Abstract
The invasive nature of glioblastoma renders them incurable by current therapeutic interventions. Using a novel invasive human glioma model, we previously identified the neurotrophin receptor p75NTR (aka CD271) as a mediator of glioma invasion. Herein, we provide evidence that preventing phosphorylation of p75NTR on S303 by pharmacological inhibition of PKA, or by a mutational strategy (S303G), cripples p75NTR-mediated glioma invasion resulting in serine phosphorylation within the C-terminal PDZ-binding motif (SPV) of p75NTR. Consistent with this, deletion (ΔSPV) or mutation (SPM) of the PDZ motif results in abrogation of p75NTR-mediated invasion. Using a peptide-based strategy, we identified PDLIM1 as a novel signaling adaptor for p75NTR and provide the first evidence for a regulated interaction via S425 phosphorylation. Importantly, PDLIM1 was shown to interact with p75NTR in highly invasive patient-derived glioma stem cells/tumor-initiating cells and shRNA knockdown of PDLIM1 in vitro and in vivo results in complete ablation of p75NTR-mediated invasion. Collectively, these data demonstrate a requirement for a regulated interaction of p75NTR with PDLIM1 and suggest that targeting either the PDZ domain interactions and/or the phosphorylation of p75NTR by PKA could provide therapeutic strategies for patients with glioblastoma.
Introduction
Human malignant glioma is one of the most common adult primary central nervous system tumors with a median survival of only 14.6 months after diagnosis.1, 2 A major barrier to effective treatment of glioblastoma is their highly invasive nature; they extend tendrils several centimeters away from the main tumor mass rendering them incurable by localized therapy including surgery or radiotherapy.3, 4 Invading malignant glioma cells comprise a cell population that are genotypically and phenotypically distinct from their noninvasive counterparts, activating a number of coordinate cellular programs including those necessary for migration, invasion and survival.4, 5, 6, 7, 8, 9, 10, 11, 12 Many individual genes have been implicated in glioma invasion and recently studies identified a subclass of glioma-expressing genes involved in cell migration and invasion that strongly correlate with poor patient survival.13, 14, 15, 16, 17
We previously found that the neurotrophin receptor, p75NTR, was upregulated in invasive glioma cells18, 19 and established p75NTR as a major contributor to their invasive nature.18, 19 p75NTR is a transmembrane glycosylated receptor expressed by neurons, neural stem cells, astrocytes, oligodendrocytes precursors and Schwann cells20 where it functions through interactions with several ligands and co-receptors21, 22, 23 to mediate cell death, survival, migration and axonal growth inhibition (reviewed in Reichardt23 and Kraemer et al.24). Using an unbiased approach, we found that p75NTR through a neurotrophin-dependent mechanism regulates the invasive behavior of glioma cells,18, 19 a function also observed in Schwann cells, neural crest cells, melanocytes and melanoma.25, 26, 27, 28, 29, 30, 31
Although the precise elucidation of p75NTR downstream signaling remains unknown, a number of effector pathways have been uncovered including RhoA,32 JNK,33 MAPK,34 NFκB,26 ceramide35 and HIF.36 Although the exact biological implications of these signaling events are largely undefined, cellular context, localization and proteolytic processing are major determinants for p75NTR-mediated events.24, 37, 38 p75NTR itself does not contain intrinsic kinase activity and instead requires association with other proteins,24 some of which require the phosphorylation of the receptor by kinases such as the cAMP-dependent protein kinase PKA, where in cerebellar neuron phosphorylation by PKA regulates the translocation of p75NTR to lipid rafts.39 p75NTR also contains protein interaction domains including a PDZ-binding motif at the extreme C-terminus of the receptor shown to interact with the Fas-associated phosphatase (FAP-1) in 293T cells40 and Par3 during Schwann cell myelination.41 PDZ domains have been found in a number of intracellular signaling proteins42, 43 and, in the context of a LIM domain, can interact with the actin cytoskeleton.44, 45, 46, 47, 48 PDLIM1 is a member of the ALP subfamily of PDZ/LIM proteins, which are postulated to have a role in actin cytoskeleton architecture.46, 49, 50, 51, 52, 53, 54
There is mounting evidence that the neurotrophin receptor family and their respective ligands are rationale therapeutic targets. For example, p75NTR (CD271) has been shown to be a crucial molecule driving melanoma initiation, self-renewal, invasion and metastasis,30, 55, 56 features also associated with glioma.18, 19, 57, 58, 59, 60 We have previously provided evidence for the targeting of p75NTR using clinically applicable γ-secretase inhibitors that prevent intramembrane proteolysis of p75NTR, a prerequisite for glioma invasion.18, 19 Use of γ-secretase inhibitors have, however, generated considerable debate owing to the numerous cellular targets and unforeseen clinical complications.61, 62 Thereby, the identification of pathways utilized by p75NTR to drive cancer invasion and tumorigenesis would provide alternate avenues for therapeutic targeting of this signaling axis. Thus, as an alternative approach, we undertook defining the molecular signaling downstream of p75NTR that are required to mediate glioma invasion. Herein, we provide evidence that phosphorylation of p75NTR by PKA modulates the phosphorylation of specific amino acid residues within the cytoplasmic domain of p75NTR resulting in a regulated interaction with PDLIM1, a novel signaling adaptor required for p75NTR-mediated glioma invasion.
Results
PKA inhibition abrogates p75NTR-induced glioma invasion
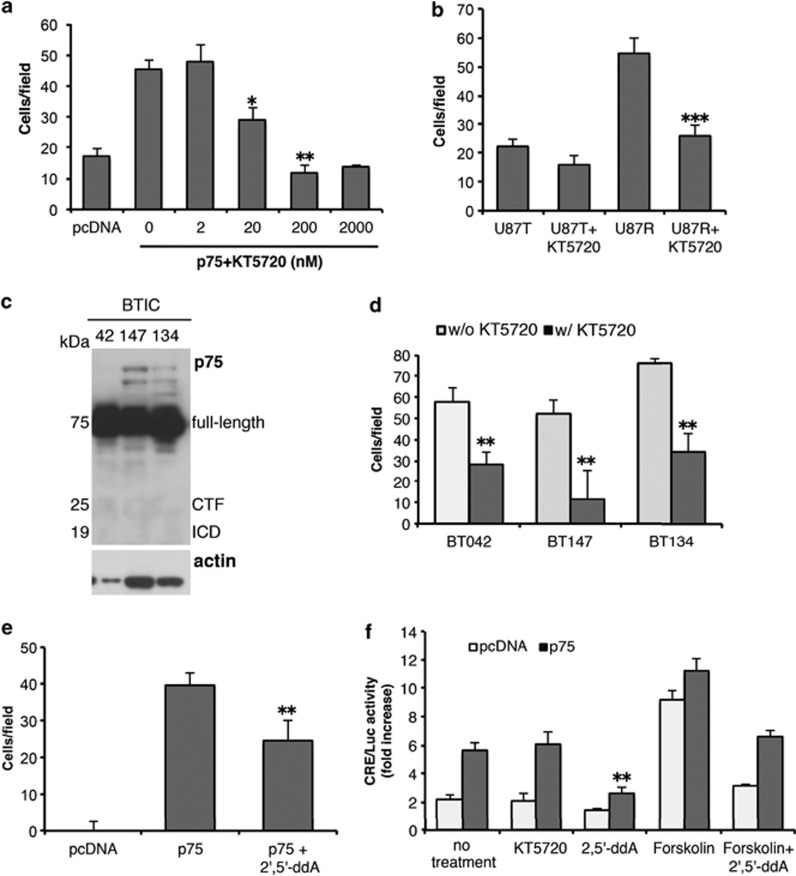
PKA has been shown to induce serine phosphorylation of the cytoplasmic tail of p75NTR, an event required for lipid-raft localization and downstream signaling in cerebellar neurons.39 Consistent with a role for compartmentalized signaling cholesterol depletion by cholesterol oxidase or methyl-β-cyclodextrin abolished the ability to mediate glioma invasion in vitro (Supplementary Figure S1) suggesting a role for lipid rafts and perhaps PKA for p75NTR-mediated glioma invasion. To determine whether PKA activity is important for p75NTR-regulated activities in glioma, we checked whether PKA activation could promote glioma invasion. Using a PKA-selective pharmacological inhibitor KT5720, invasive U87 human glioma cells stably expressing p75NTR (CD271) (U87p75NTR) were assessed for their ability to invade collagen. Treatment of U87p75NTR cells with KT5720 resulted in a dose-dependent inhibition of invasion as compared with noninvasive U87pcDNA cells (Figure 1a). This inhibitory effect was also observed in a highly invasive glioma cell line established by serial in vivo selection (U87R) in which p75NTR also regulates its invasive behavior18 (Figure 1b), and importantly, in three independent p75NTR expressing patient-derived primary cultures, herein referred to as brain tumor-initiating cells ((BT042, BT134, BT147) Figures 1c and d). This decrease in invasion was also observed following treatment with the adenylyl cyclase inhibitor 2', 5'-dideoxyadenosine which inhibits the production of cAMP (Figure 1e). Moreover, glioma cells expressing p75NTR had significantly higher levels of cAMP as assessed using a transcriptional CRE-reporter assay, and transcriptional activity was further augmented by the adenylyl cyclase activator forskolin (1 μM) (Figure 1f). Taken together, these data suggest that cAMP/PKA-induced p75NTR phosphorylation is required for glioma invasion.
Figure 1.
PKA inhibition significantly abrogated p75NTR-induced glioma invasion. (a) Treatment with increasing concentrations of the PKA inhibitor KT5720 (KT) inhibited invasion of U87 cells expressing full-length p75NTR (p75). Following pre-treatment with KT5720 for 1 h, the invasive ability of the U87 cells expressing full-length p75NTR (p75) or empty vector (pcDNA; control) were determined using collagen-coated transwells. Similar results were observed in two independent experiments. Asterisk (*) indicate P<0.05 and double asterisks (**) indicate P<0.01 versus non-treated U87p75 cells (one-way ANOVA with the Newman-Keuls post-test). (b) Highly invasive U87R and noninvasive U87T cells were isolated by serial in vivo selection and assessed for their invasive ability in the absence or presence of KT5720 (200 nM). Asterisks (***) indicate P<0.001 as compared with non-treated cells. (c) Western blot shows the expression of p75NTR in patient-derived glioma cells BT042, BT147 and BT134. (d) Histogram shows that the invasion of the patient-derived glioma cells (BTIC) in the presence of fetal bovine serum was suppressed by KT5720 (200 nM). Asterisks (**) indicate P<0.01 as compared with the corresponding non-treated BTIC. (e) p75NTR-mediated glioma invasion is dependent on cAMP. U87pcDNA or p75 cells were treated for 1 h with an adenylyl cyclase inhibitor (2', 5'-dideoxyadenosine(2', 5'-ddA)) and invasion assays (described above) were performed. Asterisk (**) indicate P<0.01 versus non-treated U87p75 cells. (f) U87p75NTR treated with the PKA inhibitor KT5720 (1 h, 200 nM), 2', 5'-ddA (1 h, 30 μM) or the adenylyl cyclase activator forskolin (1 μM) in the presence or absence of 2', 5'-ddA were assessed for cAMP activity using a CRE-luciferase reporter construct. Histogram shows the fold increases in luciferase activity as compared with vehicle-treated cells. Asterisk (**) indicate P<0.01 versus non-treated U87p75 cells. Data represent the mean±s.e.m. of triplicates, representative of at least three independent experiments.
Mutation of the PKA phosphorylation site (S303G) abrogates p75NTR-mediated invasion
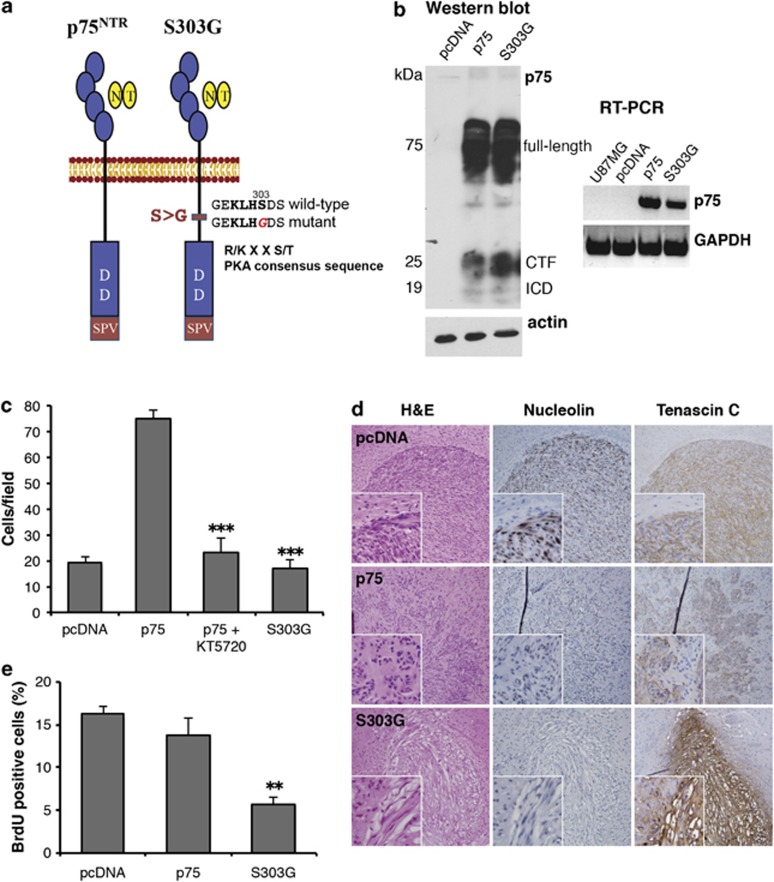
In mouse cerebellar neurons, PKA phosphorylates the cytoplasmic tail of p75NTR on serine 304 (human equivalent S303).39 To assess the importance of S303 phosphorylation by PKA and its potential role in p75NTR-mediated glioma invasion, U87 cells stably expressing a mutant construct of p75NTR/CD271 with a single point mutation at S303 were generated (p75S303G; Figure 2a) and assessed for receptor localization, topology and ability to bind neurotrophin. Previously, we demonstrated that glioma cells secrete high levels of the p75NTR ligand BDNF and upon p75NTR expression the ligand becomes cell associated, presumably bound to p75NTR.18 Flow cytometry analysis using a monoclonal antibody specific to the extracellular domain of p75NTR confirmed plasma membrane expression and correct topography of p75S303G (Supplementary Figure S2). In addition, glioma-derived BDNF was localized to the cell membrane, similar to the wild-type receptor, suggesting that BDNF binding was not compromised in the mutant (Supplementary Figure S3).
Figure 2.
Mutation of the PKA phosphorylation site (S303G) abrogates p75NTR-mediated invasion in vitro and in vivo. (a) Schematic diagram of wild-type and mutant p75NTR constructs. p75NTR has one putative PKA recognition consensus site (R/K X X S/T; KLHS) within the intracellular domain (ICD) of the protein. A PKA phosphorylation-defective mutant was generated by substitution of amino acid S303 by glycine (S303G). (b) U87MG glioma cells were stably transfected with pcDNA3.1 (pcDNA), pcDNA 3.1 containing wild-type p75NTR (p75) or pcDNA 3.1 containing the S303G mutant of p75NTR. RT–PCR and western blot confirm the expression of the wild-type and mutant receptors. RT–PCR analysis of GAPDH and western blot analysis of actin were included as loading controls. Western blot analysis detected the full-length p75NTR, the 25-kDa C-terminal fragment (CTF) and the 19-kDa ICD of p75NTR. (c) Invasion of S303G cells in collagen-coated transwell chambers was significantly decreased compared with wild-type p75NTR-expressing cells (p75). Noninvasive U87pcDNA were used for comparison. Histogram shows the mean±s.e.m. from three independent experiments; asterisks (***) indicate P<0.001 as compared with non-treated U87p75 (one-way ANOVA with the Neuman-Keuls post-test). (d) pcDNA, p75 and S303G cells (5 × 104 cells) were implanted intracerebrally into SCID mice and allowed to grow for 28 days. The mice were killed, and paraffin brain sections were stained with Hematoxylin & Eosin (H&E) or antibodies against human nucleolin or tenascin-C (brown) to visualize the tumors. Sections were counterstained with hematoxylin (blue). (e) In vivo tumor proliferation was determined by injecting bromodeoxyuridine (BrdU) into tumor-bearing mice 24 h prior to killing of the mice. Frozen brain sections were stained with an antibody against BrdU and counterstained with toluidine blue to visualize the cell nucleus. Cells that had divided during the 24 h prior to killing of the mice stained positively for BrdU, and the percentage of BrdU-positive cells was counted. Histogram represents the percentage of BrdU-positive cells in five consecutive fields. Values shown are the mean±s.e.m. from five independent mice; asterisks (**) indicate P<0.01 as compared with control U87pcDNA (one-way ANOVA with the Neuman-Keuls post-test).
The p75S303G cells were then assessed for their invasive ability. Mutation of p75NTR at S303 resulted in a significant decrease in invasion compared with the wild-type p75NTR despite these constructs being expressed at comparable levels in vitro (Figures 2b and c). To confirm whether the loss of invasive ability in vitro was maintained in vivo, p75S303G cells were implanted into the brains of immunocompromised (SCID) mice. U87p75NTR or U87pcDNA were used for comparison. Twenty-eight days after implantation, mice were killed and formalin-fixed paraffin-embedded brain sections were stained with Hematoxylin & Eosin (Figure 2d, left panel) or antibodies against human nucleolin or the extracellular matrix protein tenascin C63 to visualize the tumor (Figure 2d, center and right panel). Implantation of U87pcDNA glioma cells formed well-circumscribed tumors, while U87p75NTR glioma cells formed tumors with highly infiltrative edges. In striking contrast to the invasive U87p75NTR and consistent with the in vitro data, p75S303G-expressing tumors were well circumscribed, similar to the U87pcDNA. Comparable results were seen in three independent experiments. Taken together, these data suggest that phosphorylation of p75NTR at S303 is required for p75NTR mediated-glioma invasion.
As we have demonstrated that p75NTR undergoes γ-secretase-mediated processing in human glioma, and this was required for p75NTR-induced invasion,19 the possibility existed that mutation of the PKA phosphorylation site would affect regulated intramembrane proteolysis of p75NTR. However, as observed in Figure 2b, both the C-terminal fragment (α-secretase generated) and intracellular domain fragment (γ-secretase generated) were detected in p75S303G-expressing cells in vitro. These data suggest that in addition to intramembrane proteolysis of p75NTR, other elements are required for the invasive behavior, such as posttranslational modifications and/or specific subcellular localization. In addition, although no difference was observed in the overall growth or viability in the presence of the PKA inhibitor KT5720 or in the PKA-defective p75NTR-expressing glioma cells in vitro, a reproducible reduction in bromodeoxyuridine incorporation was observed in vivo (Figure 2e) suggesting that the brain microenvironment has an impact on the growth rate or the cellular viability of these cells.
PKA regulates the phosphorylation of Serine425 in the p75NTR-PDZ-binding motif
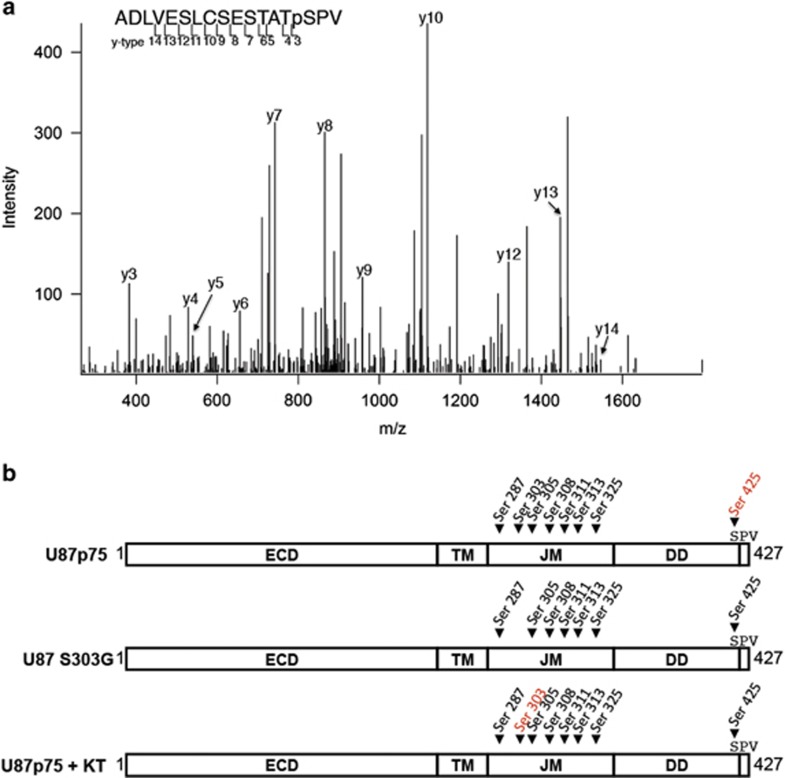
To confirm that S303 on p75NTR is phosphorylated in human glioma, we assessed endogenous phosphorylation by nano-liquid chromatography–tandem mass spectrometry. Phosphorylation of S303 was detected on the wild-type receptor but no phosphorylation was detected at amino acid 303 in p75S303G cells (Figure 3a). Phosphorylation was, however, detected on seven other serine residues (287, 303, 305, 308, 311, 313, 325 and 425) including a previously unidentified phosphorylation event on S425 of p75NTR (Figure 3b). This newly identified phosphorylation site is located within the extreme C-terminus of the protein and is part of a PDZ-binding motif.40, 64 Phosphorylation of S425 was also observed in cells treated with the PKA inhibitor KT5720 suggesting a regulatory role for this specific phosphorylation event (Figure 3b).
Figure 3.
PKA regulates the phosphorylation of S425 within the p75NTR-PDZ binding motif. p75NTR (U87p75), mutant S303G p75NTR or cells expressing p75NTR that were treated with the PKA inhibitor KT5720 (200 nM) were lysed, immunoprecipitated with anti-p75NTR, run on 10% sodium dodecyl sufate–polyacrylamide gel electrophoresis and proteins were visualized by silver stain. Protein bands representing p75NTR were removed from the gel and in-gel trypsin digestion was performed. Digested peptides and phosphorylated serine residues were identified using nano-liquid chromatography–tandem mass spectrometry (LC/MS/MS) on a LTQ Orbitrap velos (Thermo Scientific; Southern Alberta Mass Spectrometry Center). (a) Modification and peptide search were performed with Mascot software v2.3. The MS/MS spectrum for the phospho-peptide ADLVESLCSESTAT(pS)PV, with annotated b and y ions, is shown. Phosphorylation at serine -1 is indicated by presence of y3 ion (+80Da for the mass of the phosphate group). (b) Shown is a schematic representation of p75NTR indicating serine phosphorylated residues (black text) within the intracellular domain of p75NTR isolated from U87 cells expressing wild-type p75NTR (U87p75), mutant S303G p75NTR or cells expressing p75NTR that were treated with the PKA inhibitor KT5720 (200 nM). Red text indicates unphosphorylated serine residues.
Mutation of the p75NTR-PDZ-binding motif reduces glioma invasion
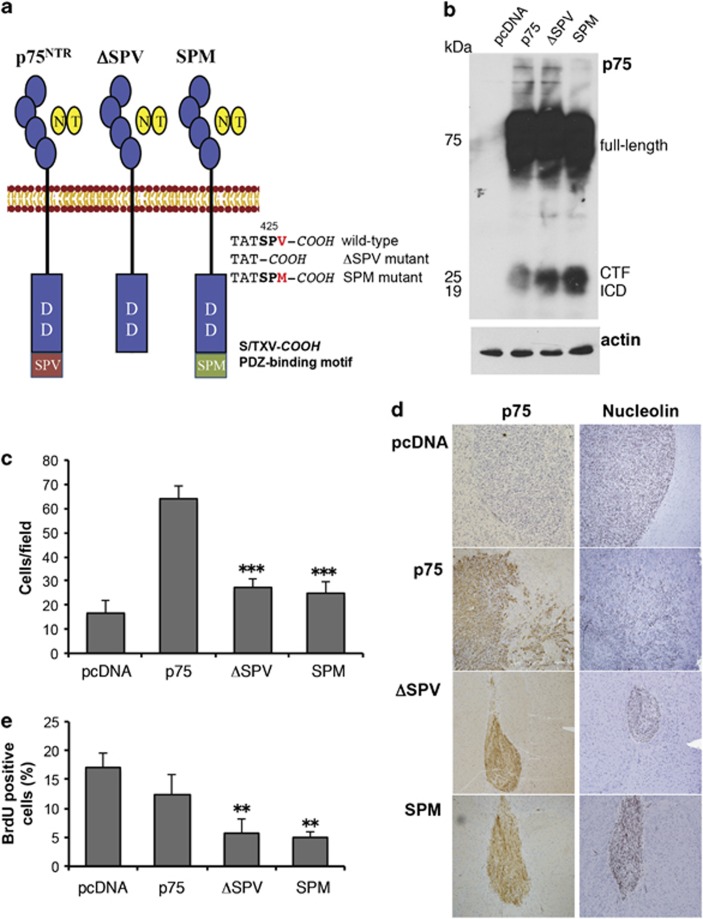
To assess the physiological relevance of phosphorylation at S425 and the importance of the C-terminal PDZ-binding domain of p75NTR, we generated two specific p75NTR mutants, both predicted to compromise p75NTR binding to PDZ-containing effector proteins.40, 64 The first construct was generated by deleting the entire PDZ-binding motif (SPV) at the extreme C-terminal of p75NTR (ΔSPV; Figure 4a). In the second the C-terminal valine was substituted with methionine (SPM; Figure 4a). U87 glioma cells were stably transfected with ΔSPV or SPM constructs and receptor expression (Figure 4b; Supplementary Figure S4) and BDNF binding (Supplementary Figure S5) was confirmed. Disruption of the PDZ-binding domain resulted in a significant decrease in p75NTR-mediated invasion in vitro (Figure 4c). To confirm this in vivo, these cells were implanted into the brains of SCID mice and allowed to grow for 28 days. The mice were killed, and paraffin-embedded brain sections were stained with antibodies against human nuclei (Figure 4d; brown, center panel) and p75NTR (Figure 4d; brown, right panel). Disruption of the PDZ-binding motif of p75NTR resulted in tumors with distinctly demarcated edges. Moreover, tumors generated from glioma cells expressing either SPM or ΔSPV p75NTR mutants were smaller and showed significant reduction of bromodeoxyuridine incorporation in vivo (Figure 4e), a result analogous to tumors generated using the PKA phosphorylation-defective U87 cells. These data suggest that the PDZ-binding domain of p75NTR is required for p75NTR-mediated glioma invasion.
Figure 4.
p75NTR C-terminal PDZ-binding motif is required for glioma invasion in vitro and in vivo. (a) Schematic diagram of the p75NTR PDZ-binding motif mutants (ΔSPV; SPM). (b) Western blot analysis using a C-terminal domain antibody to p75NTR confirms the expression of the wild-type (p75) and mutant (ΔSPV; SPM) p75NTR constructs. Western blot for actin was used as a loading control. (c) Histogram shows the number of U87 p75NTR wild-type and PDZ-binding mutants (ΔSPV; SPM) that migrated through a collagen-coated transwell. Values are the mean±s.e.m. from three independent experiments; asterisks (***) indicate P<0.001 as compared with p75NTR (one-way ANOVA with the Neuman-Keuls post-test). (d) U87pcDNA, p75NTR, ΔSPV and SPM glioma cells (5 × 104 cells) were implanted intracerebrally into SCID mice and allowed to grow for 28 days. The mice were killed, and paraffin brain sections were stained with human nucleolin and p75NTR (brown) to visualize the tumors. Sections were counterstained with hematoxylin (blue). (e) Bromodeoxyuridine (BrdU) was injected into the tumor-bearing mice 24 h prior to sacrifice. Frozen brain sections were stained with an antibody against BrdU and counterstained with toluidine blue to visualize the cell nucleus. Histogram represents the percentage of BrdU-positive cells in five consecutive fields. Values shown are the mean±s.e.m. from five independent mice; asterisks (**) indicate P<0.01 as compared with U87pcDNA (one-way ANOVA with the Neuman-Keuls post-test).
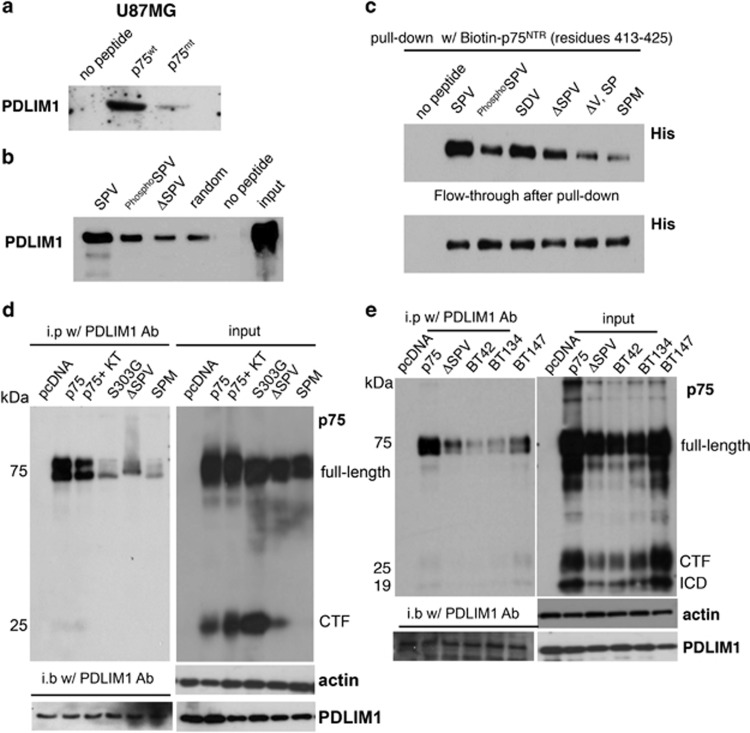
PDLIM1 directly interacts with the PDZ-binding motif of p75NTR an interaction that is regulated by phosphorylation of S425
As the PDZ-binding domain was important for p75NTR-mediated glioma invasion, we sought to identify the PDZ domain-containing protein that binds p75NTR to regulate invasion. Using biotinylated peptides corresponding to the C-terminus of either p75NTR (p75WT peptide, Biotin-N-LCSESTATSPV-COOH, residues 417-427), or a valine deletion mutant of p75NTR (p75MT peptide, Biotin-N- LCSESTATSP-COOH, residues 417–426) as affinity reagents, we identified PDLIM1 as a potential p75NTR-interacting protein using nano-liquid chromatography–tandem mass spectrometry (Supplementary Figure S6). PDLIM1 (aka CLP-36, elfin, hCLIM1) was selected for further investigation based on the presence of an N-terminal PDZ domain and a C-terminal LIM domain that has been shown to associate with α-actinin-1 and α-actinin-446, 65 and implicated in cell motility, endocytosis and cancer invasion.66, 67, 68, 69, 70, 71 To assess the specificity of the PDLIM1-p75NTR interaction in glioma cells, we first demonstrated that PDLIM1 bound the wild-type p75NTR peptide but not the PDZ-binding motif mutant (Figure 5a). Next, based on the identification of a PKA-regulated phosphorylation on S425 (Figure 3), we determined whether the interaction of PDLIM1 with p75NTR was regulated by phosphorylation of S425. Therefore, p75NTR C-terminal peptides containing an unphosphorylated S425 (SPV, Biotin-N-LCSESTATSPV-COOH, residues 417–427), a phosphorylated S425 (PhosphoSPV, Biotin-N-LCSESTATPhosphoSPV-COOH, residues 417–427) or deleted for the PDZ-binding motif (ΔSPV, Biotin-N-VESLCSESTAT-COOH, residues 414–424) were assessed for their ability to interact with PDLIM1. As shown in Figure 5b, both the PhosphoSPV and ΔSPV peptide were compromised for their ability to bind PDLIM1 suggesting that PDLIM1 interacts with the C-terminal cytoplasmic domain of p75NTR when S425 is in an unphosphorylated state.
Figure 5.
PDLIM1 associates with p75NTR via the C-terminal PDZ-binding motif, an interaction regulated by phosphorylation of S425. (a) Using a biotinylated peptide to the C-terminus of either p75NTR (p75wt peptide, Biotin-N-LCSESTATSPV-COOH, residues 417-427) or a Valine deletion mutant of p75NTR (p75MT peptide, biotin-N- LCSESTATSP-COOH, residues 417-426) as affinity reagents, PDLIM1 was confirmed as a p75NTR-interacting protein by western blot analysis. (b) To assess the physiological relevance of the phosphorylation of S425 within the PDZ-binding motif, biotin-labeled peptides corresponding to the p75NTR wild-type C-terminal (SPV), the S425 phosphorylated C-terminal (PhosphoSPV) and the PDZ-binding motif deleted C-terminal (ΔSPV) were used as affinity reagents. PDLIM1 was shown to interact with maximum affinity to the wild-type unphosphorylated peptide. (c) Biotin-labeled p75NTR peptides (SPV, PhosphoSPV, ΔSPV, ΔV, SP and SPM) were incubated with His-tagged recombinant PDLIM1 protein, respectively. Pull-down assays were performed to determine the amount of PDLIM1 protein bound to each peptide. Flow-through shows unbound proteins after incubation. (d) Immunoprecipitation with PDLIM1 antibodies co-precipitated p75NTR and PDLIM1 while the mutant alleles of p75NTR (S303G, ΔSPV and SPM) or p75NTR cells treated with the PKA inhibitor KT5720 (KT; 200 nM) were severely compromised for PDLIM1 binding. (e) Interaction of PDLIM1 with p75NTR was confirmed in patient-derived glioma stem cells/tumor-initiating cells (BT42, BT134, BT147) by immunoprecipitation using anti-PDLIM1. Input cell lysates were assessed by western blot analysis for expression of p75NTR and PDLIM1.
To determine whether there is a direct interaction between PDLIM1 and p75NTR, in vitro pull-down experiments were performed using His-tagged PDLIM1 and a biotinylated-p75NTR C-terminal peptide (Biotin-N-LCSESTATSPV-COOH, residues 417–427) (Figure 5c). As predicted, His-tagged PDLIM1 was found to directly bind the p75NTR peptide containing an unphosphorylated serine at position 425 (SPV), an interaction that was dramatically reduced when S425 was phosphorylated (PhosphoSPV), when the C-terminal valine was either substituted by methionine (SPM, Biotin-N-LCSESTATSPM-COOH, residues 417-427) or deleted (ΔV, Biotin-N-SLCSESTATSP-COOH, residues 416-426) or when the entire PDZ-motif (ΔSPV) was removed. No change in binding was observed when proline within the PDZ-binding domain was substituted with aspartate (SDV, Biotin-N-LCSESTATSDV-COOH, residues 417-427). Furthermore, and consistent with these data, immunoprecipitation of lysates from p75NTR-expressing glioma cells with either p75NTR or PDLIM1 antibodies resulted in the co-precipitation of PDLIM1 and p75NTR, respectively (Figures 5d and e, Supplementary Figure S7). Similar results were observed using lysates from U87 cells expressing only the intracellular domain of p75NTR or other glioma cell lines (U251, U118) expressing the p75NTR (Supplementary Figures S7 and S8). In contrast, the ability to co-precipitate PDLIM1 with the mutant alleles of p75NTR (S303G, ΔSPV and SPM) or p75NTR cells treated with the PKA inhibitor KT5720 was severely compromised (Figure 5d). Most importantly, the interaction of PDLIM1 with p75NTR was confirmed in genetically diverse patient-derived glioma cells (Figure 5e).
PDLIM1 is critical for glioma cell invasion both in vitro and in vivo
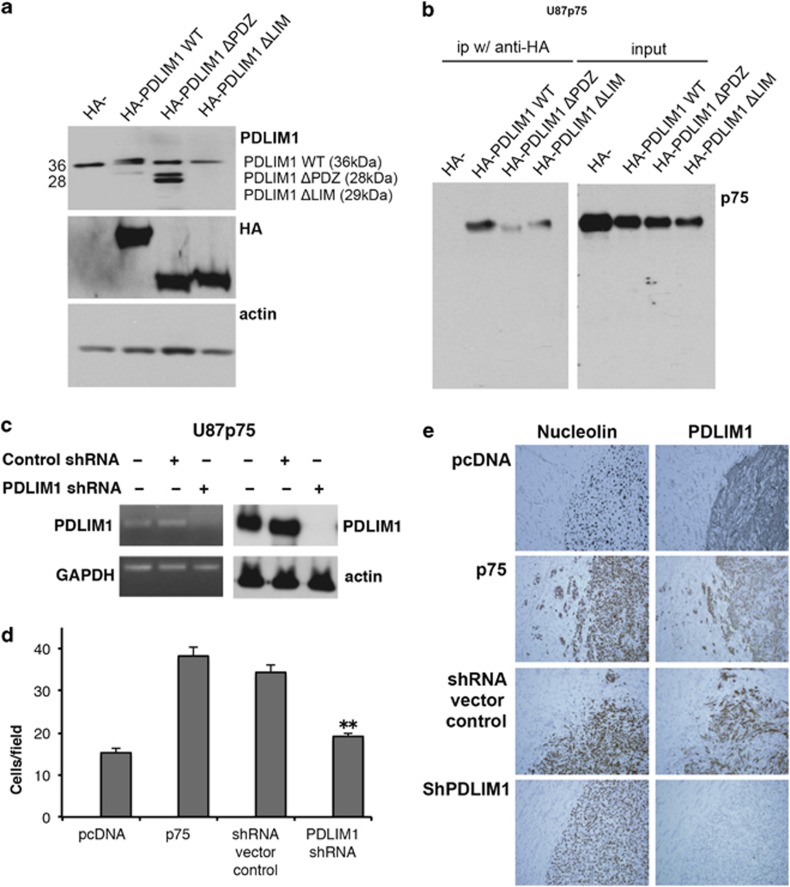
As PDLIM1 possesses two functional domains, PDZ and LIM, we next assessed the requirement of these domains for the interaction with p75NTR. To address this, HA-tagged PDLIM1 constructs lacking either the PDZ domain (ΔPDZ) or the LIM domain (ΔLIM) were generated. U87p75 glioma cells were transfected with HA-PDLIM1-ΔPDZ, HA-PDLIM1-ΔLIM or full-length HA-PDLIM1 (Figure 6a) and assessed for their ability to interact with p75NTR. Although removal of either the PDZ or LIM domain of PDLIM1 compromised the ability of the protein to interact with p75NTR, the removal of the PDZ domain resulted in a nearly complete loss of p75NTR binding (Figure 6b).
Figure 6.
PDLIM1 is critical for glioma cell invasion. (a) U87p75 cells were transfected with full-length HA-PDLIM1 or HA-PDLIM1 lacking either the PDZ domain (ΔPDZ) or the LIM domain (ΔLIM). Shown are western blot analyses for PDLIM1 (top), HA- (middle) and actin (bottom). The PDLIM1 antibody was developed against the LIM domain and thus does not recognize the HA-PDLIM1- ΔLIM therefore the expression of the ΔLIM construct was confirmed by the HA-specific antibody. HA and actin were used to confirm similar protein expression and protein loading, respectively. (b) Shown is p75NTR western blot analysis of cell lysates from U87p75 cells expressing HA-PDLIM1-WT, HA-PDLIM1-ΔPDZ or HA-PDLIM1-ΔLIM immunoprecipitated with anti-HA (left panel). Right panel shows the level of p75NTR in each input cell lysate. Immunoprecipitation with HA antibodies co-precipitated p75NTR. (c) To generate PDLIM1 knockdown cells, U87p75 cells were transfected with PDLIM1shRNA or a control shRNA. RT–PCR (left panels) and western blot analysis (right panels) show reduced or absent PDLIM1 expression when cells are expressing PDLIM1 shRNA. GAPDH and actin were used as loading controls. (d) Inhibition of PDLIM1 expression in U87p75 human glioma cells by shRNA results in a significant decrease of p75NTR-mediated invasion through collagen-coated transwells. Histogram shows the mean±s.e.m. from five independent experiments; asterisks (**) indicate P<0.01 as compared with U87 shRNA vector control (one-way ANOVA with the Neuman-Keuls post-test). (e) U87pcDNA, p75NTR and p75NTR shPDLIM1 glioma cells (5X104 cells) were implanted intracerebrally into SCID mice and allowed to grow for 28 days. The mice were killed, and paraffin brain sections were stained with human nucleolin (brown) to visualize the tumors (left panel) or PDLIM1 (brown, right panel). Sections were counterstained with hematoxylin (blue).
Next, to directly assess the role of PDLIM1 in glioma behavior, U87p75 cells were stably transfected with PDLIM1 shRNA (pLK0.1-puro shPDLIM1) or a control shRNA (pLK0.1-puro). Western blot analysis and RT–PCR showed that PDLIM1 expression was dramatically reduced in U87p75 cells transfected with PDLIM1 shRNA as compared with the control shRNA (Figure 6c). The cells stably expressing PDLIM1 shRNA were then assessed for their invasive ability in vitro where knockdown of PDLIM1 was found to significantly reduce p75NTR-mediated invasion (Figure 6d). To confirm this in vivo, cells were implanted into the brains of SCID mice and allowed to grow for 4 weeks. The mice were killed, and paraffin-embedded brain sections were stained with antibodies against human nuclei (Figure 6e; brown, left panel) or PDLIM1 (Figure 6e; brown, right panel). As predicted, knockdown of PDLIM1 resulted in complete abrogation of p75NTR-mediated glioma invasion in vivo.
These data identify the PDZ-domain containing protein PDLIM1 as a novel p75NTR-interacting protein that is essential for p75NTR-dependent glioma invasion, an interaction regulated by phosphorylation of S425 within the PDZ-binding domain.
Discussion
The highly invasive nature of glioblastoma renders them incurable by surgery and radiotherapy, thus warranting attention to the molecular determinants regulating their invasive behavior. We previously demonstrated that the p75NTR signaling axis is one such determinant in glioma invasion.18, 19 Using pharmacological and loss-of-function approaches, we defined a specific role for serine phosphorylation of p75NTR (CD271) in glioblastoma tumorigenesis and invasion. We demonstrate that phosphorylation of p75NTR on S303 is required for glioma invasion both in vitro and in vivo and that this regulation requires the subsequent dephosphorylation of a second serine site present in the PDZ-binding motif, S425. We provide evidence that dephosphorylation of S425 is essential for the interaction of p75NTR with the PDZ and LIM domain containing protein PDLIM1. Moreover, we show for the first time that S425 within the canonical PDZ-binding motif of p75NTR is phosphorylated and this regulates the interaction of p75NTR with PDLIM1. Although it has been shown in other proteins that phosphorylation within the PDZ-binding motif increases the interaction with PDZ-containing proteins,72, 73 we provide evidence that phosphorylation of p75NTR on S425 prevents its binding to PDLIM1, a negative regulation that has also been observed between the β2-adrenergic receptor and NHERF1 to control recycling of the receptor.74 In addition, we demonstrate that crippling the PDZ-binding domain, or downregulating PDLIM1, abrogates p75NTR-mediated invasion. Importantly, our study was not limited to established cell lines, as a role for PDLIM1 in glioma invasion was also confirmed in primary patient-derived cultures grown and maintained under neurosphere conditions, an environment that promotes the isolation of glioma cells with a cancer stem-like phenotype60, 75, 76 and maintains the highly invasive nature of the patient tumor.
It is now clear that the spatial localization of receptors and/or downstream effector molecules is necessary for appropriate recognition and subsequent transmission of extracellular signals (for example, see reviews by Gauthier and Robbins,77 Casaletto and McClatchey,78 Gold et al.79 and Hupalowska and Miaczynska80). For instance, the localization of p75NTR to lipid rafts has been shown to be important for at least some aspects of its biological activity.39, 81, 82, 83, 84, 85 We have previously shown that regulated intramembrane proteolysis of p75NTR is required for p75NTR-mediated glioma invasion.19 Herein, we found that although disruption of phosphorylation at S303, or mutation of the PDZ-motif (ΔSPV or SPM) results in abrogation of p75NTR-mediated invasion, it does not compromise regulated intramembrane proteolysis of the receptor as these mutant receptors still undergo cleavage to generate both C-terminal fragment and intracellular domain fragments (Figures 2b, 3b, 4d and e). These data therefore suggest that processing of the receptor, although previously shown to be necessary, is not sufficient to mediate the invasive behavior of glioma cells and supports a model where either the localization of the receptor and/or its cleavage products (intracellular domain), or the posttranslational modification of these products, may be required to induce glioma invasion, possibilities that are not mutually exclusive and all may be involved in some context.
The role of PDLIM1 in invasion is not limited to glioma and is involved in breast cancer cell migration, invasion and metastasis through interaction with the actin cytoskeleton,70 a key function of many LIM domain-containing proteins.44, 45, 46, 47, 48 Furthermore a role for p75NTR has been established in melanoma cell migration and tumorigenesis28, 30, 55, 56 for which we have shown an interaction of PDLIM1 with p75NTR (Supplementary Figure S8). In addition PDLIM1 is highly expressed in dorsal root ganglion neurons especially after nerve injury and may regulate the actin cytoskeleton during neurite outgrowth86, 87 a function also shown to involve neurotrophins and p75NTR.23, 24
Although there are numerous LIM domain-containing proteins in the mammalian genome,88 there are only 10 proteins that contain a PDZ and LIM domain. It remains to be determined whether other PDZ/LIM family members have a role in glioma invasion. Furthermore, whether other PDZ domain proteins interact with the C-terminus of p75NTR in glioma cells is undetermined, however, it is interesting that the PDLIM PDZ domains may have unique binding specificity with their respective substrates.89 Of note, a recent study suggests that MDA-9/syntenin a PDZ domain-containing protein, is important in glioma invasion and pathogenesis.90
The requirement for PDLIM1 in p75NTR-mediated glioma invasion with its regulated binding to the receptor suggests a logical therapeutic axis for treatment of glioblastoma. Development of reversible small-molecule inhibitors that target neuron-specific PDZ domains have been proposed as a means to specifically target compartmentalized signaling within the CNS.91 In addition, peptides targeting the PDZ domain of PTPN4, a tyrosine phosphatase, are efficient inducers of glioma cell death.92 Thus, targeting the PDZ domain interaction of p75NTR may provide a tissue and receptor specific therapeutic approach for patients with malignant glioma and may have global application in other cancers (that is, metastatic melanoma and breast) where p75NTR and/or PDLIM1 have also been implicated.28, 29, 30, 55, 93, 94
Materials and methods
Cell culture and treatments
U87 glioma cell lines were grown in Dulbecco's modified eagle's medium (Gibco-BRL, Bethesda, MD, USA) supplemented with 10% fetal bovine serum (Gibco-BRL) at 37 °C in a humidified 5% CO2 incubator. Stable transfectants were maintained in identical media with 400 μg/ml of G418 (Sigma, St. Louis, MO, USA). Brain tumor-initiating cells were established within the Brain Tumor Stem Cell Core at the University of Calgary and maintained as previously described.19, 60 Human Metastatic melanoma cells 3S5 and 70W97, 98 were obtained from Dr Robert Kerbel Mount Sinai Research Institute, Toronto Canada. U87 or brain tumor-initiating cells were treated with the indicated concentrations of KT5720 (1h; KT, Calbiochem, Gibbstown, NJ, USA), 2'5'-dideoxyadenosine (1 h, 30 μM; Sigma) or forskolin (30mins, 1 μM, Calbiochem), and cultured at 37 °C. Control cells were treated with 1% dimethyl sulfoxide (vehicle). All cell lines were tested for mycoplasma contamination.
Mutagenesis and transfection
The human p75NTR expression vector was constructed as described previously.19 To generate the p75NTR serine phosphorylation site mutant (p75S303G), the p75NTR C-terminal PDZ-binding motif truncation mutant (p75ΔSPV) and the p75NTR C-terminal PDZ-binding motif substitution mutant (p75SPM), site-directed mutagenesis was performed (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions. Constructs were inserted into pcDNA 3.1 vectors (Invitrogen, Carlsbad, CA, USA). Human PDLIM1 cDNA was purchased from Open Biosystems (Cat. no. MHS6278-202755473, Huntsville, AL, USA). Vectors encoding HA-tagged full-length PDLIM1 (residues1–329), ΔPDZ (residues 68-329) or ΔLIM mutant (residues 1–250) were cloned into the HA-pcDNA3.1 vector. ΔPDZ or ΔLIM mutants were obtained by PCR using the pcDNA 3.1 HA-PDLIM1plasmid as a template and verified by DNA sequencing. Transfection and selection of glioma cell lines was performed as described previously.19 Flow cytometry and western blotting confirmed appropriate expression.
Invasion assay
Cells (2.5 × 104) were seeded on collagen-coated transwell inserts (8 μm pore size; Corning Costar, Corning, NY, USA) and incubated for 3 h at 37 °C in a 5% CO2 incubator. Media was aspirated and the cells were stained in 1% crystal violet in 95% ethanol for 1 min, rinsed in phosphate-buffered saline (PBS) and cells on the upper membrane surfaces were removed with a cotton swab. The number of cells that migrated were counted by light microscopy, and the sum of cells in five microscopic fields was recorded.
Reporter gene assay
To examine cAMP activity, transcription of a CRE-promoter was evaluated in U87 cells transfected with a CRE-luciferase reporter plasmid. U87 cells were plated in a six-well tissue culture plate at 1 × 104 cells per well and transfected with a pCRE-Luc reporter plasmid using lipofectamine 2000 (Invitrogen). Three days after transfection, cells were treated as outlined in each experiment. For analyses, cells were lysed in 0.5 ml of lysis buffer (Dual-Luciferase Reporter Assay System, Promega, Madison, WI, USA) and cell lysate (10 μl) was combined with 50 μl of luciferase assay reagent. Luciferase activity was measured with a GloMax-multi detection system (Promega).
Immunoprecipitation and immunoblot
Cells were lysed with RIPA buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% (v/v) NP-40, protease inhibitor cocktail (Roche, Indianapolis, IN, USA), phosphatase inhibitor (Sigma)), protein quantification was determined, and proteins were resolved by sodium dodecyl sufate–polyacrylamide gel electrophoresis. Western blot analysis was performed using rabbit polyclonal anti-human p75NTR intracellular domain (Promega), mouse monoclonal anti-β-actin (Millipore, Billerica, MA, USA) or mouse monoclonal PDLIM1 (Abcam, Cambridge, MA, USA) with the appropriate HRP-conjugated secondary antibodies (SantaCruz Biotechnology, Santa Cruz, CA, USA). For immunoprecipitation experiments, whole-cell lysates were incubated with the indicated antibody in the presence of Sepharose-Protein A/G (Sigma). Immunoprecipitates were boiled with 2 × Laemmli buffer, resolved by sodium dodecyl sufate–polyacrylamide gel electrophoresis and western blotting were performed using anti-p75NTR (Promega), anti-PDLIM1 (Abcam) or anti-HA (Abcam) antibodies.
Reverse transcription–polymerase chain reaction (RT–PCR)
RNA from U87 cells was purified using Trizol (Invitrogen) according to the manufacturer's instructions. cDNA synthesis and analysis by PCR were performed as previously described.18, 53 These references also list primer sequences.
Peptide affinity assay and mass spectrometric analysis
For the detection of p75NTR-interacting proteins, wild-type and mutant p75NTR peptides, or a random peptide (control), were labeled on the amino terminus with biotin (synthesized by CanPeptide, Montréal, QC, Canada). U87 glioma cells were lysed as described above and incubated with biotin-labeled peptides for 1 h at 4 °C. NeutrAvidin agarose (Thermo Scientific, Rockford, lL, USA) was added and incubated for 1 h. Peptide bound to the agarose beads was centrifuged, washed with PBS and boiled with 2 × Laemmli buffer. Proteins that bound to each peptide were resolved on a 10% sodium dodecyl sufate–polyacrylamide gel electrophoresis gel and silver-stained as described previously95 or transferred to nitrocellulose for western blot analysis. Proteins that bound to the wild-type p75NTR peptide and not to the mutant peptides were excised from the gel, and processed LC-MS/MS analysis as described.95 A similar procedure was followed for the detection of serine phosphorylation on the p75NTR mutant and wild-type receptors following immunoprecipitation with anti-p75NTR (Promega).
Animals
Six- to 8-week-old female SCID mice (Charles River Laboratories, Shrewsbury, MA, USA) were housed in groups of three to five and maintained on a 12-h light/dark schedule with a temperature of 22 °C±1 °C and a relative humidity of 50%±5%. Food and water were available ad libitum. All procedures were reviewed and approved by the University of Calgary Animal Care Committee.
In vivo studies using an intracranial glioma model
Glioma cells harvested using Puck's EDTA were resuspended in PBS and implanted intracerebrally into SCID mice (5 × 104 cells/mouse) as previously described.18, 19, 96 Animals were randomized prior to the injection of tumor cells. At 4 weeks, BrdU was given by intraperitoneal injection 24 h prior to killing of the mice, brains were removed, fixed in 4% formalin and paraffin-embedded. Whole brain tissue sections from all animals in each group were examined by immunohistochemistry and assessed by two independent investigators, including one blinded assessment. To ensure adequate power to detect invasion, all analysis included five animals per experimental group and three independent experiments were performed.
Immunohistochemistry
Whole brain sections were fixed with cold acetone, and rehydrated through an ethanol gradient. Endogenous peroxidases were inactivated with 0.075% H2O2/methanol, and nonspecific binding blocked with Rodent Block M (Biocare Medical, Concord, CA, USA). Sections were incubated with 1:100 diluted rabbit polyclonal anti-p75NTR antibody (Promega), mouse monoclonal anti-human nucleolin (Abcam) or anti-Tenascin C (Novus Biologicals, Littleton, CO, USA) in antibody diluent (Biocare Medical). Following washing with PBS, the appropriate biotinylated secondary antibody (Vector Laboratories, Burlingame, CA, USA) was applied and detected using the VECTASTAIN Elite ABC kit (Vector Laboratories) and SIGMAFAST DAB (Sigma). Hematoxylin was used as a counterstain and sections were mounted with Entellan (Electron Microscopy Sciences, Hatfield, PA, USA).
Pull-down analysis of p75NTR-PDLIM1 direct interaction
One microgram of C-terminal His-tagged recombinant PDLIM1 (Fitzgerald, Acton, MA, USA) and biotin-labeled p75NTR C-terminal peptides (1mg/ml) (described above) were incubated in 300 μl of binding buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40) overnight at 4 °C. NeutrAvidin agarose (Thermo Scientific) was added for 1 h, beads were centrifuged, washed with PBS and boiled with 2 × Laemmli buffer. Proteins bound to each peptide were resolved by sodium dodecyl sufate–polyacrylamide gel electrophoresis and bound PDLIM1 was detected by western blot analysis using anti-His (Abcam) or anti-PDLIM1.
PDLIM1 shRNA
pLKO.1-shPDLIM1 vector TRCN0000161271 (shPDLIM1; GCCTTGGTTAATTGACTCACA) as well as pLKO.1-shControl SHC002 were purchased from Sigma and used according to the manufacturer's instructions. shPDLIM1 and shControl were transfected into U87p75NTR cells using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions and cells expressing the shRNA were selected in media containing Puromycin (5 μg/ml). Inhibition of PDLIM1 expression was confirmed by RT–PCR and western blot analysis.
Statistical analysis
Data were analyzed by one-way ANOVA and Student's t-test comparison, using GraphPad Instat3 software (La Jolla, CA, USA). Statistical significance was reached at P<0.05.
Acknowledgments
We would like to thank the Brain Tumor Stem Cell Core supported by funds from the Hotchkiss Brain Institute, the Alberta Cancer Foundation and the Terry Fox Research Institute. We also thank the Calgary Brain Tumour and Tissue Bank generously supported by funds from the Clark H. Smith Family and the Southern Alberta Mass Spectrometry (SAMS) Centre. This work was supported by grants to SMR and DLS from the Alberta Children's Hospital Foundation, the Alberta Cancer Foundation and Alberta Innovates Health Solutions. RSG acknowledges fellowship support from Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES), Ministério da Educação - Brasil.
Author Contributions
BYA, RSG, SMR and DLS conceived the overall project and designed all experiments. BYA, RSG, JJR, XH, JZ, N-HD and MA performed all of the experiments. BYA, RSG, SMR and DLS analyzed the data. BYA, RSG, SMR and DLS wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008 Jul 31; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol 2006; 1: 97–117. [DOI] [PubMed] [Google Scholar]
- Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 2003; 21: 1624–1636. [DOI] [PubMed] [Google Scholar]
- Mariani L, McDonough WS, Hoelzinger DB, Beaudry C, Kaczmarek E, Coons SW et al. Identification and validation of P311 as a glioblastoma invasion gene using laser capture microdissection. Cancer Res 2001; 61: 4190–4196. [PubMed] [Google Scholar]
- Joy AM, Beaudry CE, Tran NL, Ponce FA, Holz DR, Demuth T et al. Migrating glioma cells activate the PI3-K pathway and display decreased susceptibility to apoptosis. J Cell Sci 2003; 116: 4409–4417. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 2009; 4: 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin Ensign SP, Mathews IT, Symons MH, Berens ME, Tran NL. Implications of Rho GTPase signaling in glioma cell invasion and tumor progression. Front Oncol 2013; 3: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska A, Symons M. Signaling determinants of glioma cell invasion. Adv Exp Med Biol 2013; 986: 121–141. [DOI] [PubMed] [Google Scholar]
- Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci 2007; 64: 458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko PG, Ilina O, Friedl P. Interstitial guidance of cancer invasion. J Pathol 2012; 226: 185–199. [DOI] [PubMed] [Google Scholar]
- Teodorczyk M, Martin-Villalba A. Sensing invasion: cell surface receptors driving spreading of glioblastoma. J Cell Physiol 2010; 222: 1–10. [DOI] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res 2004; 64: 6503–6510. [DOI] [PubMed] [Google Scholar]
- Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 2005; 102: 5814–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Walling J, Ahn S, Kotliarov Y, Su Q, Quezado M et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res 2009; 69: 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006. 157–173. [DOI] [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AL, Lun X, Rahn JJ, Liacini A, Wang L, Hamilton MG et al. The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol 2007; 5: e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rahn JJ, Lun X, Sun B, Kelly JJ, Weiss S et al. Gamma-secretase represents a therapeutic target for the treatment of invasive glioma mediated by the p75 neurotrophin receptor. PLoS Biol 2008; 6: e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragnolini AB, Friedman WJ. The function of p75NTR in glia. Trends Neurosci 2008; 31: 99–104. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD. Neurobiology: a move to sort life from death. Nature 2004; 427: 798–799. [DOI] [PubMed] [Google Scholar]
- Barker PA. p75NTR is positively promiscuous: novel partners and new insights. Neuron 2004; 42: 529–533. [DOI] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 2006; 361: 1545–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer BR, Yoon SO, Carter BD. The biological functions and signaling mechanisms of the p75 neurotrophin receptor. Handb Exp Pharmacol 2014; 220: 121–164. [DOI] [PubMed] [Google Scholar]
- Anton ES, Weskamp G, Reichardt LF, Matthew WD. Nerve growth factor and its low-affinity receptor promote Schwann cell migration. Proc Natl Acad Sci USA 1994; 91: 2795–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BD, Kaltschmidt C, Kaltschmidt B, Offenhauser N, Bohm-Matthaei R, Baeuerle PA et al. Selective activation of NF-kappa B by nerve growth factor through the neurotrophin receptor p75. Science 1996; 272: 542–545. [DOI] [PubMed] [Google Scholar]
- Bentley CA, Lee KF. p75 is important for axon growth and schwann cell migration during development. J Neurosci 2000; 20: 7706–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JL, Menter DG, Hamada J, Marchetti D, Nakajima M, Nicolson GL. Mediation of NGF-stimulated extracellular matrix invasion by the human melanoma low-affinity p75 neurotrophin receptor: melanoma p75 functions independently of trkA. Mol Biol Cell 1993; 4: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menter DG, Herrmann JL, Marchetti D, Nicolson GL. Involvement of neurotrophins and growth factors in brain metastasis formation. Invasion Metastasis 1994; 14: 372–384. [PubMed] [Google Scholar]
- Shonukan O, Bagayogo I, McCrea P, Chao M, Hempstead B. Neurotrophin-induced melanoma cell migration is mediated through the actin-bundling protein fascin. Oncogene 2003; 22: 3616–3623. [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell 1992; 71: 973–985. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron 1999; 24: 585–593. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Carter BD, Dobrowsky RT, Chao MV. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature 1996; 383: 716–719. [DOI] [PubMed] [Google Scholar]
- Volonte C, Angelastro JM, Greene LA. Association of protein kinases ERK1 and ERK2 with p75 nerve growth factor receptors. J Biol Chem 1993; 268: 21410–21415. [PubMed] [Google Scholar]
- Dobrowsky RT, Werner MH, Castellino AM, Chao MV, Hannun YA. Activation of the sphingomyelin cycle through the low-affinity neurotrophin receptor. Science 1994; 265: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Le Moan N, Houslay DM, Christian F, Houslay MD, Akassoglou K. Oxygen-dependent cleavage of the p75 neurotrophin receptor triggers stabilization of HIF-1alpha. Mol Cell 2011; 44: 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood CK, Reid K, May LM, Bartlett PF, Coulson EJ. Palmitoylation of the C-terminal fragment of p75(NTR) regulates death signaling and is required for subsequent cleavage by gamma-secretase. Mol Cell Neurosci 2008; 37: 346–358. [DOI] [PubMed] [Google Scholar]
- Skeldal S, Matusica D, Nykjaer A, Coulson EJ. Proteolytic processing of the p75 neurotrophin receptor: A prerequisite for signalling?: Neuronal life, growth and death signalling are crucially regulated by intra-membrane proteolysis and trafficking of p75(NTR). Bioessays 2011; 33: 614–625. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Yamashita T, Yoshikawa H, Tohyama M. PKA phosphorylates the p75 receptor and regulates its localization to lipid rafts. Embo J 2003; 22: 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie S, Hachiya T, Rabizadeh S, Maruyama W, Mukai J, Li Y et al. Functional interaction of Fas-associated phosphatase-1 (FAP-1) with p75(NTR) and their effect on NF-kappaB activation. FEBS Lett 1999; 460: 191–198. [DOI] [PubMed] [Google Scholar]
- Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK et al. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science 2006; 314: 832–836. [DOI] [PubMed] [Google Scholar]
- Teyra J, Sidhu SS, Kim PM. Elucidation of the binding preferences of peptide recognition modules: SH3 and PDZ domains. FEBS Lett 2012; 586: 2631–2637. [DOI] [PubMed] [Google Scholar]
- Ivarsson Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett 2012; 586: 2638–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaavuniemi T, Ylanne J. Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with alpha-actinin–analysis of patient mutations. Exp Cell Res 2006; 312: 1299–1311. [DOI] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 1998; 393: 809–812. [DOI] [PubMed] [Google Scholar]
- Vallenius T, Luukko K, Makela TP. CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J Biol Chem 2000; 275: 11100–11105. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun 2000; 272: 505–512. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 1998; 393: 805–809. [DOI] [PubMed] [Google Scholar]
- Bashirova AA, Markelov ML, Shlykova TV, Levshenkova EV, Alibaeva RA, Frolova EI. The human RIL gene: mapping to human chromosome 5q31.1, genomic organization and alternative transcripts. Gene 1998; 210: 239–245. [DOI] [PubMed] [Google Scholar]
- Wang H, Harrison-Shostak DC, Lemasters JJ, Herman B. Cloning of a rat cDNA encoding a novel LIM domain protein with high homology to rat RIL. Gene 1995; 165: 267–271. [DOI] [PubMed] [Google Scholar]
- Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol 1997; 139: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrado M, Senatorov VV, Trivedi R, Fariss RN, Tomarev SI. Pdlim2, a novel PDZ-LIM domain protein, interacts with alpha-actinins and filamin A. Invest Ophthalmol Vis Sci 2004; 45: 3955–3963. [DOI] [PubMed] [Google Scholar]
- Kotaka M, Kostin S, Ngai S, Chan K, Lau Y, Lee SM et al. Interaction of hCLIM1, an enigma family protein, with alpha-actinin 2. J Cell Biochem 2000; 78: 558–565. [DOI] [PubMed] [Google Scholar]
- Kotaka M, Ngai SM, Garcia-Barcelo M, Tsui SK, Fung KP, Lee CY et al. Characterization of the human 36-kDa carboxyl terminal LIM domain protein (hCLIM1). J Cell Biochem 1999; 72: 279–285. [DOI] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 2010; 466: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmer T, Welte Y, Behrens D, Fichtner I, Przybilla D, Wruck W et al. The nerve growth factor receptor CD271 is crucial to maintain tumorigenicity and stem-like properties of melanoma cells. PLoS One 2014; 9: e92596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T et al. Identification of human brain tumour initiating cells. Nature 2004; 432: 396–401. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760. [DOI] [PubMed] [Google Scholar]
- Kelly JJ, Stechishin O, Chojnacki A, Lun X, Sun B, Senger DL et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells 2009. 1722–1733. [DOI] [PubMed]
- Eli Lilly and Company. Lilly halts development of semagacestat for Alzheimer's disease based on preliminary results of phase III clinical trials 2010.
- Hopkins CR. ACS chemical neuroscience molecule spotlight on ELND006: another gamma-secretase inhibitor fails in the clinic. ACS Chem Neurosci 2011; 2: 279–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Zemp FJ, Senger D, Robbins SM, Yong VW. ADAM-9 is a novel mediator of tenascin-C-stimulated invasiveness of brain tumor-initiating cells. Neuro Oncol e-pub ahead of print 1 February 2015. [DOI] [PMC free article] [PubMed]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 1997; 275: 73–77. [DOI] [PubMed] [Google Scholar]
- Bauer K, Kratzer M, Otte M, de Quintana KL, Hagmann J, Arnold GJ et al. Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 2000; 96: 4236–4245. [PubMed] [Google Scholar]
- Quick Q, Skalli O. Alpha-actinin 1 and alpha-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res 2010; 316: 1137–1147. [DOI] [PubMed] [Google Scholar]
- Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F et al. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 2005; 128: 51–62. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T et al. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin Cancer Res 2008; 14: 5348–5356. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Tsuda H, Honda K, Onozato K, Takano M, Tamai S et al. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod Pathol 2009; 22: 499–507. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhan Y, Tu Y, Chen K, Wu C. PDZ and LIM domain protein 1(PDLIM1)/CLP36 promotes breast cancer cell migration, invasion and metastasis through interaction with alpha-actinin. Oncogene 2014; 34: 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, King BL, Milan G, Maffei P, Jagger DJ et al. The Alstrom syndrome protein, ALMS1, interacts with alpha-actinin and components of the endosome recycling pathway. PLoS One 2012; 7: e37925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Nandelstadh P, Ismail M, Gardin C, Suila H, Zara I, Belgrano A et al. A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol Cell Biol 2009; 29: 822–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann CL, Martinez E, Geiger JC, Zimmer M, Traut MH, Stein V et al. Serine phosphorylation of ephrinB2 regulates trafficking of synaptic AMPA receptors. Nat Neurosci 2008; 11: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature 1999; 401: 286–290. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci 1992; 12: 4565–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992; 255: 1707–1710. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Robbins SM. Ephrin signaling: One raft to rule them all? One raft to sort them? One raft to spread their call and in signaling bind them? Life Sci 2003; 74: 207–216. [DOI] [PubMed] [Google Scholar]
- Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer 2012; 12: 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MG, Gonen T, Scott JD. Local cAMP signaling in disease at a glance. J Cell Sci 2013; 126: 4537–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalowska A, Miaczynska M. The new faces of endocytosis in signaling. Traffic 2012; 13: 9–18. [DOI] [PubMed] [Google Scholar]
- Bilderback TR, Gazula VR, Lisanti MP, Dobrowsky RT. Caveolin interacts with Trk A and p75(NTR) and regulates neurotrophin signaling pathways. J Biol Chem 1999; 274: 257–263. [DOI] [PubMed] [Google Scholar]
- Bilderback TR, Grigsby RJ, Dobrowsky RT. Association of p75(NTR) with caveolin and localization of neurotrophin-induced sphingomyelin hydrolysis to caveolae. J Biol Chem 1997; 272: 10922–10927. [DOI] [PubMed] [Google Scholar]
- Fujitani M, Kawai H, Proia RL, Kashiwagi A, Yasuda H, Yamashita T. Binding of soluble myelin-associated glycoprotein to specific gangliosides induces the association of p75NTR to lipid rafts and signal transduction. J Neurochem 2005; 94: 15–21. [DOI] [PubMed] [Google Scholar]
- Gil C, Cubi R, Aguilera J. Shedding of the p75NTR neurotrophin receptor is modulated by lipid rafts. FEBS Lett 2007; 581: 1851–1858. [DOI] [PubMed] [Google Scholar]
- Huang CS, Zhou J, Feng AK, Lynch CC, Klumperman J, DeArmond SJ et al. Nerve growth factor signaling in caveolae-like domains at the plasma membrane. J Biol Chem 1999; 274: 36707–36714. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Ohno K, Funahashi S, Miyazaki K, Nagano A, Sato K. CLP36 interacts with palladin in dorsal root ganglion neurons. Neurosci Lett 2010; 476: 53–57. [DOI] [PubMed] [Google Scholar]
- Ohno K, Kato H, Funahashi S, Hasegawa T, Sato K. Characterization of CLP36/Elfin/PDLIM1 in the nervous system. J Neurochem 2009. 790–800. [DOI] [PubMed]
- Smith MA, Hoffman LM, Beckerle MC. LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol 2014; 24: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JM, Papagrigoriou E, Berridge G, Yang X, Phillips C, Gileadi C et al. Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions. Protein Sci 2007; 16: 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegelman TP, Das SK, Hu B, Bacolod MD, Fuller CE, Menezes ME et al. MDA-9/syntenin is a key regulator of glioma pathogenesis. Neuro Oncol 2014; 16: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer LL, Neubig RR. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 2009; 34: 126–141. [DOI] [PubMed] [Google Scholar]
- Babault N, Cordier F, Lafage M, Cockburn J, Haouz A, Prehaud C et al. Peptides targeting the PDZ domain of PTPN4 are efficient inducers of glioblastoma cell death. Structure 2011; 19: 1518–1524. [DOI] [PubMed] [Google Scholar]
- Verbeke S, Meignan S, Lagadec C, Germain E, Hondermarck H, Adriaenssens E et al. Overexpression of p75(NTR) increases survival of breast cancer cells through p21(waf1). Cell Signal 2010; 22: 1864–1873. [DOI] [PubMed] [Google Scholar]
- Verbeke S, Tomellini E, Dhamani F, Meignan S, Adriaenssens E, Xuefen, le B. Extracellular cleavage of the p75 neurotrophin receptor is implicated in its pro-survival effect in breast cancer cells. FEBS Lett 2013; 587: 2591–2596. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 2006; 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Robbins SM, Senger DL. Assessing the mechanisms of glioma invasion. In: Martinez Murillo R, Martinez A (eds). Neuromethods. Springer: New York, USA, 2013, pp 275–298. [Google Scholar]
- Kerbel RS, Man MS. Single-step selection of unique human melanoma variants displaying unusually aggressive metastatic behavior in nude athymic mice. Invasion Metastasis 1984; 4: 31–43. [PubMed] [Google Scholar]
- Kerbel RS, Man MS, Dexter D. A model of human cancer metastasis: extensive spontaneous and artificial metastasis of a human pigmented melanoma and derived variant sublines in nude mice. J Natl Cancer Inst 1984; 72: 93–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.