampelopsis brevipedunculata, Vitaceae, chloroplast genome, basal lineage of rosids
Introduction
Flowering plants are the largest clade in the land plants, containing more than 250,000 species (Friis et al., 2006). Within the land plants, the eudicot clade comprises approximately 75% of all flowering plant species, including several major subclades; namely, rosids, asterids, Saxifragales, Santalales, and Caryophyllales (The angiosperm phylogeny group II [APG II], 2003; Judd and Olmstead, 2004; Soltis et al., 2005). Among these, rosid is the largest major clade of core eudicots, comprising 140 families and 70,000 species that include nearly one third of all angiosperms (Magallón-Puebla et al., 1999; Soltis et al., 2005; Jansen et al., 2006). Due to rapid radiation, angiosperms show extraordinary diversity in habit, morphology, anatomy, physiology, and reproductive biology (Friis et al., 2006). This distinction in flowering plants has offered key challenges to evolutionary biologists investigating the origin and evolution of their traits, and determining these issues precisely depends on having a well agreed upon and strongly supported phylogenetic framework. Over the past three decades, several morphological and molecular phylogenetic studies have been used to access the relationships among the major clades, resulting in angiosperms being classified into 59 orders and 413 families (The angiosperm phylogeny group III [APG III], 2009).
In earlier studies, molecular phylogenetic analyses of flowering plants were analyzed based on one to several genes of the chloroplast (cp), mitochondrial, and nuclear genomes, but most of this analysis was based on chloroplast marker genes (Jansen et al., 2006). The relationships among many of the major lineages of angiosperms have been resolved by these efforts; however, the relationship between angiosperms and seed plants are still unclear (Friis et al., 2006). Burleigh and Mathews (2004) reported that based on rootings of phylogenetic tree analysis of DNA sequences data suggested that angiosperms are sister group to all other seed plants, whereas, Ginkgo and cycads separate angiosperm groups. Though, it is not straight forward to identify the position of the root in this tree. So, this pattern of relationships is difficult to interpret in evolutionary terms as it conflicts with stratigraphic evidence (Burleigh and Mathews, 2004). The complete cp genome sequence analyses have resolved problematic deep level relationships in the angiosperms (Goremykin et al., 2003, 2004, 2005; Leebens-Mack et al., 2005; Chang et al., 2006; Jansen et al., 2006), implying that Chloranthaceae and magnoliids are sister to a clade of monocots and eudicots plus Ceratophyllaceae (Jansen et al., 2007; Moore et al., 2007). Similarly, improvements have been made in elucidating relationships within the larger monocot (Graham et al., 2006) and asterid (Bremer et al., 2002) clades. Also, cp genome sequences have been widely used for plant identification, phylogenetic studies and to increase phylogenetic resolution at low taxonomic levels (Parks et al., 2009). Hence, there is rising interest in increasing the analysis of complete chloroplast genome sequences and emerging evolutionary models for phylogenetic analysis of cp sequences to address these problems (Ané et al., 2005; Jansen et al., 2006).
Despite these achievements, the position of rosids is still the least resolved major clade in the angiosperms (Soltis and Soltis, 2004). The relationships among Vitaceae are indistinct in angiosperms, and the family does not appear to have any close relatives to other families of rosids (Soltis et al., 2000). In the Cronquist system, the Vitaceae family was placed near Rhamnaceae (Cronquist, 1988). Previous studies also reported that basal nodes with the core eudicot clade have constantly received low internal support (Judd and Olmstead, 2004; Soltis et al., 2005, 2007; Schönenberger and von Balthazar, 2006), and the phylogenetic position of Vitaceae has been controversial for many years. Earlier molecular phylogenetic analyses were conducted using one to four genes, suggesting weak support for the placement of Vitaceae as a sister to asterids (Chase et al., 1993), Caryophylales (Chase et al., 1993), Dilleniaceae (Hilu et al., 2003), rosids (Savolainen et al., 2000; Soltis et al., 2000, 2003), or Saxifragales (Savolainen et al., 2000). Moreover, the latest Angiosperm Phylogeny Group (APG) III in 2009 reported that Vitaceae has its own order, Vitales (Green and Martin, 2013). Jansen et al. (2006) reported that Vitaceae was a sister-group to all other rosids based on phylogenetic analyses. Although, analyses agree on the composition of the rosid clade, the relationships within the rosids remain unclear (Wang et al., 2009).
Here, we report the complete cp genome sequence of Ampelopsis brevipedunculata for the first time. In addition to describing the structure of the cp genome, we provide the comparative analyses of the cp genome sequences of its closely related species of rosids. We also present the results of phylogenetic analyses of DNA sequences for 70 protein-coding genes from Ampelopsis and 32 angiosperm cp genomes, including 18 in the rosids clade. The phylogenetic analyses enabled elucidation of the relationships and placement of Vitaceae to other major lineages of rosids and show the importance of taxon sampling. The complete cp genome sequence of Ampelopsis also provides valuable data useful to chloroplast genetic engineering of this economically important medicinal and ornamental plant.
Materials and Methods
DNA Extraction and Sequencing
Total genomic DNA was extracted from fresh young leaves of the A. brevipedunculata plant using a modified CTAB Method (Doyle and Doyle, 1990). The high quality DNA was sequenced using an Illumina NextSeq 500 (LabGenomics, South Korea). The pair-end library was constructed with an insert size of ∼101 bp. Sequence trimming, assembly and mapping were performed using Genious v7.1.9 (Biomatters, New Zealand). The chloroplast genome reads were aligned to its closest cpDNA sequence of Vitis (GenBank accession number: NC_007957). The consensus sequences were extracted and gaps were filled by polymerase chain reaction (PCR) amplification using specific primers based on the gap between sequences. The PCR products were purified and sequenced by conventional Sanger sequencing. The sequencing data and gene annotation were submitted to GenBank and assigned an accession number of KT831767.
Annotation and Genome Analysis of the Ampelopsis Chloroplast Genome
The initial annotation of the chloroplast genome was conducted using a Dual Organeller GenoMe Annotator (DOGMA; Wyman et al., 2004). From this initial annotation, putative starts, stops, and intron positions were identified based on comparisons to homologous genes of Vitis, Liquidambar, and Nicotiana tabacum. Further, the identified tRNAs were confirmed with tRNAscan-SE 1.21 (Schattner et al., 2005). A circle cp genome map was drawn using the OGDRAW program (Lohse et al., 2009).
Comparative Genome Analysis of the Ampelopsis cp Genome
The complete cp genome of Ampelopsis was compared with that of four other species, Vitis, Liquidambar, Penthorum and N. tabacum, using the mVISTA program in a Shuffle-LAGAN mode (Frazer et al., 2004). Ampelopsis was set as a reference.
Analysis of Tandem Repeats and Single Sequence Repeats (SSRs)
The presence of tandem repeats with more than 30 bp and a minimum of 90% sequence identity was also analyzed. PHOBOS v3.3.12 was used to identify tandem repeats and single sequence repeats (SSRs). The analysis parameters of alignment scores for the match, mismatch, gap, and N positions were set as 1, -5, -5, and 0, respectively (Mayer et al., 2010).
Synonymous (KS) and Non-synonymous (KA) Substitution Rate Analysis
The Ampelopsis cp genome sequence was compared with the cp genome sequences of Vitis, Liquidambar, and Penthorum. To analyze synonymous (KS) and non-synonymous (KA) substitution rates, the same individual functional protein-coding exons were extracted and aligned separately using Geneious v7.1.9. These aligned sequences were translated into protein sequences and analyzed. The synonymous (KS) and non-synonymous (KA) substitution rates for each protein-coding exon were estimated in DnaSP (Librado and Rozas, 2009).
Phylogenetic Analysis
In this study, genome model have been selected based on closely related to each other families of eudicots and also from previously reported studies in the literatures. A molecular phylogenetic tree was constructed using 70 protein-coding genes of 33 angiosperms. Among these 33 taxa, Nelumbo was set as the outgroup. The 33 completed cp genome sequences representing the lineages of angiosperms were downloaded from the NCBI Organelle Genome Resource database (Supplementary Table S1). The 70 protein-coding gene sequences were aligned using MAFFT v7.017 (Katoh and Standley, 2013) through Geneious v7.1.9. The aligned protein-coding gene sequences were saved in PHYLIP format using Clustal X v2.1 (Larkin et al., 2007) and used to generate a phylogenetic tree. Phylogenetic analysis was conducted based on maximum likelihood (ML) analysis using the general time-reversible invariant-sites (GTRI) nucleotide substitution model with the default parameters in RAxML v. 7.2.6 (Stamatakis et al., 2008). The bootstrap probability of each branch was calculated by 1000 replications.
Results
Ampelopsis cp Genome Assembly, Organization, and Gene Content
Overall, 50,269,822 paired-end reads (101 × 2) with an insert size of ∼101 bp were constructed and 2,688,617 reads were generated using 1,646,907,308 base pairs. De novo assembly was performed using Geneious v7.1.9. The generated contigs were assembled using the cpDNA genome sequence of Vitis vinifera as a reference and gaps were filled by Sanger sequencing.
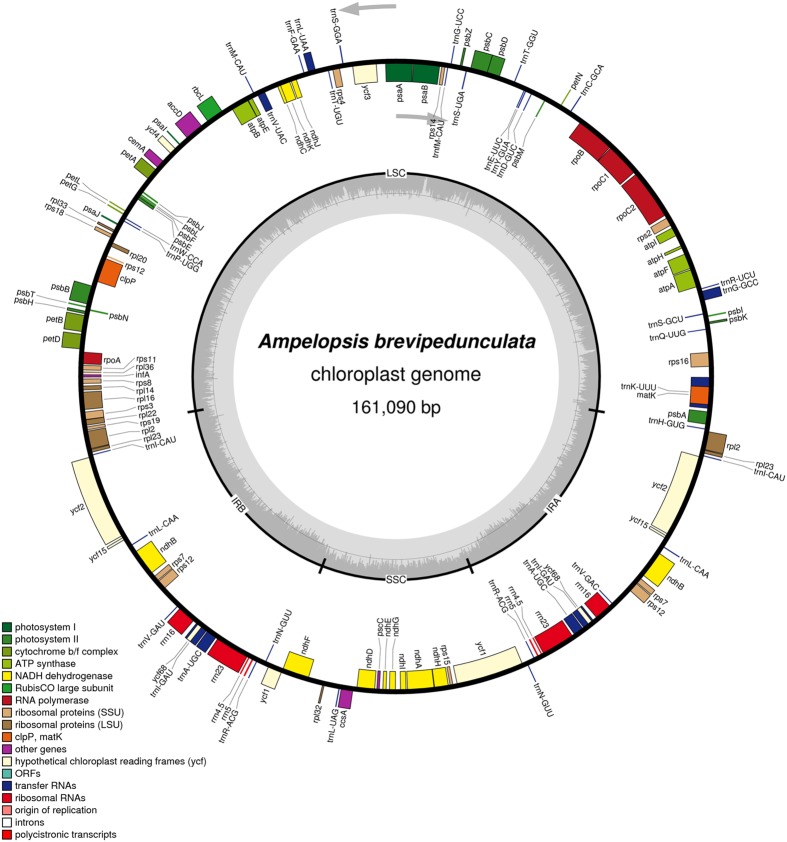
The complete cp genome sequence of A. brevipedunculata (KT831767) is 161,090 bp and shows a characteristic circular structure, including a pair of IRs (26,394 bp each) that divide the genome into two single-copy regions (LSC 89,266; SSC 19,036 bp; Figure 1). Coding regions (92,772 bp), comprising protein-coding genes (80,943 bp), tRNA genes (2,795 bp) and rRNA genes (9,034 bp) account for 57.59% of the genome, whereas non-coding regions (68,318 bp), including introns (16,931 bp) and intergenic spacers (51,387 bp), account for the remaining 42.41% of the genome. The overall A+T content of the whole genome is 62.6% (Table 1).
FIGURE 1.
Gene map of the Ampelopsis brevipedunculata. Genes lying outside of the outer layer circle are transcribed in the counterclockwise direction, whereas genes inside of this circle are transcribed in the clockwise direction. The colored bars indicate known protein-coding genes, tRNA genes and rRNA genes. The dashed darker gray area in the inner circle denotes GC content, while the lighter gray area indicates the AT content of the genome. LSC, large-single-copy; SSC, small-single-copy; IR, inverted repeat.
Table 1.
Summary of chloroplast genome characteristics of Vitaceae.
| Genome features | Ampelopsis brevipedunculata | Vitis vinifera |
|---|---|---|
| Size (bp) | 161,090 | 160,928 |
| LSC length (bp) | 89,266 | 89,147 |
| SSC length (bp) | 19,036 | 19,065 |
| IR length (bp) | 26,394 | 26,358 |
| Number of genes | 113 | 113 |
| Protein-coding genes | 79+6 | 79+6 |
| tRNA genes | 30+7 | 30+7 |
| rRNA genes | 4+4 | 4+4 |
| Number of genes duplicated in IR | 18 | 18 |
| GC content (%) | 37.4 | 37.4 |
There are a total of 131 genes in the genome, including 79 protein-coding genes, 30 tRNA genes, four ribosomal RNA genes and 18 duplicated genes (Figure 1; Table 2). Of the 18 duplicated genes in the IR region, seven are protein-coding, seven are tRNA and four are rRNA genes. Eighteen genes contain introns (one class I intron, trnLUAA and 17 class II introns), and three of these genes clpP, rps12, and ycf3, contain two introns (Table 3). The 5′-end exon of the rps12 gene is located in the LSC region, and the intron 3′-end exon of the gene is situated in the IR region. Overall, the gene order in the Ampelopsis chloroplast genome is identical to that of Vitis and tobacco.
Table 2.
List of genes present in Ampelopsis chloroplast genome.
| Category | Gene group | Gene name | ||||
|---|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5a | rrn5a | rrn16a | rrn23a | |
| Transfer RNA genes | trnA-UGCa,b | trnC-GCA | trnD-GUC | trnE-UUC | trnF-GAA | |
| trnfM-CAU | trnG-GCC | trnG-UCCb | trnH-GUG | trnI-CAUa | ||
| trnI-GAUa,b | trnK-UUUb | trnL-CAAa | trnL-UAAb | trnL-UAG | ||
| trnM-CAU | trnN-GUUa | trnP-UGG | trnQ-UUG | trnR-ACGa | ||
| trnR-UCU | trnS-GCU | trnS-GGA | trnS-UGA | trnT-GGU | ||
| trnT-UGU | trnV-GACa | trnV-GAU | trnV-UACb | trnW-CCA | ||
| trnY-GUA | ||||||
| Small subunit of ribosome | rps2 | rps3 | rps4 | rps7a | rps8 | |
| rps11 | rps12a,c,d | rps14 | rps15 | rps16b | ||
| rps18 | rps19 | |||||
| Large subunit of ribosome | rpl2a | rpl14 | rpl16b | rpl20 | rpl22 | |
| rpl23 | rpl32 | rpl33 | rpl36 | |||
| DNA-dependent RNA polymerase | rpoA | rpoB | rpoC1b | rpoC2 | ||
| Translational initiation factor | infA | |||||
| Genes for photosynthesis | Subunits of photosystem I | psaA | psaB | psaC | psaI | psaJ |
| ycf3c | ycf4 | |||||
| Subunits of photosystem II | psbA | psbB | psbC | psbD | psbE | |
| psbF | psbH | psbI | psbJ | psbK | ||
| psbL | psbM | psbN | psbT | psbZ | ||
| Subunits of cytochrome | petA | petBb | petDb | petG | petL | |
| petN | ||||||
| Subunits of ATP synthase | atpA | atpB | atpE | atpFb | atpH | |
| atpI | ||||||
| Large subunit of Rubisco | rbcL | |||||
| Subunits of NADH dehydrogenase | ndhAb | ndhBa,b | ndhC | ndhD | ndhE | |
| ndhF | ndhG | ndhH | ndhI | ndhJ | ||
| ndhK | ||||||
| Other genes | Maturase | matK | ||||
| Envelope membrane protein | cemA | |||||
| Subunit of acetyl-CoA | accD | |||||
| C-type cytochrome synthesis gene | ccsA | |||||
| Protease | clpPc | |||||
| Component of TIC complex | ycf1a | |||||
aTwo gene copies in IRs; bgene containing a single intron; cgene containing two introns; dgene divided into two independent transcription units.
Table 3.
Location and length of intron-containing genes in the Ampelopsis chloroplast genome.
| Gene∗ | Location | Exon I | Intron I | Exon II | Intron II | Exon III |
|---|---|---|---|---|---|---|
| Nucleotides in base pairs | ||||||
| atpF | LSC | 144 | 747 | 414 | ||
| clpP | LSC | 71 | 817 | 292 | 634 | 228 |
| ndhA | SSC | 552 | 1132 | 540 | ||
| ndhB | IR | 777 | 679 | 756 | ||
| petB | LSC | 6 | 695 | 642 | ||
| petD | LSC | 8 | 731 | 475 | ||
| rps12# | LSC | 114 | – | 232 | 536 | 26 |
| rpl2 | IR | 390 | 674 | 435 | ||
| rpl16 | LSC | 9 | 1068 | 399 | ||
| rpoC1 | LSC | 432 | 763 | 1617 | ||
| rps16 | LSC | 40 | 909 | 236 | ||
| trnG-GCC | LSC | 23 | 707 | 37 | ||
| trnA-UGC | IR | 38 | 803 | 35 | ||
| trnI-GAU | IR | 42 | 950 | 35 | ||
| trnK-UUU | LSC | 37 | 2512 | 29 | ||
| trnL-UAA | LSC | 37 | 516 | 50 | ||
| trnV-UAC | LSC | 39 | 574 | 37 | ||
| ycf3 | LSC | 126 | 739 | 228 | 745 | 153 |
∗Identical duplicate gene containing introns in the IR region are not included. #The rps12 is a trans-spliced gene with the 5′ end located in the LSC region and duplicated in the 3′ end in the IR regions.
Comparative Analysis of the Ampelopsis Chloroplast Genome
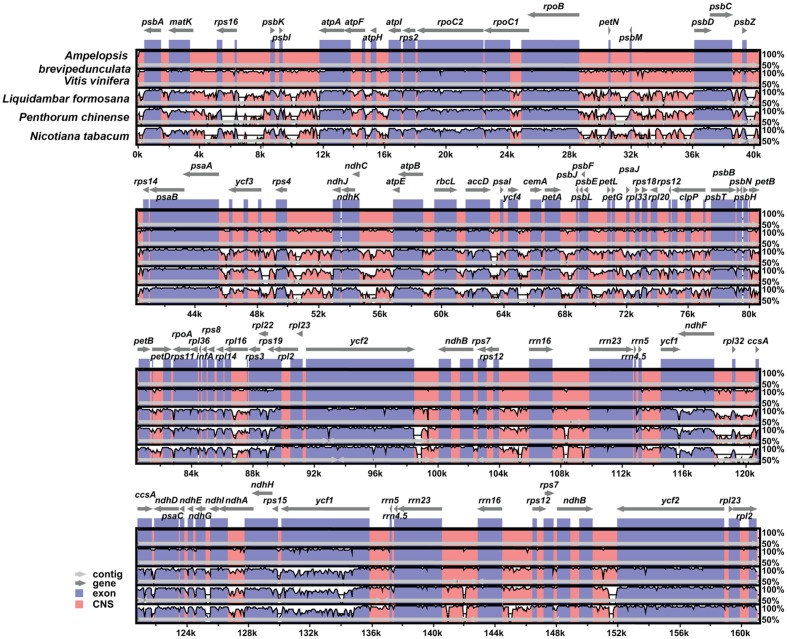
mVISTA was used to study the cp genome sequence variations in the orders of Vitales and Saxifragales, as well as in Nicotiana. The coding region was found to be more highly conserved than the non-coding regions (Figure 2), and the most dissimilar coding regions of the five chloroplast genomes were rpl22, rps19, and ycf1.
FIGURE 2.
Comparison of the cp genome sequence of Ampelopsis brevipedunculata, Vitis vinfera, Liquidambar formosana, Penthorum chinense, and Nicotiana tabacum generated with mVista. Gray arrows indicate the position and direction of each gene. Red and blue areas indicate intergenic and genic regions, respectively. Black lines define regions of sequence identity with A. brevipedunculata, using a 50% identity cutoff. Dashed rectangles denote highly divergent regions of A. brevipedunculata compared to V. vinfera, L. formosana, P. chinense, and N. tabacum.
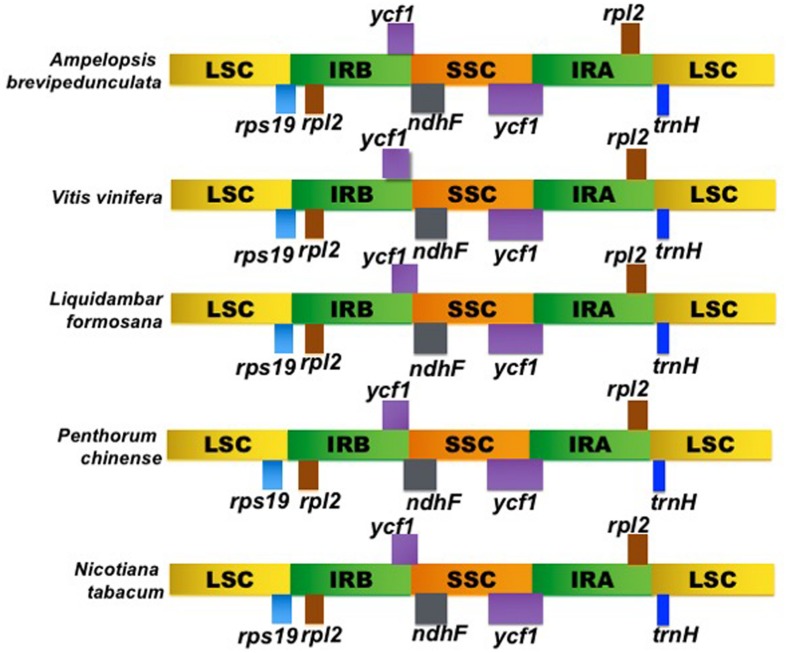
The LSC/IRB/SSC/IRA boundary regions of the Ampelopsis cp genome were compared to the corresponding regions of its four closely related cp genomes, Vitis, Liquidambar, Penthorum, and Nicotiana (Figure 3). The rps19 gene of Ampelopsis, Vitis and Liquidambar was extended from the IRB to the LSC region with 7–37 bp variability. However, the rps19 gene of Penthorum and Nicotiana was shifted to an LSC region with a 2–15 bp gap. At the IRB/SSC boundary, the ycf1 and ndhF genes were overlapped in Ampelopsis, Vitis, Liquidambar, Penthorum, and Nicotiana. Expansion, contraction, and shifting of the ycf1 gene was observed in the boundary regions of the SSC/IRA. The size variation of ycf1 from 5172 to 5682 bp was identified in all cp genomes. The trnH gene was located in the LSC region of all genomes, but varied from 0 to 21 bp apart from the IRA/LSC junctions. When compared with other closely related cp genomes of Vitis, the Ampelopsis was found to have very little size differences in the LSC, IR, and SSC regions.
FIGURE 3.
Comparison of the borders of the LSC, SSC, and IR regions of Ampelopsis brevipedunculata, Vitis vinfera, Liquidambar formosana, Penthorum chinense, and Nicotiana tabacum cp genomes.
Repeat Sequence Analysis
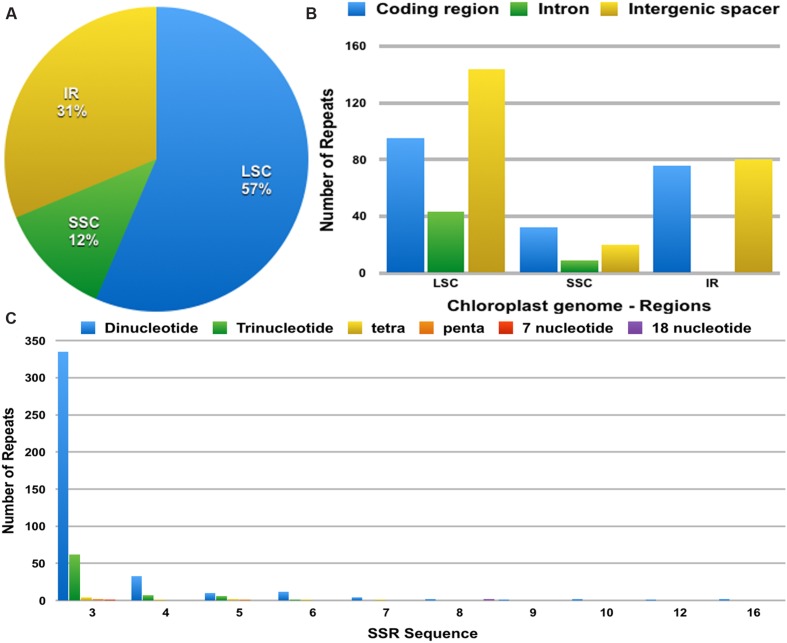
The distribution, type and presence of simple sequence repeats (SSR) or microsatellites was studied in the cp genome of Ampelopsis. A total of 493 SSRs were identified (Figure 4). Of these, 282 were found in the LSC regions, whereas 156 and 61 were in the IR and SSC regions, respectively (Figure 4A). Moreover, 203 SSRs were found in the protein-coding regions, 235 were in intergenic spacers and 61 in the introns of the Ampelopsis cp genome (Figure 4B). Among these SSRs, dipolymers were most common, accounting for 81.5%, while tripolymers accounted for 15.42%, and tri-, tetra-, penta-, 7-nucleotide, and 18-nucleotide polymers occurred with less frequency (Figure 4C). Moreover, three penta-, one 7-nuclelotide and two 18-nucleotide polymers were detected in the cp genome. The size and location of tetra-, penta-, 7-nucleotide and 18-nucleotide polymers are shown in Table 4. A total of 17 polymers was identified in the genome, whereas 11 were localized in intergenic spacers, six in coding regions and none in introns.
FIGURE 4.
The distribution, type and presence of simple sequence repeats (SSRs) in the cp genome of Ampelopsis brevipedunculata. (A) Presence of SSRs in the LSC, SSC, and IR regions. (B) Presence of SSRs in the protein-coding regions, intergenic spacers and introns of LSC, SSC, and IR regions. (C) Presence of polymers in the cp genome of A. brevipedunculata.
Table 4.
Distribution of tetra, penta, and hexapolymer single sequence repeats (SSRs) in Ampelopsis chloroplast genome.
| SSR type | SSR sequence | SSR size (bp) | Start | End | Location |
|---|---|---|---|---|---|
| Tetra | (AAAT)3 | 12 | 52,799 | 52,810 | trnF-GAA/ndhJ (IGS) |
| Tetra | (AAAT)3 | 12 | 126,739 | 126,750 | ndhA (CDS) |
| Tetra | (AATC)3 | 12 | 127,487 | 127,498 | ndhA (CDS) |
| Tetra | (AGAT)3 | 13 | 31,776 | 31,788 | petN/psbM (IGS) |
| Tetra | (AAAG)3 | 13 | 127,046 | 127,058 | ndhA (CDS) |
| Tetra | (AATC)3 | 14 | 68,471 | 68,484 | petA/psbJ (IGS) |
| Tetra | (AGAT)4 | 17 | 1627 | 1643 | psbA/matK (IGS) |
| Tetra | (AAAT)5 | 22 | 104,318 | 104,339 | rps12/trnV-GAU (IGS) |
| Tetra | (AATT)5 | 23 | 54,955 | 54,977 | ndhC/trnV-UAC (IGS) |
| Tetra | (AAAT)6 | 26 | 146,014 | 146,039 | trnV-GAC/rps12 (IGS) |
| Tetra | (AAAG)7 | 29 | 47,086 | 47,114 | ycf3 (CDS) |
| Penta | (AATAT)3 | 15 | 55,700 | 55,714 | ndhC/trnV-UAC (IGS) |
| Penta | (AAAAT)3 | 15 | 70,508 | 70,522 | psbE/petL (IGS) |
| Penta | (AATAT)5 | 27 | 31,699 | 31,725 | petN/psbM (IGS) |
| 7-nucleotide | (AAAAAAT)3 | 21 | 14,750 | 14,770 | atpF/atpH (IGS) |
| 18-nucleotide | (AATATCGTCACTAGCATC) | 78 | 96,562 | 96,639 | ycf2 (CDS) |
| 18-nucleotide | (AATATCGTCACTAGCATC) | 78 | 153,718 | 153,795 | ycf2 (CDS) |
A total of 11 tandem repeats were identified in the cp genome of Ampelopsis (Table 5). Of these, six were present in the intergenic spacers of atpH/atpI (1), rpoB/trnC-GCA (1), psbM/trnD-GUA (1), trnE-UUC/trnT-GGU (1), psaA/ycf3 (1) and ndhF-rpl32 (1), three were located in the protein-coding regions of accD (1) and ycf2 (2) and two were present in the intron and exon of ycf3.
Table 5.
Distribution of tandem repeats in Ampelopsis chloroplast genome.
| S. no. | Repeat length (bp) | Consensus size × copy number | Start | End | Location |
|---|---|---|---|---|---|
| 1 | 30 | 15 × 2 | 15,791 | 15,832 | atpH/atpI (IGS) |
| 2 | 40 | 20 × 2 | 28,961 | 29,000 | rpoB/trnC-GCA (IGS) |
| 3 | 45 | 21 × 2 | 32,738 | 32,782 | psbM/trnD-GUA (IGS) |
| 4 | 40 | 16 × 2 | 34,723 | 34,762 | trnE-UUC/trnT-GGU (IGS) |
| 5 | 30 | 15 × 2 | 45,933 | 45,962 | psaA/ycf3 (IGS) |
| 6 | 35 | 16 × 2 | 47,890 | 47,924 | ycf3 (exon and intron) |
| 7 | 35 | 17 × 2 | 61,967 | 62,014 | accD (CDS) |
| 8 | 48 | 24 × 2 | 46,377 | 46,406 | ycf3 (exon and intron) |
| 9 | 78 | 18 × 4 | 96,562 | 96,639 | ycf2 (CDS) |
| 10 | 37 | 17 × 2 | 119,000 | 119,036 | ndhF-rpl32 (IGS) |
| 11 | 78 | 18 × 4 | 153,718 | 153,795 | ycf2 (CDS) |
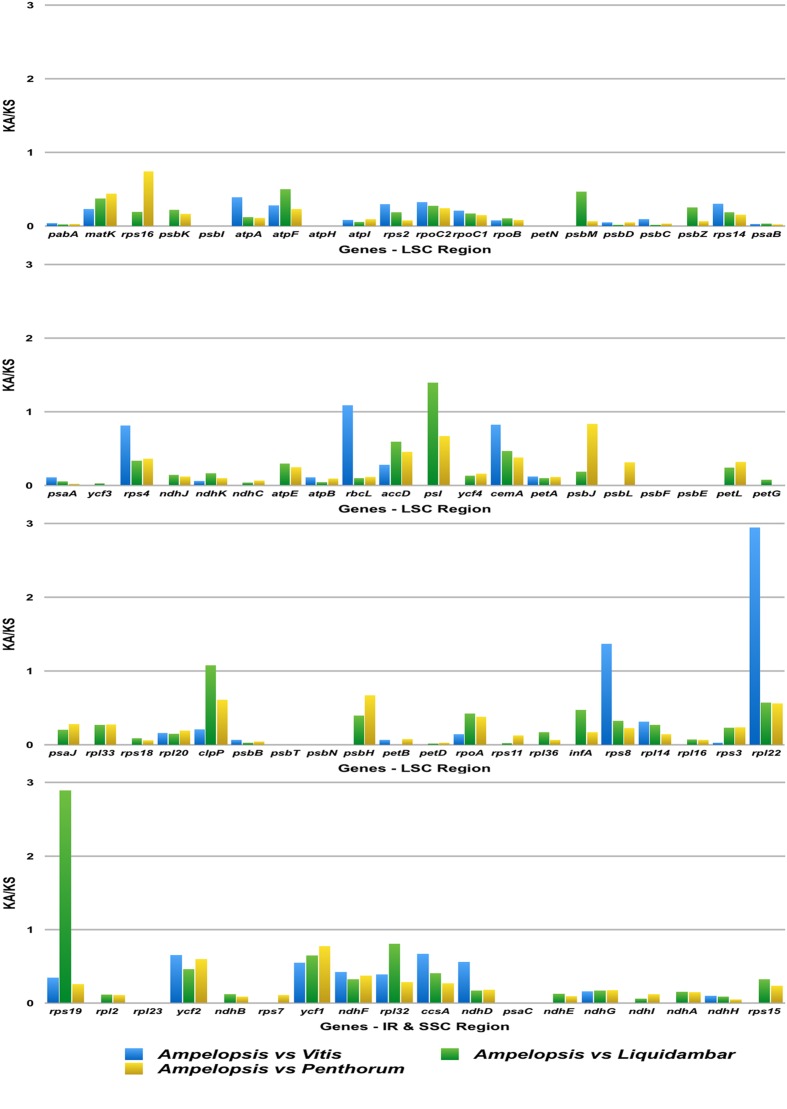
Synonymous (KS) and Non-synonymous (KA) Substitution Rate Analysis
A total of 78 genes encoding 91 protein-coding exons in the cp genome of Ampelopsis were used to analyze synonymous and non-synonymous rates against Vitis, Liquidambar, and Penthorum (Figure 5). The KA/KS ratio of all genes was less than 1, except for rpl22 of Lychnis. The KA/KS ratio of rpl22 of Ampelopsis vs. Vitis was 2.95, while rps19 of Ampelopsis vs. Liquidambar was 2.89.
FIGURE 5.
KA/KS values of 78 protein-coding genes of Ampelopsis brevipedunculata, Vitis vinfera, Liquidambar formosana, and Penthorum chinense. Blue color boxes indicate the KA/KS ratio of the A. brevipedunculata vs. V. vinfera, Green color boxes indicate A. brevipedunculata vs. L. formosana and Yellow color boxes indicate A. brevipedunculata vs. P. chinense.
Phylogenetic Analysis
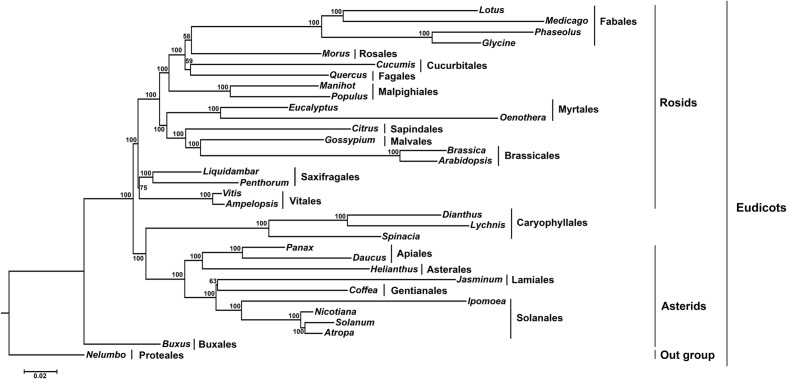
In this study, we analyzed the relationship between Vitales and Rosids. The reconstructed phylogeny showed that it divided into two clades, rosids, and asterids (Figure 6). Within asterids, Caryophyllales (core eudicots) deviated from asterids and formed two sister clades with a 100% bootstrap (BS) value. In another major rosid clade, Saxifragales, and Vitaceae diverged from rosids and formed two sister clades with 100% BS value. These two clades are the earliest diverging lineages of rosids, and Saxifragales forms a sister clade to Ampelopsis and Vitis (Vitaceae) with 75% BS value.
FIGURE 6.
Molecular phylogenetic tree of 33 angiosperms based on 70 protein-coding genes in the cp genome. The tree was constructed by maximum likelihood (ML) analysis of the conserved regions using the RaxML program and the GTR+I nucleotide model. The stability of each tree node was tested by bootstrap analysis with 1000 replicates. Bootstrap values are indicated on the branches, and the branch length reflects the estimated number of substitutions per 1000 sites. Nelumbo was set as the outgroup.
Discussion
Most angiosperms commonly encode 74 protein-coding genes, while an additional five are present in only some species (Millen et al., 2001). However, the Ampelopsis cp genome has 79 protein-coding genes, 30 tRNA genes and four ribosomal RNA genes, which is similar to Vitis and Saxifragales. This might have been because the genome shares its gene contents with the Vitaceae family and its closest relative, the Saxifragales family. Moreover, the total numbers of introns in the plastid are the same in this family and Saxifragales. Several lineages of angiosperms have lost introns from the rpl2 gene independently (Downie et al., 1991), which could also be considered a characteristic feature of the core members of Caryophyllales (Logacheva et al., 2008). However, the Vitaceae family has not lost any introns in the genes.
The cp genome size of Ampelopsis was compared with that of V. vinifera. Both genomes showed a similar genome size. The detected variation in sequence length between these two genomes is only 162 bp, which might be due to the insertion of sequences in the non-coding region of Ampelopsis. Among plant lineages, the genome size varies due to the expansion and contraction of the border regions between the IR regions and the single-copy regions (Wang and Messing, 2011). Hence, in this study, the exact IR border positions and their adjacent genes of one Vitaceae, two Saxifrageles and one Nicotiana cp genomes were compared. The organization of the Ampelopsis genome with a pair of IR regions separated by the SSC and LSC regions is identical to most sequenced angiosperm chloroplast genomes. The photosynthetic dicot cp genome size of angiosperms varies from 150,519 bp (Lotus; Kato et al., 2000) to 162,686 bp (Amborella; Goremykin et al., 2003), whereas the size of the Ampelopsis cp genome (161,090 bp) is also within this size range. The size of the IR region (26,394 bp) of the Ampelopsis is also well within the size range of other sequenced dicot genomes that range from 23,302 (Calycanthus; Goremykin et al., 2003) to 27,807 bp (Oenothera; Hupfer et al., 2000). However, the gene content and order of the Ampelopsis cp genome is exactly the same as that of Vitis, tobacco and many other unreorganized angiosperm cp genomes. Previous studies also reported that several rosid cp genomes have lost the rpl22 gene, including legumes (Spielmann et al., 1988; Milligan et al., 1989; Gantt et al., 1991; Doyle et al., 1995; Saski et al., 2005). At least two independent losses of rpl22 have occurred in the rosids (Jansen et al., 2006). However, multiple independent genes infA (Millen et al., 2001), rps16 (Doyle et al., 1995), and accD (Downie and Palmer, 1992; Cosner et al., 1997) have also been lost from the angiosperms. Taken together, these results indicate that gene losses are not always dependable markers of phylogenetic relationships.
Remarkably, the ACG start codon was found in ndhD and psbL of the Ampelopsis cp genome. Earlier studies also showed that, due to RNA editing during the translation process, the ACG start codon of rps19 has been converted into an initiation codon, AUG, in Nicotiana and Dianthus (Neckermann et al., 1994; Raman and Park, 2015). The same process might also have occurred in these two genes of the Ampelopsis cp genome. This evidence indicates that the evolutionary rates of cp genomes in Vitaceae are comparatively mild based on the relatively minor variations in the IR regions. High sequence polymorphisms are frequently observed in closely related species of land plants and considered as it is highly conserved regions in the chloroplast genome (Wicke et al., 2011). The occurrence of several SSR sites in the Ampelopsis cp genome showed that these sites can be used to estimate the intraspecific level of polymorphism leading to very sensitive phylogeographic and population structure studies of this species.
The synonymous and non-synonymous nucleotide substitution patterns are very important markers in gene evolution studies (Kimura, 1983). Non-synonymous nucleotide substitutions have occurred less frequently than synonymous substitutions, and the ratio of KA/KS was less than one in most protein-coding regions (Makalowski and Boguski, 1998). In this study, the ratio of KA/KS was significantly less than one in all protein-coding regions of Ampelopsis, except for two genes. Nevertheless, the KA/KS ratio of rpl22 Ampelopsis vs. Vitis and rps19 of Ampelopsis vs. Liquidambar was 2.95 and 2.89, respectively. When compared with gene rpl22 Ampelopsis vs. Vitis, 10 amino acids were changed, as were nine amino acids in rps19 of Ampelopsis vs. Liquidambar. Most of the changes occurred in the second and third position of the codon rather than the first position. This fluctuation might have been due to non-synonymous substitution in the rpl22 and rps19 genes and is the result of silent mutation.
The eudicots are considered the largest clade of angiosperms, containing over 75% of the extant species (Soltis et al., 2005). Within eudicots, Nelumbo diverges first and forms a clade with Buxus. Buxus is a sister to a strongly supported eudicot clade that includes two discreetly to well-supported groups encompassing the rosids and asterids. Previous studies have clearly indicated that Ranunculus, proteales and Buxales are early diverging lineages of eudicots (Friis et al., 2011). While, recent molecular phylogenetic analysis revealed that Caryophyllales belongs to sister relationship with asterids (Jansen et al., 2006; Raman and Park, 2015).
In core eudicots, the rosid clade is well-supported, but the least resolved major clade, comprising more than 25% of all angiosperm species (Schönenberger and von Balthazar, 2006). Soltis et al. (2003) reported that Saxifragales are generally associated with rosids, though support is not high and the order has been linked with caryophyllids. Saxifragales are clearly the earliest diverging lineage of core eudicots (Soltis et al., 2005; Magallon, 2007). Recently, based on 24 plastid inverted repeats, 10 plastid, and 2 nuclear genes analysis clarified the internal relationships of rosids. This study suggested that rosids formed two major clades, Fabidae and Malvidae, and the addition of Saxifragales as a basal order and Vitaceae as sister to all other rosid clade (Wang et al., 2009). Based on these considerations, phylogenetic analysis was conducted based on 70 protein-coding genes of 33 angiosperms to understand the position of Ampelopsis in the eudicots. Molecular phylogenetic analysis showed that both Saxifragales and Vitaceae diverged from rosids and formed two separate clades within rosids with 100% BS value. Vitals (Ampelopsis and Vitis) are a sister clade to Saxifragales with a 75% moderate bootstrap value. However, according to The angiosperm phylogeny group III [APG III] (2009), the placement of Vitaceae is in its own order, while Vitales is in the eudicots (Green and Martin, 2013). Although phylogenetic studies support that Vitaceae is an early diverging member and forms a sister-group to all other rosids (Jansen et al., 2006), the analysis conducted in this study shows that Saxifragales and Vitaceae are the ancient early diverging members of the rosid clade and Saxifragales formed a sister relationship with Viataceae. The phylogenetic analysis based on these results strongly supports that Vitaceae is a sister to Saxifragales and that both are early diverging clades within the rosids. However, the relationship of Vitaceae with Saxifragales is equivocal.
Conclusion
In summary, the chloroplast genome of Ampelopsis was sequenced and characterized for the first time. The Ampelopsis genome shares the same overall organization and gene contents of most of the unreorganized angiosperm chloroplast genomes, including its closest species, Vitis. The LSC/IRB/SSC/IRA boundary regions of the Ampelopsis cp genome were compared to its closely related genomes and no intense variations were identified in Vitaceae. A phylogenetic tree constructed with 70 protein-coding genes of 33 angiosperms revealed strong support for the position of Saxifragales and Vitaceae as the basal and earliest diverging lineages. Moreover, the analysis indicated that Saxifragales forms a sister to Vitaceae of the rosids. Overall, the results of this study provide better support of the evolution and molecular biology of the plant, Ampelopsis, and will enable its genetic improvement.
Author Contributions
Conceived and designed the experiments: SP. Performed the experiments, analyzed the data, and wrote the paper: GR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a The National Institute of Biological Resources of Korea grant funded by the Korean Government (No. NIBR201631201), South Korea and the Research Center for Policy Suggestion of the Ministry of Educational Science (2014).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00341
References
- Ané C., Burleigh J. G., McMahon M. M., Sanderson M. J. (2005). Covarion structure in plastid genome evolution: a new statistical test. Mol. Biol. Evol. 22 914–924. 10.1093/molbev/msi076 [DOI] [PubMed] [Google Scholar]
- Bremer B., Bremer K., Heidari N., Erixon P., Olmstead R. G., Anderberg A. A., et al. (2002). Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol. Phylogenet. Evol. 24 274–301. 10.1016/S1055-7903(02)00240-3 [DOI] [PubMed] [Google Scholar]
- Burleigh J. G., Mathews S. (2004). Phylogenetic signal in nucleotide data from seed plants: implications for resolving the seed plant tree of life. Am. J. Bot. 91 1599–1613. 10.3732/ajb.91.10.1599 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Lin H. C., Lin I. P., Chow T. Y., Chen H. H., Chen W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23 279–291. 10.1093/molbev/msj029 [DOI] [PubMed] [Google Scholar]
- Chase M., Soltis D., Olmstead R., Morgan D., Les D., Mishler B., et al. (1993). Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann. Missouri Bot. Gard. 80 528–580. 10.2307/2399846 [DOI] [Google Scholar]
- Cosner M. E., Jansen R. K., Palmer J. D., Downie S. R. (1997). The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet. 31 419–429. 10.1007/s002940050225 [DOI] [PubMed] [Google Scholar]
- Cronquist A. (1988). The Evolution and Classification of Flowering Plants, 2nd Edn. Bronx, NY: New York Botanical Garden. [Google Scholar]
- Downie S. R., Olmstead R. G., Zurawski G., Soltis D. E., Soltis P. S., Watson J. C., et al. (1991). Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution 45 1245–1259. 10.2307/2409731 [DOI] [PubMed] [Google Scholar]
- Downie S. R., Palmer J. D. (1992). “Use of chloroplast DNA rearrangements in reconstructing plantphylogeny,” in Molecular Systematics of Plants, eds Soltis P. S., Soltis D. E., Doyle J. J. (New York, NY: Chapman and Hall; ), 14–35. [Google Scholar]
- Doyle J. J., Doyle J. L. (1990). Isolation of plant DNA from fresh tissue. Focus 12 13–15. [Google Scholar]
- Doyle J. J., Doyle J. L., Palmer J. D. (1995). Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst. Bot. 20 272–294. 10.2307/2419496 [DOI] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32 W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis E. M., Crane P. R., Pedersen K. R. (2011). Early Flowers and Angiosperm Evolution. Cambridge: University of Cambridge. [Google Scholar]
- Friis E. M., Pedersen K. R., Crane P. R. (2006). Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Paleogeogr. Paleoclimatol. Paleoecol. 232 251–293. 10.1016/j.palaeo.2005.07.006 [DOI] [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J., Weeden N. F., Palmer J. D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin V. V., Hirsch-Ernst K. I., Wolfl S., Hellwig F. H. (2003). Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol. Biol. Evol. 20 1499–1505. 10.1093/molbev/msg159 [DOI] [PubMed] [Google Scholar]
- Goremykin V. V., Hirsch-Ernst K. I., Wolfl S., Hellwig F. H. (2004). The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol. Biol. Evol. 21 1445–1454. 10.1093/molbev/msh147 [DOI] [PubMed] [Google Scholar]
- Goremykin V. V., Holland B., Hirsch-Ernst K. I., Hellwig F. H. (2005). Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol. Biol. Evol. 22 1813–1822. 10.1093/molbev/msi173 [DOI] [PubMed] [Google Scholar]
- Graham S. W., Zgurski J. M., McPherson M. A., Cherniawsky D. M., Saarela J. M., Horne E. S. C., et al. (2006). Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. Aliso 22 3–20. [Google Scholar]
- Green R., Martin G. (2013). A rosid is a rosid is a rosid. or not. Adv. Classification Res. Online 23 9–16. 10.7152/acro.v23i1.14228 [DOI] [Google Scholar]
- Hilu K. W., Borsch T., Muller K., Soltis D. E., Soltis P. S., Savolainen V., et al. (2003). Inference of angiosperm phylogeny based on matK sequence information. Amer. J. Bot. 90 1758–1776. 10.3732/ajb.90.12.1758 [DOI] [PubMed] [Google Scholar]
- Hupfer H., Swaitek M., Hornung S., Herrmann R. G., Maier R. M., Chiu W. L., et al. (2000). Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome 1 of the five distinguishable Euoenthera plastomes. Mol. Gen. Genet. 263 581–585. 10.1007/s004380051204 [DOI] [PubMed] [Google Scholar]
- Jansen R. K., Cai Z., Daniell H., Raubeson L., DePamphilis C. W., Leebens-Mack J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Kaittanis C., Saski C., Lee S. B., Tomkins J., Alverson A. J., et al. (2006). Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol. Biol. 6:32 10.1186/1471-2148-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd W. S., Olmstead R. G. (2004). A survey of tricolpate (eudicot) phylogeny. Amer. J. Bot. 91 1627–1644. 10.3732/ajb.91.10.1627 [DOI] [PubMed] [Google Scholar]
- Kato T., Kaneko T., Sato S., Nakamura Y., Tabata S. (2000). Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 7 323–330. 10.1093/dnares/7.6.323 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1983). The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J., Raubeson L. A., Cui L., Kuehl J., Fourcade M., Chumley T., et al. (2005). Identifying the basal angiosperms in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol. Biol. Evol. 22 1948–1963. 10.1093/molbev/msi191 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Logacheva M. D., Samigullin T. H., Dhingra A., Penin A. A. (2008). Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale -a wild ancestor of cultivated buckwheat. BMC Plant Biol. 8:59 10.1186/1471-2229-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Bock R. (2009). Organellar genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 25 1451–1452. [DOI] [PubMed] [Google Scholar]
- Magallon S. (2007). From fossils to molecules: phylogeny and the core eudicot floral ground plan in Hamamelidoideae (Hamamelidaceae, Saxifragales). Syst. Bot. 32 317–347. 10.1600/036364407781179617 [DOI] [Google Scholar]
- Magallón-Puebla S., Crane P. R., Herendeen P. (1999). Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Missouri Bot. Gard. 86 297–372. 10.2307/2666180 [DOI] [Google Scholar]
- Makalowski W., Boguski M. S. (1998). Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. U.S.A. 95 9407–9412. 10.1073/pnas.95.16.9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Leese F., Tollrian R. (2010). Genome-wide analysis of tandem repeats in Daphnia pulex–a comparative approach. BMC Genomics 11:277 10.1186/1471-2164-11-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen R. S., Olmstead R. G., Adams K. L., Palmer J. D., Lao N. T., Heggie L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13 645–658. 10.1105/tpc.13.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan B. G., Hampton J. N., Palmer J. D. (1989). Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol. Biol. Evol. 6 355–368. [DOI] [PubMed] [Google Scholar]
- Moore M. J., Bell C. D., Soltis P. S., Soltis D. E. (2007). Using plastid genome scale-data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. U.S.A. 104 19363–19368. 10.1073/pnas.0708072104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckermann K., Zeltz P., Igloi G. L., Kössel H., Maier R. M. (1994). The role of RNA editing in conservation of start codons in chloroplast genomes. Gene 146 177–182. 10.1016/0378-1119(94)90290-9 [DOI] [PubMed] [Google Scholar]
- Parks M., Cronn R., Liston A. (2009). Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84 10.1186/1741-7007-7-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman G., Park S. (2015). Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS ONE 10:e0141329 10.1371/journal.pone.0141329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C., Lee S., Daniell H., Wood T., Tomkins J., Kim H. G., et al. (2005). Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59 309–322. 10.1007/s11103-005-8882-0 [DOI] [PubMed] [Google Scholar]
- Savolainen V., Chase M. W., Morton C. M., Soltis D. E., Bayer C., Fay M. F., et al. (2000). Phylogenetics of flowering plants based upon a combined analysis of plastid atpB and rbcL gene sequences. Syst. Biol. 49 306–362. 10.1080/10635159950173861 [DOI] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33 W686–W689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönenberger J., von Balthazar M. (2006). Reproductive structures and phylogenetic framework of the rosids - progress and prospects. Plant Syst. Evol. 260 87–106. [Google Scholar]
- Soltis D. E., Gitzendanner M. A., Soltis P. S. (2007). A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int. J. Plant Sci. 168 137–157. 10.1086/509788 [DOI] [Google Scholar]
- Soltis D. E., Senters A. E., Zanis M. J., Kim S., Thompson J. D., Soltis P. S., et al. (2003). Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. Amer. J. Bot. 90 461–470. 10.3732/ajb.90.3.461 [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S. (2004). Amborella not a “basal angiosperm”? Not so fast. Am. J. Bot. 91 997–1001. 10.3732/ajb.91.6.997 [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Chase M. W., Mort M. E., Albach D. C., Zanis M., et al. (2000). Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 133 381–461. 10.1111/j.1095-8339.2000.tb01588.x [DOI] [Google Scholar]
- Soltis D. E., Soltis P. S., Endress P. K., Chase M. W. (2005). Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer Associates Inc. [Google Scholar]
- Spielmann A., Roux E., von Allmen J., Stutz E. (1988). The soybean chloroplast genome: completed sequence of the rps19 gene, including flanking parts containing exon 2 of rpl2 (upstream), but lacking rpl22 (downstream). Nucl. Acids Res. 16:1199 10.1093/nar/16.3.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web-server. Syst. Biol. 75 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- The angiosperm phylogeny group II [APG II] (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 141 399–436. [Google Scholar]
- The angiosperm phylogeny group III [APG III] (2009). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 161 105–121. [Google Scholar]
- Wang H., Moore M. J., Soltis P. S., Bell C. D., Brockington S. F., Alexandre R., et al. (2009). Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. U.S.A. 106 3853–3858. 10.1073/pnas.0813376106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Messing J. (2011). High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE 6:e24670 10.1371/journal.pone.0024670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G. M., dePamphilis C. W., Muller K. F., Quandt D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76 273–297. 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Boore J. L., Jansen R. K. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20 3252–3255. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.