Abstract
Whole-genome analysis was applied to investigate atypical point-source transmission of 2 invasive group A streptococcal (GAS) infections. Isolates were serotype M4, ST39, and genetically indistinguishable. Comparison with MGAS10750 revealed nonsynonymous polymorphisms in ropB and increased speB transcription. This study demonstrates the usefulness of whole-genome analyses for GAS outbreaks.
Keywords: group A Streptococcus, invasive disease, speB, Streptococcuspyogenes, whole-genome sequencing
Streptococcus pyogenes or group A Streptococcus (GAS) is a major cause of mortality from bacterial infections globally, causing invasive disease as well as superficial infections [1]. Genetic analysis of GAS transmission during a point-source outbreak has been previously limited to single-gene approaches, such as sequencing the gene encoding the hypervariable M protein (emm-typing), or methods such as multilocus sequence typing and pulsed-field gel electrophoresis [1]. In recent research, several studies have reported increased resolution using whole-genome sequencing over conventional transmission analysis techniques [2, 3]. In contrast to prior reports, we document 2 invasive GAS disease cases occurring after exposure to a single subclinical source patient. Whole-genome sequencing confirmed that the isolates were genetically indistinguishable and identified unique polymorphisms that potentially contributed to the unusual transmission observed.
CASE REPORT
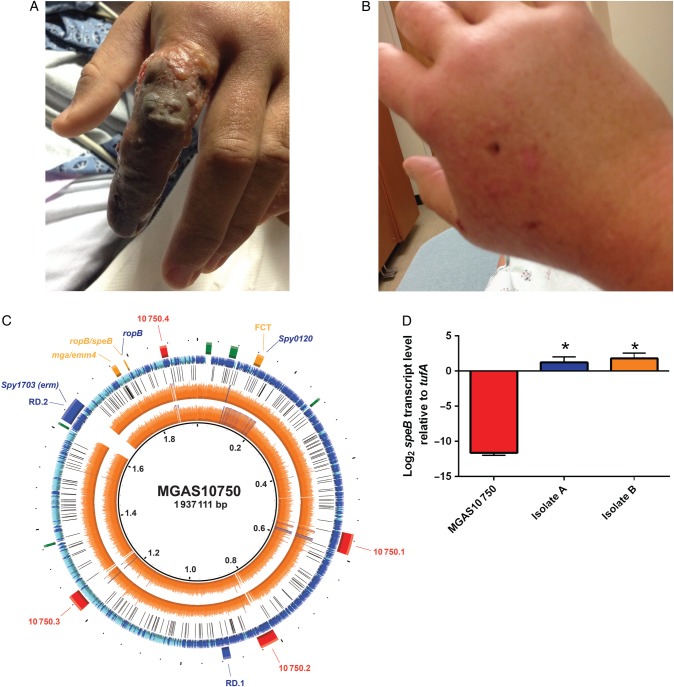
Patient A is a 30-year-old healthy man who suffered a scratch injury from a 17-year-old autistic student on his left index finger during an altercation. He presented to our hospital with fever (39.7°C), tachycardia, swelling, pain, and erythema of the left hand as well as numbness in the distal phalanx. Despite broad-spectrum antibiotics, the patient exhibited progressive swelling and blistering of the digit, lymphangitic spread involving his forearm, and severe pain (Figure 1A). He then underwent surgical debridement; operative cultures grew GAS (isolate A). Patient A improved postoperatively and was discharged 1 week later on oral clindamycin.
Figure 1.
Comparative genomic analyses of invasive clinical isolates (A and B) show genetic identity to each other, but genetic variability compared with M4 type strain MGAS10750, resulting in increased speB transcript levels. (A) Clinical pictures of patient (A) (Isolate A) showing inoculation sites and subsequent clinical sequelae. (B) Same as panel (A) except for patient (B) (isolate B). (C) Genome sequencing coverage map of isolates (A and B) vs the serotype M4 reference genome MGAS10750. Ring legend (inside to outside): genome coordinates in Mbp (Ring 1); Isolate (B) genome sequencing coverage (Ring 2); isolate (A) genome sequencing coverage (Ring 3); location of polymorphisms in isolate A and B strains relative to MGAS10750 (Ring 4); location of coding sequences (CDS) in the reference genome MGAS10750 with forward (blue) and reverse (aqua) directions indicated (Ring 5); selected landmarks in the reference genome MGAS10750 including phage (red), regions of difference ([RD] blue), ribosomal operons (green), and selected genes (Ring 6). (D) TaqMan real-time quantitative reverse transcription-polymerase chain reaction for speB was performed on cells grown to stationary phase for the strains indicated. Data graphed are mean ± standard deviation of 4 biological replicates done on 2 separate occasions (ie, 8 samples). The * indicates P < .05 compared with MGAS10750 as determined by one-way analysis of variance followed by Tukey's post hoc test.
Patient B is a 48-year-old woman with a history of breast cancer status postbilateral mastectomy and right-sided lymph node dissection. She also suffered a scratch wound from the child during the same altercation. That night she became febrile (40°C), hypotensive, and developed swelling in her right hand, which progressed to erythema extending to her upper arm (Figure 1B). She presented to an outside hospital where she underwent debridement of the right hand. Both blood (isolate B) and surgical cultures grew GAS. She was treated with clindamycin and meropenem with gradual clinical improvement.
Rapid streptococcal testing of the oropharynx of the seemingly asymptomatic, assumed source subject returned positive. He was treated with benzathine penicillin.
MATERIALS AND METHODS
Deoxyribonucleic Acid Typing
Genomic deoxyribonucleic acid (DNA) was extracted and sequenced using an Illumina MiSeq platform 250-base pair paired-end protocol to an average depth of approximately 1600×. The sequences were assembled and comparative genomics were performed. In brief, paired-end sequencing reads were assembled de novo using CLC Bio Genomics Workbench version 8.0.3 (QIAGEN) and the SPAdes genome assembler (http://bioinf.spbau.ru/spades) and then mapped to the previously sequenced MGAS10750 serotype M4 reference genome (NC_008024.1). Polymorphisms in the core genome were called against MGAS10750 using CLC Bio Genomics Workbench, version 8.0.3. Genomic visualizations were generated using BLAST Ring Image Generator. Core genomic differences between the 38 completed GAS genomes, isolate A, and isolate B strains were determined using MUMmer, and neighbor network phylogenetic relationships were visualized using SplitsTree (http://splitstree.org/). BioProject (PRJNA300859) and BioSample numbers for the sequence reads of the 2 strains were generated (SAMN04230426 and SAMN04230441 for isolate A and isolate B, respectively).
Transcript Analysis
Isolate A, B, and MGAS10750 were grown in duplicate on 2 separate days to stationary phase in Todd-Hewitt broth with 0.2% yeast extract (THY), and ribonucleic acid (RNA) was extracted using the RNeasy Mini Kit (QIAGEN). Complementary DNA was reverse transcribed from RNA using Superscript III (Invitrogen). TaqMan real-time quantitative reverse transcription-polymerase chain reaction was performed with an Applied Biosystems 7500 system using the ΔCT method of analysis. Determination of speB transcript levels was performed (oligonucleotides 5′-CGCACTAAACCCTTCAGCTCTT-3′, 5′-ACAGCACTTTGGTAACCGTTGA-3′ and probe 6FAM-5′-GCCTGCGCCGCCACCAGTA-3′-TAMRA) using the previously validated tufA as an internal control (oligonucleotides 5′-CTACTTTAACAGCTGCAATCACA-3′, 5′-AGAAGCGTAATCTTTT-3′ and probe 6FAM- TATTGGCACGTCGCTTGCCTTCATC-TAMRA).
RESULTS AND DISCUSSION
Transmission of GAS resulting in subsequent invasive disease has been well described following contact with a person with invasive GAS infection or asymptomatic GAS-colonized healthcare workers [4]. Because our cases did not fit either paradigm, a literature search confirmed the lack of previous point-source transmission studies demonstrating an invasive GAS outbreak in the community setting arising from an asymptomatic pharyngeal GAS-positive individual (Supplementary Table 1).
Given the unusual transmission circumstances and limited resolution of traditional epidemiological methodologies, we sought to characterize this GAS outbreak using a whole-genome approach (Supplementary Tables 2 and 3). Consistent with a common point-source and close temporal relationship, the 2 isolates were genetically indistinguishable at the whole-genome level (Figure 1C). Although we cannot be certain that the GAS strain from the 17-year-old student was identical to the 2 case isolates due to lack of the probable source patient's clinical isolate, the likelihood that both patients acquired an identical isolate almost simultaneously from another person or intermediary is exceedingly low.
Both isolates were serotype M4, ST-39, and phylogenetic analyses comparing the invasive isolates to 38 completed GAS genomes showed distinct clustering from other serotypes (Supplementary Figure 1). Both strains lacked the hasABC genes necessary for hyaluronic acid (HA) capsule biosynthesis, a finding consistent with recent reports identifying serotype M4 GAS, as well as M22 and M89, as lacking capsule [5–7]. Although the GAS HA capsule is a virulence factor and important for resisting phagocytosis [8, 9], these cases clearly show that acapsular GAS strains have the capacity to cause severe infections. Because experimental data suggest that capsule interferes with GAS adherence to epithelial cells [10], it is possible that the lack of capsule may have facilitated the efficient transmission seen in our cases.
Mapping of the genomic sequences of invasive isolates A and B to the previously sequenced reference serotype M4 strain MGAS10750 revealed 252 genetic variations in the core (ie, nonphage) genome (Figure 1C, Supplementary Tables 2 and 4). It is important to note that we identified 2 nonsynonymous single-nucleotide polymorphisms (SNPs) in the regulator of proteinase B gene (ropB; also rgg), resulting in amino acid changes T104I and S116L relative to MGAS10750 (Supplementary Table 4). A diverse array of SNPs in ropB have been previously shown to influence disease manifestations, transcript levels, and strain virulence in GAS, presumably due to alterations in production of the streptococcal cysteine protease, SpeB (streptococcal pyrogenic exotoxin B), which is the main regulatory target of RopB [11, 12]. The combination of T104I/S116L polymorphisms observed in the outbreak strains have not previously been described in systematic RopB studies performed in serotype M1 and M3 GAS; however, no systematic study of RopB in serotype M4 strains has been performed to date. Thus, to determine whether the T104I/S116L polymorphisms impacted speB expression, RNA was isolated from the 2 outbreak strains as well as MGAS10750, and transcript analyses were performed. We found nearly 2000-fold higher speB transcript levels in the 2 outbreak isolates (Figure 1D) compared with MGAS10750. Because speB expression is known to vary by site as well as at different points during course of infection, expression measured in growth culture may not exactly reflect what is occurring in vivo. Despite this minor limitation, inasmuch as SpeB is a potent virulence factor implicated in tissue destruction and systemic dissemination, the high levels of speB expression observed in our outbreak strains may have contributed to the invasive phenotype [11, 13].
In addition to the novel ropB allele observed our strains, both strains contained an insertion resulting in a frame-shift mutation in a gene encoding a putative sortase (MGAS10750_spy0120) (Supplementary Table 4) located in the fibronectin, collagen-binding, T antigen (FCT) region [14]. Group A Streptococcus FCT sortases are critical for assembly of pili, which are important for the initial stages of GAS adhesion to epithelial cells. However, GAS pili may also reduce systemic dissemination by increasing susceptibility of the organism to neutrophil extracellular traps [15]. Thus, the observed putative sortase mutation may have also contributed to the rapid dissemination of infection in our patients. Finally, both A and B isolates lacked the mobile genetic element (RD.2) found in MGAS10750 encoding for macrolide resistance (ermT/R) (Figure 1D). Consistent with genetic findings, both strains were erythromycin susceptible.
CONCLUSIONS
In summary, we describe the application of whole-genome sequencing to confirm an atypical point-source transmission scenario in 2 patients with invasive GAS acquired from a mutual, asymptomatic contact. This approach allowed for genetic identity confirmation of the 2 outbreak isolates and identification of unique SNPs potentially involved in the severe clinical disease observed in our patients. In addition, it is evident from the literature that the continued falling costs and reduction in sequencing turnaround time has led to increasing utility and impact of whole-genome sequencing in routine diagnostic and public health microbiology. Thus, application of whole-genome sequencing to the investigation of invasive GAS outbreaks could be widely used to clarify the circumstances underlying these potentially devastating events.
Supplementary Data
Supplementary material is available online at Open Forum Infectious Diseases online (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
We thank Lara Weaver (North Carolina State Laboratory of Public Health) for handling the isolates. We also thank Sue Lynn Ledford, Kim McDonald, and Mark Werdel (Wake County Human Services) for conducting the public health investigation and providing infection control measures. We also acknowledge the case patients, both of whom gave written informed consent.
Financial support. This work was supported by National Institute of Allergy and Infectious Diseases (grant R01AI089891 [to S. A. S.]; grants K24-AI093969 and R01-AI068804 [to V. G. F.]) and MD Anderson Cancer Center (MDACC) Bioinformatics Shared Resource support grant P30 CA016672 from the National Cancer Institute. J. G. P. is supported by the MDACC Odyssey Program and CFP Foundation.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Bessen DE, McShan WM, Nguyen SV et al. . Molecular epidemiology and genomics of group A Streptococcus. Infect Genet Evol 2015; 33:393–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben Zakour NL, Venturini C, Beatson SA et al. . Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing. J Clin Microbiol 2012; 50:2224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tse H, Bao JY, Davies MR et al. . Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J Infect Dis 2012; 206:341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Working Group on Prevention of Invasive Group A Streptococcal Infections. Prevention of invasive group A streptococcal disease among household contacts of case-patients: is prophylaxis warranted? JAMA 1998; 279:1206–10. [PubMed] [Google Scholar]
- 5.Flores AR, Jewell BE, Fittipaldi N et al. . Human disease isolates of serotype M4 and M22 group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio 2012; 3:e00413–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu L, Olsen RJ, Nasser W et al. . Trading capsule for increased cytotoxin production: Contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. MBio 2015; 6:e01378–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashbaugh CD, Alberti S, Wessels MR. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J Bacteriol 1998; 180:4955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wessels MR, Moses AE, Goldberg JB et al. . Hyaluronic acid capsule is a virulence factor for mucoid group A Streptococci. Proc Natl Acad Sci U S A 1991; 88:8317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foley MJ, Wood WB Jr. Studies on the pathogenicity of group A Streptococci. II. The antiphagocytic effects of the M protein and the capsular gel. J Exp Med 1959; 110:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollands A, Pence MA, Timmer AM et al. . Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A Streptococcus M1T1 clone. J Infect Dis 2010; 202:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen RJ, Laucirica DR, Watkins ME et al. . Polymorphisms in regulator of protease B (RopB) alter disease phenotype and strain virulence of serotype M3 group A Streptococcus. J Infect Dis 2012; 205:1719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll RK, Shelburne SA III, Olsen RJ et al. . Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest 2011; 121:1956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukomski S, Montgomery CA, Rurangirwa J et al. . Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun 1999; 67:1779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessen DE, Kalia A. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect Immun 2002; 70:1159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotty Alexander LE, Maisey HC, Timmer AM et al. . M1T1 group A Streptococcal pili promote epithelial colonization but diminish systemic virulence through neutrophil extracellular entrapment. J Mol Med 2010; 88:371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.