Abstract
Fusion of donor mesenchymal stem cells with parenchymal cells of the recipient can occur in the brain, liver, intestine and heart following transplantation. The therapeutic benefit or detriment of resultant hybrids is unknown. Here we sought a global view of phenotypic diversification of mesenchymal stem cell-cardiomyocyte hybrids and associated time course. Using single-cell RNA-seq, we found hybrids consistently increase ribosome components and decrease genes associated with the cell cycle suggesting an increase in protein production and decrease in proliferation to accommodate the fused state. But in the case of most other gene groups, hybrids were individually distinct. In fact, though hybrids can express a transcriptome similar to individual fusion partners, approximately one-third acquired distinct expression profiles in a single day. Some hybrids underwent reprogramming, expressing pluripotency and cardiac precursor genes latent in parental cells and associated with developmental and morphogenic gene groups. Other hybrids expressed genes associated with ontologic cancer sets and two hybrids of separate experimental replicates clustered with breast cancer cells, expressing critical oncogenes and lacking tumor suppressor genes. Rapid transcriptional diversification of this type garners consideration in the context of cellular transplantation to damaged tissues, those with viral infection or other microenvironmental conditions that might promote fusion.
Mesenchymal/multipotent stem/stromal cells (MSCs) can fuse with parenchymal cells of the brain1, liver2, small intestine3 and heart1,4,5,6,7,8,9 following transplantation. Fusion of this type might be tightly controlled and restricted to certain cell types as with sperm-egg fusion and skeletal myoblast fusion. However, it is more likely that regulation of fusion is bypassed in the context of transplantation and the altered tissue environment of damaged or diseased tissue. So-called “accidental cell fusion” can result from cell stress including nutrient deprivation and hypoxia which can render cell membranes leaky or unstable10,11. Unstable cell membranes are biophysically susceptible to membrane fusion12. It may be for this reason that cell fusion appears to occur more readily in the context of hypoxia than normoxia13,14. Accidental cell fusion can also be mediated by viral fusogenic proteins of an active virus or activated elements of an endogenous virus15,16,17,18,19,20,21,22,23. It is estimated that more than 17 of 29 virus families that infect human cells have elements capable of fusing cells15.
We and others have proposed that accidental cell fusion can give rise to fusion products capable of acquiring phenotypic and functional properties of either or both fusion partners2,24,25,26,27,28,29,30,31,32. The beneficial effects of such an outcome include cell fusion between myeloma cells and B cells to form hybridomas and associated monoclonal antibodies33. Similarly, fusion between dendritic cells and tumor cells augments secretion of paracrine factors and can be used as anti-tumor immunotherapy34. Return of liver function has been reported after fusion between transplanted bone marrow MSCs and diseased hepatocytes2,25,35 and endogenous c-kit+ cells can form cardiomyocytes in an infarcted murine heart as a result of cell fusion36.
Accidental cell fusion might also enable catastrophic events including the development of tumor cells and/or metastatic spread of tumor cells. Spontaneous fusion has been reported between normal breast epithelium and breast cancer cells37,38, among breast tumor cells themselves39,40, and between breast cancer epithelium and tumor stromal cells including MSCs41,42. In vitro studies of hybrids formed between normal breast epithelium (M13SV1-EGFP-Neo) and breast cancer cells (HS578T-Hyg) showed increased “locomotory activity” compared to the normal parental line43. Fusion-enhanced migration was associated with altered CCL21/CCR7 signaling, which was previously linked to metastatic spreading of breast cancer to lymph nodes. Increased metastatic potential of hybrids was also observed in vivo when breast cancer cell variants (MDA-MB-231) with tropism for either lung or bone injected in nude mice gave rise to hybrids capable of metastases to both organs39.
Here we probe the extent of transcriptional diversification of hybrids formed between MSCs and cardiomyocytes, and the beneficial or detrimental outcomes of diversification at the single cell level. We probe this particular cell pairing as hybrids of this type have been most frequently reported in the context of cell transplantation to the heart. We utilize single-cell RNA-seq since each hybrid is predicted to be transcriptionally distinct and hence population analyses may mute unique expression profiles.
Results and Discussion
Accidental Fusion via Measles Virus Fusogens
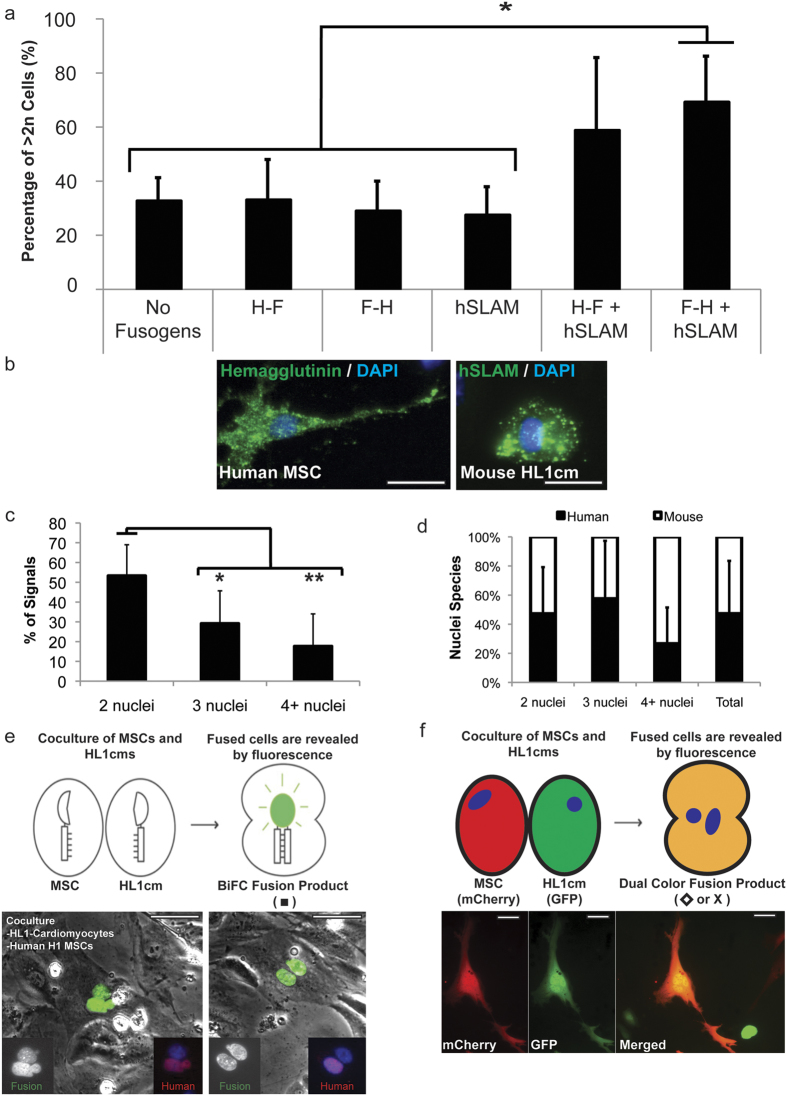
Here we take the case of fusion of MSCs with cardiomyocytes, which has been detected by mulitiple investigators in vivo1,4,5,6,7,8,9. Single-cell transcriptome analysis of hybrids by RNA-seq should enable direct comparison of individual fusion products with parental cells to determine the degree of programming/reprogramming in hybrid cells toward one or both of the parental cells, the rapidity with which programming/reprogramming occurs and the extent to which unique advantageous or deleterious transcriptome features emerge. To this end we developed a means to efficiently induce accidental cell fusion between MSCs and cardiomyocytes in vivo9 and in vitro (shown here) via expression of viral fusogens. The in vitro system mimics the measles virus and associate receptor and enables fusion only when the hemagglutinin (H) protein binds to the human signaling lymphocytic activation molecule (hSLAM), which then forms a trimeric complex with the fusion protein (F) to initiate fusion. To test the specificity of the system, two separate populations of HL-1 cardiomyocytes (HL1cm) were transfected with a bicistronic H–F, bicistronic F–H, hSLAM, or no construct. Co-cultures were generated containing HL1cm transfected with each combination (i.e., H–F/hSLAM, F–H/hSLAM, H–F/no construct, etc.) and monitored for seven days. When all three parts of the fusion system were delivered (either H–F/hSLAM or F–H/hSLAM, Fig. 1a,b), the percentage of cells with DNA content greater than 2n increased from about 30% in controls to 59.1% ± 26.6% (P = 0.23) and 69.5% ± 16.7% (*P < 0.05) respectively, which confirms the specificity of the system.
Figure 1. Induction and detection of cell fusion via the measles virus system and bimolecular fluorescence complementation (BiFC) and dual-color fluorescence, respectively.
(a) DNA content of HL1cm was analyzed via DAPI staining with flow cytometry and DNA content of experimental co-cultures (H–F/hSLAM and F–H/hSLAM) was compared to co-culture controls (no fusogen/no fusogen, H–F/no fusogen, F–H/no fusogen, and hSLAM/no fusogen) (*P < 0.05). Data are represented as mean ± standard deviation (SD). (b) Immunocytochemistry for hemagglutinin (H) on hMSC (green) and hSLAM on HL1cm (green). Nuclei stained with DAPI (blue). Scale bar = 50 μm. (c) Greater than 50% of detected fusion products contained two nuclei. Data are represented as mean ± SD. (*P < 0.05, **P < 0.01). (d) Approximately 50% of the total nuclei of fusion products were of human origin, supporting the specificity of the system. (e) Schematic for the bimolecular fluorescence complementation (BiFC) system. Fusion products were detected with fluorescence microscopy for BiFC (green). Cells were then labeled for human nuclear antigen (HNA, red, only present in the hMSC nuclei). Representative fusion products were detected with BiFC (green), HNA (red), and nuclei detected with DAPI (blue). Scale bar = 50 μm. (f) Schematic for the dual color fluorescence system. Fusion products were detected with fluorescence microscopy for dual expression of GFP from HL1cm and mCherry from mMSC. Representative fusion product is shown below schematic. Scale bar = 50 μm.
To ensure that fusion occurred between two discrete fusion partners, we induced fusion between mouse HL1cm and human H1 MSCs (hMSC) and detected fusion products using a bimolecular complementation (BiFC)-based reporter assay44 (Fig. 1e). This technique is a powerful tool for detecting fusion in vitro and its use was essential for these studies to ensure that single-cell transcriptomes emerged from bona fide fusion products due to the inducible nature of the signal. After identifying fusion products (Fig. 1e), cells were stained for human nuclear antigen (HNA; present on hMSCs and not HL1cm). Over half of the detected fusion products had two nuclei (53.3% ± 15.7%, *P < 0.05 vs. three nuclei and **P < 0.01 vs. 4+ nuclei) (Fig. 1c). Of the fusion products with two nuclei, 47.2% ± 32.0% were of human origin confirming the ability of the fusion system to merge at least one cell of each type (i.e., MSCs and HL1cm) in any given fusion product (Fig. 1d). These data confirm the ability of the measles virus-based system to promote accidental cell fusion between hMSCs and cardiomyocytes and of the BiFC-based detection method to robustly identify hybrids. To ensure that the BiFC complex did not introduce transcriptional bias unrelated to fusion, we also used a dual color (DC) approach wherein different fluorescence reporter genes were expressed in each fusion partner (Fig. 1f).
Transcriptome Diversification of MSC-Cardiomyocyte Fusion Products
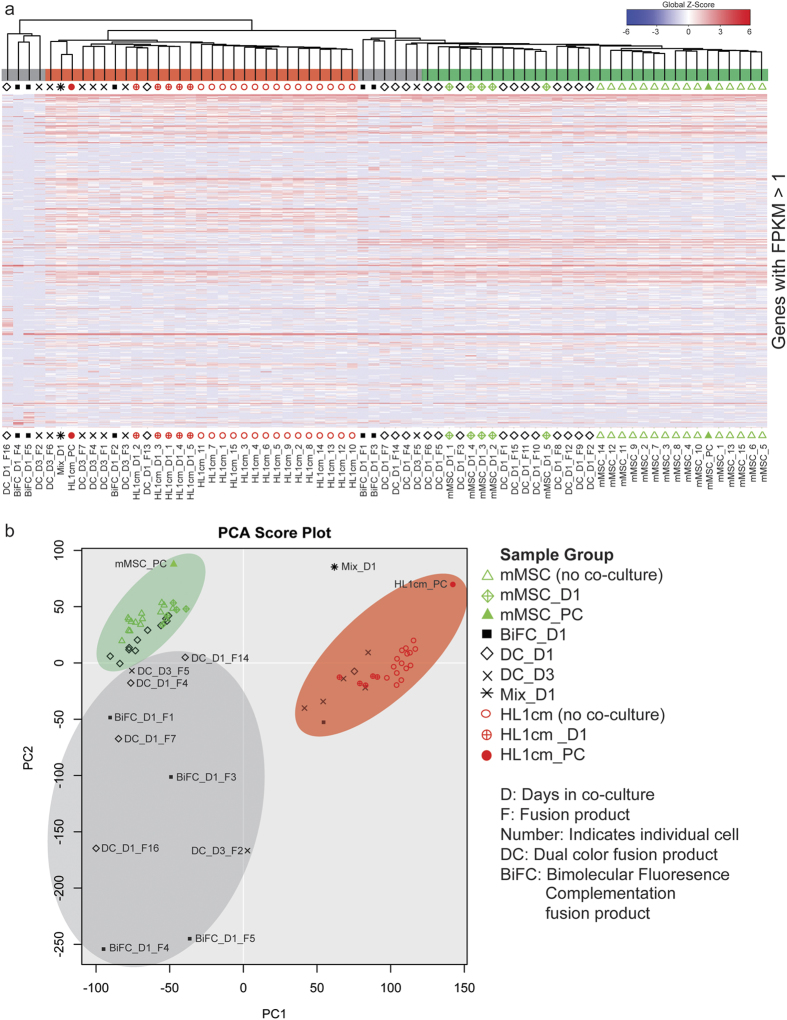
Single-cell RNA-seq was performed on isolated mouse MSCs (mMSC) HL1cm hybrids. We switched to an autologous murine system as we no longer needed the xenogeneic system to validate the BiFC approach and because most reports of MSC-CM fusion reflect autologous systems1,4,8. Hierarchical clustering (HC) and principal component analysis (PCA) were executed to compare transcripts of five fusion products (BiFC_D1_F1–5, 24 h) identified using BiFC, twenty-three fusion products (DC_D1_F1–16, 24 h; DC_D3_F1–7, 72 h) identified using dual color (DC) expression of GFP and mCherry, the parental controls, and the population controls (mMSC_PC and HL1cm_PC). Parental controls included 15 cells of each parental type isolated prior to co-culture (mMSC_1–15 and HL1cm_1–15) and 5 cells of each parental cell type isolated 24 h after co-culture (mMSC_D1_1–5 and HL1cm_D1_1–5). In addition, a population containing a mixture of both parental cells and fusion products obtained 24 h after co-culture was included (Mix_D1). Resulting HC (Fig. 2a, shown are all genes with Fragments Per Kilobase of transcript per Million mapped reads (FPKM) >1 for any sample45) and corresponding PCA plots (Fig. 2b) showed population-level controls correlated with the average of the single cells of that population supporting the accuracy of the single-cell data. The Mix_D1 population control was positioned approximately midway between parental cells reflecting the higher relative fraction of parental cells to fusion products in this population. Analysis of fusion products revealed extensive heterogeneity with ten expressing a unique transcriptome, seven clustering closely with cardiomyocytes and eleven clustering more closely with mMSCs (Fig. 2a,b). Gain or loss of gene expression did not favor particular chromosomes in a substantive way, though minor nuances were noted (Supplementary Fig. S2 and S3).
Figure 2. Hierarchical clustering (HC) and principal component analysis (PCA) of all genes (with FPKM >1) of mMSC-cardiomyocyte fusion products and parental controls.
(a) A global view of gene expression of hybrids (BiFC_D1_F1–5, DC_D1_F1–16, and DC_D3_F1–7), parental cells (mMSC_1–15, mMSC_D1_1–5, HL1cm_1–15, and HL1cm_D1_1–5) and population controls (mMSC_PC, HL1cm_PC, and Mix_D1). Global Z-Score reflects the number of standard deviations away from the mean of expression of all genes in the display. Gene expression is shown in fragments per kilobase of exon per million fragments mapped (FPKM). (b) PCA score plot of hybrids, parental cells and population controls.
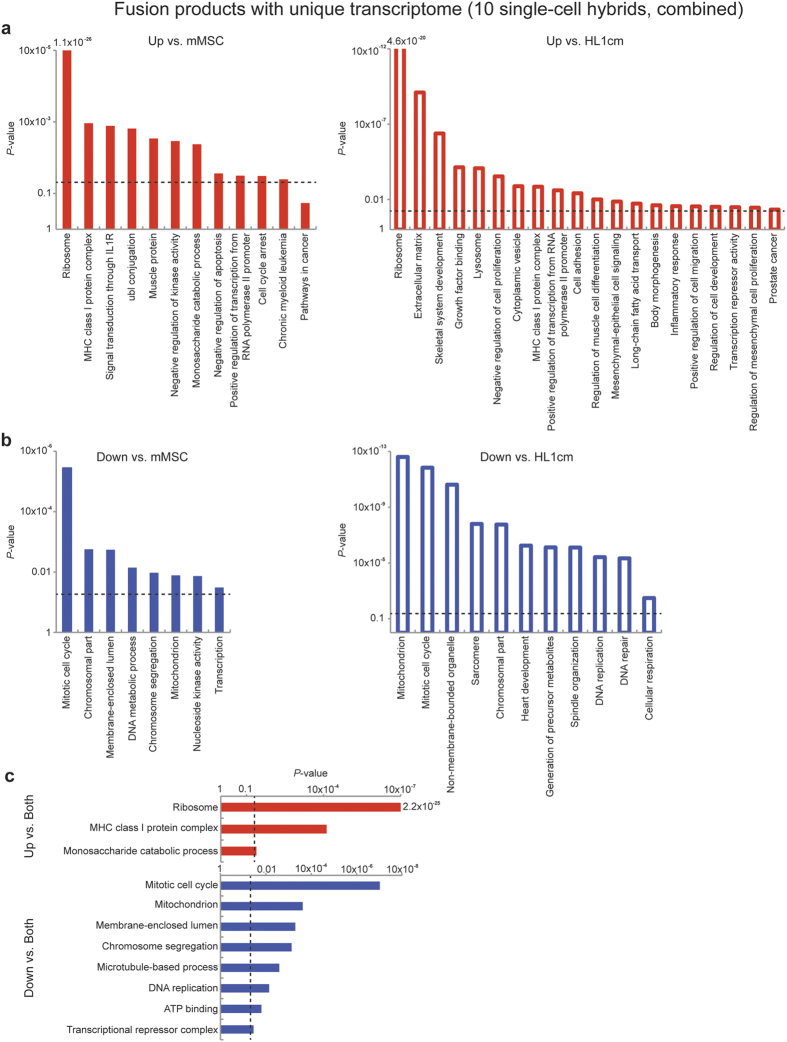
To identify enriched, function-related gene groups and to isolate interacting proteins of the ten hybrids with unique transcriptomes (combined) relative to parental cells, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources. Significant genes were identified relative to parental controls using the Single Cell Differential Expression (SCDE) toolkit and separated according to increased or decreased FPKM values46. From the large list of functional clusters that emerged from our analysis (Supplementary Table S3) certain trends were identified. All hybrids with unique transcriptomes realized a significant increase of ribosomal genes relative to both fusion partners (14 Rpl family genes, 9 Rps family genes and Ubb, Fig. 3a,c). Ribosomal biogenesis is tightly coupled to cell growth and proliferation47 and thus we expected to see a corresponding increase in genes associated with the cell cycle. Instead, hybrids with unique transcriptomes exhibited a significant decrease in FPKM values of the genes associated with the mitotic cell cycle, chromosome segregation and DNA replication compared to both fusion partners. Recent studies suggest hyperactive ribosomal biogenesis in the absence of proliferation can be associated with cellular stress including proteotoxic stress, translational infidelity, expression of oncogenes and loss of tumor suppressors48. We would add the possibility that cellular stress associated with cell fusion might stimulate ribosomal biogenesis as the cell attempts to recalibrate protein synthesis. Recalibration efforts of the cell might also account for decreased FPKM values associated with non-membrane bounded organelles and membrane enclosed lumen (Fig. 3b). Most other significant changes in gene groups of hybrids occurred relative to one or the other fusion partner. For example, when compared to parental cardiomyocytes the fusion products with unique transcriptomes combined showed a decrease in genes associated with heart development and sarcomere formation suggesting a loss of cardiomyocyte contractile function, which was similar to the results found in an in vitro functional study of hybrids formed between human MSCs and neonatal rat ventricular myocytes32.
Figure 3. Gene ontology of hybrids with unqiue transcriptomes.
P-value of functional annotation for differentially expressed genes identified by SCDE for ten hybrids (combined) with unique transcriptomes relative to either fusion partner (see hybrids associated with gray shaded region of Fig. 2a,b). Increased FPKM values are denoted in red (a) and decreased FPKM values are denoted in blue (b) compared to each fusion partner. Versus HL1cm: open bars, versus mMSC: closed bars. (c) P-value of functional annotation for differentially expressed genes identified by SCDE for ten hybrids with a unique transcriptome (combined, see hybrids associated with gray shaded region of Fig. 2a,b) relative to both fusion partners (15 of each). Increased FPKM values relative to both fusion partners are denoted in red and decreased FPKM values are denoted in blue. The dashed line represents a P-value of 0.05. See also Supplementary Table 3.
Function-related Gene Groups of Hybrids Can Mirror Parental Groups
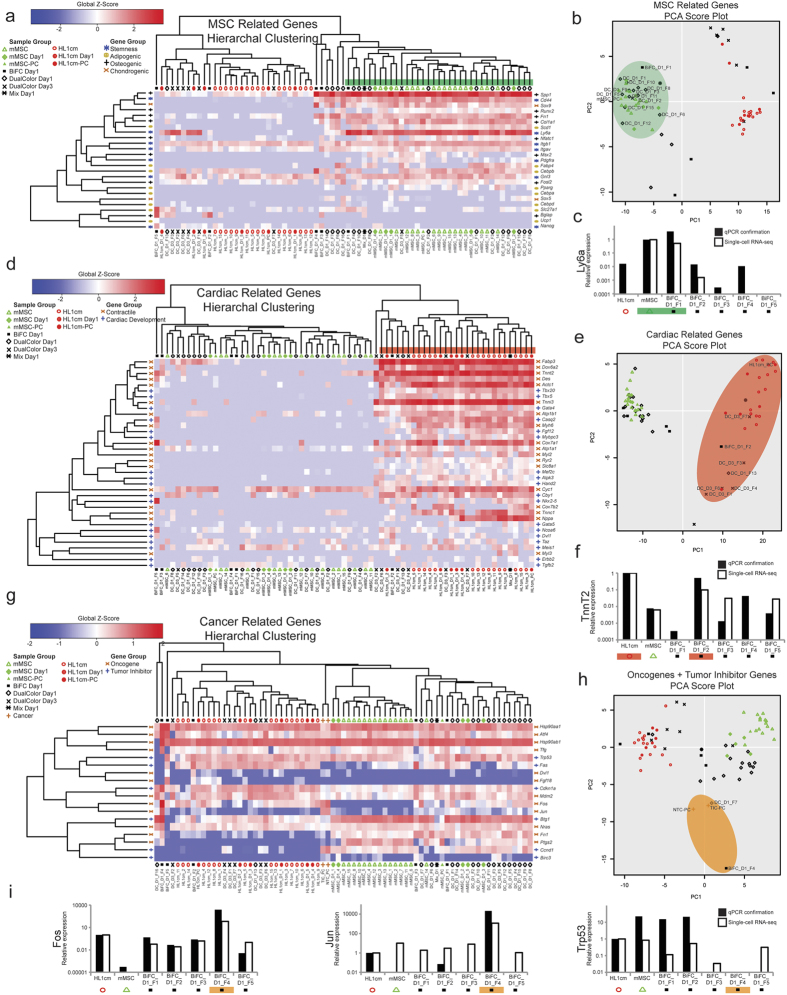
To probe more deeply the contribution of parental lines to programming/reprogramming of hybrid transcriptomes, we defined specific gene sets relevant to each parental cell type including genes related to the differentiation of each parental cell (i.e., mesoderm and cardiac precursors for cardiomyocytes and pluripotent cells for MSCs) and, in the case of MSCs, genes related to mesodermal cell types that can arise from MSCs (i.e., adipocytes, osteoblasts, chondrocytes, Supplementary Table S4). For the MSC gene set, thirteen of twenty-eight hybrids (BiFC_D1_F1, DC_D1_1–5, 8–12, 15, DC_D3_F5) had a profile quite similar to mMSCs (Fig. 4a,b). These thirteen expressed genes related to stemness (Ly6a, CD44, Itgb1, Itgav, and Gnl3) and mesodermal differentiation (Spp1, Sox9, Col1a1, Scd1 and Fn1). RNA-seq data was confirmed with qPCR analysis of Ly6a (Fig. 4c). BiFC_D1_F3 and BiFC_D1_F5 (which do not cluster with mMSCs according to this gene set and clustered with neither mMSC nor HL1cm according to the entire gene set, Fig. 2) expressed high levels of the pluripotency gene, Nanog (68.8 and 124.5 FPKM in BiFC_D1_F3 and BiFC_D1_F5 respectively). This was unexpected since neither mMSCs (average 1.92 ± 5.34 FPKM) nor HL1cm (average 3.57 ± 6.03 FPKM) express Nanog at a high level. Nanog is a transcription factor, known for its control of proliferation and self-renewal in the inner cell mass, supporting a reversion or reprogrammed cell state for BiFC_D1_F3 and BiFC_D1_F5. Of note, twelve of the thirteen hybrids that clustered with mMSCs were obtained one day following co-culture as opposed to those obtained three days after co-culture which either clustered with HL1cm or expressed a unique transcriptome. To explore this trend we next focused on genes associated with cardiomyocyte determination and contractility.
Figure 4. mMSC-cardiomyocyte fusion products can express a cardiomyocyte cell-like, stem cell-like or distinct transcriptome.
(a) HC of fusion products in relation to a set of genes related to stemness (*), adipogenic differentiation (o), osteogenic differentiation (+), or chondrogenic differentiation (x). (b) PCA analysis of fusion products and controls for the MSC gene set. (d) HC of fusion products for a set of genes related to cardiac development (+) or contractile ability (x). (e) PCA analysis of fusion products and controls for the cardiac gene set. (g) HC of fusion products, a population of tumor initiating cancer cells (TIC-PC), a population of nontumorigenic cancer cells (NTC-PC), and controls for a set of oncogenes and a set of tumor suppressor genes. (h) PCA analysis of fusion products, cancer populations and controls for the combined oncogene tumor suppressor gene set. (c,f,i) qPCR confirmation of RNA-seq data. See also Supplementary Table 4 and Supplementary Fig. 4.
Seven (BIFC_D1_F2, DC_D1_F13, DC_D3_F1, 3, 4, 6, 7) of twenty-eight hybrids had a transcriptome quite similar to HL1cm (Fig. 4d,e). Of the seven, five corresponded to hybrids three days after co-culture. These seven hybrids expressed genes related to cardiac development to a lesser extent (Hand2, Cby1, Tbx5, Tbx20, and Gata4) and to contractile machinery to a greater extent (Actc1, Tnni3, Cox6a2, fabp3, Cyc1, Atp1a1, Atp1b1, Des, Tnnt2, and Myh6). RNA-seq data was confirmed with qPCR analysis (Fig. 4f). Of particular interest, BiFC_D1_F5 (which clustered far from HL1cm according to this gene set and clustered with neither mMSC nor HL1cm according to the entire gene set, Fig. 2), expressed a vastly different cardiac gene profile overall and unexpectedly expressed mesodermal precursor gene Nkx2.5 (2545 FPKM) at a level far exceeding the cardiomyocyte controls (average Nkx2–5 expression 12.6 ± 18.2 FPKM) (Fig. 4d). This is consistent with the ontology of hybrids with unique transcriptomes showing increased regulation of muscle cell differentiation when compared to HL1cm (Fig. 3a). Of note, BiFC_D1_F5 also expressed other developmental (Ncoa6, Dvl1) and contractile genes (Cox7a1, Des, Tnnt2 and Myh6) at levels comparable to cardiomyocyte controls. Enticing is the possibility that fusion can enable reprogramming to an early mesodermal state although it should be made clear that single-cell sequencing technologies sample only approximately 5–10% of the total transcripts of a cell, and therefore low-abundance genes including transcription factors associated with cell differentiation may have been missed49, especially in hybrids where bona fide replicates are challenging to obtain and more challenging to confirm.
Cancer-related Gene Groups Prevalent in Some Hybrids
To this point, analyses of transcriptomes of fusion products have been biased toward beneficial outcomes of fusion. Since ten hybrids expressed dramatically diverse and distinct transcriptional features, we reassessed the ontologic outcomes related to cancer fates. Recall from Fig. 3a, that cancer gene clusters (Chronic myeloid leukemia vs. mMSC and Prostate cancer vs. HL1cm) were identified when compared with both mMSC and HL1cm at statistically significant levels. As neither parental cell type had a cancer-like phenotype, we conducted further analysis on genes in these sets to discern the prevalence (i.e., number of hybrids) and degree of tumor susceptibility of hybrids (i.e., clustering with bona fide tumor cells). These genes were classified into one list containing two gene groups, oncogenes or tumor inhibitors (Supplementary Table S4). HC and PCA were performed (Fig. 4g,h, Supplementary Fig. S4). In addition, we included RNA-seq data from two populations (tumor initiating cells (TIC-PC) and nontumorigenic cancer cells (NTC-PC)) of MMTV-Wnt-1 murine tumor cells (previously reported50). We selected these breast cancer cells because they were processed in silico in a manner similar to our approach and therefore FPKMs could be directly compared. Preferable may have been prostate or leukemia cells, but we were unable to find an appropriate single-cell RNA-seq data set. Despite this, the PCA plot showed BiFC_D1_F4 and DC_D1_F7 clustered with both TIC-PC and NTC-PC. Of note, BiFC_D1_F4 and DC_D1_F7 showed decreased FPKM values for the tumor suppressor gene, p53 (Trp53, no detectable FPKM and 6.65 FPKM, respectively) and increased levels of the proto-oncogenes Fos and Jun. Fos levels were 16952.1 FPKM for BiFC_D1_F4 and 754.0 FPKM for DC_D1_F7. Jun levels were 1581.5 FPKM for BiFC_D1_F4 and 14.1 FPKM for DC_D1_F7. Average oncogene levels for parental cell with highest expression (Fos, 128.2 ± 111.2 FPKM, HL1cm and Jun 3.49 ± 3.62 FPKM, mMSC) were less than levels observed in these two hybrids and Trp53 expression (231.5 ± 165.8 FPKM, mMSC; 232.1 ± 144.7 FPKM HL1cm) is reliably detected as opposed to undetected/low levels of these two hybrids. These RNA-seq trends were confirmed with qPCR analysis of Fos, Jun, and Trp53 (Fig. 4i). BiFC_D1_F4 even had higher levels of Fos (550.3 and 1201.5 FPKM for TIC-PC and NTC-PC, respectively) and Jun (421.1 and 477.0 FPKM for TIC-PC and NTC-PC, respectively) expression than the breast cancer cell populations (accounting in large part for the spread between BiFC_D1_F4 and the TIC/NTC in the PCA score plot, Fig. 4h, Supplementary Fig. S4). Thus the increased FPKM values of oncogenes, clustering with tumor-forming cancer cell populations, and decreased or undetected FPKM values of tumor suppressors in some fusion products suggests transcriptome diversity associated with accidental cell fusion has potential to support the emergence of a cancer phenotype.
Conclusions
Thus, here we utilize a robust means to increase accidental cell fusion in vitro and an inducible detection system to reliably identify and isolate fusion products subsequently analyzed using single-cell RNA-seq. We found ten hybrids expressed a unique transcriptome, seven clustered closely with cardiomyocytes and eleven clustered more closely with mMSCs supporting the hypothesis that cell fusion is an ingenious means to generate transcriptional diversity. Grouping of hybrids might reflect, in part, the ratio of parental nuclei51. Figure 1 indicates approximately 47% of hybrids have more than two nuclei and interestingly 64% of hybrids showed significant similarity to one or the other fusion partner. However, at the later time point (day 3) hybrids appear to tend toward a cardiomyocyte-like phenotype compared to day 1 (Chi-square statistic, 10.73; P < 0.005 of HC, Fig. 2a) suggesting such hybrids are more apt to survive or that numbers of nuclei alone do not dictate phenotypic outcomes. Gene ontology analysis of differentially expressed genes showed that the fusion products with unique transcriptomes consistently recalibrate protein synthesis and limit proliferation as indicated by increased FPKM values of genes associated with ribosomal proteins and decreased FPKM values of genes associated with cytoplasmic organelles and the cell cycle, respectively. Some fusion products underwent considerable reprogramming expressing pluripotency and cardiac precursor genes at high levels relative to parental cells. Also of interest was the observation that two hybrids appeared to produce a transcriptome, which aligned closely to that of breast cancer cells, increasing critical oncogenes and decreasing tumor suppressor genes. For these reasons, it will be important to monitor stem cell transplantation and other scenarios that enable membrane instability for the emergence of tumors.
Methods
Human MSCs (hMSC), mouse bone marrow MSCs (mMSC) and mouse HL-1 cardiomyocytes (HL1cm) were expanded and cultured as previously described52,53. hMSCs were used only in Fig. 1 to confirm specificity of the measles virus system. mMSCs were used for single-cell RNA-seq experiments. The first five fusion products were detected via the inducible bimolecular fluorescence complementation (BiFC) system44. The remaining twenty-three hybrids were detected via the more commonly used dual-color (DC) fluorescence methodology. Briefly, GFP was constitutively expressed in HL1cms and mCherry in mMSCs. Hybrids in this case are dual-labeled with both GFP and mCherry. BiFC positive (GFP) cells or dual-labeled cells were sorted using FACS. Sorted, single-cell, fusion products were captured on chips using the Fluidigm C1 system. mRNA libraries were constructed using the Illumina Nextera XT according to the manufacturer’s protocol and sequenced on the Illumina Miseqv3 (Supplementary Fig. S1, Supplementary Tables S1,S2,S3,S4). RNA-seq data has been uploaded to the Gene Expression Omnibus (GEO) database (GSE69926). RNA-seq data was confirmed with qPCR analysis on the cDNA from the Fluidigm C1 chip for selected genes.
A detailed methods section is available in the Online Data Supplement.
Additional Information
How to cite this article: Freeman, B. T. et al. Single-cell RNA-seq reveals activation of unique gene groups as a consequence of stem cell-parenchymal cell fusion. Sci. Rep. 6, 23270; doi: 10.1038/srep23270 (2016).
Supplementary Material
Acknowledgments
This study was funded by NIH, NHLBI, HL089679; University of Minnesota Genomics Center New Investigator Award; NSF-GRFP, 2010105691. We also thank Drs. Joshua Baller and John Garbe of the University of Minnesota Supercomputing Institute for ongoing informatics support.
Footnotes
Author Contributions B.T.F., conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.P.J., collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; B.M.O., collection and assembly of data, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
References
- Alvarez-Dolado M. et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature 425, 968–973 (2003). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature 422, 897–901 (2003). [DOI] [PubMed] [Google Scholar]
- Rizvi A. Z. et al. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci USA 103, 6321–6325 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J. et al. Predominant fusion of bone marrow-derived cardiomyocytes. Cardiovasc Res 68, 387–393 (2005). [DOI] [PubMed] [Google Scholar]
- Kruglyakov P. V. et al. In vitro and in vivo differentiation of mesenchymal stem cells in the cardiomyocyte direction. B Exp Biol Med 142, 503–506 (2006). [DOI] [PubMed] [Google Scholar]
- Noiseux N. et al. Mesenchymal stem cells overexpressing Akt dramatically repair infarcted myocardium and improve cardiac function despite infrequent cellular fusion or differentiation. Mol Ther 14, 840–850 (2006). [DOI] [PubMed] [Google Scholar]
- Nygren J. M. et al. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat Cell Biol 10, 584–592 (2008). [DOI] [PubMed] [Google Scholar]
- Oh H. et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA 100, 12313–12318 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. T., Kouris N. A. & Ogle B. M. Tracking fusion of human mesenchymal stem cells after transplantation to the heart. Stem Cells Transl Med 4, 685–694 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D. M. & Foss A. J. Endothelial cells of tumor vessels: abnormal but not absent. Cancer Metastasis Rev 19, 109–120 (2000). [DOI] [PubMed] [Google Scholar]
- Morikawa S. et al. Abnormalities in pericytes on blood vessels and endothelial sprouts in tumors. Am J Pathol 160, 985–1000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal Roy S. & Sarkar M. Membrane fusion induced by small molecules and ions. J Lipids 2011, 528784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltsyn A. A. et al. Intermittent hypoxia stimulates formation of binuclear neurons in brain cortex- a role of cell fusion in neuroprotection? Exp Biol Med (Maywood) 239, 595–600 (2014). [DOI] [PubMed] [Google Scholar]
- Lazova R., Chakraborty A. & Pawelek J. M. Leukocyte-cancer cell fusion: initiator of the warburg effect in malignancy? Adv Exp Med Biol 714, 151–172 (2011). [DOI] [PubMed] [Google Scholar]
- Duelli D. & Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nature Rev Cancer 7, 968–976 (2007). [DOI] [PubMed] [Google Scholar]
- Kouris N. A. et al. Directed Fusion of Mesenchymal Stem Cells with Cardiomyocytes via VSV-G Facilitates Stem Cell Programming. Stem Cells Int 2012, 414038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouris N. A. et al. A nondenatured, noncrosslinked collagen matrix to deliver stem cells to the heart. Reg Med 6, 569–582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F. & Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol 68, 1179–1185 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joag S. V. et al. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J Virol 70, 3189–3197 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson G. B. et al. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J Exp Med 188, 1159–1171 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Marin M., Kim J. H., Desai T. M. & Melikyan G. B. Distinct requirements for HIV-cell fusion and HIV-mediated cell-cell fusion. J Biol Chem 290, 6558–6573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P. & Zheng J. High-risk HPV E5-induced cell fusion: a critical initiating event in the early stage of HPV-associated cervical cancer. Virology J 7, 238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. et al. Human papillomavirus 16 E5 induces bi-nucleated cell formation by cell-cell fusion. Virology 384, 125–134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagasse E. et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med 6, 1229–1234 (2000). [DOI] [PubMed] [Google Scholar]
- Willenbring H. et al. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med 10, 744–748 (2004). [DOI] [PubMed] [Google Scholar]
- Miller R. A. & Ruddle, F. H. Pluripotent teratocarcinoma-thymus somatic cell hybrids. Cell 9, 45–55 (1976). [DOI] [PubMed] [Google Scholar]
- Cowan C. A., Atienza J., Melton D. A. & Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373 (2005). [DOI] [PubMed] [Google Scholar]
- Tada M., Tada T., Lefebvre L., Barton S. C. & Surani M. A. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J 16, 6510–6520 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo A. et al. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J 23, 1431–1440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N. et al. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature 416, 542–545 (2002). [DOI] [PubMed] [Google Scholar]
- Wei H. J. et al. FOXF1 mediates mesenchymal stem cell fusion-induced reprogramming of lung cancer cells. Oncotarget 5, 9514–9529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrin I. Y., Yoon W., Li L., Shepherd N. & Bursac N. Rapid fusion between mesenchymal stem cells and cardiomyocytes yields electrically active, non-contractile hybrid cells. Sci Rep 5, 12043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C. & Cuello A. C. Hybrid hybridomas and their use in immunohistochemistry. Nature 305, 537–540 (1983). [DOI] [PubMed] [Google Scholar]
- Lindner M. & Schirrmacher V. Tumour cell-dendritic cell fusion for cancer immunotherapy: comparison of therapeutic efficiency of polyethylen-glycol versus electro-fusion protocols. Eur J Clin Invest 32, 207–217 (2002). [DOI] [PubMed] [Google Scholar]
- Willenbring H. & Grompe M. Delineating the hepatocyte’s hematopoietic fusion partner. Cell Cycle 3, 1489–1491 (2004). [DOI] [PubMed] [Google Scholar]
- van Berlo J. H. et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509, 337–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar T. et al. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin Exp Metastasis 28, 75–90 (2011). [DOI] [PubMed] [Google Scholar]
- Ozel C. et al. Hybrid cells derived from breast epithelial cell/breast cancer cell fusion events show a differential RAF-AKT crosstalk. Cell Comm Signal 10, 10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. & Kang Y. Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc Natl Acad Sci USA 106, 9385–9390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Y., Ha S. A., Yang Y. S. & Kim J. W. p-Glycoprotein ABCB5 and YB-1 expression plays a role in increased heterogeneity of breast cancer cells: correlations with cell fusion and doxorubicin resistance. BMC Cancer 10, 388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen B. M. et al. Spontaneous fusion with, and transformation of mouse stroma by, malignant human breast cancer epithelium. Cancer Res 66, 8274–8279 (2006). [DOI] [PubMed] [Google Scholar]
- Noubissi F. K., Harkness T., Alexander C. M. & Ogle B. M. Apoptosis-induced cancer cell fusion: a mechanism of breast cancer metastasis. FASEB J 29, 4036–45 (2015). [DOI] [PubMed] [Google Scholar]
- Berndt B. et al. Fusion of CCL21 non-migratory active breast epithelial and breast cancer cells give rise to CCL21 migratory active tumor hybrid cell lines. PLoS One 8, e63711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. P., Vincenz C., Eliceiri K. W., Kerppola T. K. & Ogle B. M. Bimolecular fluorescence complementation analysis of eukaryotic fusion products. Biol Cell 102, 525–537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O., Teichmann S. A. & Marioni J. C. Computational and analytical challenges in single-cell transcriptomics. Nature Rev Genetics 16, 133–145 (2015). [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Silberstein L. & Scadden D. T. Bayesian approach to single-cell differential expression analysis. Nat Methods 11, 740–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J., Yetil A. & Felsher D. W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10, 301–309 (2010). [DOI] [PubMed] [Google Scholar]
- Orsolic I. et al. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Sem Cancer Bio S1044–579X (2015). [DOI] [PubMed] [Google Scholar]
- Grun D., Kester L. & van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods 11, 637–640 (2014). [DOI] [PubMed] [Google Scholar]
- Feng W. et al. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells 32, 1734–1745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlath G. K. & Blau H. M. Expression of muscle genes in heterokaryons depends on gene dosage. J Cell Biol 102, 124–130 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P. & Hematti P. Derivation and immunological characterization of mesenchymal stromal cells from human embryonic stem cells. Exp Hematol 36, 350–359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhelper M. E. et al. Proliferation in vivo and in culture of differentiated adult atrial cardiomyocytes from transgenic mice. Am J Physiol 259, H1826–1834 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.