Abstract
The misfolding and aggregation of proteins into amyloid fibrils characterizes many neurodegenerative disorders such as Parkinson’s and Alzheimer’s diseases. We report here a method, termed SAVE (single aggregate visualization by enhancement) imaging, for the ultrasensitive detection of individual amyloid fibrils and oligomers using single-molecule fluorescence microscopy. We demonstrate that this method is able to detect the presence of amyloid aggregates of α-synuclein, tau, and amyloid-β. In addition, we show that aggregates can also be identified in human cerebrospinal fluid (CSF). Significantly, we see a twofold increase in the average aggregate concentration in CSF from Parkinson’s disease patients compared to age-matched controls. Taken together, we conclude that this method provides an opportunity to characterize the structural nature of amyloid aggregates in a key biofluid, and therefore has the potential to study disease progression in both animal models and humans to enhance our understanding of neurodegenerative disorders.
Keywords: Parkinson’s, CSF, biomarkers, single-molecule
Many neurodegenerative diseases share a common underlying process which involves the misfolding and aggregation of a native soluble protein into insoluble deposits that can accumulate in the brain and are accompanied by progressive neuronal damage.1 Alzheimer’s disease (AD) and Parkinson’s disease (PD) are the most common of these disorders, together affecting up to 8% of people over the age of 65 years. In AD, the amyloid-β (Aβ) peptide accumulates as extracellular deposits, or plaques, and the protein tau as intracellular inclusions, whereas in PD, α-synuclein (αS) accumulates as Lewy bodies within neuronal cells. Despite plaques and deposits being the most clearly evident characteristics of the diseases, much evidence now points toward soluble aggregate species as being the key cytotoxic agents.2−7 Due to the highly heterogeneous nature and complexity of such species, single-molecule fluorescence techniques have proved to be extremely useful for studying both the oligomers and fibrils of amyloid proteins, as such methodologies can probe below the ensemble average of the system, and characterize even the rarest of species.3,6,8,9 Such techniques have, however, until now, required the protein of interest to be covalently labeled with an organic fluorophore, which is not always straightforward and requires carefully controlled reactions to ensure that it has no significant effect on the behavior of the protein.10,11 The need for labeling makes it very challenging to apply single-molecule fluorescence techniques to samples of proteins extracted from in vivo sources. To address these issues, we have developed an approach to detect aggregates that does not rely on covalently labeling proteins with organic fluorophores, but instead relies on addition of a dye that noncovalently binds the oligomers and fibrils, and not the monomeric protein. This method, that we term SAVE (single aggregate visualization by enhancement) imaging, uses highly sensitive single-molecule instrumentation to detect the fluorescence emission signal from dye molecules that interact with the common motif of extended β-sheet structure that is present in amyloid-related protein aggregates.
The most commonly used assays for detecting amyloid fibrils exploits the binding of the benzothiazole salt thioflavin-T (ThT) (Figure 1A) to amyloid structures, which leads to an increase in its fluorescence intensity by several orders of magnitude, thereby making it an unusually sensitive and efficient reporter of extended β-sheet structure.12,13 Indeed, the fluorescence quantum yield, Φfl, of ThT in water at room temperature is 1 × 10–4, but when docked between surface side-chains in grooves running parallel to the long axis of the cross β-sheets, the fluorescence quantum yield increases by many orders of magnitude, for example, Φfl = 0.43 for ThT bound to amyloid fibrils formed from insulin.14,15 Previous work16,17 has used total internal reflection microscopy (TIRF) combined with ThT fluorescence to follow the elongation of fibrils in real-time. In this case, the species were several micrometers in length and appeared as clear fibril-like structures in the images. It is generally thought that only fibrils contribute significantly to an increase in the fluorescence intensity of ThT, since the smaller aggregates (particularly those below the diffraction limit of light (∼250 nm)) have fewer binding-sites. However, here we show that the high detection efficiency of single-molecule techniques enables the observation of an enhancement in fluorescence through binding both to fibrils, and oligomers. Not only are we able to observe the much smaller species in vitro, but similar aggregates can also be detected in the cerebrospinal fluid samples from people both with and without Parkinson’s disease. This is the first time, to our knowledge, that ThT-active aggregates have been detected in human CSF and opens up new possibilities to understanding the cause of Parkinson’s disease and could also be used as a method for early diagnosis.
Figure 1.
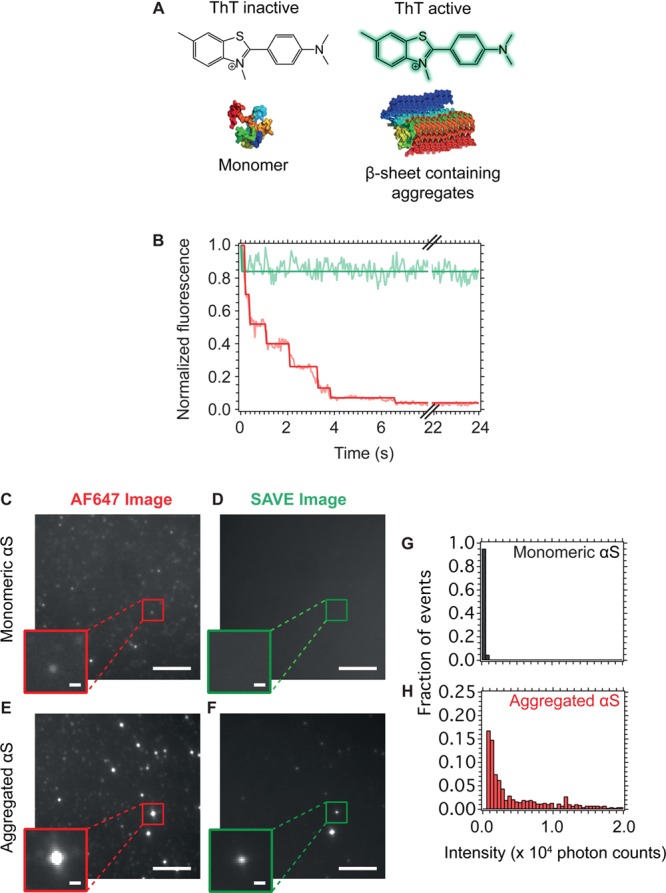
(A) Molecular structure of ThT. In the unbound state, ThT has a low fluorescence intensity; however, when bound to extended β-sheet containing oligomers or fibrils, the fluorescence quantum yield increases significantly (more then 4000 fold). (B) Fluorescence intensity of an individual αS aggregate. The red trace shows the AF647 fluorescence, which decreases over time in stepwise drops, due to photobleaching of each dye molecule. The green trace shows the intensity of ThT, which remains constant over time, since exchange of ThT occurs at a much higher rate than photobleaching of the individual ThT molecules. (C) AF647 fluorescence image of monomeric αS. (D) SAVE image of monomeric αS, there are no visible puncta, since ThT does not bind with an increase in fluorescence to monomeric protein. (E) AF647 image of aggregated αS. The brighter puncta correspond to aggregates, and the dimmer puncta monomeric protein. (F) SAVE images of the same regions as in (E). Only the aggregates (bottom) give rise to fluorescent signal, and these correlate well with the brighter puncta in (E). The scale bars in (C)–(F) are 5 μm and in the zoom are 500 nm in length. AF647 intensity histograms of labeled monomeric and aggregated αS are shown in (G) and (H), respectively. As expected, the aggregated αS has an intensity distribution shifted to higher values than for monomeric αS.
Results and Discussion
Monitoring the Aggregation of αS
To detect individual protein aggregates, we have merged traditional bulk techniques with single-molecule instrumentation, and have made use of TIRF microscopy to detect and count individual ThT-bound aggregates, one-by-one, generated from both in vitro and in vivo sources. The success of this approach can be attributed to a number of factors. First, as ThT is only significantly fluorescent when bound to the underlying cross β-sheet structure of amyloid, it can be added to a sample at high concentrations to promote binding, but without generating high background fluorescence that prevents single-molecule detection. Second, the unusually large Stokes shift (i.e., the wavelength difference between the excitation and emission spectral maxima) of ThT is exploited in the design of our instrumentation; excitation at 405 nm is coupled with detection of fluorescence emission at wavelengths greater than 500 nm. This strategy effectively filters out nonspecific sources of autofluorescence that generally have a smaller Stokes’ shift, while still allowing the detection of even low amounts of ThT fluorescence. In addition, the use of a high numerical aperture objective lens, and a highly sensitive electron multiplied charge coupled device (EMCCD) camera, allows even those aggregates with extremely low fluorescence intensity to be successfully imaged.
The ability of SAVE imaging to detect protein aggregates is first demonstrated using αS covalently labeled with Alexa Fluor 647 (AF647) after incubation under conditions we have previously shown to favor the formation of oligomers and fibrils.3 The use of labeled monomeric protein allows all αS-containing species to be specifically imaged through excitation and detection of AF647 fluorophores. Figure 1B shows a typical intensity trace of a single aggregate of AF647-labeled αS; the AF647 intensity profile (red) shows a series of stepwise decreases, characteristic of photobleaching of the single fluorophores. In contrast, there is no noticeable photobleaching of ThT (green trace) as the dye binding is transient, and so there is a rapid turnover of fluorescent molecules bound to the aggregates. Therefore, even though the ThT signal is lower than that from the AF647 fluorescence, the lack of noticeable photobleaching enables the imaging of the aggregates for effectively any length of time without being subjected to a decrease in intensity. SAVE imaging takes advantage of this by integrating the fluorescence from the samples over several seconds, during which time the aggregates remain bright, whereas unwanted sources of fluorescence photobleach.
Figure 1C and D shows the AF647 and SAVE images (same region) of fluorescently labeled monomeric αS. Although multiple distinct puncta can be observed in the AF647 image, no such signals are visible in the SAVE image, since monomeric αS does not bind and enhance the fluorescence of ThT. Figure 1E and F shows the AF647 and SAVE images of a mixture of aggregated and monomeric αS. The brighter puncta in the AF647 image appear as discrete puncta in the corresponding SAVE image, and these can be attributed to αS aggregates. Once again, the dimmer puncta in the AF647 image, which correspond to monomeric αS, have no coincident puncta in the SAVE image. One of the advantages of SAVE is that discrete species can be identified and characterized, and Figure 1G and H shows the intensity distribution from individual puncta detected in the AF647 image of the monomeric and aggregated αS, respectively. The red bars indicate the species detected in which there is a coincident fluorescence spot in the SAVE image, whereas the gray are those in which there is no coincident spot. As expected, the intensity distribution is shifted to higher values for the coincident species, as these have multiple labeled αS monomeric units present in them.
To test the ability of SAVE imaging to detect different αS species, wild-type αS without an added label was incubated under conditions favoring its aggregation (Materials and Methods), and samples were imaged at a range of time-points (Figure 2). During the first 2 h of the aggregation process, the protein sample consisted mainly of monomeric species, and so no fluorescent puncta were observed. However, from 4 to 18 h, fluorescent puncta could be identified, and their numbers increased over time. Due to the diffraction-limit of light, the size of the oligomers cannot be deduced; however the puncta must correspond to species <250 nm in size, and so are likely to be either small extended β-sheet containing fibrils, or oligomers. These have been shown in previous work to be highly cytotoxic to neuronal cells, resulting in increased reactive oxygen species generation.3 The number of fluorescent puncta can be seen to increase over time such that, after 21 h, fibrils larger than 250 nm, and brighter than those at earlier time-points are observed.
Figure 2.
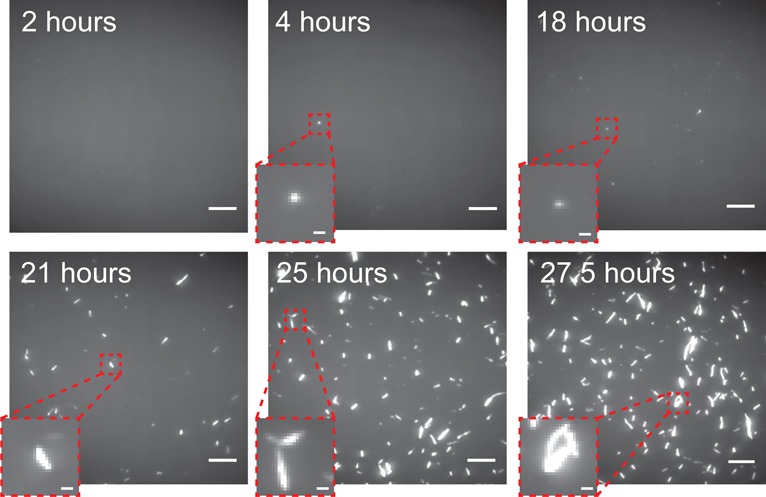
SAVE images of αS species incubated under aggregating conditions. At 2 h, no fluorescent puncta were visible, since only monomeric protein is present. However, after 4 h, diffraction-limited puncta corresponding to oligomers or small fibrils (<250 nm) become visible. Eventually, after 21 h, larger fibrillar species are present. Scale bars are 5 μm and 500 nm in the zoomed insets.
In addition to visualizing aggregates of αS, we explored the ability of SAVE imaging to observe aggregates of the peptides associated with AD, namely, Aβ and tau. Figure 3 shows the AF647 and SAVE images when Aβ and tau were left to aggregate for 3 h and 45 minutes, respectively. At early times, the species are imaged as distinct puncta and correspond to either small fibrils (<250 nm), or oligomers. In the case of Aβ, these oligomers have been shown to be highly cytotoxic to neuronal cells.18
Figure 3.
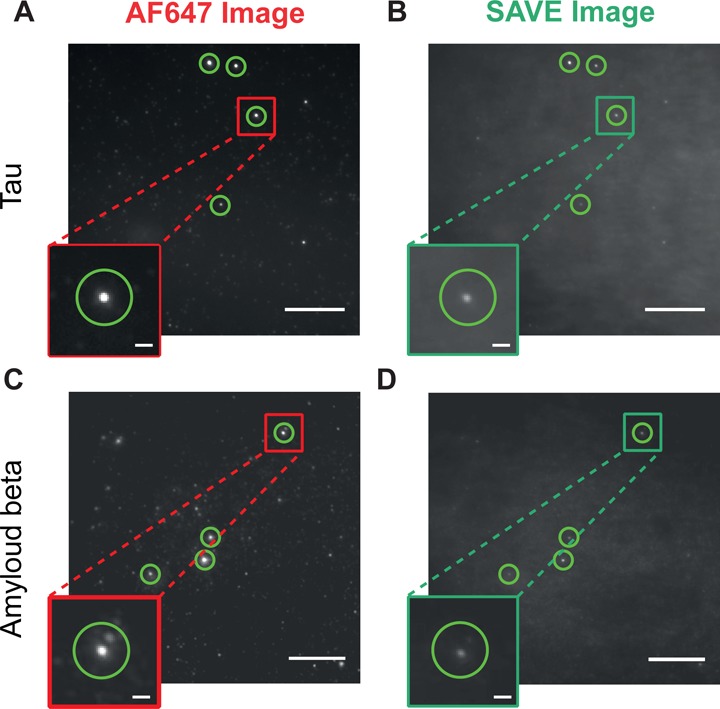
SAVE images of AF647 labeled Aβ and tau aggregates. Aβ was imaged via its AF647 fluorescence (A) and using SAVE (B) after 3 h of aggregation at a concentration of 100 pM, and Tau via AF647 fluorescence (C) and SAVE (D) after 45 minutes at a concentration of 37.5 pM. The brighter puncta in (A) and (C) are coincident with the puncta in (B) and (D), respectively. Scale bars are 10 μm and 1 μm in the zoomed insets. The green circles highlight example aggregated species.
Limit of Detection of Oligomers
SAVE imaging was evaluated using a concentrated sample of unlabeled αS oligomers generated using a protocol that gives rise to >90% of the monomer being incorporated into the oligomeric species.19 The sizes and structural characteristics of the oligomers were determined using the same set of established biophysical techniques, described by Chen et al.,19 and repeated here using atomic force microscopy (AFM) (Figure 4A), dynamic light scattering (DLS) (Figure 4B), and circular dichroism (CD) (Figure 4C) before being characterized by SAVE imaging (Figure 5). The DLS data show that the dominant species have hydrodynamic radii with a mean value of ∼13 nm, significantly larger than that of a monomer (∼4 nm) and the CD spectrum is intermediate between that of a monomer and a β-sheet rich fibrillar species. The AFM images of the oligomers show only the presence of spherical species, and no signs of fibrillar fragments. An approximate concentration of the oligomers was determined by measuring the absorbance of the monomer, and by using the fact that, on average, the oligomers contain 25 monomer units.19
Figure 4.
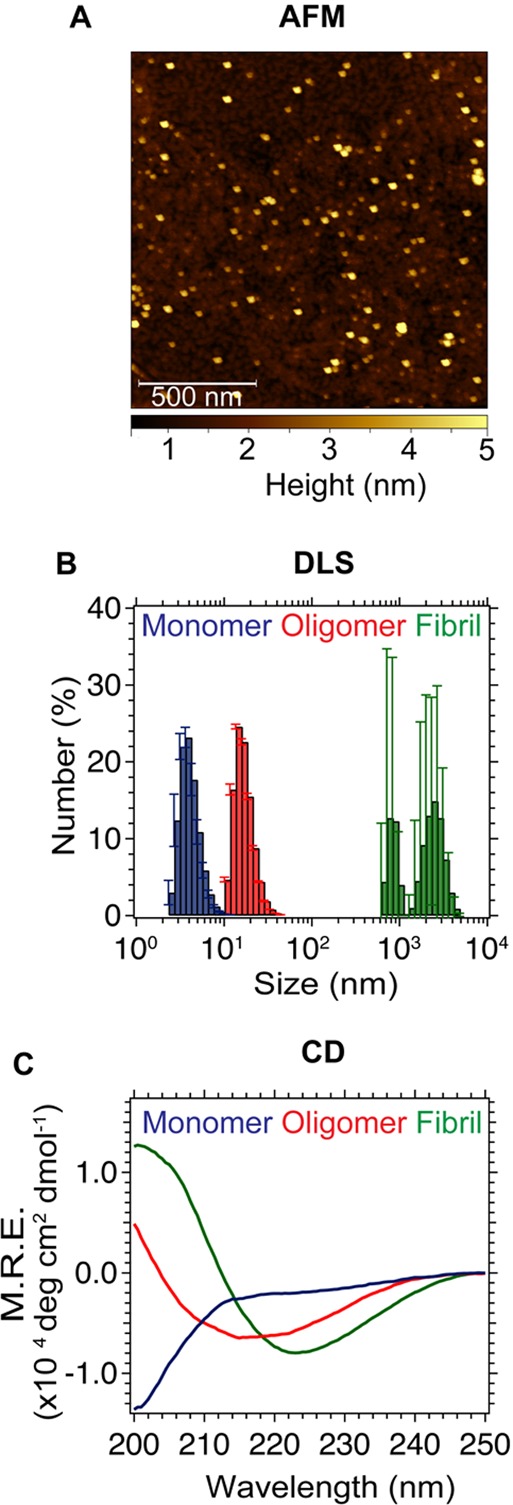
(A) AFM image of enriched oligomers. (B) DLS peak for monomer (blue), enriched oligomers (red), and fibrils (green) (mean from three measurements, error bars are from the standard deviation). (C) CD spectra of monomer, enriched oligomers, and fibrils.
Figure 5.
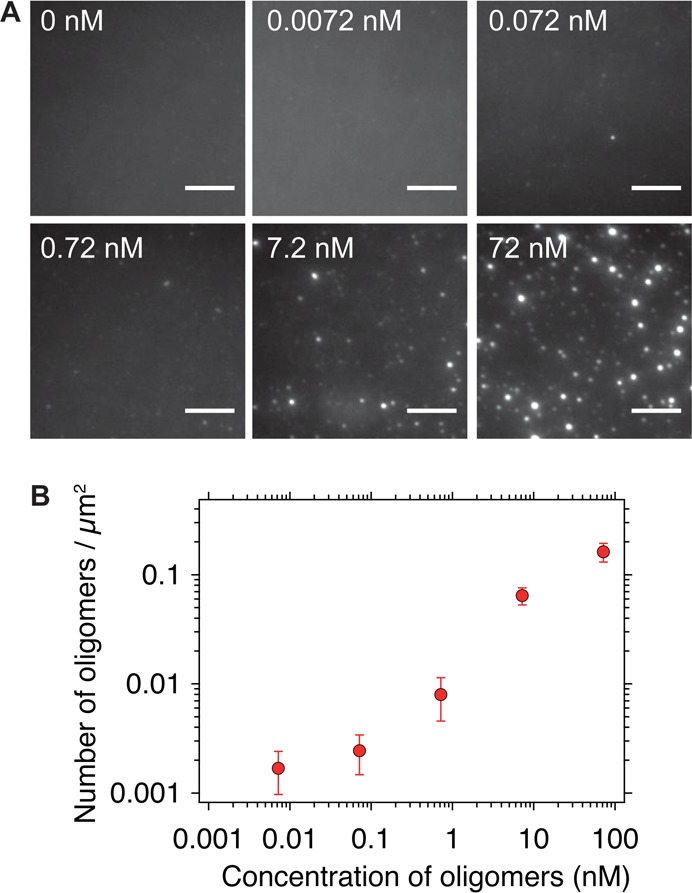
(A) SAVE images of enriched oligomers at a range of concentrations in 5 μM ThT. Scale bars are 5 μm B. Number of events detected when a range of concentrations of enriched oligomers (mean ± SD, n = 30 images). There is a linear detection regime between 0.1 and 10 nM. At lower concentrations, the number of oligomers detected is similar to the number of events detected from buffer alone, while at higher concentrations, the field-of-view is saturated, and counting individual puncta becomes problematic.
To determine the limit of detection by SAVE imaging, samples with a range of concentrations of the enriched oligomers were diluted into 5 μM ThT solutions and the resultant numbers of oligomers quantified by SAVE imaging (Figure 5). At oligomer concentrations spanning 0.1–10 nM, a linear relationship was observed between the concentration of oligomers and the number of detected events. At low concentrations (∼7 pM), the number of events detected (1.69 × 10–3 ± 0.72 × 10–3 counts/μm2) is similar to that from the background alone (0.72 × 10–3 ± 0.38 × 10–3 counts/μm2) and so detecting oligomers becomes challenging. The limit of blank (LoB) is the highest apparent number of counts expected to be found when replicates of a sample containing no analyte are detected, and is given by the expression:20
The limit of detection (LoD) was determined by utilizing both the measured LoB and test replicates of a sample known to contain a low concentration of analyte, and is defined as
The LoB was determined to be 1.35 × 10–3 oligomers/μm2, and the LoD to be 2.52 × 10–3 counts/μm2. Therefore, this method can accurately detect oligomers at concentrations as low as ∼10 pM. At higher concentrations (∼100 nM), the spatial separation of puncta in the field of view becomes too low, and resolving individual oligomers becomes problematic, although this issue can be rectified through prior dilution of the sample.
Detection of Aggregates in Biofluids
The results presented so far confirm that SAVE imaging is able to detect aggregates present in samples formed in vitro, and thus we explored whether extended β-sheet containing aggregates could be detected in human biofluids formed in vivo. Single-molecule detection of species in biofluids is made difficult as a result of the inability to label specifically the biomolecule of interest, and the high background fluorescence that typically results from the multiple components of the samples. CSF, however, is usually free of any cells, and has a very low concentration of protein, and so we observe negligible background signal. Additionally, ThT becomes fluorescent only when bound to molecules containing an extended β-sheet structure typical of aggregates. It is a combination of these factors that allows SAVE imaging to specifically detect amyloid-like aggregates, and not monomeric protein in CSF samples. CSF samples are likely to be the most informative, since CSF is in closer contact with the central nervous system than other more accessible fluids, such as plasma, or urine, and so are more likely to reflect the neurodegenerative processes relating to PD.
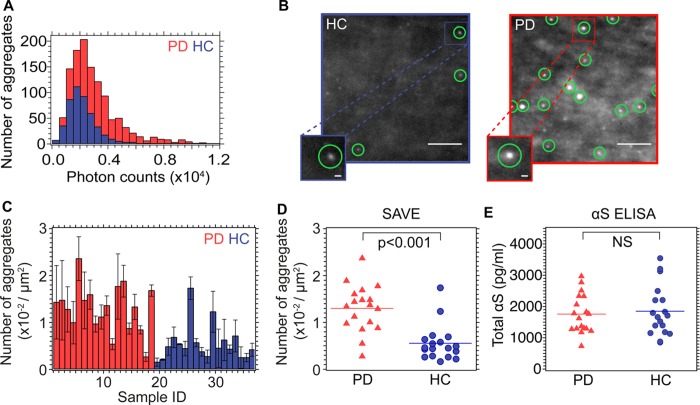
We have examined a series of CSF samples obtained via lumbar puncture from healthy controls (HCs) and from patients suffering from PD (the HC and PD samples were previously obtained for a CSF biomarker study21). Samples from the latter have previously been shown to contain both monomeric and aggregated forms of αS,22,23 and we first measured the total concentration of αS in each of the samples using ELISA methods (Figure 6E). Some reports in the literature have suggested that the total concentration of αS is lower in the CSF of PD patients compared to that from HCs;24,25 our results, however, are in agreement with other reports26−28 showing no significant differences between the sample sets. The total αS concentration, however, does not necessarily reflect differences in the concentrations of specific structural conformations of αS, including the potentially damaging aggregated forms, limiting the utility on total αS as a biomarker. We therefore used SAVE imaging to determine whether or not the total aggregate load was different between samples from healthy controls and PD patients. ThT was added to a final concentration of 5 μM in samples of CSF diluted 1:10 in PBS, it was possible to observe fluorescence signals in samples from both PD patients (n = 18) and healthy controls (HC) (n = 18) (Figure 6) that can be identified as aggregates (Supporting Information Tables S2 and S3 provide sample information, aggregate counts and intensities, and Figure 6 show the results from the analysis of the CSF samples). The data also show that significantly more puncta are, however, detected in the SAVE images from the PD samples (0.013 ± 0.005 aggregates/μm2, mean ± SD) compared to those from HCs (0.006 ± 0.004 aggregates/μm2, mean ± SD) giving rise to a 95% confidence interval: −0.01 to −0.004, p < 0.0001. In addition, the distribution of integrated fluorescence intensity (or brightness) of the aggregates (Figure 6A) could be determined by the technique. This distribution was found to be shifted to higher values for the PD compared to the HC samples, indicating either that the aggregates contained more monomer units, or had a more ordered extended β-sheet structure.
Figure 6.
(A) Photon count intensity distribution histograms from two representative samples of CSF (PD (red), sample 10; controls (blue), sample 23); these cases give rise to counts closest to mean for the PD and HC samples, respectively. (B) Representative TIRF images from sample 10 (red) and sample 23 (blue). For each sample, 27 images were taken (three 3 × 3 grid scans), and the number of puncta counted. For sample 10, 1201 oligomers were detected, whereas for the HC, only 348 oligomers were observed. Green circles show detected oligomers. Scale bar is 5 μm in the main images, and 500 nm in the zoomed image. (C) Histogram of mean number of oligomers for each sample, red bars are from PD CSF samples, and blue from HC samples. (D) Box plots of the same data for the number of oligomers detected. Horizontal lines show the mean counts for PD and HC samples. (E) Box plots of total αS concentrations from ELISA measurement.
Previous studies of CSF samples using a dual-ELISA method indicate that aggregated αS is significantly more abundant in CSF from PD patients than in those from healthy individuals, or indeed, from those suffering from Alzheimer’s disease (AD), or from progressive supranuclear palsy (PSP).29 Our results are in broad agreement with these latter studies, as we have also detected a higher concentration of extended β-sheet containing aggregates in the CSF of PD patients compared to healthy controls. These results indicate that the ultrasensitive detection of aggregated protein in CSF is possible, and may, in addition, differentiate between disease and nondisease states. In addition, single-molecule methods have been combined with fluorescently tagged antibodies to detect amyloid aggregates;31,32 however, as with ELISA studies, these methods rely on immunocapture of the protein of interest, which can be problematic. Single-molecule fluorescence has also previously been used to detect the number of β-amyloid seeds present in CSF, using seeded amplification.33 In contrast to these previous studies, SAVE imaging relies on the structural motif of the extended crossed β-sheet, and so can directly detect aggregates regardless of their protein composition and without amplification, providing a complementary measurement to ELISA. It may be the case that this is a more relevant measure, since neurodegenerative diseases may be due to a more general loss of protein homeostasis,34 and so aggregates of a range of proteins may result. SAVE imaging therefore provides a sensitive measurement of the entire aggregate load in biofluids, and simultaneously allows the in-depth characterization of the nature of these aggregates. The ability to visualize aggregates directly in accessible in vivo fluids provides the opportunity of tracking the relationship between aggregate formation and disease onset. This approach may be a valuable tool in future studies of the progression of disease, therapeutics, or even early diagnosis, all of which remain major hurdles to neurodegenerative diseases. A limitation of this technique is that it is unable to determine which proteins the aggregates are composed of
Conclusion
In this paper, we have discussed the development of a method for directly visualizing unlabeled aggregates with extended cross β-sheet structure at the single-molecule level and that we can straightforwardly detect enriched oligomers, containing on average 25 monomers. In addition to imaging in vitro oligomer formation, we have also shown that SAVE imaging can be used to characterize aggregates present in human biofluids, and we demonstrate that CSF from PD patients contains a significantly higher number of aggregates than CSF from healthy controls. Unlike ELISA-based methods, SAVE imaging detects a generic structural motif rather than a specific epitope of a particular proteins. As we analyze aggregates one-by-one, differences in their fluorescence intensities can be measured, and this may provide insights into their structural nature, information that is very difficult to very to obtain using traditional ensemble-based techniques. The results show that this method has the potential to be used to study, in great detail, unlabeled aggregates formed in vitro, and hence to define the intrinsic kinetics and mechanism of aggregate formation, in addition to providing new insights into the role of oligomers in the onset and progression of disease. Furthermore, this method may act as a diagnostic tool for neurodegeneration, an advance that is critical for the implementation of clinical trials, and for monitoring the effectiveness of potential therapies.
Methods
Preparation of αS
Either the wild-type or A90C variant (for labeled αS) of monomeric αS was purified from Escherichia coli as described previously.3,35 For A90C, the single cysteine was labeled with maleimide-modified Alexa Fluor 647 (AF647) as previously reported.36 The excess dye was removed by passing the labeled protein through a P10 desalting column containing Sephadex G25 matrix (GE Healthcare, Waukesha, WI). The protein was concentrated using Amicon Ultra Centricons (Millipore, Billerica, MA), divided into aliquots, before being flash-frozen in liquid nitrogen and stored at −80 °C. Each aliquot was only thawed once prior to use.
Aggregation of αS
For the aggregation reactions, a 70 μM solution of either wild-type or AF647 labeled αS in 25 mM Tris buffer (pH 7.4) and 0.1 M NaCl (with 0.01% NaN3 to prevent bacterial growth during the experiments) was incubated in the dark at 37 °C, with constant agitation at 200 rpm for 5 days, during which time aliquots were taken.
Aggregation of Amyloid Beta and Tau
To demonstrate the ability to visualize aggregates from other proteins that form extended β-sheet aggregates, we also incubated AF647 labeled cys-Aβ1–42 (500 nM in SSPE buffer (Fluka), pH 7.4, at 37 °C with constant agitation at 200 rpm), and AF647 labeled K18 tau ΔK (10 μM in 50 mM ammonium acetate buffer, pH 7, with 2 μM heparin and 1 mM DTT at 37 °C) as described previously.9,37
Preparation of Enriched Oligomers
A solution of monomeric αS at ca. 800 μM (12 mg/mL) was incubated in PBS at 37 °C without agitation for 20–24 h. The solution was then passed through a 0.22 μm cutoff filter (Millipore, product number SLGP033RK) to remove any particles of dust and/or large protein aggregates that may have been present. The monomeric fraction of the solution (>95%) is removed from the oligomers of αS by passing the solution through a series of 100 kDa cutoff filters (Millipore, product number UFC5100BK). The concentration of the final oligomeric solution was estimated from the absorbance at 275 nm, using an extinction coefficient of 5600 M–1 cm–1.
Characterization of Enriched Oligomers
AFM
AFM images of αS oligomers were taken using a Nanowizard II atomic force microscope (JPK, Berlin, Germany). Images were acquired at room temperature in air using tapping mode. αS oligomers (∼1 μM, 10 μL) were deposited on freshly cleaved mica, incubated for 10 minutes, followed by washes with deionized water, and left to dry in air.
CD
Far-UV CD spectra of the αS oligomers were acquired in PBS at 20 °C. Proteins were diluted to a final concentration of ∼10 μM, and the spectra were acquired using a 1 mm path length cuvette and a J-810 Jasco spectropolarimeter (Tokyo, Japan), equipped with a thermostatic cell holder.
DLS
The intensity-weighted mean hydrodynamic diameter with polydispersity index PdI (a parameter used to describe the width of the size distribution) and the diameter number distribution profile of oligomeric αS (35 μM) in PBS at 25 °C were determined by DLS using a Zetasizer Nano ZSP (Malvern Instruments) at a back scattering angle of 173°.
Preparation of ThT Solution
ThT stock solutions were prepared by diluting ThT (Sigma-Aldrich, product number T3516) into neat ethanol (Sigma-Aldrich, product number 459836) to give a final concentration of ∼1 M. Following this, a 100-fold dilution into prefiltered PBS (0.02 μm syringe filter, Whatman, product number 6809–2101) was performed to give an ∼100 μM stock solution of ThT. It was important to ensure that the ThT solutions were freshly filtered each day, and that the concentrations of the stocks were not too high, since insoluble ThT can give rise to fluorescent puncta. The final concentration was determined from the absorbance at 412 nm, using an extinction coefficient of 36 000 M–1 cm–1. The stock solution was stored in the dark at 4 °C, and was only used for a maximum of 2 weeks after preparation.
Preparation of Slides for Single-Molecule Analysis
Borosilicate glass coverslips (VWR international, 20 × 20 mm, product number 63 1-0122) were cleaned using an argon plasma cleaner (PDC-002, Harrick Plasma) for at least 1 h to remove any fluorescent residues. Frame-Seal slide chambers (9 × 9 mm2, Biorad, Hercules, CA, product number SLF-0601) were affixed to the glass, and 50 μL of poly-l-lysine (70 000–150 000 molecular weight, Sigma-Aldrich, product number P4707-50 ML) was added to the coverslide on the inside of the chamber and incubated for at least 30 minutes, before being washed with 20 nm filtered PBS buffer (phosphate buffered saline tablets, Sigma-Aldrich, product number P4417) which had been prebleached in a bespoke solvent bleacher set at 470 nm light overnight. Prior to use, each batch of coverslides were tested for fluorescent artifacts (i.e., false positives) by analyzing a solution of 5 μM ThT only. We found that the poly-l-lysine solution degraded over time, leading to fluorescent artifacts, and so only fresh solutions of this were used.
Single-Molecule Imaging
Imaging was performed using a home-built total internal reflection fluorescence microscope. This imaging mode restricts detectable fluorescence signal to within 200 nm from the sample slide. For imaging of AF647 labeled αS in the presence of ThT, the output from two lasers operating at 405 nm (Oxxius LaserBoxx, product number LBX-405-100-CIR-PP) and 640 nm (Coherent Cube, product number 1150205) were aligned and directed parallel to the optical axis at the edge of a 1.49 NA TIRF objective (APON60XO TIRF, Olympus, product number N2709400), mounted on an inverted Olympus IX-71 microscope. Fluorescence was collected by the same objective was separated from the returning TIR beam by a dichroic (Di01-R405/488/561/635, Semrock), and passed through appropriate filters (BLP01-488R and BLP01-635R, Semrock). The fluorescence was then passed through a 2.5× beam expander and dual-view (Photometrics). The images were recorded on an EMCCD camera (Evolve 512, Photometrics) operating in frame transfer mode (EMGain of 11.5 e–/ADU and 250 ADU/photon). Each pixel was 109 nm in length. For each data set, 3 × 3 image grids were measured in three different regions of the coverslide. The distance between the nine images measured in each grid was set to 350 μm, and was automated (bean-shell script, Mircomanager) to prevent user bias. Images were recorded at 33 frames s–1 for 150 frames first from the red channel (AF647 emission) with 640 nm illumination (10–50 W/cm2), followed by 150 frames in the blue channel (ThT emission) with 405 nm illumination (150–200 W/cm2). For unlabeled oligomers, either from the enriched preparation, or those present in CSF, the sample was only excited with 405 nm illumination, and images were recorded from the blue channel only. All samples were stored and diluted in LoBind microcentrifuge (Eppendorf, Hamburg, Germany) to limit surface adsorption, as shown to be successful in our previous work.38,39
CSF Samples
CSF samples were collected from 18 patients with clinically defined PD (aged 51–85 years, mean ± SD = 66 ± 8) and 18 normal healthy individuals (aged 46–71 years, mean ± SD = 61 ± 9) by lumbar puncture (sample information is given in Supporting Information Figures S4 and S5). The HC and PD samples were previously obtained for a CSF biomarker study.21 A standardized protocol for the collection and storage of CSF (www.neurochem.gu.se/TheAlzAssQCProgram) was followed. Briefly, lumbar puncture was performed between 9 and 12 a.m. A volume of 15 mL of CSF was collected in sterile polypropylene tubes. The collected CSF samples were gently mixed to avoid gradient effects and centrifuged in the original tube at 4000 rpm for 10 minutes at +4 °C. It was then divided into 0.5 mL aliquots that were frozen on dry ice and stored at −80 °C in LoBind microcentrifuge tubes (Eppendorf, Hamburg, Germany) pending analysis. Blood-contaminated samples (>500 red blood cells per μL) were excluded. Time between sample collection, centrifugation, and freezing was maximum 1 h. The patients were diagnosed according to the UK PD Society Brain Bank Criteria. The study was conducted in accordance with local clinical research regulations and with the provisions of the Helsinki declaration. An informed consent was obtained from all subjects, including access to their clinical data. CSF samples were analyzed for total αS using the αS ELISA Kit (Covance, Berkeley, CA) according to kit insert instructions.
Prior to SAVE imaging, the CSF samples were diluted 10-fold into prefiltered PBS (0.02 μm syringe filter, Whatman, product number 6809-2101) and ThT was added to give a final concentration of 5 μM. The sample was then added to the preprepared borosilicate slides, and incubated for 10 minutes before being imaged as described previously.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.5b00324.
Data analysis; optimum concentration for SAVE imaging; optimum threshold for spot counting; patient sample information (PDF)
Author Present Address
∇ M.K.: iNANO, Aarhus University, Gustav Wieds Vej 14, DK-8000 Aarhus C, Denmark.
Author Contributions
⊥ M.H.H. and S.F.L. contributed equally to this work.
⊥ M.H.H. and S.F.L. performed experiments and wrote the manuscript. S.G., M.J.D., L.T., M.K., J.S.B., and M.I. aided with experiments and contributed to writing the manuscript. S.W.C. provided enriched oligomer samples and performed the biophysical characterization of them. N.K.M. performed CSF sample collection/preparation and helped with the initial analysis of the data. H.Z. performed the ELISAs on the CSF samples. M.H.H., S.F.L., D.K., N.W., N.C., and C.M.D. designed experiments.
This research study was funded in part by the Wellcome Trust/MRC Joint Call in Neurodegeneration award (WT089698) to the UK Parkinson’s Disease Consortium (UKPDC) and the NIHR rare disease translational research collaboration and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. We are also grateful to the Augustus Newman and Wolfson Foundations for their support. We thank the Royal Society for the University Research Fellowship of Dr. Steven F. Lee (UF120277).
The authors declare no competing financial interest.
Supplementary Material
References
- Chiti F.; Dobson C. M. (2006) Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 75, 333–366. 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; et al. (2011) In Vivo Demonstration That Alpha-Synuclein Oligomers Are Toxic. Proc. Natl. Acad. Sci. U. S. A. 108, 4194–4199. 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremades N.; Cohen S. I. A.; Deas E.; Abramov A. Y.; Chen A. Y.; Orte A.; Sandal M.; Clarke R. W.; Dunne P.; Aprile F. A.; et al. (2012) Direct Observation of the Interconversion of Normal and Toxic Forms of α-Synuclein. Cell 149, 1048–1059. 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro T. F.; Putcha P.; Tetzlaff J. E.; Spoelgen R.; Koker M.; Carvalho F.; Hyman B. T.; McLean P. J. (2008) Formation of Toxic Oligomeric Alpha-Synuclein Species in Living Cells. PLoS One 3, e1867. 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings L. M.; Oddo S.; Green K. N.; McGaugh J. L.; LaFerla F. M. (2005) Intraneuronal Abeta Causes the Onset of Early Alzheimer’s Disease-Related Cognitive Deficits in Transgenic Mice. Neuron 45, 675–688. 10.1016/j.neuron.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Cleary J. P.; Walsh D. M.; Hofmeister J. J.; Shankar G. M.; Kuskowski M. A.; Selkoe D. J.; Ashe K. H. (2005) Natural Oligomers of the Amyloid-Beta Protein Specifically Disrupt Cognitive Function. Nat. Neurosci. 8, 79–84. 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- Koffie R. M.; Meyer-Luehmann M.; Hashimoto T.; Adams K. W.; Mielke M. L.; Garcia-Alloza M.; Micheva K. D.; Smith S. J.; Kim M. L.; Lee V. M.; et al. (2009) Oligomeric Amyloid Beta Associates with Postsynaptic Densities and Correlates with Excitatory Synapse Loss near Senile Plaques. Proc. Natl. Acad. Sci. U. S. A. 106, 4012–4017. 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra N.; Blum C.; Segers-Nolten I. M. J.; Claessens M. M. A. E.; Subramaniam V. (2012) Molecular Composition of Sub-Stoichiometrically Labeled α-Synuclein Oligomers Determined by Single-Molecule Photobleaching. Angew. Chem., Int. Ed. 51, 8821–8824. 10.1002/anie.201200813. [DOI] [PubMed] [Google Scholar]
- Narayan P.; Meehan S.; Carver J. A.; Wilson M. R.; Dobson C. M.; Klenerman D. (2012) Amyloid-β Oligomers Are Sequestered by Both Intracellular and Extracellular Chaperones. Biochemistry 51, 9270–9276. 10.1021/bi301277k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina-Ishii C.; Chan Y.-H. M.; Johnson J. M.; Kung L. A.; Lenz P.; Boxer S. G. (2006) Diffusive Dynamics of Vesicles Tethered to a Fluid Supported Bilayer by Single-Particle Tracking. Langmuir 22, 5682–5689. 10.1021/la0534219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M. J.; Jacobson K. (1997) Single-Particle Tracking: Applications to Membrane Dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399. 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Maskevich A. A.; Stsiapura V. I.; Kuzmitsky V. A.; Kuznetsova I. M.; Povarova O. I.; Uversky V. N.; Turoverov K. K. (2007) Spectral Properties of Thioflavin T in Solvents with Different Dielectric Properties and in a Fibril-Incorporated Form. J. Proteome Res. 6, 1392–1401. 10.1021/pr0605567. [DOI] [PubMed] [Google Scholar]
- LeVine H. 3rd (1993) Thioflavine T Interaction with Synthetic Alzheimer’s Disease Beta-Amyloid Peptides: Detection of Amyloid Aggregation in Solution. Protein Sci. 2, 404–410. 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulatskaya A. I.; Maskevich A. A.; Kuznetsova I. M.; Uversky V. N.; Turoverov K. K. (2010) Fluorescence Quantum Yield of Thioflavin T in Rigid Isotropic Solution and Incorporated into the Amyloid Fibrils. PLoS One 5, e15385. 10.1371/journal.pone.0015385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancalana M.; Koide S. (2010) Molecular Mechanism of Thioflavin-T Binding to Amyloid Fibrils. Biochim. Biophys. Acta, Proteins Proteomics 1804, 1405–1412. 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T.; Hamada D.; Hasegawa K.; Naiki H.; Goto Y. (2003) Direct Observation of Amyloid Fibril Growth Monitored by Thioflavin T Fluorescence. J. Biol. Chem. 278, 16462–16465. 10.1074/jbc.C300049200. [DOI] [PubMed] [Google Scholar]
- Ban T.; Hoshino M.; Takahashi S.; Hamada D.; Hasegawa K.; Naiki H.; Goto Y. (2004) Direct Observation of Aβ Amyloid Fibril Growth and Inhibition. J. Mol. Biol. 344, 757–767. 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- Narayan P.; Holmström K. M.; Kim D.-H.; Whitcomb D. J.; Wilson M. R.; St. George-Hyslop P.; Wood N. W.; Dobson C. M.; Cho K.; Abramov A. Y.; et al. (2014) Rare Individual Amyloid-β Oligomers Act on Astrocytes to Initiate Neuronal Damage. Biochemistry 53, 2442–2453. 10.1021/bi401606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. S.; Drakulicb S.; Deas E.; Cremades N. (2015) Structural Characterization of Toxic Oligomers That Are Kinetically Trapped during α-Synuclein Fibril Formation. Proc. Natl. Acad. Sci. U.S.A. 112, E1994–E2003. 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster D. A.; Pry T. (2008) Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 29, S49–S52. [PMC free article] [PubMed] [Google Scholar]
- Magdalinou N. K.; Paterson R. W.; Schott J. M.; Fox N. C.; Mummery C.; Blennow K.; Bhatia K.; Morris H. R.; Giunti P.; Warner T. T.; et al. (2015) A Panel of Nine Cerebrospinal Fluid Biomarkers May Identify Patients with Atypical Parkinsonian Syndromes. J. Neurol., Neurosurg. Psychiatry 86, 1240–1247. 10.1136/jnnp-2014-309562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda T.; Qureshi M. M.; Ardah M. T.; Varghese S.; Shehab S. a. S.; Kasai T.; Ishigami N.; Tamaoka A.; Nakagawa M.; El-Agnaf O. M. A. (2010) Detection of Elevated Levels of α-Synuclein Oligomers in CSF from Patients with Parkinson Disease. Neurology 75, 1766–1772. 10.1212/WNL.0b013e3181fd613b. [DOI] [PubMed] [Google Scholar]
- El-Agnaf O. M. A.; Salem S. A.; Paleologou K. E.; Curran M. D.; Gibson M. J.; Court J. A.; Schlossmacher M. G.; Allsop D. (2006) Detection of Oligomeric Forms of Alpha-Synuclein Protein in Human Plasma as a Potential Biomarker for Parkinson’s Disease. FASEB J. 20, 419–425. 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B.; Cullen V.; Kahn I.; Krastins B.; Outeiro T. F.; Pepivani I.; Ng J.; Schulz-Schaeffer W.; Kretzschmar H. A.; McLean P. J.; et al. (2008) Direct Quantification of CSF Alpha-Synuclein by ELISA and First Cross-Sectional Study in Patients with Neurodegeneration. Exp. Neurol. 213, 315–325. 10.1016/j.expneurol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Tokuda T.; Salem S. A.; Allsop D.; Mizuno T.; Nakagawa M.; Qureshi M. M.; Locascio J. J.; Schlossmacher M. G.; El-Agnaf O. M. A. (2006) Decreased Alpha-Synuclein in Cerebrospinal Fluid of Aged Individuals and Subjects with Parkinson’s Disease. Biochem. Biophys. Res. Commun. 349, 162–166. 10.1016/j.bbrc.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Noguchi-Shinohara M.; Tokuda T.; Yoshita M.; Kasai T.; Ono K.; Nakagawa M.; El-Agnaf O. M. A.; Yamada M. (2009) CSF Alpha-Synuclein Levels in Dementia with Lewy Bodies and Alzheimer’s Disease. Brain Res. 1251, 1–6. 10.1016/j.brainres.2008.11.055. [DOI] [PubMed] [Google Scholar]
- Reesink F. E.; Lemstra A. W.; van Dijk K. D.; Berendse H. W.; van de Berg W. D. J.; Klein M.; Blankenstein M. A.; Scheltens P.; Verbeek M. M.; van der Flie W. M. (2010) CSF α-Synuclein Does Not Discriminate Dementia with Lewy Bodies from Alzheimer’s Disease. Alzheimer's Dementia 22, 87–95. [DOI] [PubMed] [Google Scholar]
- Spies P. E.; Melis R. J. F.; Sjögren M. J. C.; Rikkert M. G. M. O.; Verbeek M. M. (2009) Cerebrospinal Fluid Alpha-Synuclein Does Not Discriminate between Dementia Disorders. Alzheimer's Dementia 5, P342. 10.1016/j.jalz.2009.04.1181. [DOI] [PubMed] [Google Scholar]
- Tokuda T.; Salem S. A.; Allsop D.; Mizuno T.; Nakagawa M.; Qureshi M. M.; Locascio J. J.; Schlossmacher M. G.; El-Agnaf O. M. A. (2006) Decreased Alpha-Synuclein in Cerebrospinal Fluid of Aged Individuals and Subjects with Parkinson’s Disease. Biochem. Biophys. Res. Commun. 349, 162–166. 10.1016/j.bbrc.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Bieschke J.; Giese A.; Schulz-Schaeffer W.; Zerr I.; Poser S.; Eigen M.; Kretzschmar H. (2000) Ultrasensitive Detection of Pathological Prion Protein Aggregates by Dual-Color Scanning for Intensely Fluorescent Targets. Proc. Natl. Acad. Sci. U. S. A. 97, 5468–5473. 10.1073/pnas.97.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke S. A.; Birkmann E.; Henke F.; Görtz P.; Lange-Asschenfeldt C.; Riesner D.; Willbold D. (2007) Single Particle Detection of Abeta Aggregates Associated with Alzheimer’s Disease. Biochem. Biophys. Res. Commun. 364, 902–907. 10.1016/j.bbrc.2007.10.085. [DOI] [PubMed] [Google Scholar]
- Pitschke M.; Prior R.; Haupt M.; Riesner D. (1998) Detection of Single Amyloid Beta-Protein Aggregates in the Cerebrospinal Fluid of Alzheimer’s Patients by Fluorescence Correlation Spectroscopy. Nat. Med. 4, 832–834. 10.1038/nm0798-832. [DOI] [PubMed] [Google Scholar]
- Ciryam P.; Tartaglia G. G.; Morimoto R. I.; Dobson C. M.; Vendruscolo M. (2013) Widespread Aggregation and Neurodegenerative Diseases Are Associated with Supersaturated Proteins. Cell Rep. 5, 781–790. 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W.; Cherny D.; Subramaniam V.; Jovin T. M. (2004) Rapid Self-Assembly of Alpha-Synuclein Observed by in Situ Atomic Force Microscopy. J. Mol. Biol. 340, 127–139. 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Thirunavukkuarasu S.; Jares-Erijman E. A.; Jovin T. M. (2008) Multiparametric Fluorescence Detection of Early Stages in the Amyloid Protein Aggregation of Pyrene-Labeled Alpha-Synuclein. J. Mol. Biol. 378, 1064–1073. 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Shammas S. L.; Garcia G. A.; Kumar S.; Kjaergaard M.; Horrocks M. H.; Shivji N.; Mandelkow E.; Knowles T. P. J.; Mandelkow E.; Klenerman D. (2015) A Mechanistic Model of Tau Amyloid Aggregation Based on Direct Observation of Oligomers. Nat. Commun. 6, 7025. 10.1038/ncomms8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks M. H.; Tosatto L.; Dear A. J.; Garcia G. A.; Iljina M.; Cremades N.; Dalla Serra M.; Knowles T. P. J.; Dobson C. M.; Klenerman D. (2015) Fast Flow Microfluidics and Single-Molecule Fluorescence for the Rapid Characterization of α-Synuclein Oligomers. Anal. Chem. 87, 8818–8826. 10.1021/acs.analchem.5b01811. [DOI] [PubMed] [Google Scholar]
- Tosatto L.; Horrocks M. H.; Dear A. J.; Knowles T. P. J.; Dalla Serra M.; Cremades N.; Dobson C. M.; Klenerman D. (2015) Single-Molecule FRET Studies on Alpha-Synuclein Oligomerization of Parkinson’s Disease Genetically Related Mutants. Sci. Rep. 5, 16696. 10.1038/srep16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.