Abstract
While reef degradation is occurring worldwide, it is not uncommon to see phase shifts from coral to macroalgal dominated reefs. Numerous studies have addressed the mechanisms by which macroalgae may outcompete corals and a few recent studies highlighted the putative role of bacteria at the interface between macroalgae and corals. Some studies suggest that macroalgae may act as vectors and/or foster proliferation of microorganisms pathogenic for corals. Using a combination of high throughput sequencing, bacterial culturing, and in situ bioassays we question if the adversity of macroalgal-associated bacteria to corals is mediated by specific bacterial taxa. Using Illumina sequencing, we characterized and compared the bacterial community from two Lobophora (Dictyotales, Phaeophyceae) species. The two species presented distinctive bacterial communities. Both species shared approximately half of their OTUs, mainly the most abundant bacteria. Species-specific OTUs belong to Planctomycetes, Proteobacteria, and Bacteroidetes. In total, 16 culturable bacterial strain were isolated and identified from the Lobophora surface, consisting of 10 genera (from nine families, four classes, and three phyla), some of which are not known as, but are related to pathogens involved in coral diseases, and others are naturally associated to corals. When patches of marine agar with 24 h cultures of each of these bacteria were placed in direct contact with the branches of the scleractinian coral Acropora muricata, they caused severe bleaching after 24 h exposure. Results suggest that regardless of taxonomic affinities, increase in density of these bacteria can be adverse to corals. Nevertheless, the microbial community associated to macroalgal surface may not represent a threat to corals, because the specific bacterial screening and control exerted by the alga preventing specific bacterial proliferation.
Keywords: coral bleaching, Illumina sequencing, in situ bioassay, Lobophora, macroalgal–coral interaction, macroalgal bacterial assemblage, macroalgal culturable epibacteria
Introduction
Competition between benthic macroalgae and corals, two ecosystem engineers of tropical reefs, is a key process shaping the structure of reef communities. Contrary to healthy reefs where macroalgae and corals maintain a stable coexistence, in disturbed reef ecosystems, macroalgae often gain dominance over scleractinian corals. While declines in coral cover are generally associated with increases in the abundance of fleshy (Hughes, 1994; McClanahan et al., 1999), and crustose coralline algae (Antonius and Afonso-Carillo, 2001; Pueschel and Saunders, 2009), in many cases it remains unclear whether the algae are directly or indirectly responsible for coral death or whether they simply settle on dead coral surfaces which are newly open substrate (McCook et al., 2001). In the pursuit of deciphering the mechanisms by which macroalgae may outcompete corals, the first studies focused on effects directly attributable to the alga, e.g., overgrowth, shading, abrasion, recruitment barriers, and allelopathic interactions (see McCook et al., 2001 for review). The concept of holobiont initially proposed for corals (Rohwer et al., 2002) and more recently adopted for algae (Barott et al., 2011) raised the awareness that the microbial component may play a significant ecological role in biotic interactions. As shown for corals (Rohwer et al., 2002; Reshef et al., 2006; Kvennefors et al., 2010; Mouchka et al., 2010), there is increasing evidence suggesting that algal-associated microbiota are species-specific (Sapp et al., 2007; Barott et al., 2011; Hollants et al., 2013a,b) and play an important role in the normal functioning of the algal host (Provasoli and Pintner, 1980; Keshtacher-Liebso et al., 1995; Nakanishi et al., 1996; Matsuo et al., 2003; Croft et al., 2005, 2006; Joint et al., 2007). By extension, the microbiota may also partake important roles in the ecological interactions (e.g., herbivory, competition, etc.) of the algal hosts with other organisms. The microbiota associated to corals and algae feasibly play a role in the outcome of the competition between these major coral reef components. A series of studies indicate (1) that macroalgae can act as reservoirs and vectors of coral pathogens (Nugues et al., 2004; Barott et al., 2011; Sweet et al., 2013) and (2) that macroalgal diffusible compounds can lead to changes in coral microbial assemblages resulting in coral vulnerability or even mortality (Smith et al., 2006; Morrow et al., 2011, 2012). Macroalgae may thus disturb microbial communities of the corals, and convey and/or foster the development of pathogenic bacteria to corals. These results lead to question whether only the known coral-pathogenic bacteria cause bleaching of coral, or if any proliferating bacteria may cause harm to corals.
In coral reef ecosystems the brown algal order Dictyotales (Phaeophyceae) represents one of the most important algal groups, and the genus Lobophora (Dictyotales, Phaeophycea) plays a particularly important ecological role in algal–coral–grazing interactions and competition (Coen and Tanner, 1989; Nugues and Bak, 2008; Diaz-Pulido et al., 2009; Anthony et al., 2011; Slattery and Lesser, 2014). Lobophora can outcompete coral species (Jompa and McCook, 2002a,b; Nugues and Bak, 2006) and it can produce allelopathic compounds acting against corals (Rasher and Hay, 2010; Slattery and Lesser, 2014; Vieira et al., 2016). Nevertheless, association with live corals is restricted to only specific Lobophora species (Vieira et al., 2014, 2016). However, in these studies it is difficult to demonstrate whether the allelochemicals responsible for coral bleaching, are produced by the alga or the associated microbiota. Kubanek et al. (2003) isolated and characterized a complex molecule with antifungal properties from a Lobophora species and discussed the possibility that the new compounds could be the product of a microbial symbiont. It therefore seems imperative that future studies need to specifically address the ecological roles that microbial communities associated with macroalgal have.
The present study focuses on Lobophora-associated bacteria, characterizes the bacterial community associated to the macroalgae, and assesses the adversity of bacterial isolates of this macroalgal-holobiont on the Scleractinian coral Acropora muricata Linnaeus (1758).
Materials and methods
Study area and collection of samples
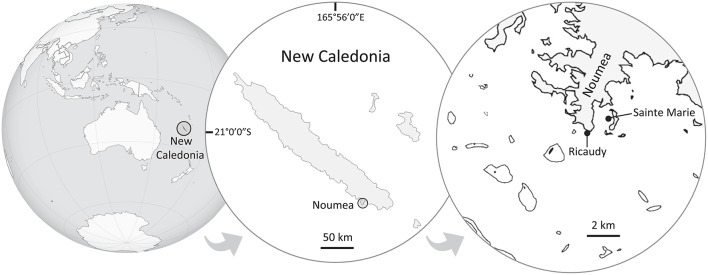
This study was performed in Noumea lagoon, New Caledonia, where two species of Lobophora were collected in two different sites, both fringing reefs, located 1.2 km apart (Figure 1). Lobophora monticola was collected in Sainte Marie Bay at 5 m depth (latitude: −22.297713°, longitude: 166.481639°) and Lobophora rosacea in Ricaudy at 2 m depth (latitude: −22.315317°, longitude: 166.457717°; Figure 1). The two species are naturally found growing associated with corals. L. monticola is generally found growing attached to Acropora corals, and L. rosacea is commonly found growing niched within branching corals such as Acropora. The site in Sainte Marie Bay is located several meters away from the mangrove shore, and is protected from wind exposure, resulting in turbid and still waters. The site in Ricaudy is exposed to dominant wind and thus experiences strong wave action. Algal thalli were collected right off their coral substrates (i.e., Acropora, Figures 2A,B); epiphyte-free (i.e., algal epiphytes) specimens were specifically selected. For Illumina sequencing, specimens were put in ziploc bags under water, stored in a cooler, frozen at −80°C, and freeze-dried. For the bacterial isolation and culture, algal thalli were rinsed three times consecutively with sterile seawater right after collection on the boat, placed in sterile vials, and stored in a cooler. Once in the laboratory, thalli were rinsed one more time with a sterile saline solution (36 g.L−1) under a biological safety cabinet.
Figure 1.
Map of New Caledonia showing the sampling and bioassays sites. Lobophora rosacea was collected in Ricaudy and L. monticola in Sainte Marie Bay.The distance between the two sites is ~1.2 km.
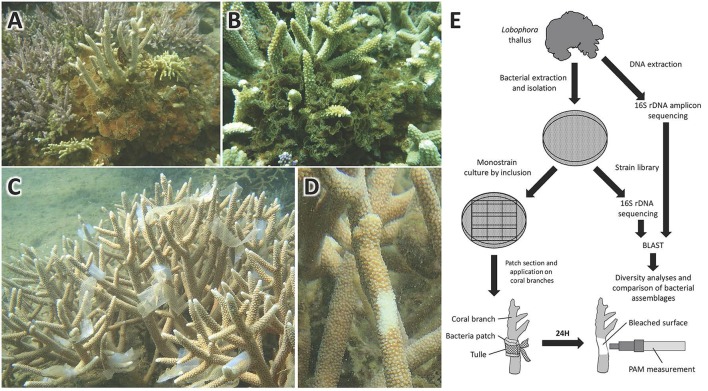
Figure 2.
Pictures of (A) L. rosacea and (B) L. monticola growing on Acropora colonies. Pictures of in situ bioassays on Acropora muricata colonies, showing (C) the marine agar patches with mono-specific bacterial cultures applied onto A. muricata colonies branches, and (D) the bleaching induced by the bacteria after 24 h exposure. Flowchart of the method deployed from the bacterial extraction on the algal-surface to the bioassays and bacterial diversity characterization (E).
Bacterial community barcoding and characterization
Bacterial DNA extraction and amplification
DNA was extracted from five freeze-dried replicates (a replicate = a unique specimen) of each species (L. rosaceae and L. monticola) using the Quick-gDNA kit (Zymo Research™) applying the manufacturer protocol.
The total 16S rDNA region was amplified using the universal primers 27F and 1494R (Lane, 1991) with the following changes to the original protocol: after an initial denaturation at 95°C for 2 min, conditions were 35 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 20 s, and extension at 72°C for 90 s. The final extension was at 72°C for 3 min. The 25 μl reaction mixture contained 250 μM dNTPs, 0.6 μM of each primer, 1 × 2PCR buffer mix, 2 μl of template DNA (with a final concentration of about 10 ng μl–1), and 0.3 μl of Taq polymerase (Advantage® 2 Clontech). PCR products were cleaned using ExoFastAP enzyme following the manufacturer protocol (Thermo Scientific™) and amplified DNA was submitted to Molecular Research (MR DNA), Shallowater, Texas where a nested-PCR was performed prior to sequencing. Modified 8 bp key-tagged primer 799F along with the reverse primer 1193R (fragment ~400 bp), which avoid chloroplast cross amplification (Bodenhausen et al., 2013), were used and PCR conditions were as follow: 95°C for 3 min, 30 cycles of 95°C for 20 s, 50°C for 30 s, 72°C for 30 s, and a final elongation of 72°C for 3 min. Samples were pooled together in equal proportions based on their molecular weight and DNA concentrations and purified using calibrated Ampure XP beads. DNA libraries were prepared by using a Illumina TruSeq DNA library preparation protocol and paired-end (2 × 250 bp) sequencing performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on a MiSeq following the manufacturer's guidelines.
16S rRNA analysis and bacterial community diversity
All the diversity analyses were performed using the program QIIME 1.8.0: Quantitative Insights Into Microbial Ecology (Caporaso et al., 2010). Sequences were screened for a minimum read length of 350 bp and < 2 or more undetermined nucleotides. The filtered dataset, containing only high quality sequences, was submitted to a conservative chimera detection filter using the ChimeraSlayer method (Haas et al., 2011). Selected high quality chimera-free sequences were clustered into Operational Taxonomic Units (OTUs) using the UCLUST (Edgar, 2010) with a pairwise identity threshold of 0.97.
Representative sequences for each OTU were picked using the “most-abundant” method and OTU sequence alignment was performed with Pynast (Caporaso et al., 2010). The Ribosomal Database Project (RDP; Wang et al., 2007) classifier was used for taxonomic assignment with a 95% confidence threshold. Sequences with the best match for eukaryotes (i.e., chloroplasts and mitochondria) were excluded from the OTU table in downstream analyses. To assign each OTU to the closest matching described taxon, searches were performed against the Greengenes reference database (version 12_10; McDonald et al., 2012) with a maximum e-value to record an assignment of 0.001. The degree of relatedness of the subsets of the most common sequences was inferred using the phylogenetic reconstruction with Qiime's script make_phylogeny.py and using, by default, FastTree (Price et al., 2010) from Qiime.
Diversity estimation and comparison of bacterial assemblages
In order to obtain direct descriptors of the diversity of bacterial assemblages, we calculated two widely used diversity indices, the Shannon (H′) and the Simpson indices of diversity (1 – D). The non-parametric ACE and the Chao 1 richness index were calculated with the software estimates (Version 9; Colwell, 2013) to estimate microbial diversity. Principal component and cluster analyses were used to determine the similarity between the samples and the species. To compare the difference of community composition between the two Lobophora species an analysis of similarities (ANOSIM) was performed in R (R Development Core Team, 2013) using the “vegan” package. Finally, to identify which bacterial groups mostly contribute to the difference between the two Lobophora species, a Similarity Percentages (SIMPER) analysis was performed in R using the “vegan” package.
Bacterial culture and bioassays
Bacterial extraction
Bacteria were extracted from the plant surface by implementing three independent extraction methods on different thalli. For the first method, the bacteria were extracted by swapping the thallus surface with a cotton swab and streaking it on a marine agar (MA, Laboratorios Conda) plate. Then the swab cotton was introduced into a sterile Falcon tube with 10 mL of Marine Broth (MB, Laboratorios Conda) and shaken vigorously for 1 min and incubated 48 h at 30°C, under shaking conditions (120 rpm). The second method consists of inoculating a MA petri dish by directly placing an algal thallus on it. After firmly pressing it against the agar, the thallus was left 5 min on it. The process was renewed with the other side of the thallus on the same petri dish and the plates were incubated at 30°C during 48 h. In the third method, a thallus was placed into a 15 mL sterile tube with 10 mL of sterile saline water and shaken for 1 min. Three MA plates were inoculated by spreading, respectively, 100 μL of the 1/10, 1/100 dilutions and pure suspension. Plates were then incubated 48 h at 30°C.
Bacterial isolation
After the 48 h incubation, bacteria presenting visually diverse colony morphologies were selected for isolation. A small portion of a bacterial colony was gently scooped with a disposable sterile inoculating loop and streaked into a MA plate, which was then incubated 48 h at 30°C. This process was renewed two more times to assure pure isolates. Several colonies of a given bacteria were then placed into 10 mL of MB within a 50 mL sterile tube and incubated 24 h at 30°C. Hundred microliters of the resulting culture was spread onto a MA plate and incubated 24 h at 30°C. Finally, colonies picked from the marine agar plates and introduced in 800 μL of MB or 800 μL of bacteria from the liquid culture were put together with 800 μL sterile 25% glycerol solution into a cryotube and stored at −80°C.
Bacterial DNA extraction and identification
Bacterial DNA was extracted with the DNeasy® Blood and Tissue Kit (Qiagen, California, USA), following the manufacturer's instructions. The 16S rRNA gene (1500 pb) was used for bacterial identification. Amplification of the 16S rRNA gene by PCR, using the universal 16S rRNA primers (Table S1), was followed by sequencing of the resulting PCR amplicons. Identification to the species level was performed by comparison to a Basic Local Alignment Search Tools (BLAST) database comprising over 1 million entries of bacteria.
In situ bioassays
A flowchart illustrating the methodology is given in Figure 2E. Isolated bacteria were put in direct contact with the coral branches by means of marine agar patches. The marine agar was inoculated by inclusion in order to have the bacteria growing on and within the marine agar. To do so, 100 μL of bacteria (concentration of ca. 30 bacteria/mL) from the marine broth culture were put directly on the empty petri dish after which the marine agar, which had cooled down to room temperature, was poured on the petri dish. The mix, marine agar with the 100 μL bacteria solution, was then gently mixed to homogenize the bacteria into the marine agar. The plates were then incubated 24 h at 30°C. Ten agar strips of 2 cm2 were cut from the petri dish under the biosafety cabinet and put into sterile re-sealable zipper storage bags and stored into a cooler until application. The agar strips were then applied on the coral branches and fixed to it with sterile labeled tulle bands (Figures 2C–E). Seawater bacteria were used as control in addition to sterile marine agar patches. After 24 h application on the coral branches, the agar strips were removed. Bioassays were performed in situ directly on coral colonies. A total of 10 replicas were used, with one coral colony representing one replica, on which the 16 isolated strains were tested separately, resulting in a total of 160 bioassays.
Coral bleaching measurement and statistical analyses
Pulse Amplitude Modulated (PAM) fluorometry was used to assess the effects of bacteria on coral health (effective quantum yield). PAM fluorometry measurements were carried out with a Diving-PAM (Walz) directly after removal of the strips. PAM fluorometry measures the photosynthetic efficiency of photosystem II within the endosymbiotic Symbiodinium spp. that may be used as a quantitative measure of photoinactivation during coral bleaching (Warner et al., 1999). PAM fluorometry values of healthy corals range between 0.5 and 0.7, depending on the coral species and time of the day. Values between 0 and 0.2 are indicative of severe bleaching or mortality (Fitt et al., 2001). PAM fluorometry measurements were made right where the strips were applied and 5 cm next to it, as a spatial control in order to have a coral health baseline for comparison. Data were tested for normality with a Shapiro–Wilk test. Since the data respected the parametric assumptions, a one-way ANOVA was subsequently performed, followed by the Tukey post-hoc honestly significant difference (HSD) test. Statistical analyses were performed using R.
Results
Bacterial community characterization through 16S rRNA NGS and culturing
Characterization through 16S rRNA NGS
In a dataset of more than 644,377 sequences obtained from the 10 Lobophora specimens studied, 3247 and 3313 OTUs were identified from L. rosacea and L. monticola, respectively. On average each sample had 1386 ± 162 OTUs (Table S2). Shannon and Simpson diversity indices were calculated for each sample and were used to compare the gross structure of bacterial assemblages among our samples. Both indices were rather stable across replicate samples, with an average value of 5.14 ± 0.32 and 5.67 ± 0.19 (H′), and 0.977 ± 0.010 and 0.989 ± 0.003 (1 – D), for L. monticola and L. rosacea, respectively. The Shannon (F = 10.0372, p = 0.1675) and Simpson (F = 6.2934, p = 0.2321) indices were not statistically significant different between the two species. The non-parametric estimators predicted an average number per sample of 2777 ± 606 and 2839 ± 386 OTUs (ACE), and of 2805 ± 646 and 2881 ± 604 (Chao1) OTUs for L. monticola and L. rosacea, respectively. Chao1 (F = 0.0374, p = 0.9857) and ACE (F = 0.0269, p = 0.9897) both richness estimators were not statistically significant different between the two species. Comparison of the core OTUs shared by at least four replicas per species between the two Lobophora species is presented in Figure 3. The sequences are publicly available in MG-RAST with the accession numbers 467520195.3–4675204.3.
Figure 3.
Relative abundance of the bacterial taxa, present in at least four replicates per species, associated with Lobophora rosacea (light gray bubbles on the left side) and L. monticola (dark gray bubbles on the right side), at different taxonomic levels (Phylum, Class, Order, Family).
Comparison between Lobophora species bacterial assemblages
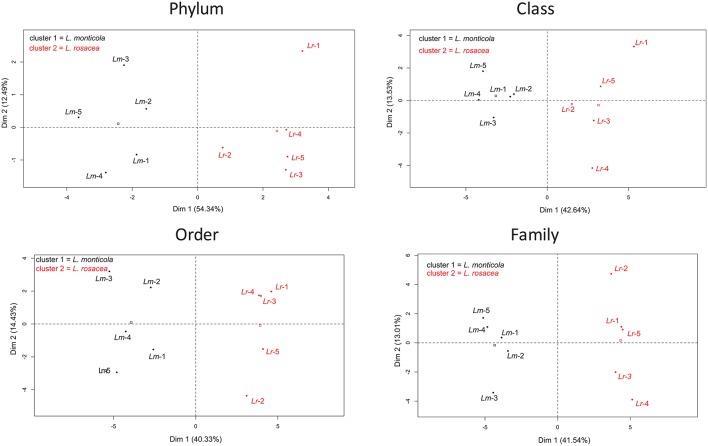
The ANOSIM analysis showed a statistically significant difference in the community composition at all taxonomic levels (R = 0.488−0.94; p = 0.006−0.012) between the two Lobophora species. The most influential taxa, determined with the SIMPER analysis, are given in Table S3. We performed PCA analysis to determine if the bacterial assemblage associated to the Lobophora species is species-specific. The four analyses resulted in the classification of the ten samples into two distinct clusters corresponding to the two Lobophora species. The first component that contributed significantly to explaining the relationships among the samples (eigen-values > 1), by clearly disjointing the two species, accounted for 40–54% of the variance depending on the taxonomic level, with a stronger value at the phylum level (54%; Figure 4). The most abundant phyla were shared by both species (Proteobacteria, Bacteroidetes, Planctomycetes, and Verrucomicrobia).
Figure 4.
Principal Component Analysis (PCA) for the bacterial assemblages from 10 samples of two Lobophora species, L. monticola (Lm-x), and L. rosacea (Lr-x).
Characterization through culturing
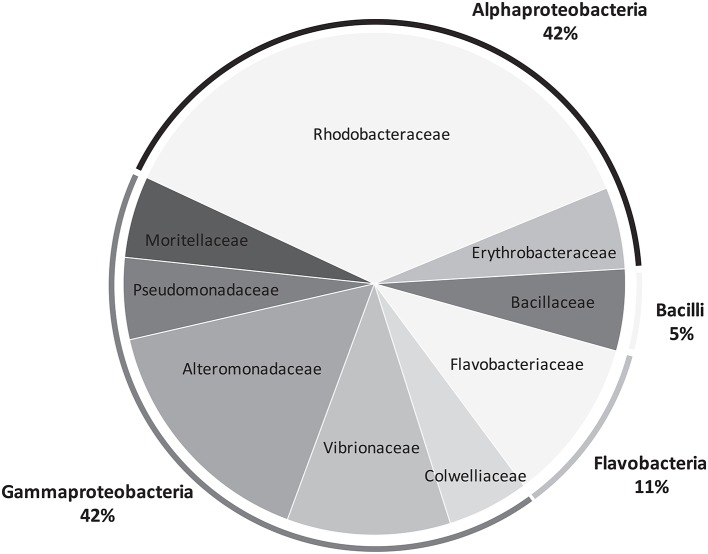
Bacteria grew quite well after 48 h of incubation, as visually assessed. We managed to culture 20 strains, 10 for each species of Lobophora. BLAST-search revealed 16 different strains (Table 1) with similarity to GenBank sequences ranging from 95 to 100% (Table S4). The two species of Lobophora were composed of similar percentages of surface-associated cultivable bacteria per phylum, with 85 and 90% Proteobacteria for L. rosacea and L. monticola, respectively, and 10% Bacterioides for both species, with an additional 5% Firmicutes for L. rosacea (Table 1). The isolated and successfully cultured 16 strains belonged to ten different genera: Bacillus, Erythrobacter, Microbulbifer, Muricauda, Paramoritella, Ruegeria, Shimia, Tenacibaculum, Thalassomonas, and Vibrio (Figure 5). Out of the nine families isolated, three (Colwelliaceae, Bacillaceae, Moritellaceae) were not detected and four of these families were abundantly characterized in the Illumina 16S rRNA amplicon sequencing (Alteromonadacea, Rhodobacteracea, Flavobacteriaceae, and Erythrobacteriaceae).
Table 1.
List of the bacterial strains, with their GenBank accession numbers, isolated from Lobophora rosacea and L. monticola using culture approaches, identified with the BLAST-search.
| Algal host | Strain | Isolated bacterial species | Family | Phylum | Accession number |
|---|---|---|---|---|---|
| L. monticola | LMB | Ruegeria sp.2 | Rhodobacteraceae | α-Proteobacteria | KU560505 |
| LMC | Thalassomonas sp. | Colwelliaceae | γ-Proteobacteria | KU560494 | |
| LMD | Ruegeria sp.3 | Rhodobacteraceae | α-Proteobacteria | KU560501 | |
| LME | Ruegeria sp.1 | Rhodobacteraceae | α-Proteobacteria | KU560502 | |
| LMF | Vibrio sp.2 | Vibrionaceae | γ-Proteobacteria | KU560496 | |
| LMG | Microbulbifer sp.1 | Alteromonadaceae | γ-Proteobacteria | KU560493 | |
| LMH | Tenacibaculum sp. | Flavobacteriaceae | Bacteroidetes | KU560487 | |
| LMI | Ruegeria sp.2 | Rhodobacteraceae | α-Proteobacteria | KU560506 | |
| LMJ | Paramoritella sp. | Moritellaceae | γ-Proteobacteria | KU560495 | |
| LMM | Ruegeria sp.4 | Rhodobacteraceae | α-Proteobacteria | KU560504 | |
| L. rosacea | LR1 | Shimia sp.1 | Rhodobacteraceae | α-Proteobacteria | KU560499 |
| LR11 | Shimia sp.1 | Rhodobacteraceae | α-Proteobacteria | KU560500 | |
| LR2 | Erythrobacter sp. | Sphingonmonadaceae | α-Proteobacteria | KU560498 | |
| LR3 | Muricauda sp. | Flavobacteriaceae | Bacteroidetes | KU560488 | |
| LR4 | Ruegeria sp.1 | Rhodobacteraceae | α-Proteobacteria | KU560503 | |
| LR5 | Vibrio sp.1 | Vibrionaceae | γ-Proteobacteria | KU560497 | |
| LR6 | Microbulbifer sp.1 | Alteromonadaceae | γ-Proteobacteria | KU560492 | |
| LR7 | Microbulbifer sp.2 | Alteromonadaceae | γ-Proteobacteria | KU560491 | |
| LR8 | Bacillus sp. | Bacillaceae | Firmicutes | KU560489 | |
| LR9 | Microbulbifer sp.3 | Alteromonadaceae | γ-Proteobacteria | KU560490 |
Figure 5.
Pie chart representing the bacterial diversity, at the family and phylum levels, recovered from Lobophora surface using culture approaches.
In situ bioassays
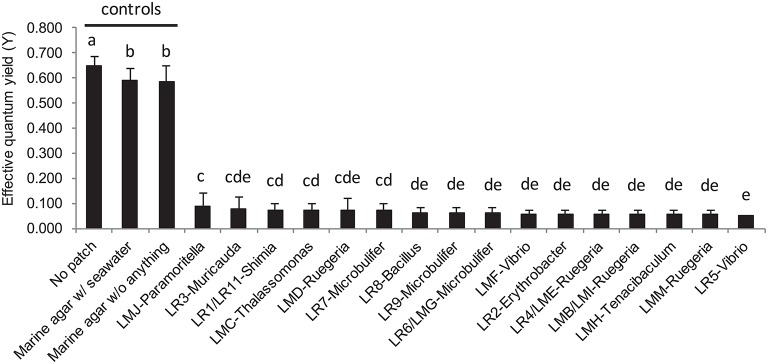
After 24 h exposure, the surface area of the coral A. muricata in direct contact with each of the macroalgae-associated culturable bacterial patches, showed severe visual bleaching (Figure 2C) and an almost complete suppression of coral photosynthetic efficiency across all tested strains, with a relative average quantum yield decrease to 0.064 ± 0.051 (± S.D.), (p < 0.001; Figure 6). Nevertheless, coral tissue on which agar patches were applied was left intact. Seawater bacterial patches, used as control in addition to “sterile” marine agar patches, did not cause coral bleaching.
Figure 6.
Barplot representation of the photosynthetic efficiency from Acropora muricata measured by Pulse Amplitude Modulated (PAM) fluorometry following the in situ bioassays with mono-specific bacterial inclusion culture with the 16 strains isolated from L. rosacea and L. monticola. The statistical analyses, comparing the bacterial culture patches to control patches, were performed using one-way ANOVA and Tukey's HSD post-hoc test. Letters indicate distinct groupings based on post-hoc statistical comparison among sub-fractions. n = 10 assays. Error bars represent standard deviation of the mean.
Discussion
This study shows that all culturable bacteria isolated from macroalgae of the genus Lobophora, pertaining to various taxa, caused severe bleaching, and significantly suppressed photosynthetic efficiency of the coral A. muricata. Our experiments show that macroalgae can indirectly cause coral mortality by means of their surface-associated microbiota. These results suggest also that any bacterial biofilms might be capable of bleaching corals but, as discussed hereafter, a set of ecological factors are generally preventing specific bacterial growth.
Bacterial assemblage diversity
This is the first bacterial community characterization of species from the genus Lobophora using Illumina metabarcoding sequencing. Our 16S rRNA comparative analyses of the bacterial communities between the two Lobophora species showed that the two diversity indices, one more sensitive to richness (Shannon) and the other to evenness (Simpson) were not significantly different between the two species. In contrast, multivariate analyses conducted on Illumina sequencing results clearly showed the species-specificity of the bacterial assemblages associated to individuals of L. rosacea and L. monticola. The two species mainly differ in the most abundant bacterial taxa among those shared by both species. Results indicate that the two Lobophora species have comparable richness of bacterial assemblages but contrasting composition. The two Lobophora species grow in habitats with contrasting environmental conditions. Considering the habitat specificity of these two species, it is questionable whether the bacterial assemblage species-specificity could be linked to the different niches that the two species inhabit. Transplant or common garden experiments will allow concluding whether/to what extent Lobophora species bacterial community composition is controlled by ecological factors or the evolutionary history of the host.
Similar phyla were found with relatively similar percentages between the two Lobophora species. They also shared four bacterial families (i.e., Flavobacteriaceae, Rhodobacteraceae, Alteromonadaceae, Vibrionaceae), three genera (i.e., Ruegeria, Microbulbifer, Vibrio), and two OTUs (Microbulbifer sp. and Ruegeria sp.1). Barott et al. (2011) performed pyrosequencing on the microbial communities of the major ecological functional algal groups in Curaçao (The Netherlands Antilles) including the genus Dictyota (Dictyotales, Phaeophyceae), and established a microbial fingerprint for each group. When comparing their results with ours it is clear that our Lobophora fingerprints (relative abundance percentage) do not resemble any of the algal groups studied by Barott et al. (2011; Figure S1). To what extent technical (marker), region (Caribbean vs. Pacific) or host specificity play a role is uncertain. When considering the major phyla, the two Lobophora species had three major phyla in common with the other algal groups (Proteobacteria, Bacteoidetes, and Planctomycetes), but did not present Firmicutes. Furthermore, Lobophora presented one additional major phylum (Verrucomicrobia) not detected in the other algal groups.
Potential coral pathogens?
The present bioassay results evoke symptoms of “white” diseases, such as the white plague syndrome affecting massive and encrusting corals, the white band disease affecting Acropora spp., and the Acroporid white syndrome affecting A. hyacinthus. While none of the isolated bacterial OTUs have yet been reported in the literature as coral pathogens the majority of the isolated genera (Vibrio, Ruegeria, Thalassomonas, Shimia, Microbulbifer) (Table 1) were documented either as pathogens of corals or other organisms, or associated to coral diseases. Several species of the genus Vibrio are known as agents responsible for coral diseases such as the “yellow blotch/band disease” (Cervino et al., 2004; Rosenberg et al., 2007; Bourne et al., 2008, 2009; Sunagawa et al., 2009). Some members of the genus Thalassomonas were reported as the causative agent of the “white plague” (Rosenberg et al., 2007). Species of the genus Ruegeria presented antibacterial properties (Porsby et al., 2008) and were found associated to the “yellow band disease” (Apprill et al., 2013). The genus Shimia was also found associated to corals affected with the “Porites White Patch Syndrome” (Séré et al., 2013). Finally the genera Microbulbifer and Bacillus presented antimicrobial activities (Kim et al., 2008; Nithyanand and Pandian, 2009). When looking at the list of coral-associated bacteria provided by Mouchka et al. (2010) established at the ordinal level, we notice that five of the orders that are more prevalent in infected or bleached corals, were present in Lobophora, two relatively abundant orders, Rhodobacterales (8 and 23% for L. rosacea and L. monticola, respectively) and Alteromonadales (10 and 9%), and three rare orders (Chromatiales, Clostridiales, and Vibrionales). Similarly, Barott et al. (2011) showed that two Dictyota species harbored the highest percentages (39.6 and 40.8%) of potential pathogens associated with coral diseases, which supports the idea that members of the Dictyotales hold a reservoir of potential coral pathogens.
Ecological insight
It is only recently that the microbial component has been considered in the interactions between algae and corals, and we have only begun to scratch the surface of understanding the complex interactions between coral, algae, and microbes (Table 2). Recent studies showed that macroalgae may alter microbial communities of corals, and convey and/or foster the development of pathogenic bacteria to corals (Table 2).
Table 2.
Comparison of studies on microbial mediation in macroalgal–coral interaction.
| Reference | Objectives and methods | Results and conclusion | Algae | Corals |
|---|---|---|---|---|
| Barott et al., 2011 | Microbial diversity analysis; 16S rDNA tag pyrosequencing | Algae serve as reservoirs for potential coral pathogens | CCA, Dictyota bartayresiana, Halimeda opuntia, Turf | Montastra annularis |
| Barott et al., 2012 | Microbial diversity analysis; 16S rDNA tag pyrosequencing | Algae caused hypoxia on adjacent coral tissue and shifts in the bacterial communities at the interaction zones | CCA, Dictyota bartayresiana, Halimeda opuntia, Turf | Montastra annularis |
| Barott and Rohwer, 2012, a review | DAM [dissolved organic matter (DOM), direct contact, disease, algae and microbes] model | Macroalgae promote heterotrophic microbial overgrowth of coral | – | – |
| Morrow et al., 2011 | Effects of allelochemicals from macroalgae and cyanobacteria on coral microorganisms; bacterial bioassays and 16S rDNA sequencing | Alter coral microbiome | Acanthophora spicifera, Lobophora variegata, Dictyota sp., D. pulchella, Lyngbya polychroa, L. majuscula | Montastraea faveolata, Porites astreoides |
| Morrow et al., 2012 | Effects of algal extracts on coral bacterial assemblage; 16S rRNA DGGE | Algal extracts induce bacterial assemblage shifts | Dictyota sp., Halimeda opuntia, Lobophora variegata | Montastraea faveolata, Porites astreoides |
| Morrow et al., 2013 | Effects of algal contact on coral bacterial assemblage; 16S rRNA DGGE | Algal contact induce bacterial assemblage shifts | Dictyota menstrualis, Halimeda opuntia | Montastraea faveolata, Porites astreoides |
| Nugues et al., 2004 | Effects of algal contact on coral; field experiment | Transmission of coral disease | Halimeda opuntia | Montastraea faveolata |
| Smith et al., 2006 | Effects of dissolved compounds from algae on corals; laboratory experiments | Dissolved compounds from algae | Caulerpa, CCA, Cyanobacteria, Dictyosphaeria cavernosa, Halimeda, Microdictyon, Peysonnellia, Turf mixed | Acropora, Favia, Fungia, Hydnophora, Montastrea, Montipora, Pavona, Pocillopora verrucosa, Porites, Stylophora |
| Sweet et al., 2013 | Original source of coral pathogens | Algae serve as reservoirs for a variety of different potential coral pathogens. Algal-associated microbes alone are unlikely to cause coral death | Caulerpa cupressoides, C. racemosa, Chlorodesmis fastigiata, Dictyota frabilis, Halimeda macroloba, Hincksia sp., Hydroclathrus clathrus, Hypnea sp., Laurencia sp., Padina australis, Sargassum polycystum | Acropora muricata, Montastraea faveolata |
| Thurber et al., 2012 | Effects of macroalgae on coral growth and microbial community structure | Algae caused the disappearance of a γ-proteobacterium; increases or decreases in microbial taxa already present in corals; establishment of new taxa to the coral microbiome; vectoring and growth of microbial taxa from the macroalgae to the coral | Dictyota menstrualis, Galaxuara obtusata, Halimeda tuna, Lobophora variegata, Sargassum polyceratium | Porites astreoides |
| This study | Effects of macroalgae-associated bacteria on corals; bacterial bioassays and 16S rDNA Illumina sequencing | Macroalgae-associated bacteria induce quick and strong coral bleaching | Lobophora monticola, L. rosacea | Acropora muricata |
| Vermeij et al., 2009 | Effects of macroalgae and microbes on survival and settlement success of coral planulae | Macroalgae indirectly cause planular mortality by enhancing microbial concentrations or by weakening the coral's resistance to microbial infections | Ulva fasciata, Acanthophora spicifera, Pterocladiella caerulescens, Sargassum polyphyllum | Montipora capitata |
Our findings suggest that (1) Lobophora hosts surface and core bacterial orders present in diseased and bleached corals, and epibacteria experimentally capable of bleaching corals, and that (2) all tested bacteria can equally induce strong and quick coral bleaching. The bleaching potential from the tested bacteria is apparently not restricted to bacteria from the genus Lobophora considering the panel of bacteria isolated which belong to a variety of taxa but also the natural presence of some of these taxa in healthy corals. In other words, regardless of their taxonomic affinity, dense mature bacterial films can intrinsically bleach corals. But while these results seem to concur with recent findings on the role of macroalgae as conveyors and fosterers of coral pathogens, this does not imply necessarily that macroalgae represent a threat to corals. The mere presence of bacteria is not a threat, as the consequences depend on conditions allowing them to proliferate. In fact, corals themselves also naturally host pathogenic bacteria that can be detrimental to the health of the host under certain conditions. Nonetheless, only specific bacteria have actually demonstrated the capacity to efficiently proliferate and bleach corals. Smith et al. (2006) previously showed that macroalgal diffusible compounds enhanced the activity of coral- or seawater-associated bacteria, leading to coral mortality. These latter results support the idea that bacterial proliferation can generally be adverse to coral. Consequently, although it is true that macroalgae may harbor coral pathogens (Nugues et al., 2004; Barott et al., 2011; Sweet et al., 2013), bacterial adversity is not restricted to the pathogenic strains, but appears related to bacterial density. The natural presence of potentially pathogenic species within coral microbial communities (Mouchka et al., 2010; Barott et al., 2011) supports the idea that adversity toward corals is linked to microbial density. This implies that these specific pathogenic strains have not evolved the capacity to bleach corals but the capacity to take advantage over the other bacteria associated to the host when the necessary conditions are in place. Consequently, in healthy coral reefs, the bacterial community associated to macroalgal surfaces may not represent a threat. Although, macroalgae may act as reservoirs for coral pathogens, there has only been anecdotal reports of bacterial infections in corals attributed to macroalgal contact (e.g., Nugues et al., 2004), and the algal triggered coral disease hypothesis still remains mainly unexplored.
While any bacteria may potentially be adverse to corals, a combination of biotic (e.g., allelopathy) and abiotic (e.g., temperature) factors is regulating microbial composition and abundance on both the coral and the alga (Ritchie, 2006; Mao-Jones et al., 2010; Stratil et al., 2013). Comparably to corals (Ritchie, 2006), algae have the capacity to control the density of specific strains, which coexist in the algal surface biofilm (Barott et al., 2011; Egan et al., 2013). Bacterial regulation may therefore be a key factor preventing bacterial adversity toward corals. Lobophora natural products have experimentally demonstrated a broad-spectrum of antibacterial activities (see Vieira et al., 2015 for review). These compounds potentially act as regulators controlling the algal–bacterial communities. Disruption in the coral or algal microbial community equilibrium may result in adverse conditions for corals.
Future studies should be directed at exploring and clearly identifying factors that lead to changes in the microbial community composition.
Author contributions
CV, AE, OD, and CP designed research; CV, LG, FH, and JG performed the bacterial culture and the in situ bioassays; AE, ES, and TA performed the Illumina sequencing analyses; AE, ES, TA, and CV analyzed the Illumina sequencing data; CV and FH analyzed the bioassays data; and CV, AE, TA, LG, ES, FH, CP, and OD co-wrote the paper. All authors critically revisited this work for important intellectual content and approved the final version to be published. All authors agree to be accountable for all aspects of the work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by UMR ENTROPIE (UR, IRD, CNRS). CV was a Ph.D. fellow of the University of Pierre and Marie Curie and Ghent University and was part of MARES, a Joint Doctorate program selected under Erasmus Mundus coordinated by Ghent University (FPA 2011-0016). AE was supported by postdoctoral fellowship SFRH/BPD/63703/2009 and SFRH/BPD/107878/2015 of the Portuguese National Science Foundation (Fundacão para a Ciência e a Tecnologia, FCT) and all CCMAR members by FCT program UID/Multi/04326/2013.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00316
16S rRNA primers and PCR conditions.
List of the OTUs identified from L. rosacea and L. monticola.
The most influential taxon determined with the SIMPER analysis. The values are the average contributions of each species to the average overall Bray–Curtis dissimilarity.
BLAST-search results for the isolated bacteria strains from L. rosacea and L. monticola.
Barplot representation of the relative abundance of phyla associated with the Lobophora monticola and L. rosacea characterized by Illumina sequencing in comparison with Barott et al. (2011) results with other macroalgae.
References
- Anthony K., Maynard J. A., Diaz−Pulido G., Mumby P. J., Marshall P. A., Cao L., et al. (2011). Ocean acidification and warming will lower coral reef resilience. Global Change Biol. 17, 1798–1808. 10.1111/j.1365-2486.2010.02364.x [DOI] [Google Scholar]
- Antonius A., Afonso-Carillo J. (2001). Pneophyllum conicum killing reef-corals in Mauritius: a new Indo-Pacific syndrome? Bull. Mar. Sci. 69, 613–618. [Google Scholar]
- Apprill A., Hughen K., Mincer T. (2013). Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ. Microbiol. 15, 2063–2072. 10.1111/1462-2920.12107 [DOI] [PubMed] [Google Scholar]
- Barott K. L., Rodriguez−Brito B., Janouškovec J., Marhaver K. L., Smith J. E., Keeling P., et al. (2011). Microbial diversity associated with four functional groups of benthic reef algae and the reef−building coral Montastraea annularis. Environ. Microbiol. 13, 1192–1204. 10.1111/j.1462-2920.2010.02419.x [DOI] [PubMed] [Google Scholar]
- Barott K. L., Rodriguez-Mueller B., Youle M., Marhaver K. L., Vermeij M. J., Smith J. E., et al. (2012). Microbial to reef scale interactions between the reef-building coral Montastraea annularis and benthic algae. Proc. Biol. Sci. 279, 1655–1664. 10.1098/rspb.2011.2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott K. L., Rohwer F. L. (2012). Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. 10.1016/j.tim.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Bodenhausen N., Horton M. W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8:e56329. 10.1371/journal.pone.0056329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. 10.1016/j.tim.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Bourne D., Iida Y., Uthicke S., Smith-Keune C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. 10.1038/ismej.2007.112 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervino J. M., Hayes R. L., Polson S. W., Polson S. C., Goreau T. J., Martinez R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864. 10.1128/AEM.70.11.6855-6864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen L., Tanner C. (1989). Morphological variation and differential susceptibility to herbivory in the tropical brown alga Lobophora variegata. Mar. Ecol. Prog. Ser. 54, 287–298. 10.3354/meps054287 [DOI] [Google Scholar]
- Colwell R. (2013). EstimateS: Statistical Estimation of Species Richness and Shared Species From Samples. Version IX. Available online at: http://purloclcorg/estimates
- Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. 10.1038/nature04056 [DOI] [PubMed] [Google Scholar]
- Croft M. T., Warren M. J., Smith A. G. (2006). Algae need their vitamins. Eukaryotic Cell 5, 1175–1183. 10.1128/EC.00097-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pulido G., McCook L. J., Dove S., Berkelmans R., Roff G., Kline D. I., et al. (2009). Doom and boom on a resilient reef: climate change, algal overgrowth and coral recovery. PLoS ONE 4:e5239. 10.1371/journal.pone.0005239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. 10.1111/1574-6976.12011 [DOI] [PubMed] [Google Scholar]
- Fitt W. K., Brown B. E., Warner M. E., Dunne R. P. (2001). Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51–65. 10.1007/s003380100146 [DOI] [Google Scholar]
- Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollants J., Leliaert F., Clerck O., Willems A. (2013a). What we can learn from sushi: a review on seaweed–bacterial associations. FEMS Microbiol. Ecol. 83, 1–16. 10.1111/j.1574-6941.2012.01446.x [DOI] [PubMed] [Google Scholar]
- Hollants J., Leliaert F., Verbruggen H., Willems A., De Clerck O. (2013b). Permanent residents or temporary lodgers: characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc. Biol. Sci. 280, 20122659. 10.1098/rspb.2012.2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. P. (1994). Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551. 10.1126/science.265.5178.1547 [DOI] [PubMed] [Google Scholar]
- Joint I., Tait K., Wheeler G. (2007). Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos. Trans. R. Soc. B Biol. Sci. 362, 1223–1233. 10.1098/rstb.2007.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jompa J., McCook L. J. (2002a). Effects of competition and herbivory on interactions between a hard coral and a brown alga. J. Exp. Mar. Biol. Ecol. 271, 25–39. 10.1016/S0022-0981(02)00040-0 [DOI] [Google Scholar]
- Jompa J., McCook L. J. (2002b). The effects of nutrients and herbivory on competition between a hard coral (Porites cylindrica) and a brown alga (Lobophora variegata). Limnol. Oceanogr. 47, 527–534. 10.4319/lo.2002.47.2.0527 [DOI] [Google Scholar]
- Keshtacher-Liebso E., Hadar Y., Chen Y. (1995). Oligotrophic bacteria enhance algal growth under iron-deficient conditions. Appl. Environ. Microbiol. 61, 2439–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-K., Ravichandran Y. D., Khan S. B., Kim Y. T. (2008). Prospective of the cosmeceuticals derived from marine organisms. Biotechnol. Bioprocess Eng. 13, 511–523. 10.1007/s12257-008-0113-5 [DOI] [Google Scholar]
- Kubanek J., Jensen P. R., Keifer P. A., Sullards M. C., Collins D. O., Fenical W. (2003). Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. U.S.A. 100, 6916–6921. 10.1073/pnas.1131855100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvennefors E. C. E., Sampayo E., Ridgway T., Barnes A. C., Hoegh-Guldberg O. (2010). Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site-and species-specificity of common bacterial associates. PLoS ONE 5:e10401. 10.1371/journal.pone.0010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. (1991). 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (Chichester: Wiley; ), 125–175. [Google Scholar]
- Linnaeus C. (1758). Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis. Editio Decima, Reformata. Holmiae (Stockholm): Laurentii Salvii. [Google Scholar]
- Mao-Jones J., Ritchie K. B., Jones L. E., Ellner S. P. (2010). How microbial community composition regulates coral disease development. PLoS Biol. 8:e1000345. 10.1371/journal.pbio.1000345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Suzuki M., Kasai H., Shizuri Y., Harayama S. (2003). Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ. Microbiol. 5, 25–35. 10.1046/j.1462-2920.2003.00382.x [DOI] [PubMed] [Google Scholar]
- McClanahan T., Aronson R., Precht W., Muthiga N. (1999). Fleshy algae dominate remote coral reefs of Belize. Coral Reefs 18, 61–62. 10.1007/s003380050155 [DOI] [Google Scholar]
- McCook L., Jompa J., Diaz-Pulido G. (2001). Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19, 400–417. 10.1007/s003380000129 [DOI] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., Desantis T. Z., Probst A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K. M., Liles M. R., Paul V. J., Moss A. G., Chadwick N. E. (2013). Bacterial shifts associated with coral-macroalgal competition in the Caribbean Sea. Mar. Ecol. Prog. Ser. 488, 103–117. 10.3354/meps10394 [DOI] [Google Scholar]
- Morrow K. M., Ritson-Williams R., Ross C., Liles M. R., Paul V. J. (2012). Macroalgal extracts induce bacterial assemblage shifts and sublethal tissue stress in Caribbean corals. PLoS ONE 7:e44859. 10.1371/journal.pone.0044859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K., Paul V., Liles M., Chadwick N. (2011). Allelochemicals produced by Caribbean macroalgae and cyanobacteria have species-specific effects on reef coral microorganisms. Coral Reefs 30, 309–320. 10.1007/s00338-011-0747-1 [DOI] [Google Scholar]
- Mouchka M. E., Hewson I., Harvell C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. 10.1093/icb/icq061 [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Nishijima M., Nishimura M., Kuwano K., Saga N. (1996). Bacteria that induce morphogenesis in Ulva pertusa (Chlorophyta) grown under axenic conditions. J. Phycol. 32, 479–482. 10.1111/j.0022-3646.1996.00479.x [DOI] [Google Scholar]
- Nithyanand P., Pandian S. K. (2009). Phylogenetic characterization of culturable bacterial diversity associated with the mucus and tissue of the coral Acropora digitifera from the Gulf of Mannar. FEMS Microbiol. Ecol. 69, 384–394. 10.1111/j.1574-6941.2009.00723.x [DOI] [PubMed] [Google Scholar]
- Nugues M., Bak R. (2008). Long-term dynamics of the brown macroalga Lobophora variegata on deep reefs in Curacao. Coral Reefs 27, 389–393. 10.1007/s00338-007-0346-3 [DOI] [Google Scholar]
- Nugues M. M., Bak R. P. (2006). Differential competitive abilities between Caribbean coral species and a brown alga: a year of experiments and a long-term perspective. Mar. Ecol. Prog. Ser. 315, 75–86. 10.3354/meps315075 [DOI] [Google Scholar]
- Nugues M. M., Smith G. W., Hooidonk R. J., Seabra M. I., Bak R. P. (2004). Algal contact as a trigger for coral disease. Ecol. Lett. 7, 919–923. 10.1111/j.1461-0248.2004.00651.x [DOI] [Google Scholar]
- Porsby C. H., Nielsen K. F., Gram L. (2008). Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74, 7356–7364. 10.1128/AEM.01738-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L., Pintner I. J. (1980). Bacteria induced polymorphism in an axenic laboratory strain of Ulva lactuca (Chlorophyceae). J. Phycol. 16, 196–201. 10.1111/j.1529-8817.1980.tb03019.x [DOI] [Google Scholar]
- Pueschel C. M., Saunders G. W. (2009). Ramicrusta textilis sp. nov. (Peyssonneliaceae, Rhodophyta), an anatomically complex Caribbean alga that overgrows corals. Phycologia 48, 480–491. 10.2216/09-04.1 [DOI] [Google Scholar]
- Rasher D. B., Hay M. E. (2010). Seaweed allelopathy degrades the resilience and function of coral reefs. Commun. Integr. Biol. 3, 564–566. 10.4161/cib.3.6.12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2013). R: a Language and Environment for Statistical Computing. Vienna: The R Foundation for Statistical Computing, Institute for Statistics and Mathematics; Available online at: http://cran.r-project.org [Google Scholar]
- Reshef L., Koren O., Loya Y., Zilber−Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. 10.1111/j.1462-2920.2006.01148.x [DOI] [PubMed] [Google Scholar]
- Ritchie K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. 10.3354/meps322001 [DOI] [Google Scholar]
- Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. 10.3354/meps243001 [DOI] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Sapp M., Schwaderer A. S., Wiltshire K. H., Hoppe H.-G., Gerdts G., Wichels A. (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699. 10.1007/s00248-006-9162-5 [DOI] [PubMed] [Google Scholar]
- Séré M. G., Tortosa P., Chabanet P., Turquet J., Quod J.-P., Schleyer M. H. (2013). Bacterial communities associated with Porites white patch syndrome (PWPS) on three Western Indian Ocean (WIO) coral reefs. PLoS ONE 8:e83746. 10.1371/journal.pone.0083746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Lesser M. P. (2014). Allelopathy in the tropical alga Lobophora variegata (Phaeophyceae): mechanistic basis for a phase shift on mesophotic coral reefs? J. Phycol. 50, 493–505. 10.1111/jpy.12160 [DOI] [PubMed] [Google Scholar]
- Smith J. E., Shaw M., Edwards R. A., Obura D., Pantos O., Sala E., et al. (2006). Indirect effects of algae on coral: algae−mediated, microbe−induced coral mortality. Ecol. Lett. 9, 835–845. 10.1111/j.1461-0248.2006.00937.x [DOI] [PubMed] [Google Scholar]
- Stratil S. B., Neulinger S. C., Knecht H., Friedrichs A. K., Wahl M. (2013). Temperature−driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2, 338–349. 10.1002/mbo3.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Desantis T. Z., Piceno Y. M., Brodie E. L., Desalvo M. K., Voolstra C. R., et al. (2009). Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521. 10.1038/ismej.2008.131 [DOI] [PubMed] [Google Scholar]
- Sweet M. J., Bythell J. C., Nugues M. M. (2013). Algae as reservoirs for coral pathogens. PLoS ONE 8:e69717. 10.1371/journal.pone.0069717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R. V., Burkepile D. E., Correa A. M., Thurber A. R., Shantz A. A., Welsh R., et al. (2012). Macroalgae decrease growth and alter microbial community structure of the reef-building coral, Porites astreoides. PLoS ONE 7:e44246. 10.1371/journal.pone.0044246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij M., Smith J., Smith C., Thurber R. V., Sandin S. (2009). Survival and settlement success of coral planulae: independent and synergistic effects of macroalgae and microbes. Oecologia 159, 325–336. 10.1007/s00442-008-1223-7 [DOI] [PubMed] [Google Scholar]
- Vieira C., D'hondt S., De Clerck O., Payri C. E. (2014). Toward an inordinate fondness for stars, beetles and Lobophora? Species diversity of the genus Lobophora (Dictyotales, Phaeophyceae) in New Caledonia. J. Phycol. 50, 1101–1119. 10.1111/jpy.12243 [DOI] [PubMed] [Google Scholar]
- Vieira C., Gaubert J., De Clerck O., Payri C., Culioli G., Thomas O. P. (2015). Biological activities associated to the chemodiversity of brown algae belonging to the genus Lobophora (Dictyotales, Phaeophyceae). Phytochem. Rev. 10.1007/s11101-015-9445-x [DOI] [Google Scholar]
- Vieira C., Thomas O. P., Culioli G., Genta-Jouve G., Houlbreque F., Gaubert J., et al. (2016). Allelopathic interactions between the brown algal genus Lobophora (Dictyotales, Phaeophyceae) and scleractinian corals. Sci. Rep. 6, 18637. 10.1038/srep18637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M. E., Fitt W. K., Schmidt G. W. (1999). Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc. Natl. Acad. Sci. U.S.A. 96, 8007–8012. 10.1073/pnas.96.14.8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
16S rRNA primers and PCR conditions.
List of the OTUs identified from L. rosacea and L. monticola.
The most influential taxon determined with the SIMPER analysis. The values are the average contributions of each species to the average overall Bray–Curtis dissimilarity.
BLAST-search results for the isolated bacteria strains from L. rosacea and L. monticola.
Barplot representation of the relative abundance of phyla associated with the Lobophora monticola and L. rosacea characterized by Illumina sequencing in comparison with Barott et al. (2011) results with other macroalgae.