Abstract
Fragment-based lead discovery has proved to be an effective alternative to high-throughput screenings in identifying chemical matter that can be developed into robust lead compounds. The search for optimal combinations of biophysical techniques that can correctly and efficiently identify and quantify binding can be challenging due to the physicochemical properties of fragments. In order to minimize the time and costs of screening, optimal combinations of biophysical techniques with maximal information content, sensitivity, and robustness are needed. Here we describe an approach utilizing automated microscale thermophoresis (MST) affinity screening to identify fragments active against MEK1 kinase. MST identified multiple hits that were confirmed by X-ray crystallography but not detected by orthogonal methods. Furthermore, MST also provided information about ligand-induced aggregation and protein denaturation. The technique delivered a large number of binders while reducing experimentation time and sample consumption, demonstrating the potential of MST to execute and maximize the efficacy of fragment screening campaigns.
Keywords: binding affinity, biophysical screening, drug discovery, protein aggregation, surface plasmon resonance
Introduction
Over the last decade, fragment-based lead discovery (FBLD) has been adopted throughout academia and industry as a tool to identify viable starting points from which lead compounds can be generated. To date, at least nine FBLD projects have led to compounds in phase II or III studies, including one approved drug.1 The FBLD approach aims to identify very small molecular fragments (150–300 Da) that bind to specific sites of target proteins and are subsequently grown, merged, or linked to produce more potent drug leads.
A variety of biophysical techniques have been applied in FBLD campaigns, ranging from protein and ligand-observed nuclear magnetic resonance (NMR)–based approaches, X-ray crystallography, surface plasmon resonance (SPR), and mass spectrometry (MS) to isothermal titration calorimetry (ITC) and thermal shift assays.2,3 Each method has advantages and disadvantages in terms of sample consumption, degree of automation, and assay complexity. To fully exploit the benefits of FBLD, these biophysical methods should be fast, efficient, and precise in their ability to characterize binding fragments, while also reducing the number of false positives and false negatives. Microscale thermophoresis (MST) is an emerging technique that has been broadly applied to investigate biomolecular interaction of a variety of drug targets. MST detects the directed movement of fluorescent molecules in microscopic temperature gradients in microliter-volume capillaries to quantify interaction affinities4,5 (Suppl. Fig. 1). Each molecule has distinct thermophoretic properties, which are determined by its size, charge, and hydration shell. Binding of ligands typically change at least one of these parameters, resulting in changes in the thermophoretic movement of the molecule. The change in thermophoresis can be used to derive dissociation constants (Kd) within minutes by sequentially scanning capillaries with varying ligand concentrations. MST has been shown to be well suited to detect binding of small molecules, fragments, or even ions to biomolecules,6–8 thus making this technique applicable to FBLD. Moreover, since movement of fluorescent molecules through a detection volume is monitored over time, additional information about protein aggregation and denaturation can be derived from the shape of MST traces.9
Here we report an automated, affinity-based FBLD approach by MST to screen a library containing 193 fragments targeted against mitogen/extracellular signal-regulated kinase (MEK1) from the MAPK signaling cascade. MEK1 is responsible for the phosphorylation and activation of downstream ERK proteins, thereby regulating proliferation, differentiation, transcription regulation, and development. Aberrant signaling in this pathway is associated with unregulated cell growth, and targeting this cascade has become a viable means of developing anticancer therapies.10 The library used for screening was designed by a target-restrained virtual screening campaign, so that a large number of binding fragments was expected.11 The results presented here demonstrate that MST generates quantitative data for affinity rankings in a rapid and precise manner. Fourteen out of 19 fragments subjected to crystallization studies within the MST top-25 affinity ranking were shown to bind to MEK1 by X-ray crystallography. In addition, MST identified protein aggregating effects asserted by a number of fragments, thereby preventing a carryover of false positive into later stages of lead development. Thus, the use of MST in FBLD represents a significant advance toward a fast, reliable, and cost-effective approach to identify and develop fragment-based leads.
Material and Methods
Protein Labeling
The protein construct of MEK1 (residues 37–383) was purchased from Crelux (Crelux GmbH, Martinsried, Germany). Fluorescence labeling of MEK1 was performed following the protocol for N-hydroxysuccinimide (NHS) coupling of the dye NT647 (NanoTemper Technologies, Munich, Germany) to lysine residues. Briefly, 100 µL of a 20 µM solution of MEK1 protein in labeling buffer (130 mM NaHCO3, 50 mM NaCl, pH 8.2) was mixed with 100 µL of 60 µM NT647-NHS fluorophore (NanoTemper Technologies) in labeling buffer and incubated for 30 min at room temperature (RT). Unbound fluorophores were removed by size-exclusion chromatography with MST buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 10 mM MgCl2) as running buffer. The degree of labeling was determined using extinction coefficients ε280 = 20,800 M−1 cm−1 for MEK1 and ε653 = 250,000 M−1 cm−1 for the NT647 fluorophore, with a correction factor Ccorr of 0.031 at 280 nm, using Cprotein = A280 – (A653*Ccorr)/ε280 and Cfluorophore = A653/ε653, and was between 0.9 and 1.1 throughout all labeling reactions. For storage, NT647-MEK1 was frozen in 8 µL aliquots at −80 °C. Prior to MST experiment, aliquots of NT647-MEK1 were thawed on ice and centrifuged for 15 min at 4 °C and 23,000 × g to remove protein aggregates.
Protein Thermal Stability Measurements
To compare stability of NT647-MEK1 and unmodified MEK1 protein, thermal unfolding profiles of the proteins were recorded using the Prometheus NT.48 instrument (NanoTemper Technologies). For this, 30 µL of a 2 µM solution of each protein in assay buffer was prepared, and 3 × 10 µL was loaded into nanoDSF grade standard capillaries (NanoTemper Technologies) for triplicate measurements. Thermal unfolding of triplicates was analyzed in a thermal ramp from 25 to 80 °C with a heating rate of 1 °C/min. Unfolding transition temperatures (Tm) were automatically determined by the software and represented as mean ± SD.
Assay Development for MST Screening
Pretests using premium-coated and standard treated MST capillaries (NanoTemper Technology) were performed to test for adsorption of NT647 MEK1 to capillary walls by analyzing capillary scans recorded by the Monolith NT.115 prior to MST experiments. MEK1 did not adsorb to capillary walls in MST buffer, including 0.05% Pluronic F127 (Sigma-Aldrich, St. Louis, MO), 5 mM DTT, and 5% DMSO, but strongly adsorbed to hydrophobic and standard treated capillaries in the absence of Pluronic F127. Moreover, in the absence of Pluronic F127, reproducibility of MST signals was low, and aberrant MST traces occurred, pointing toward aggregation of the protein. For subsequent experiments, standard treated capillaries and MST buffer with 0.05% Pluronic F127, 5% DMSO, and 5 mM DTT (assay buffer) were used.
The interaction between adenosine triphosphate (ATP) and NT647-MEK1 was established on a Monolith NT.115 instrument (NanoTemper Technologies) and was used as a positive control throughout the screening. For this, ATP serial dilutions where prepared in assay buffer and mixed 1:1 with a solution of 30–50 nM NT647-MEK1 to yield a final volume of 20 µL per dilution. The reaction mixtures were loaded into standard treated capillaries and subsequently analyzed by MST at 20% and 80% MST power, respectively, and a light-emitting diode (LED) intensity of 30%. Analysis of the interaction by thermophoresis after either 30 s laser-on time at MST 20% or 5 s laser-on time at MST 80% yielded similar Kd values with similar signal-to-noise levels, so that a measurement protocol for the screening with 80% and analysis of binding after 5 s laser-on time was chosen to minimize measurement time. Stability and reproducibility of the interaction were tested by remeasuring ATP binding experiments after a 2h incubation time in capillaries at RT. Here, no change in fluorescence intensity, protein adsorption, binding amplitude, or Kd value was observed, showing that the interaction was robust, and thus a suitable positive control for the screening campaign.
MST Fragment Screening
Fragment stocks (100 mM) in DMSO were diluted into assay buffer to reach a final concentration of 10 mM. Subsequent liquid handling steps were carried out using a Microlab Starlet liquid handling system (Hamilton Robotics, Bonaduz, Switzerland), modified with a multititer plate (MTP) turn-and-tilt station (NanoTemper Technologies) and CoRe and iSWAP grippers (Hamilton Robotics, Bonaduz, Switzerland) for capillary chip and MTP handling. Fragment predilutions were prepared for MST experiments by 12-fold 1:2 serial dilutions in assay buffer containing 10% DMSO in Greiner White nonbinding 384-well plates (Greiner Bio-One, Frickenhausen, Germany) to yield final volumes of 10 µL. NT647-MEK1 stocks were centrifuged for 15 min at 23,000 × g to remove aggregates, and the supernatant was subsequently transferred to the liquid handling system and diluted into assay buffer without DMSO to reach a final NT647-MEK1 concentration of 60 nM. NT647-MEK1 solution (10 µL) was then added to the fragment dilutions in the plate and mixed carefully by pipetting up and down five times to reach a final NT647-MEK1 concentration of 30 nM, a final DMSO concentration of 5%, and a final reaction volume of 20 µL. For MST experiments, four rows with a total of eight 12-fold dilution series were prepared just in time before the measurement. From these four rows, four capillary chips with standard treated capillaries (NanoTemper Technologies) were filled with two dilution series per chip by automated dipping of the capillaries into each row of the multiwell plates. Four loaded chips were then transferred to a Monolith NT.Automated instrument (NanoTemper Technologies), and the MST for each capillary was recorded at MST 80% with a laser-on time of 5 s, thereby producing eight binding curves per run, with run times of ~16 min. Over the time course of the screening, a total of 13 positive controls using the NT647-MEK1-ATP interaction was performed (approximately 1 positive control every two runs). All measurements were performed at a fixed temperature of 25 °C. Additionally, a subset of fragments was investigated with laser-on times of 20 s (shown in Fig. 4). Kd values were calculated from fragment concentration-dependent changes in normalized fluorescence (Fnorm) of NT647-MEK1 after 5 s of thermophoresis based on the law of mass action using the NT.Affinity Analysis software. In case fragments caused a concentration-dependent decrease in NT647-MEK1 fluorescence in the serial dilution, a “SD test” was performed to discriminate between binding-specific fluorescence quenching and loss of fluorescence due to protein precipitation. For this, mixtures containing the “unbound” and “bound” states of NT647-MEK1 (0 and 1 mM of the fragments, respectively) were first centrifuged for 10 min at 23,000 × g to remove protein precipitate, and then mixed with a 2× solution containing 4% SDS and 40 nM DTT and heated to 95 °C for 5 min to denature NT647-MEK1. Subsequently, the NT647 fluorescence of the bound and unbound samples was detected using a Monolith NT.115. If fluorescence loss in the serial dilution is caused by binding-induced quenching, fluorescence intensities of the bound and unbound samples should be identical after denaturation; in the case of protein loss due to precipitation, the fluorescence difference persists after the SD test, which was true for the tested fragments.
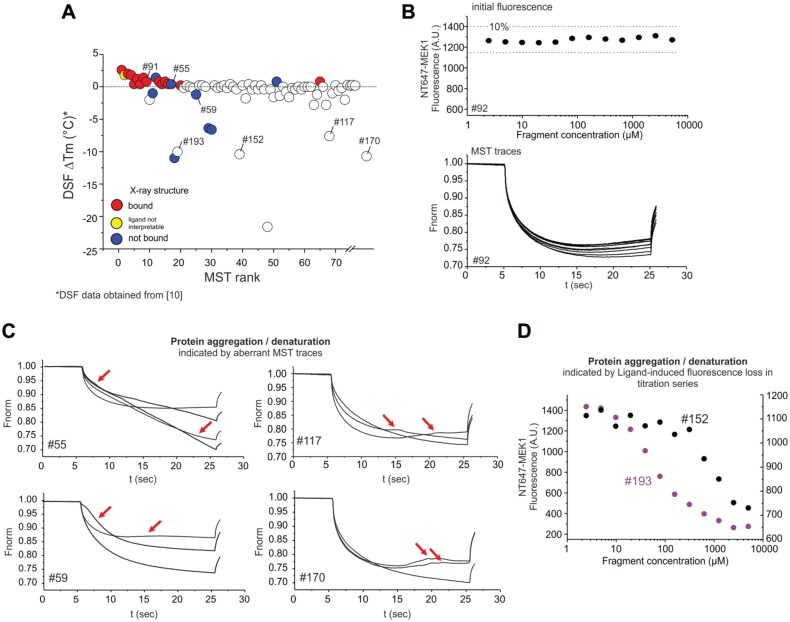
Figure 4.
Additional information derived from MST traces. (A) Plot of changes in unfolding transition temperature ΔTm by DSF vs. MST rank. (B) Example of regular fluorescence and MST traces of NT647 MEK1 in a serial dilution upon binding to fragment #92. (Upper panel) Initial fluorescence values in capillaries do not differ by more than 10% throughout the serial dilution. (Lower panel) Plot of the normalized fluorescence over time of MST experiments with NT647-MEK1 and fragment #92. MST traces from all 12 capillaries are shown. Regular MST traces have a smooth appearance and show a ligand concentration-dependent shift in magnitude. (C) Examples for fragment-induced aberrant MST traces due to protein aggregation/denaturation at elevated fragment concentration. For each fragment, three or four capillaries at the highest fragment concentrations (5–0.625 mM) are shown. Red arrows highlight aberrant MST traces. (D) Examples for fragment concentration-dependent loss of fluorescence intensity throughout the serial dilution. Fragments #152 and #193 reduce the initial fluorescence intensity by 86% and 42%, respectively, which could be attributed to protein loss.
For exemplary single-dose MST analysis, ΔFnorm values at a single fragment concentration of 150 µM were calculated by subtracting the Fnorm value at 150 µM from the averaged Fnorm values of the three lowest fragment concentrations (0.15–0.61 µM, representing the unbound state) for each serial dilution. Note that typical single-dose screenings by MST should include duplicate measurements of the fragments and DMSO references on each capillary chip.
Results and Discussion
For MST-based FBLD, we designed an automated workflow—including the preparation of serial dilutions, filling of capillary chips, and initiation of MST experiments—to quantify the binding of 193 fragments to purified MEK1 protein (Fig. 1A,B). Lysine residues on MEK1 were fluorescently labeled with the fluorophore NT647, and the protein was subsequently tested in thermal unfolding experiments, verifying that the fluorescent label did not compromise protein stability (Suppl. Fig. 2). Optimal buffer conditions were determined in pretests that evaluated MST signal reproducibility and the suppression of unspecific adsorption to capillary walls (Fig. 1C and Suppl. Fig. 3A). NT647-MEK1 functionality was confirmed by quantifying its Mg2+-dependent interaction with ATP by MST (Fig. 1D), yielding Kd values of 4.5 µM in the presence and 140 µM in the absence of MgCl2, respectively, which is in good agreement with previous results.12 Reproducibility and long-term stability of the NT647-MEK1-ATP interaction were confirmed (Suppl. Fig. 3B), and this interaction was subsequently used as a positive control throughout the MST screening.
Figure 1.
MST-based fragment screen against MEK1. (A) Schematic overview of automated Kd determination by MST. (B) Automated MST setup using a Hamilton Starlet liquid handling station in conjunction with a Monolith NT.Automated instrument. Using a standard MTP gripper (a), capillary chips can be loaded with solutions containing fluorescent target molecule and fragment dilutions in 384-well plates (b). The filled chips are then transferred to the tray of the MST instrument (c) and MST experiments are initiated. (d) Additional chips are stored in the liquid handling unit. (C) NT647-MEK1 adsorption test. Adsorption of 30 nM NT647-MEK1 to capillary walls was evaluated using capillary shape overlays. A clear adsorption to capillary walls was observed in the absence of Pluronic F127. The scheme on the right illustrates the rationale of the adsorption test, in which higher fluorescence intensity on the capillary walls results in a distorted capillary shape profile. (D) Binding of ATP to NT647-labeled MEK1 in the presence and absence of MgCl2 determined by MST. Error bars are SDs from three independent measurements. (E) Tabular overview of duration and sample consumption of the affinity screening by MST. (F) Pie chart visualization of screening results. (G) MST binding curves of the top-25 fragment hits and the internal ATP positive control (red).
For screening, 12-fold serial dilutions of fragments in assay buffer and subsequent mixing with NT647-MEK1 were prepared in 384-well plates using an automated liquid handling setup. After preparation of serial dilutions, capillary chips were automatically filled using a standard MTP gripper and then transferred to the capillary chip tray of the integrated MST instrument (Fig. 1A,B). MST experiments were initiated automatically and eight Kd values were determined per run (two serial dilutions per chip). The MST-based Kd determination for 193 fragments was completed within <7 h, consuming only 90 µg of MEK1 protein (Fig. 1E), demonstrating the fast and economical nature of this fragment screening approach. From 193 analyzed fragments, 73 (37.8%) hits were found to interact with MEK1 by MST with different affinities (Fig. 1F,G), as indicated by fragment concentration-dependent changes in the thermophoretic movement of NT647-MEK1. The relatively high hit rate reflects the preselection of the library by virtual screen for fragments that specifically bind to the ATP binding cleft of MEK1.11 The hit rate determined by MST is higher than those determined by other biophysical techniques for the same library on MEK1, highlighting the technique’s ability to identify a larger pool of chemical matter that could be explored at the start of a hit-to-lead process. From the 73 fragments that induced MST responses, 25 showed dose–response curves with clearly defined bound and unbound states and Kd values of <200 µM, and were classified as best hits (Fig. 1F,G). In addition to standard dose–response binding experiments, specificity of the interaction of the top-25 fragments with the ATP binding cleft of MEK1 was confirmed by obliteration of the binding signal in competition experiments using binding buffer with 5 mM ATP (not shown).
We next investigated how MST-derived data correlated with results derived from orthogonal methods. The MST affinity ranking showed a good correlation with a qualitative DSF ranking, which monitors shifts in the unfolding transition temperature (Tm) of MEK1 upon ligand binding. However, correlation between SPR and DSF results, as well as the correlation between SPR and MST results, was significantly lower (Fig. 2, Suppl. Fig. 4A, and Suppl. Table 1). Interestingly, a correlation trend was detected between Kd values and ΔTm values determined by DSF (Suppl. Fig. 4B), similar to previously reported observations.13 However, one potential limitation of DSF is that ligand-induced Tm shifts can be small (<1 °C), as in the present case of MEK1, and affinities have to be relatively high (Kd < 50 µM14) to register robust Tm shifts, which limits its applicability for low-affinity fragment screens. Consequently, only 12 fragments that increased the unfolding temperature of MEK by ≥0.8 °C were declared clear binders and transferred to X-ray analysis, disregarding a large number of potential false negatives. By comparison, MST enabled 73 fragments to be ranked based on their true binding affinity. An additionally performed single-dose analysis of the MST data showed that a screening with a single-fragment concentration of 150 µM would have been sufficient to robustly identify the X-ray confirmed hits (on the expense of affinity information), while reducing measurement time by sixfold to ~70 min (Suppl. Table 1 and Suppl. Fig. 5A,B). As for the affinity ranking, single-dose MST hits yielded an excellent overlap with DSF, but not with SPR hits (Suppl. Fig. 5C,D).
Figure 2.
Summary of screening results. (A) Tabular overview of the top-25 fragments from the MST-based Kd screen in comparison to results obtained by DSF, SPR, and X-ray crystallography. A full table of all fragments can be found in Suppl. Table 1. (B) Venn diagram depicting distribution and overlap of hits determined by DSF, SPR, and MST. Numbers in nonoverlapping regions of the circles indicate unique hits. Numbers in overlapping regions correspond to shared hits of two or all three methods, respectively. (C) Summary of X-ray-confirmed fragment hits determined by DSF, SPR, and MST.
Seven out of eight previously characterized X-ray positive binders11 were among the top-15 fragments ranked based on their MST affinity (Fig. 2A). In order to test whether MST-identified binders yield complex structures with MEK1, we analyzed an additional 11 MST positive fragments from the top-25 MST hits by X-ray crystallography. Binding of 7 out of these 11 fragments to MEK1 could be unambiguously confirmed, as clear electron densities with defined orientations in the ATP binding site of MEK1 were detectable (Fig. 3; other structures not shown due to proprietary reasons). Notably, even the low-affinity binder, #132,11 could be identified based on dose-dependent MST signal changes (MST rank 65; Suppl. Fig. 4C), demonstrating that MST can detect and quantify interactions over a wide concentration range if dose–response signals are detected at different fragment concentrations. The ability to detect low-affinity binders is particularly important for fragment screening campaigns, since the initial affinity of a fragment does not necessarily correlate with the potency of a mature compound, so that weak binders—which are potentially omitted by other biophysical techniques—can be of high value for the subsequent lead generation process.
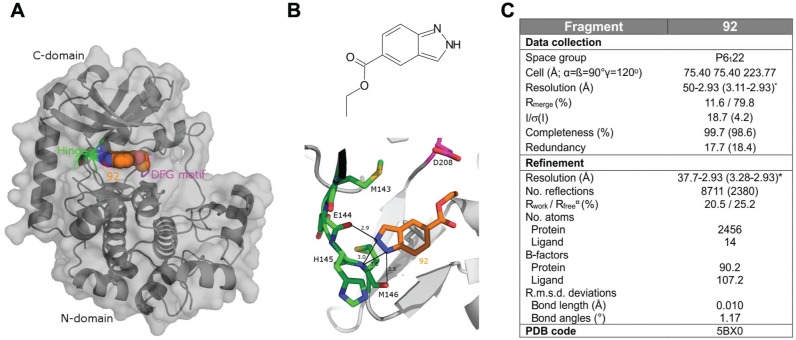
Figure 3.
Structure of MEK1 in complex with fragment 92. (A) Surface and ribbon representation of MEK1 in complex with compound 92. Protein Hinge and DFG motif are highlighted in green and pink, respectively (PDB: 5BX0). (B) Fragment 92 chemical formula and detailed view of the binding mode of compound 92. Distances indicated in the figure are measured in ångstrom. Color scheme is the same as in figure (A). (C) Summary of X-ray parameters.
Other important aspects of fragment screening campaigns comprise the exclusion of false positives and false negatives. Binding of fragments and small molecules can either stabilize or destabilize target proteins. Destabilization or denaturation of proteins is often accompanied by protein aggregation.15 MST provides a direct feedback on ligand-induced aggregation and other secondary effects that are identified based on aberrant MST traces. From the 193 investigated fragments, 81 reduced Tm by more than 0.4 °C as determined by DSF (Fig. 4A), suggesting destabilizing effects of these fragments. From those 81 fragments, MST initially identified 5 (20% of all top-25 MST hits) as potential binders, while SPR identified 14 (47% of all SPR hits) of those fragments as potential binders (Suppl. Fig. 6A). Closer inspection of MST data, however, showed that four out of the five potential hits with destabilizing properties showed clearly aberrant MST traces, pointing toward protein aggregation and/or denaturation (Fig. 2A). While MST data from regular binders show normal MST traces and a constant target fluorescence with a variance of <10% throughout the serial dilution (Fig. 4B), several fragments, including fragments #59, #106, and #186, introduced irregular, bumpy MST traces at high concentrations, which are caused by protein aggregates moving through the temperature gradient (Fig. 4C). Notably, fragment #55 stabilized MEK1 in DSF by 0.4 °C, but caused irregular MST traces and also showed no binding to MEK1 in X-ray analysis, suggesting that this fragment causes protein aggregation without destabilizing MEK1. Aggregating effects could also be identified for fragments outside of the top-25 MST ranking, which reduced Tm by DSF (Fig. 4C). The integration of algorithms that automatically detect and quantify the aggregation of target molecules from MST traces would allow for quickly identifying and excluding aggregation-inducing fragments and compounds. Besides aberrant MST traces, several fragments, including fragments #152 and #193, induced a ligand concentration-dependent drop in the fluorescence intensity of NT647-MEK1 (Fig. 4D), indicating protein loss, for example, due to protein unfolding and adsorption to multiwell plate walls or pipette tip surfaces, which could be confirmed in subsequent tests.
The exclusion of MST-identified promiscuous binders from further analysis improved the hit-to-structure ratio from the crystallized fragments from 73% to 86% (Suppl. Fig. 6B). Thus, additional MST-derived information about protein quality can be used as a filter to increase the hit rate of the screening, and to prevent destabilizing/aggregating fragments from entering the hit-to-lead stage of drug development.
In conclusion, MST robustly identified, quantified, and validated a set of fragment hits with micromolar affinity for MEK1, which could be followed up in hit-to-lead development. Moreover, MST provided additional information about unwanted secondary effects of fragments on protein integrity that were not detected by other methods, and which would have prevented false-positive hits from entering later stages of hit expansion, as well as rescuing fragments classified as false negatives. Furthermore, the primary hits identified by MST would have provided increased chemical depth of compounds forwarded for hit-to-lead expansion. Combining speed and low material requirements with the reliable identification of true binders by deriving binding affinities, MST has been demonstrated to be an effective method for screening fragments. Used in conjunction with other biophysical techniques, MST has the potential to significantly improve FBLD workflows.
Supplementary Material
Acknowledgments
We thank Lea Martin and Lisa Hanel for excellent technical assistance, and Heide Marie Resch for critical reading of the manuscript.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The project was funded by German Federal Ministry of Education and Research, funding program Photonics Research (13N11667).
References
- 1. Baker M. Fragment-Based Lead Discovery Grows Up. Nat. Rev. Drug Discov. 2013, 12 (1), 5–7. [DOI] [PubMed] [Google Scholar]
- 2. Kumar A., Voet A., Zhang K. Y. Fragment Based Drug Design: From Experimental to Computational Approaches. Curr. Med. Chem. 2012, 19 (30), 5128–5147. [DOI] [PubMed] [Google Scholar]
- 3. Murray C. W., Rees D. C. The Rise of Fragment-Based Drug Discovery. Nat. Chem. 2009, 1 (3), 187–192. [DOI] [PubMed] [Google Scholar]
- 4. Duhr S., Braun D. Why Molecules Move along a Temperature Gradient. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (52), 19678–19682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jerabek-Willemsen M., Andre T., Wanner R., et al. MicroScale Thermophoresis: Interaction Analysis and Beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar]
- 6. Parker J. L., Newstead S. Molecular Basis of Nitrate Uptake by the Plant Nitrate Transporter NRT1.1. Nature 2014, 507 (7490), 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radke M. B., Taft M. H., Stapel B., et al. Small Molecule-Mediated Refolding and Activation of Myosin Motor Function. eLife 2014, 3, e01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hussein M., Bettio M., Schmitz A., et al. Cyplecksins Are Covalent Inhibitors of the Pleckstrin Homology Domain of Cytohesin. Angew. Chemie. Int. Ed. 2013, 52 (36), 9529–9533. [DOI] [PubMed] [Google Scholar]
- 9. Mao Y., Yu L., Yang R., et al. A Novel Method for the Study of Molecular Interaction by Using Microscale Thermophoresis. Talanta 2015, 132 (0), 894–901. [DOI] [PubMed] [Google Scholar]
- 10. Saini K. S., Loi S., de Azambuja E., et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK Pathways in the Treatment of Breast Cancer. Cancer Treat. Rev. 2013, 39 (8), 935–946. [DOI] [PubMed] [Google Scholar]
- 11. Amaning K., Lowinski M., Vallee F., et al. The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against MEK1. Bioorg. Med. Chem. Lett. 2013, 23 (12), 3620–3626. [DOI] [PubMed] [Google Scholar]
- 12. Smith C. K., Windsor W. T. Thermodynamics of Nucleotide and Non-ATP-Competitive Inhibitor Binding to MEK1 by Circular Dichroism and Isothermal Titration Calorimetry. Biochemistry 2007, 46 (5), 1358–1367. [DOI] [PubMed] [Google Scholar]
- 13. Matulis D., Kranz J. K., Salemme F. R., et al. Thermodynamic Stability of Carbonic Anhydrase: Measurements of Binding Affinity and Stoichiometry Using ThermoFluor. Biochemistry 2005, 44 (13), 5258–5266. [DOI] [PubMed] [Google Scholar]
- 14. Sheth P. R., Liu Y., Hesson T., et al. Fully Activated MEK1 Exhibits Compromised Affinity for Binding of Allosteric Inhibitors U0126 and PD0325901. Biochemistry 2011, 50 (37), 7964–7976. [DOI] [PubMed] [Google Scholar]
- 15. McGovern S. L., Caselli E., Grigorieff N., et al. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem. 2002, 45 (8), 1712–1722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.