Abstract
Retroviral vector-mediated gene therapy is promising, but genotoxicity has limited its use in the clinic. Genotoxicity is highly dependent on the retroviral vector used, and foamy viral (FV) vectors appear relatively safe. However, internal promoters may still potentially activate nearby genes. We developed insulated FV vectors, using four previously described insulators: a version of the well-studied chicken hypersensitivity site 4 insulator (650cHS4), two synthetic CCCTC-binding factor (CTCF)-based insulators, and an insulator based on the CCAAT box-binding transcription factor/nuclear factor I (7xCTF/NF1). We directly compared these insulators for enhancer-blocking activity, effect on FV vector titer, and fidelity of transfer to both proviral long terminal repeats. The synthetic CTCF-based insulators had the strongest insulating activity, but reduced titers significantly. The 7xCTF/NF1 insulator did not reduce titers but had weak insulating activity. The 650cHS4-insulated FV vector was identified as the overall most promising vector. Uninsulated and 650cHS4-insulated FV vectors were both significantly less genotoxic than gammaretroviral vectors. Integration sites were evaluated in cord blood CD34+ cells and the 650cHS4-insulated FV vector had fewer hotspots compared with an uninsulated FV vector. These data suggest that insulated FV vectors are promising for hematopoietic stem cell gene therapy.
Introduction
Retroviral vector-mediated gene therapy has enormous potential to cure genetic disorders and has been used successfully to correct hematopoietic diseases in numerous clinical trials.1–6 Early SCID-X1 (X-linked severe combined immunodeficiency) clinical trials using gammaretroviral (GV) vectors were successful in treating the SCID-X1 phenotype; however, patients in two clinical trials developed leukemia as a result of vector-induced genotoxicity.7,8 Preclinical studies and additional clinical trials for SCID-X1 and Wiscott-Aldrich syndrome have continued to show similar or even higher rates of oncogenesis as a severe adverse side effect of retroviral hematopoietic stem cell (HSC) gene therapy.9–11 These side effects are caused by enhancer elements within integrated vector proviruses that activate nearby proto-oncogenes.7,11–16 Advanced designs of retroviral vectors have eliminated or greatly reduced the native retroviral enhancer elements in the long terminal repeats (LTRs), significantly increasing the safety of modern vectors.14,17–19 However, the promoters necessary for sufficient gene expression to correct a disease phenotype may contain enhancer elements with the potential to cause dysregulation, which has already been documented.4 These genotoxic side effects are a major hurdle to the safety of retroviral vector-mediated gene therapy, limiting adoption of this approach in the clinic.
Increasing the safety of retroviral vectors can be approached in several different ways, and the choice of vector parent virus is paramount. Retroviruses have specific integration site preferences that, in addition to removing the native viral enhancers, can be used to increase the safety of retroviral vectors.20 Vectors developed from foamy virus (FV), the only member of the Spumaretrovirinae subfamily of retroviruses, have an integration profile that is potentially safer than other retroviral vectors.20–24 FV vectors have a reduced preference for integration within CpG islands known to be DNA regulatory regions and near promoters compared with GV vectors, and a reduced likelihood to integrate within genes relative to lentiviral (LV) vectors.20,21 In addition to a more desirable integration profile, FV vectors also have a lower propensity to dysregulate nearby genes when directly compared with GV and LV vectors.25 These increased safety features, in addition to a large therapeutic transgene-carrying capacity17 and broad tissue tropism including human mobilized peripheral blood and cord blood-derived CD34+ cells,23,26 suggest that FV vectors are a promising vector for safer HSC gene therapy.
FV vectors appear to be relatively safe, but the internal promoter-enhancers necessary for therapeutic gene expression could potentially activate nearby genes. A proven approach to reduce this source of genotoxicity is to add chromatin insulators to retroviral vectors.18,27–29 Chromatin insulators are DNA sequences that protect genes from inappropriate transcription, often by recruiting proteins necessary for chromatin remodeling and scaffold attachment.30–32 There are two types of insulators: barrier insulators and enhancer-blocking insulators.30 Barrier insulators are sequences that allow for an abrupt change from closed to open chromatin and protect genes from being silenced. Enhancer-blocking insulators, hereafter referred to as insulators, prevent enhancer elements from acting on promoters when the insulator is between an enhancer and promoter. We hypothesized that adding insulators to FV vectors might improve their safety.
Here four previously described promising candidate insulators were evaluated for use in FV vectors. A 650-bp version of the well-studied chicken hypersensitivity site 4 (cHS4) originally described by Arumugam and colleagues33 and three synthetic insulators described by Gaussin and colleagues34 were tested. The 650-bp cHS4 (650cHS4) is a combination of the 5′ 250-bp core, which contains a CCCTC-binding factor (CTCF)-binding site, and the 3′ 400 bp of the 1.2-kbp cHS4.35,36 This 650cHS4 insulator reduces the genotoxicity of GV and LV vectors.18,33 Of the three synthetic insulators tested here, two have multiple binding sites for CTCF and the third has multiple binding sites for the CCAAT box-binding factor/nuclear factor 1 (CTF/NF1). The CTF/NF1 protein is believed to be involved in barrier insulation and its DNA-binding sequence seems to impart enhancer-blocking activity when presented in multiple repeats.34,37–39 Reduced genotoxicity of GV vectors has previously been demonstrated with all of these insulators. Using a series of assays, including a plasmid-based assay to evaluate insulator activity and a novel shuttle vector approach to assess the fidelity of insulator transfer from the vector plasmid to the integrated vector provirus, a cHS4-based insulated FV vector was found to be promising and evaluated for its relative genotoxicity.
Materials and Methods
Cell culture
All cell lines were maintained at 37°C and 5% CO2. HEK293T and HT1080 fibroblasts were cultured in HyClone high-glucose Dulbecco's modified Eagle's medium (DMEM) (SH30022.01; Thermo Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (S11550; Atlanta Biologicals, Lawrenceville, GA) and penicillin–streptomycin (50 U/ml) (17-602E; Lonza, Walkersville, MD). WEHI-3 cells (TIB-68; American Type Culture Collection [ATCC], Manassas, VA) were maintained between 2 × 105 and 2 × 106 cells/ml in Iscove's modified Dulbecco's medium (IMDM) (12-722F; Lonza) supplemented with 10% FBS, 0.05 M 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), and penicillin–streptomycin (50 U/ml). For the production of WEHI CM IL-3 tissue culture additive, 2 × 105 cells/ml were plated and cultured for 5 days. Medium was then collected, clarified by centrifugation at 400 × g, filtered through a 0.22-μm (pore size) filter (Millipore, Bedford, MA), and stored at –20°C. 32D cells (CRL-11346; ATCC) were maintained between 2 × 105 and 1 × 106 cells/ml in RPMI 1640 (SH30027.01; Thermo Scientific) supplemented with 10% FBS, 10% WEHI CM, and penicillin–streptomycin (50 U/ml). For interleukin (IL)-3 depletion experiments, 32D cells were grown in RPMI, 10% FBS, and penicillin–streptomycin (50 U/ml) after extensive washing with phosphate-buffered saline (PBS).
Plasmids and plasmid construction
Insulator elements have been previously described.33,34 Spacers in synthetic insulators were derived from mouse utrophin cDNA (GenBank accession number: BC062163.1) as previously described.34 All insulators were synthesized with flanking NsiI restriction sites. The FV vector plasmid FV-PGW was previously described40 and carries an enhanced green fluorescent protein (EGFP) reporter transgene expressed from a human phosphoglycerate kinase (PGK) promoter. FV-SGW is an FV vector plasmid containing an EGFP reporter transgene expressed from a spleen focus-forming virus (SFFV) promoter. These FV vector plasmids contain an added NsiI restriction site at the U3 deletion site of the 3′ LTR to facilitate inserting insulators. The FV-SGWKO vectors were developed by cloning an SalI restriction site-flanked R6Kγ bacterial origin of replication and kanamycin resistance cassette into the SalI restriction site within FV-SGW. The enhancer-blocking test plasmid was created by synthesizing the cytomegalovirus (CMV) enhancer, multiple cloning site with NsiI restriction site, CMV minimal promoter expressing mCherry, and poly(A) signals into a pUC57 backbone. The PGK-driven EGFP expression cassette was derived from the vector FV-PGW.40 The 1.2-kbp cHS4 insulator was amplified by PCR from chicken genomic DNA (69233; Novagen/EMD Millipore, Darmstadt, Germany), using primers 5′-ATATTCCCCCATCCTCACTGA-3′ and 5′-GAAGAAAAGAAGCAGGCTTTC-3′ and Phusion high-fidelity polymerase (Thermo Scientific). Insulators were cloned into the NsiI restriction site of all plasmids and vectors. The LTRs of FV-PGW and insulated FV constructs were cloned into the multiple cloning site of the enhancer-blocking test plasmid with ClaI and PvuII. All insulators were synthesized by Blue Heron Biotechnology (Bothell, WA) or GenScript (Piscataway, NJ).
Vector production and titer
Vectors were produced on HEK293T cells plated on poly-l-lysine-coated plates by polyethylenimine (PEI) transfection of vector plasmids with vector-packaging helper plasmids as previously described.41–43 The GV vector CL-SGN was generated from pCAG-GFP, a gift from F. Gage (plasmid no. 16664; Addgene, Cambridge, MA),44 where an SFFV EGFP neomycin cassette was cloned in between the EcoRI sites, using standard molecular biology techniques. The GV vector CL-SGN and the LV vector LV-SFFVEGFP were both pseudotyped with a vesicular stomatitis virus G glycoprotein (VSV-G) envelope. The functional titers of all vectors were determined after transduction of HT1080 fibroblasts and flow cytometric analysis for EGFP-expressing cells.
Enhancer-blocking activity assay
HEK293T cells (4 × 105) were plated on poly-l-lysine-coated wells of 12-well plates. Cells were transfected with 1 μg of control or insulator-containing enhancer-blocking test plasmid and incubated for 40–45 hr before harvest for flow cytometric analysis on an BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA). To evaluate insulating activity, the ratio of mCherry mean fluorescence intensity (MFI) to EGFP MFI was determined for the EGFP-positive cells. The control ratio was normalized to 100% and the percent expression as compared with control was determined for all inserted sequences.
Shuttle vector rescue for insulator fidelity
HT1080 fibroblasts were infected with the insulated FV-SGWKO vector, sorted for EGFP expression to greater than 95% transduction, and expanded to at least 1.5 × 106 cells. DNA was then extracted from the cells, digested with the restriction enzyme NdeI, ligated and transformed into electrocompetent TransforMax EC100D pir+ Escherichia coli (ECP09500; Epicentre, Madison, WI), and plated on kanamycin-containing plates. Colonies were isolated and sequenced by Beckman Coulter Genomics Services (Danvers, MA), using primers 5′-TATGCCTCCCGCTATGCTCG-3′ and 5′-CCTGTGGAACACCTACATCTG-3′ for the 5′ and 3′ LTRs, respectively. Retrieved sequences were aligned to vector sequence data using the BioEdit alignment tool (Ibis Biosciences, Carlsbad, CA). Retrieved sequences that did not contain chromosome junctions were further processed to verify integration. DNA was extracted from selected colonies, digested with AvrII, and sequenced (Macrogen Korea, Seoul, South Korea) using primer 5′-TAAACCGACTTGATTCGAGAA-3′.
32D genotoxicity assay
32D cells (2 × 106 or 1 × 108) were plated in log phase at 5 × 105 cells/ml in 32D WEHI CM medium and transduced with CL-SGN, FVSGW, or FVSGW-650cHS4-R vector at a multiplicity of infection (MOI) of 2 or with LV-SFFVEGFP at an MOI of 0.4, resulting in 3–5% transduction as previously described.18 CL-SGN and LV-SFFVEGFP transductions were carried out in the presence of protamine sulfate (4 μg/ml). Cells were cultured for 48 hr, to allow for transduction, and then enriched by flow sorting for EGFP-expressing cells (S3 cell sorter; Bio-Rad, Hercules, CA). Sorted cells were allowed to recover in 32D WEHI CM medium for 24 hr and then thoroughly washed with PBS and transferred to RPMI supplemented with FBS and penicillin–streptomycin. Forty-eight hours later, cells were counted and plated at a concentration of 5 × 105 cells/ml in MethoCult semisolid medium (04230; Stemcell Technologies, Vancouver, BC) containing RPMI. Colonies were scored after 4–5 weeks of incubation.
Transduction of human cord blood CD34+ cells
Human cord blood CD34+ progenitor cells (2C-101; Lonza) were stimulated and maintained between 5 × 105 and 1 × 106 cells/ml in IMDM with 10% FBS and a 100-ng/ml concentration each of recombinant human interleukin (rhIL)-3, rhIL-6, recombinant human stem cell factor (rhSCF), recombinant human thrombopoietin (rhTPO), rhFlt-3, and recombinant human granulocyte colony-stimulating factor (rhG-CSF) (ProSpec, Ness-Ziona, Israel). After 24 hr of culture, 1 × 106 cells were harvested and exposed to FVSGW or FVSGW-650cHS4-R on CH-296-coated suspension culture plates at an MOI of 10 for 16 hr. Cells were then transferred to standard tissue culture-treated plates maintained for up to 10 days. Transduced cells were sorted by EGFP at 5 days posttransduction. At 10 days, remaining cells were harvested and DNA was extracted with a Gentra Puregene tissue kit (Qiagen, Valencia, CA).
Quantitative real-time PCR for relative retroviral vector integration copy number
DNA extracted from 5-day in vitro-cultured transduced CD34+ cells was assessed for copy number by quantitative real-time PCR assay for the EGFP transgene carried by the retroviral vectors. Forty nanograms of DNA was amplified in triplicate with the EGFP primers 5′-TGAGCAAAGACCCCAACGAG-3′ and 5′-TCGTCCATGCCGAGAGTGAT-3′. As an internal control, the β-globin gene was amplified with primers 5′-ATATCCCCCAGTTTAGTAGTTGGA-3′ and 5′-CCACAAGTATCACTAAGCTCGC-3′. PCR-amplified DNA for standard amplification curves was generated with the previously described primers. Standard curves were run in triplicate with a 10-fold dilution series ranging from 100 to 107 copies. Reactions contained 10 μl of Applied Biosystems SYBR green master mix (Thermo Fisher Scientific, Grand Island, NY) and a 0.2 μM concentration of each primer in a total volume of 20 μl. Quantitative real-time PCR was performed on a CFX384 Touch (Bio-Rad) under the following thermal cycling conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 61°C for 1 min, and a final extension at 65°C for 5 min. Vector copy number per cell is expressed as the ratio of EGFP gene copies to β-globin copies.
Modified genomic sequencing-polymerase chain reaction and integration site analysis
Three micrograms of genomic DNA was analyzed by modified genomic sequencing-polymerase chain reaction (MGS-PCR) as previously described45,46 and sequenced on an Illumina MiSeq (Genomic Sequencing and Analysis Facility, University of Texas-Austin, Austin, TX). Briefly, genomic DNA was sheared to an average fragment size of 1.5 kbp with a HydroShear (Digilab, Marlborough, MA) set to a speed code of 12. Fragment ends were repaired (terminator end repair kit; Lucigen, Middleton, WI) and a linker sequence was ligated to the fragments. Fragments then underwent two sets of 30-round exponential PCR. The first, exponential PCR was done with biotin-tagged FV LTR primer 5′-ACCGACTTGATTCGAGAACC-3′ and linker primer 5′-GACCCGGGAGATCTGAATTC-3′. After MACS (magnetic-activated cell-sorting) bead purification (Dynabeads M-280 streptavidin; Thermo Fisher Scientific) to enrich for biotin-tagged fragments, samples were amplified by PCR with FV LTR primer containing a sample identifier sequence (XXXXXXX) and an Illumina MiSeq platform adaptor sequence (in boldface) 5′-AATGATACGGCGACCACCGAGATCTACAC/XXXXXXX/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/GCTAAGGGAGACATCT-3′ and a linker primer also containing a sample identifier and an Illumina MiSeq platform adaptor 5′- CAAGCAGAAGACGGCATACGAGAT/XXXXXXX/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG-3′. Samples were enriched for fragments between 400 and 800 bp by agarose gel purification (QIAquick gel extraction kit; Qiagen, Germantown, MD) and excess primers were further removed with AMPure XP beads (Beckman Coulter, Indianapolis, IN). Sequencing reads were paired with Paired-End reAd mergeR (PEAR) software,47 and processed and mapped to the Genome Reference Consortium (GRC) build GRhg38 human genome map with the Vector Integration Site Analysis (VISA) server (see https://visa.pharmacy.wsu.edu/ bioinformatics/).48,49 VISA was also used to generate a random 10,000-site data set for comparison. PERL scripts querying oncogene databases were used to assess integrations within and near known proto-oncogenes as previously described.20,41 Retroviral integration site (RIS) data sets were divided into a minimum of three nonoverlapping randomly selected matched size data sets of 1588 unique RIS for hotspot analysis.
Results
Development of insulated FV vectors
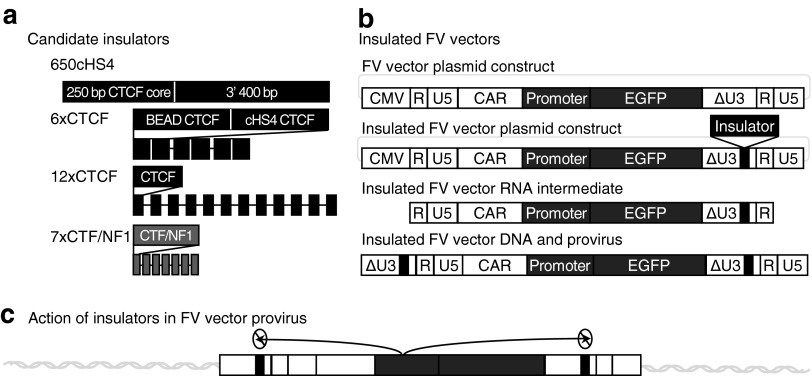
We chose four insulators, previously shown to be effective in GV or LV vectors, for our studies (Fig. 1a). The first insulator we used is derived from the well-studied chicken hypersensitivity site four (cHS4). The cHS4 insulator is a 1.2-kbp genomic region within the chicken β-globin locus that binds many proteins, including CTCF, and has been extensively used in retroviral vectors to reduce genotoxicity. Previous studies have shown that cHS4 reduces retroviral vector titers.33,50 Thus, for our studies we used the 650-bp version originally described by Arumugam and colleagues,33 which has similar activity to the full-length cHS4. We also evaluated synthetic insulators described by Gaussin and colleagues.34 These insulators have repeated CTCF-binding elements in order to increase their efficacy. 6xCTCF and 12xCTCF contain repeats of either the CTCF-binding sites from cHS4 and the blocking element α/δ (BEAD) insulator from the human α/δ T cell locus (6xCTCF) or the consensus CTCF-binding site (12xCTCF).51,52 The 7xCTF/NF1 insulator is not based on CTCF binding but has seven repeats of the binding site for CTF/NF1. This insulator was originally described as a barrier insulator but may have blocking activity as well when repeated in tandem.34,37,38
Figure 1.
Insulated foamy viral (FV) vectors. (a) Candidate insulators. The 650cHS4 insulator is derived from the 1.2-kbp chicken hypersensitivity site 4 (cHS4) from the chicken β-globin locus. This insulator contains the 5′-most 250-bp core containing the CCCTC-binding factor (CTCF)-binding domain and the terminal 400-bp portion. The 6xCTCF insulator contains alternating repeats of the CTCF-binding domain from blocking element α/δ (BEAD) and the cHS4 CTCF-binding domain. The 12xCTCF insulator contains consecutive repeats of a CTCF-binding domain consensus sequence separated by unique spacers. The 7xCTF/NF1 insulator contains consecutive repeats of the CTF/NF1-binding domain. (b) Insulated FV vector construction. Insulators were inserted into the U3 deletion site of the 3′ LTR. The FV vector DNA is transfected into HEK293 cells to make vector virions. During vector preparation the vector DNA is initially transcribed into RNA, which is encapsidated and reverse transcribed into DNA that is integrated into the host genome. During the process of reverse transcription, the 3′ LTR, including the insulator, is copied to the 5′ LTR so that the integrated provirus is flanked by the insulators. CAR is the cis-acting region containing the remaining portions of the FV gag, pol, and env sequences necessary for vector genome packaging and integration. (c) Insulators block the internal promoter from acting on adjacent host genes.
Similar to previously developed insulated retroviral vectors,53,54 the insulators were positioned in the U3 region of the 3′ LTR of replication-incompetent FV vectors (Fig. 1b).17,40 Enhancer elements function bidirectionally but insulators can prevent the enhancer activation of promoters only when between the enhancer and promoter.30 To best prevent enhancer-mediated dysregulation of nearby genes, the therapeutic transgene cassette needs to be flanked by insulators. Placing the insulator in the U3 region of the 3′ LTR uses the retroviral/vector genome replication strategy in order to accomplish this flanking. During vector production the vector genome is transcribed from a plasmid and then reverse transcribed to make integration-competent replicate vector genomes. During reverse transcription the 3′-LTR U3 region is copied to the 5′ LTR. Addition of the insulator to the U3 region of the 3′ LTR results in vector genomes and subsequent integrated vector proviruses with insulators in both the 5′ and 3′ LTRs, effectively blocking any activity from the internal promoter on the upstream or downstream flanking host genome (Fig. 1c). To reduce the potential negative impact of insulator sequences on titer, the insulators were placed at the site of a 582-bp deletion made during previous vector development.17 All developed vectors contain an EGFP gene cassette controlled by a PGK promoter or SFFV promoter in the location where a therapeutic transgene would be located.
Enhancer-blocking activity assay
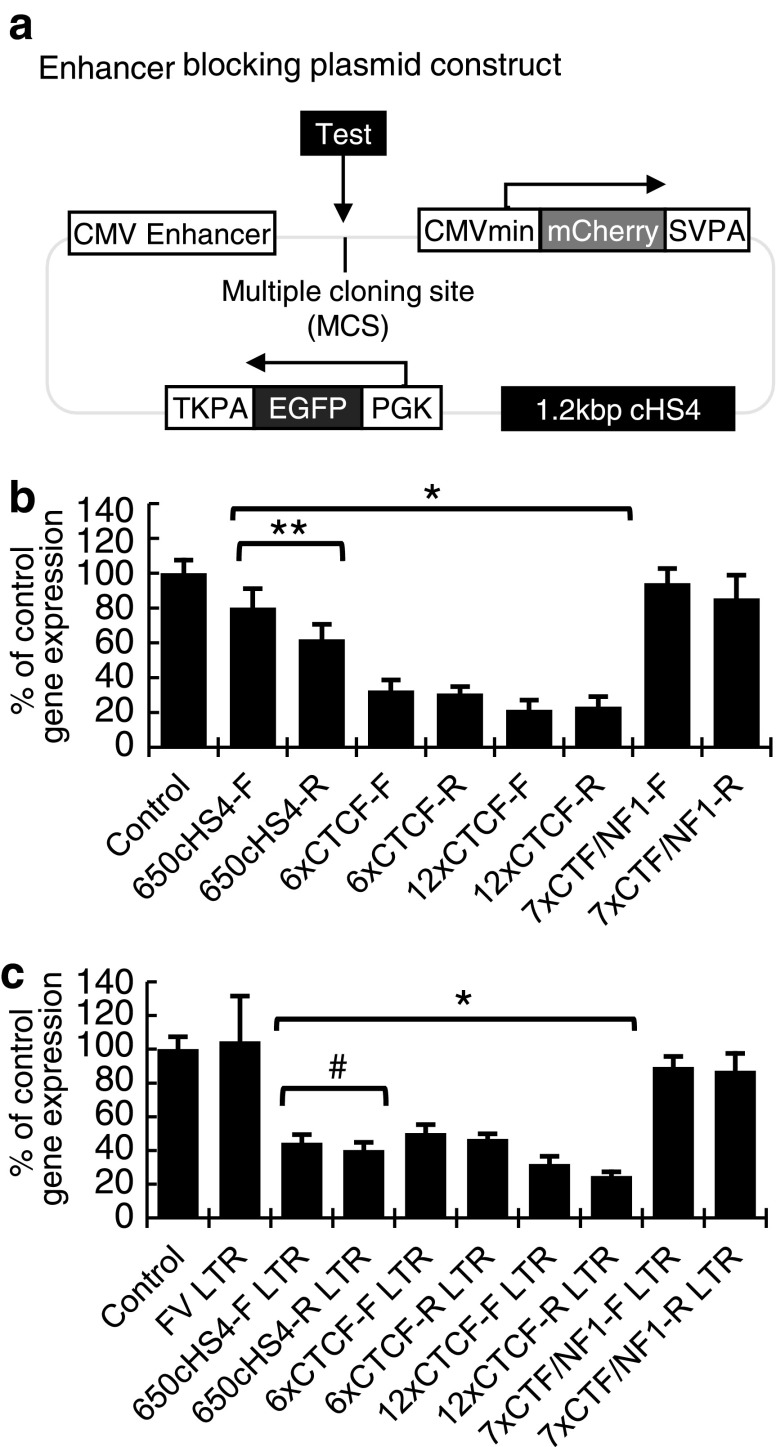
To assess the insulating activity of the candidate insulators, we designed a dual-fluorescence enhancer-blocking test plasmid that features a separated CMV enhancer and minimal promoter (CMVe/CMVmin) controlling mCherry expression and a control EGFP expression cassette (Fig. 2a). Insulators were placed between the CMVe and CMVmin and the ratio of mCherry to EGFP expression was compared (Supplementary Fig. S1; supplementary data are available online at www.liebertpub.com/hum). Because the assay is based on a circular plasmid, the CMVe can act as an enhancer on the CMVmin from either the 5′ or 3′ end of the mCherry gene cassette. To isolate the effects of the test insulator and block the CMVe from acting in the 5′ direction as a downstream enhancer, a permanent full-length cHS4 insulator was positioned at the 3′ end of the mCherry gene cassette. The full-length cHS4 insulator is present in the control as well as the plasmids containing insulators so that the reduction in mCherry gene expression normalized to EGFP observed is due to the insulator placed between the CMVe and CMVmin. This plasmid assay has some key differences from previously reported assays to improve the detection of enhancer-blocking activity. Previously described assays have used antibiotic resistance and reduction in the number of live cells to determine insulator activity, which requires selection of stably transfected cells over a few weeks.34,35 Our assay uses changes in the intensity of a fluorescence reporter, which allows for highly quantitative determination of insulator activity from transfected cells within a few days by flow cytometry. The assay also features a 400-bp spacer between the minimal CMV promoter and the enhancer element to allow for improved range of motion for the enhancer to interact with the promoter.
Figure 2.
Enhancer-blocking activity of candidate insulators. (a) Enhancer-blocking plasmid construct. Insulators or an FV vector LTR containing an insulator to be tested were placed in the multiple cloning site between the CMV enhancer and the CMV minimal promoter controlling the expression of mCherry. The ratio of mCherry expression to EGFP expression was then determined. SVPA, simian virus 40 (SV40) polyadenylation site; TKPA, thymidine kinase polyadenylation site. (b) Insulator activity. Enhancer-blocking plasmid with no insulator (Control) or with the indicated insulators was transfected into HEK293T cells and the effect of insulators on enhancers, determined by intensity of fluorescence, was evaluated by flow cytometry 40–45 hr posttransfection. Columns represent the percentage of normalized mCherry expression as compared with the uninsulated control. (c) Enhancer-blocking capacity of insulators in the context of the FV LTR. *p < 0.05 compared with control; **p < 0.05 between samples; #p < 0.05 compared with insulator alone.
Enhancer-blocking activity was found for all insulators with a broad range of activity (Fig. 2b). The 7xCTF/NF1 insulator was the least potent insulator and the 12xCTCF insulator was the most potent. Interestingly, there was a significant difference between the activities of the forward- and reverse-oriented 650cHS4 insulators, which was not observed with the synthetic insulators. To further investigate how insulators would function within the FV vector, the entire LTR from insulated FV vectors was then cloned between the CMVe and CMVmin (Fig. 2c). Inserting insulators into the FV vector LTR sequence significantly decreased the activity of the 6xCTCF insulator from 30 to 47%. For other insulators, insulator function stayed the same or was improved. Interestingly, the forward-oriented 650cHS4 insulator had observably stronger enhancer-blocking activity when tested in the presence of the FV LTR (45%) than in the absence of the LTR (80%) (Fig. 2b and c).
Effect of insulators on FV vector titer
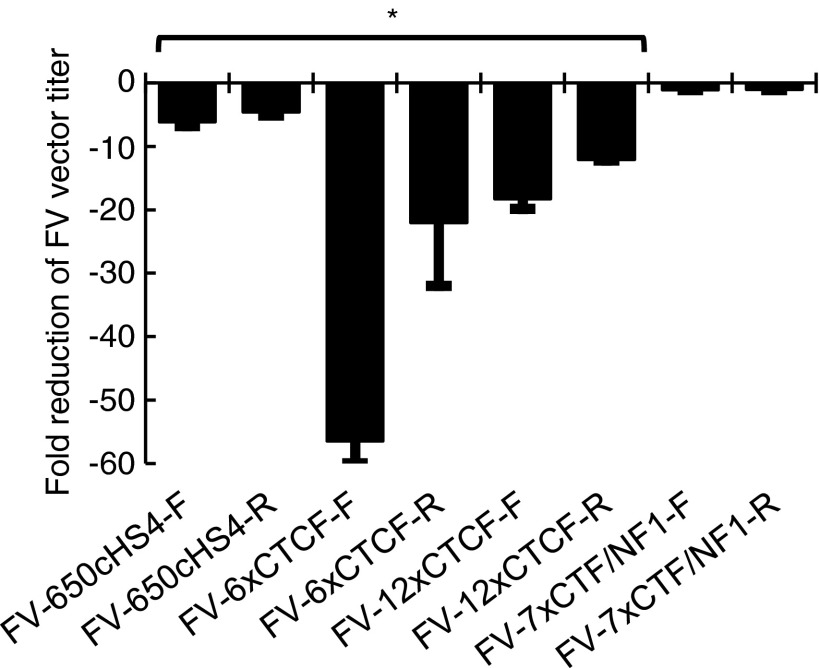
LTR-insulated FV vectors containing an EGFP expression cassette were made in HEK293T cells and titered on HT1080 human fibroblasts (Fig. 3). For these experiments EGFP acted as a marker for transduction, which could be visualized by fluorescence microscopy and quantitated by flow cytometry. The 7xCTF/NF1 insulator had a minimal effect on FV vector titer. The two CTCF-based insulators with repeated CTCF elements, 12xCTCF and 6xCTCF, dramatically reduced titers. 6xCTCF was not evaluated further because of the low titers. Neither the size of the insulator, as seen with LV vectors,50 nor the strength of the insulator had a consistent effect on titer. Interestingly, the titer of FV vectors with insulators in the forward orientation were consistently lower than FV vectors with insulators in the reverse orientation. However, the reverse-oriented 650cHS4 insulator could still be produced at a clinically relevant titer, less than 5-fold lower than the titer of an uninsulated FV vector, and more than 107 transducing units/ml after vector concentration. The choice of promoter to drive EGFP did not affect the changes in titer (data not shown).
Figure 3.
Titers of insulated FV vectors. HT1080 fibroblasts were transduced with FV vector preparations with the indicated insulators, and titers were determined by flow cytometry for EGFP expression. Columns represent the fold reduction in titer as compared with an uninsulated FV vector control. *p < 0.05 compared with control.
Fidelity of transfer of insulators to proviral LTRs
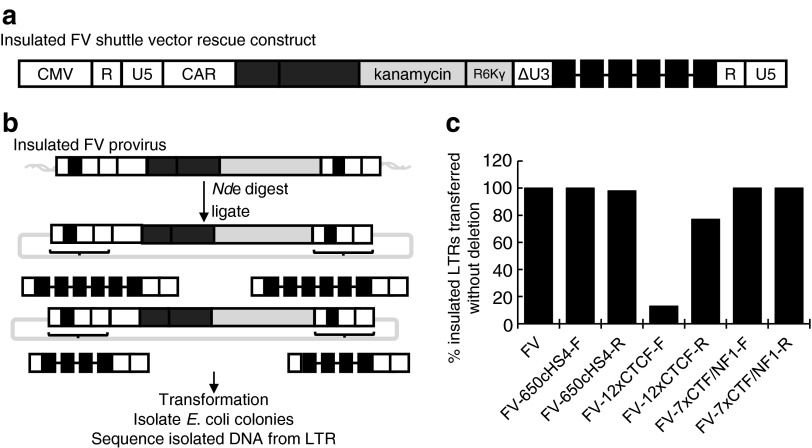
For insulators to be effective, they must be retained as the 3′ LTR of the vector plasmid is copied to the 5′ and 3′ LTRs of the integrated vector provirus during vector production, reverse transcription, and integration. Previous studies have identified problems with the retention of repeated elements within retroviruses as well as retaining a tandem repeat of the 250-bp cHS4 core.55–57 To evaluate the fidelity of transfer of insulators during vector production and transduction a shuttle vector rescue (SVR) approach was used to isolate integrated vector proviruses and sequence the LTRs (Fig. 4a and b).20 The SVR FV vector contains an R6Kγ bacterial origin of replication and a kanamycin resistance cassette between the transgene expression cassette and the FV vector LTR. After integration with the SVR FV vector, the host cell genome will contain integrated vector proviruses with bacterial origins of replication and kanamycin resistance cassettes. The DNA from the transduced cells can now be extracted and digested with a restriction enzyme that cuts the genomic DNA but does not cut the integrated provirus. Of the digested DNA, only the fragments containing integrated proviruses will have kanamycin resistance cassettes. These fragments can then be ligated and transformed into bacteria for isolation on kanamycin-containing plates. In this way, the complete provirus can be captured as a plasmid, and both LTRs can be sequenced for direct comparison. SVR thus allowed us to rapidly evaluate the fidelity of transfer of insulators from vector plasmids into both vector provirus LTRs. Both 650cHS4 and 7xCTF/NF1 were retained at high frequency regardless of orientation (Fig. 4c and Supplementary Table S1). The 12xCTCF insulator was not efficiently retained, with a dramatic decrease in fidelity when the insulator was in the forward orientation. For all captured proviruses where loss of an insulator occurred, the deletions were large (at least 150 bp) and only one to three CTCF-binding sites remained when the insulator was in the forward orientation. With few exceptions, the provirus 5′ and 3′ LTRs were identical, suggesting the errors occurred during reverse transcription. The insulator with the lowest fidelity tested in the fidelity assay, 12xCTCF in forward orientation, resulted in the lowest titer FV vector.
Figure 4.
Shuttle vector rescue for analysis of insulator fidelity. A shuttle vector rescue strategy was used to isolate whole integrated vector proviruses and evaluate the presence of insulators from both the 5′ and 3′ LTRs. (a) Schematic of the shuttle vector rescue insulated FV vector construct with an R6Kγ bacterial origin of replication and kanamycin resistance cassette between the EGFP gene cassette and the 3′ LTR. (b) Shuttle vector rescue process to rescue whole integrated proviruses from transduced HT1080 genomic DNA. Genomic DNA is isolated from transduced cells and then digested with the restriction enzyme NdeI, which does not cut within the provirus, to produce DNA fragments with an intact vector provirus. Digested DNA is then ligated and transformed into electrocompetent Escherichia coli. Plasmid DNA from transformed kanamycin-resistant colonies containing proviruses is isolated and both the 3′ and 5′ LTRs are sequenced to determine the fidelity of transfer of the insulators to each LTR. In the example, two rescued proviruses are shown; the top one has an intact 6X element, whereas the bottom one has deleted two elements in both the 5′ and 3′ LTRs. (c) Percentage of intact insulated LTRs rescued after shuttle vector rescue.
Evaluating the safety of insulated FV vectors in vitro
The FV vectors containing the 650cHS4 in reverse orientation were determined to be the most promising insulated vectors for further studies because of a clinically relevant titer, significant enhancer-blocking activity, and efficient retention of insulators in both LTRs within integrated vector proviruses. Therefore we began assessing the genotoxicity of these vectors in vitro, using a previously described assay using 32D myeloid cells.18 32D cells are an IL-3-dependent cell line where genotoxicity is measured by the frequency at which cells become IL-3 independent. IL-3 independence can be achieved by activation of autocrine IL-3 production or by activation of numerous oncogenes that negate the need for IL-3.58 As described by Li and colleagues,18 the 32D in vitro assay is 10-fold more sensitive to genotoxic events than assays using normal mouse bone marrow, and genotoxicity can be detected from GV with as few as 1 × 106 cells at 3% transduction efficiency.
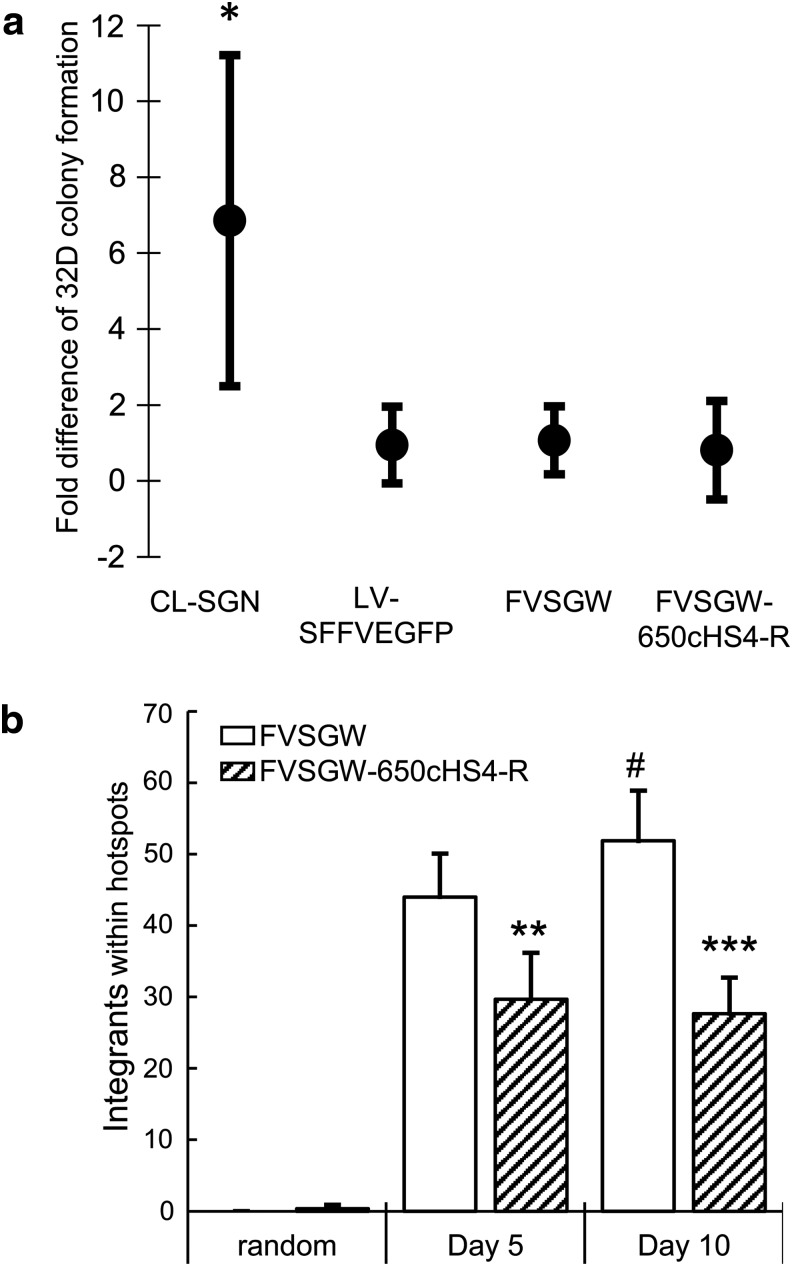
Previous studies have suggested a reduced potential for FV vector genotoxicity compared with GV vectors. In anticipation of this, we used vectors with SFFV-controlled EGFP gene cassettes and increased the number of total cells in the assay. Although not a promoter currently used in retroviral gene therapy clinical trials, the SFFV promoter represents a worse-case scenario to fully stress the action of the insulators. For FV vectors we expanded the procedure to 1 × 108 cells at 5% transduction efficiency in order to detect possible genotoxicity. The 32D cell assay shows that FVSGW and FVSGW-650cHS4-R are both significantly less genotoxic than the control GV vector (CL-SGN) (Fig. 5a). Because LV vectors are becoming more popular for gene therapy studies because of their reduced genotoxicity compared with GV vectors,28,59 1 × 108 32D cells were also transduced with an SFFV-containing LV vector (LV-SFFVEGFP). Consistent with previous findings, LV-SFFVEGFP is also significantly less genotoxic than CL-SGN with no detectable difference in the frequency of 32D colony formation compared with the FV vectors. Therefore, while CL-SGN consistently causes conversion of 32D cells to IL-3 independence, this conversion is not consistently detectable for FV or insulated FV and suggests that these vectors are both relatively safe and at least as safe as LV vectors.
Figure 5.
In vitro assessment of genotoxicity. (a) 32D cells were transduced with the indicated vectors and plated in IL-3-deficient semisolid medium for 4–5 weeks to allow for colony development. Shown is the mean fold difference with standard deviation as compared with untransduced control cells per 5 × 105 plated cells. CL-SGN, n = 4; LV-SFFVEGFP, n = 34; FVSGW, n = 45; FVSGW-650cHS4-R, n = 43. (b) CD34+ stem cells from cord blood were transduced with either FVSGW or FVSGW-650cHS4-R and cultured in vitro for 5 or 10 days before genomic DNA extraction for modified genomic sequencing-polymerase chain reaction (MGS-PCR). Captured integrations were ordered by position in the genome, and the distances between nearest integration sites were evaluated. Columns represent the percentage of total integrations within 50 kbp of two other integration sites. Each average is based on at least three randomly chosen nonoverlapping unique sets of 1588 integrations from the available MGS-PCR sequencing data. *p < 0.001 compared with LV-SFFVEGFP, FVSGW, and FVSGW-650cHS4-R; **p < 0.05 compared with FVSGW; ***p < 0.01 compared with FVSGW; #p < 0.05 compared with day 5.
Reduced genotoxicity of insulated vectors in human cord blood CD34+ cells
Ultimately, insulated FV vectors used for HSC gene therapy will need to be safe in patient cells that are used in the clinic. We thus explored potential signs of genotoxicity in transduced and in vitro-cultured human CD34+ cord blood cells by performing RIS analysis of MGS-PCR-captured integration sites. Our goal was to determine whether the 650cHS4 insulator would affect the integration profile as well as evaluate integrations near proto-oncogenes and hotspots that may have clinical relevance and warn of potential genotoxic side effects in a clinically relevant target cell. Again, FV vectors containing a highly genotoxic SFFV promoter were used to maximize potential differences between the control uninsulated vector and the insulated vector. Cord blood CD34+ cells were transduced at an MOI of 10, resulting in 12.7 and 13.2% EGFP-expressing cells, respectively, for insulated and noninsulated FV vectors (Supplementary Fig. S2a). Before DNA extraction for RIS retrieval, samples were enriched to about 80% EGFP-expressing cells and equal transduction efficiency was verified by qPCR (Supplementary Fig. S2b and c). MGS-PCR of extracted DNA resulted in the capture of 4764 unique integrations from insulated FV vector-transduced samples and 9765 unique integrations from uninsulated FV vector-transduced samples (Supplementary Table S2). The integration profiles of FV and FVSGW-650cHS4-R were assessed after 5 days of in vitro culture (Supplementary Fig. S3). Addition of the cHS4 insulator did not significantly change the distribution of integration sites in proximity to transcription start sites (TSS) or DNase hypersensitivity sites, nor did it change the frequency of integrations within genes on day 5 after vector exposure (Supplementary Table S2). We also assessed integration sites for proximity to known proto-oncogenes and extended the culture time to 10 days to evaluate any potential skewing during in vitro culture. From the 14,529 integrations sites retrieved from day 5 and the additional 5976 sites and 13,120 sites retrieved from insulated and uninsulated FV vector-transduced samples, respectively, we were able to determine that there was no significant difference in the number of FV or FV-650cHS4-R vector integrations observed near proto-oncogenes (Supplementary Table S2).
We then assessed the frequency of integration hotspots. A hotspot is defined as an area of the genome where clusters of integrations are observed more frequently than expected by chance. On the basis of previous evaluations of GV and LV vector hotspots and common insertion sites we defined three integrations in a span of 50 kbp or less as a hotspot.60,61 To avoid bias from integration site sample size differences, the MGS-PCR data sets were divided into a minimum of three nonoverlapping subsets of 1588 unique integration sites. Integrations within hotspots were then identified from these subsets. Fewer integration hotspots were observed from FVSGW-650cHS4-R-insulated FV vector-transduced CD34+ cells than from the uninsulated FV vector-transduced cells (Fig. 5b). We also observed a statistically significant increase in the number of hotspots captured from day 5 to day 10 in uninsulated FV vector-transduced samples that was not seen in the insulated counterpart. The integration hotspots were also less likely to be near known oncogene TSS, although these differences were not statistically significant (Supplementary Fig. S4). These data show that the 650cHS4 insulator reduced the frequency of integration hotspots when used in otherwise identical FV vectors.
Discussion
Retroviral vectors have enormous potential for HSC gene therapy, but genotoxicity continues to be a major concern for their clinical use. Here we describe the development of an insulated FV vector and show that in human cord blood CD34+ cells, fewer integrants occurred within hotspots compared with an uninsulated FV vector. We also describe a new rapid plasmid-based assay for insulator function and a shuttle vector assay to rapidly assess the fidelity of insulator transfer to the integrated vector provirus.
An insulated FV vector should have potent insulating activity and must be produced at high-enough titer for efficient HSC transduction. It should also maintain insulator fidelity during transfer of the insulator from the 3′ vector plasmid LTR to both LTRs of the integrated vector provirus in order to flank the transgene cassette. We found that the previously described 12xCTCF and 6xCTCF insulators were potent in a plasmid-based assay; however, they greatly reduced FV vector titers below what can be used in a clinical setting. Also, when the fidelity of transfer of the 12xCTCF insulator was evaluated, 33% (reverse orientation) to 87% (forward orientation) of integrated proviruses had deletions of the repeated CTCF elements. Taken together, these data suggest that repeated elements of CTCF mediate potent insulating activity, but future designs need to reduce recombination during vector production. This may be possible by creating repeated CTCF elements that are not based on a consensus CTCF element, but instead use different CTCF elements with mismatches to reduce the potential for recombination. Evaluation of CTCF-binding sites in the human genome has established hundreds of CTCF-binding domains, so designing repeated CTCF elements with potent insulating activity that do not reduce vector titers may be possible.62–65
Only the FV vectors with 650cHS4 in the reverse orientation fulfilled the criteria of significant insulating activity, high titer, and high fidelity of insulator transfer from vector plasmid to vector provirus. By using vectors with a strong SFFV internal promoter, we showed in the established and sensitive 32D assay that the FV vectors were significantly less genotoxic than a GV vector (CL-SGN) and similar in genotoxicity to an LV vector (LV-SFFVEGFP) (Fig. 5a). Neither the LV vectors nor the FV vectors produced consistently reproducible transformation of 32D cells to factor independence. As such, we were unable to establish a difference in genotoxicity between FVSGW and FVSGW-650cHS4-R with this assay. However, when integration sites in cord blood CD34+ cells were compared, a potential difference in safety was observed between the uninsulated and insulated FV vectors. The global distribution of captured integration sites from both vector-transduced samples were similar, as expected, with no significant difference in distribution of integrations near gene TSS, integrations within genes, or near or in known proto-oncogenes (Fig. 5b and Supplementary Table S2). We also did not identify clonal dominance within any of the samples, consistent with FV vector-transduced samples being highly polyclonal with and without an insulator.41,66 Despite these similarities, a significant reduction in the number of observed integration sites within hotspots was observed when FV vectors were insulated. The effect is seen as early as 5 days after vector exposure and increases over time as indicated by a significant increase in the number of observed integrations within hotspots in uninsulated FV vector-transduced samples (Fig. 5b). Furthermore, we did not observe a change in the number of integrations within hotspots of insulated FV vector-transduced samples over time. Therefore our data show the insulator is potentially reducing the effects of the strong promoter on the surrounding host genome, thus reducing skewing from the initial polyclonal distribution of the transduced samples and inhibiting the proliferation of cells containing integration within a hotspot.
We speculate that the initial distribution of integration sites with respect to hotspots is not significantly different for the uninsulated and insulated FV vectors. This is because the insulator element is not expected to alter the integration profile. However, as early as day 5, the uninsulated vector is potentially promoting a growth advantage of hotspots (Fig. 5b). By day 10 this difference is significantly greater, which is consistent with 5 days being sufficient time for skewing of hotspots. In this study we chose to use a highly genotoxic promoter, SFFV, in order to maximize the potential to observe a reduction of skewing by an insulator element. In the setting of gene therapy a less genotoxic promoter would be preferred, but it must still provide sufficient transgene expression to correct the disease phenotype.67,68 Also, genotoxicity has still been observed in some vectors with housekeeping promoters such as PGK, which may still benefit from the use of insulators in a clinical setting.39 Additional studies to evaluate the effect of the FV-650cHS4-R-insulated FV vector on skewing in repopulating cells are warranted, but our data in human cord blood CD34+ cells suggest that insulated FV vectors may further improve FV safety.
Evaluating insulators in both forward and reverse orientation brought to light some interesting characteristics that should be taken under consideration when developing insulated vectors. We found that insulators can have variable activity depending on their orientation and that this can be changed in the context of the vector LTR. We also found that placing an insulator in the FV vector LTR can increase or decrease the activity of an insulator (Fig. 2b and c). The direct validation and comparison of insulator activity within the context of a retroviral LTR as we have shown here has previously been understudied, and is critical to assess insulators for use in retroviral vectors. Our enhancer-blocking activity assay allowed rapid assessment of insulators in either orientation and allows rapid assessment of activity within a LTR. As new CTCF-based and also non-CTCF-based insulator sequences continue to be identified, the plasmid-based enhancer activation assay we describe here should allow high-throughput analysis of candidate insulators to efficiently identify novel promising insulators.
In summary, our findings provide additional evidence that FV vectors are relatively safe for gene therapy. We developed a high-titer 650cHS4-insulated FV vector that transfers the insulator element with high fidelity to both proviral LTRs. We also showed that this insulated FV vector may be safer than an uninsulated FV vector, based on the frequency of integrants in hotspots in cord blood CD34+ cells. Previous studies with GV and LV vectors strongly suggest that insulators add to the safety of vectors18,33,34,39 and this also seems to be the case for FV vectors. Our data strongly support further preclinical evaluation of insulated FV vectors for HSC gene therapy.
Supplementary Material
Acknowledgments
This research was supported by NIH grants AI097100 and AI102672 (G.D.T.). The authors thank Victor Bii for sharing the gammaretroviral vector CL-SGN before publication.
Author Disclosure
The authors have no competing financial interests in relation to the work described.
References
- 1.Bordignon C, Notarangelo LD, Nobili N, et al. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA-immunodeficient patients. Science 1995;270:470–475 [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, Le Deist F, Carlier F, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med 2002;346:1185–1193 [DOI] [PubMed] [Google Scholar]
- 3.Wang GP, Berry CC, Malani N, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood 2010;115:4356–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 2010;467:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santilli G, Almarza E, Brendel C, et al. Biochemical correction of X-CGD by a novel chimeric promoter regulating high levels of transgene expression in myeloid cells. Mol Ther 2011;19:122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 2003;348:255–256 [DOI] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 2008;118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Dullmann J, Schiedlmeier B, et al. Murine leukemia induced by retroviral gene marking. Science 2002;296:497. [DOI] [PubMed] [Google Scholar]
- 10.Braun CJ, Boztug K, Paruzynski A, et al. Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014;6:227–233 [DOI] [PubMed] [Google Scholar]
- 11.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis after gene therapy of SCID-X1 patients. J Clin Invest 2008;118:3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 2006;12:401–409 [DOI] [PubMed] [Google Scholar]
- 13.Zychlinski D, Schambach A, Modlich U, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther 2008;16:718–725 [DOI] [PubMed] [Google Scholar]
- 14.Bokhoven M, Stephen SL, Knight S, et al. Insertional gene activation by lentiviral and gammaretroviral vectors. J Virol 2009;83:283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosticardo M, Ghosh A, Du Y, et al. Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells. Mol Ther 2009;17:1910–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trobridge GD. Genotoxicity of retroviral hematopoietic stem cell gene therapy. Exp Opin Biol Ther 2011;11:581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trobridge G. Improved foamy virus vectors with minimal viral sequences. Mol Ther 2002;6:321–328 [DOI] [PubMed] [Google Scholar]
- 18.Li CL, Xiong D, Stamatoyannopoulos G, et al. Genomic and functional assays demonstrate reduced gammaretroviral vector genotoxicity associated with use of the cHS4 chromatin insulator. Mol Ther 2009;17:716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coci EG, Maetzig T, Zychlinski D, et al. Novel self-inactivating vectors for reconstitution of Wiskott-Aldrich syndrome. Curr Gene Ther 2015;15:245–254 [DOI] [PubMed] [Google Scholar]
- 20.Trobridge GD, Miller DG, Jacobs MA, et al. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci U S A 2006;103:1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowrouzi A, Dittrich M, Klanke C, et al. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol 2006;87:1339–1347 [DOI] [PubMed] [Google Scholar]
- 22.Deyle DR, Khan IF, Ren G, et al. Lack of genotoxicity due to foamy virus vector integration in human iPSCs. Gene Ther 2013;20:868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josephson NC, Vassilopoulos G, Trobridge GD, et al. Transduction of human NOD/SCID-repopulating cells with both lymphoid and myeloid potential by foamy virus vectors. Proc Natl Acad Sci U S A 2002;99:8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasimuzzaman M, Kim YS, Wang YD, et al. High-titer foamy virus vector transduction and integration sites of human CD34+ cell-derived SCID-repopulating cells. Mol Ther Methods Clin Dev 2014;1:14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrie PC, Huo Y, Stolitenko RB, et al. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther 2008;16:534–540 [DOI] [PubMed] [Google Scholar]
- 26.Trobridge G, and Russell DW. Cell cycle requirements for transduction by foamy virus vectors compared with those of oncovirus and lentivirus vectors. J Virol 2004;78:2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emery DW. The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum Gene Ther 2011;22:761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arumugam PI, Higashimoto T, Urbinati F, et al. Genotoxic potential of lineage-specific lentivirus vectors carrying the β-globin locus control region. Mol Ther 2009;17:1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neff T, Shotkoski F, and Stamatoyannopoulos G. Stem cell gene therapy, position effects and chromatin insulators. Stem Cells 1997;15(Suppl 1):265–271 [DOI] [PubMed] [Google Scholar]
- 30.West AG, Gaszner M, and Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev 2002;16:271–288 [DOI] [PubMed] [Google Scholar]
- 31.Burgess-Beusse B, Farrell C, Gaszner M, et al. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci U S A 2002;99:16433–16437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace JA, and Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev 2007;17:400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arumugam PI, Urbinati F, Velu CS, et al. The 3′ region of the chicken hypersensitive site-4 insulator has properties similar to its core and is required for full insulator activity. PLoS One 2009;4:e6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaussin A, Modlich U, Bauche C, et al. CTF/NF1 transcription factors act as potent genetic insulators for integrating gene transfer vectors. Gene Ther 2012;19:15–24 [DOI] [PubMed] [Google Scholar]
- 35.Chung JH, Whiteley M, and Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 1993;74:505–514 [DOI] [PubMed] [Google Scholar]
- 36.Chung JH, Bell AC, and Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci U S A 1997;94:575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pankiewicz R, Karlen Y, Imhof MO, et al. Reversal of the silencing of tetracycline-controlled genes requires the coordinate action of distinctly acting transcription factors. J Gene Med 2005;7:117–132 [DOI] [PubMed] [Google Scholar]
- 38.Ferrari S, Simmen KC, Dusserre Y, et al. Chromatin domain boundaries delimited by a histone-binding protein in yeast. J Biol Chem 2004;279:55520–55530 [DOI] [PubMed] [Google Scholar]
- 39.Cesana D, Ranzani M, Volpin M, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther 2014;22:774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trobridge GD, Beard BC, Wu RA, et al. Stem cell selection in vivo using foamy vectors cures canine pyruvate kinase deficiency. PLoS One 2012;7:e45173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olszko ME, Adair JE, Linde I, et al. Foamy viral vector integration sites in SCID-repopulating cells after MGMTP140K-mediated in vivo selection. Gene Ther 2015;22:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trobridge GD, Wu RA, Hansen M, et al. Cocal-pseudotyped lentiviral vectors resist inactivation by human serum and efficiently transduce primate hematopoietic repopulating cells. Mol Ther 2010;18:725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiem HP, Wu RA, Sun G, et al. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther 2010;17:37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C, Teng EM, Summers RG Jr, et al. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci 2006;26:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beard BC, Adair JE, Trobridge GD, et al. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol Biol 2014;1185:321–344 [DOI] [PubMed] [Google Scholar]
- 46.Rae DT, Collins CP, Hocum JD, et al. Modified genomic sequencing PCR using the MiSeq platform to identify retroviral integration sites. Hum Gene Ther Methods 2015:26:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Kobert K, Flouri T, et al. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014;30:614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921 [DOI] [PubMed] [Google Scholar]
- 49.Hocum JD, Battrell LR, Maynard R, et al. VISA—Vector integration site analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics 2015;16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbinati F, Arumugam P, Higashimoto T, et al. Mechanism of reduction in titers from lentivirus vectors carrying large inserts in the 3′LTR. Mol Ther 2009;17:1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramezani A, Hawley TS, and Hawley RG. Combinatorial incorporation of enhancer-blocking components of the chicken β-globin 5′HS4 and human T-cell receptor α/δ BEAD-1 insulators in self-inactivating retroviral vectors reduces their genotoxic potential. Stem Cells 2008;26:3257–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007;128:1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanawa H, Yamamoto M, Zhao H, et al. Optimized lentiviral vector design improves titer and transgene expression of vectors containing the chicken β-globin locus HS4 insulator element. Mol Ther 2009;17:667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emery DW, Yannaki E, Tubb J, et al. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc Natl Acad Sci U S A 2000;97:9150–9155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An W, and Telesnitsky A. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 2001;286:475–482 [DOI] [PubMed] [Google Scholar]
- 56.Nielsen TT, Jakobsson J, Rosenqvist N, et al. Incorporating double copies of a chromatin insulator into lentiviral vectors results in less viral integrants. BMC Biotechnology 2009;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breda L, Casu C, Gardenghi S, et al. Therapeutic hemoglobin levels after gene transfer in β-thalassemia mice and in hematopoietic cells of β-thalassemia and sickle cells disease patients. PLoS One 2012;7:e32345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.FitzGerald TJ, Henault S, Sakakeeny M, et al. Expression of transfected recombinant oncogenes increases radiation resistance of clonal hematopoietic and fibroblast cell lines selectively at clinical low dose rate. Radiation Res 1990;122:44–52 [PubMed] [Google Scholar]
- 59.Modlich U, Navarro S, Zychlinski D, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther 2009;17:1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cattoglio C, Facchini G, Sartori D, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 2007;110:1770–1778 [DOI] [PubMed] [Google Scholar]
- 61.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet 2002;32:166–174 [DOI] [PubMed] [Google Scholar]
- 62.Liu M, Maurano MT, Wang H, et al. Genomic discovery of potent chromatin insulators for human gene therapy. Nat Biotechnol 2015;33:198–203 [DOI] [PubMed] [Google Scholar]
- 63.Nakahashi H, Kwon KR, Resch W, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 2013;3:1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhee HS, and Pugh BF. Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 2011;147:1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H, Tian Y, Shu W, et al. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One 2012;7:e41374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiem HP, Allen J, Trobridge G, et al. Foamy-virus-mediated gene transfer to canine repopulating cells. Blood 2007;109:65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malik P, and Arumugam PI. Gene therapy for β-thalassemia. Hematology Am Soc Hematol Educ Program 2005:45–50 [DOI] [PubMed] [Google Scholar]
- 68.Hunter MJ, Zhao H, Tuschong LM, et al. Gene therapy for canine leukocyte adhesion deficiency with lentiviral vectors using the murine stem cell virus and human phosphoglycerate kinase promoters. Hum Gene Ther 2011;22:689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.