Abstract
We report a negative feedback loop between the signaling protein phospholipase D (PLD), phosphatidic acid (PA), and a specific set of microRNAs (miRNAs) during nutrient starvation of breast cancer cells. We show that PLD expression is increased in four breast cancer cell lines and that hypoxia, cell overcrowding, and nutrient starvation for 3 to 6 h increase expression even further. However, after prolonged (>12-h) starvation, PLD levels return to basal or lower levels. The mechanism for this is as follows. First, during initial starvation, an elevated PA (the product of PLD enzymatic activity) activates mTOR and S6K, known to inhibit apoptosis, and enhances cell migration especially in post-epithelial-to-mesenchymal transition (post-EMT) cancer cells. Second, continued PA production in later starvation induces expression of PLD-targeting microRNA 203 (miR-203), miR-887, miR-3619-5p, and miR-182, which reduce PLD translation. We provide direct evidence for a feedback loop, whereby PLD induction upon starvation leads to PA, which induces expression of miRNAs, which in turn inhibits PLD2 translation. The physiological relevance for breast cancer cells is that as PA can activate cell invasion, then, due to the negative feedback, it can deprive mTOR and S6K of their natural activator. It can further prevent inhibition of apoptosis and allow cells to survive nutrient deprivation, which normal cells cannot do.
INTRODUCTION
MicroRNAs (miRNAs) are short molecules of noncoding RNA, ∼22 nucleotides in length, and have crucial roles in the regulation of many cellular processes, including development, proliferation, differentiation, apoptosis, and stress response (1, 2). Mature miRNA molecules associate with the Argonaute (Ago1 and Ago2) proteins and the RNA-induced silencing complex (RISC) (3, 4). Active miRNAs regulate expression of their target genes via association of an ∼7-nucleotide-long stretch seed region with a complementary sequence in the target mRNA located in the 3′ untranslated region (UTR). Binding of miRNAs to their target mRNAs along with the RISC complex mediates inhibition of translation initiation (5). miRNA involvement in cancer development and metastasis is the subject of intense research (6–10).
Phospholipase D (PLD) has been implicated in cellular signals that suppress apoptosis and contribute to cancer cell survival (11–13). Through cell signaling, elevated PLD activity leads to activation of mammalian target of rapamycin (mTOR), a survival signal often hyperactivated in cancer (14, 15). Elevated PLD activity also subdued the tumor suppressors p53 and protein phosphatase 2A (12). Zheng et al. published a model for enhanced survival and migration signals in the developing tumor (16). In a developing tumor mass, cells inside the mass were subjected to hypoxia and nutrient and growth factor deprivation. It is proposed that cells that respond to stress by elevating PLD protein levels will survive presumably by gaining the ability to migrate. However, very little is known about PLD regulation at gene and protein levels. Our objective was to characterize a novel miRNA-mediated posttranscriptional regulation of PLD in breast cancer cells and the effect and biological function of nutrient starvation on this type of regulation.
We have identified a repertoire of miRNAs that regulate PLD translation. Biphasic PLD protein expression in response to nutrient starvation can be explained by induction of PLD-regulatory miRNA gene expression with prolonged starvation. We propose a model whereby the PLD enzymatic product phosphatidic acid (PA) induced an miRNA-mediated negative feedback on PLD protein expression in prolonged nutrient starvation of breast cancer cells. We also provide evidence of the biphasic regulation of mTOR and S6K in early and late starvation that plays into this new feedback loop.
MATERIALS AND METHODS
Cell culture and starvation.
MDA-MB-231, BT-474, and BT549 human breast cancer cells and MCF-10A human breast cells were obtained from ATCC (Manassas, VA). Human mammary epithelial cells (HMEC) were obtained from Cell Applications Inc. (San Diego, CA). MCF-7 and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS). BT-474 cells were cultured in Hybri-Care medium (ATCC) supplemented with 1.5 g/liter NaHCO3 and 10% fetal bovine serum (FBS). BT-549 cells were cultured in RPMI 1640 medium (ATCC) supplemented with 0.023 U/ml insulin and 10% FBS. HMEC and MCF-10A cells were cultured in mammary epithelial cell growth medium including bovine pituitary extract (BPE), human epidermal growth factor (hEGF), hydrocortisone, GA-1000, and insulin. HMEC were cultured on collagen-coated flasks. Cells were maintained at 37°C in an incubator with a humidified atmosphere of 5% CO2.To starve cells and render them nascent, medium was aspirated from cells, which were then washed 2× with phosphate-buffered saline (PBS) and incubated in cell starvation medium (DMEM–0.1% bovine serum albumin [BSA]) for several lengths of time as indicated in the legends of the figures.
Transfection of cells.
Cells were seeded in 6-well plates with an equal number of cells per well and were then allowed to grow for 12 to 24 h prior to transfection. Plasmid transfection reaction mixtures included 1 to 2 μg of DNA plasmid and 1 μg DNA per 2-μl volume of Transit2020 transfection reagent. Mimic transfection reaction mixtures included 100 nM the appropriate miRNA mimic and the appropriate volume of DharmaFECT 2 transfection reagent based on the cell line and number of cells seeded as recommended by the manufacturer. Cell transfections were 36 to 48 h in duration.
PLD, mTOR, and S6K activity assays.
Immunocomplex samples were processed for PLD2 activity in PC8 liposomes and n-[3H]butanol beginning with the addition of the following (final concentrations): 3.5 mM PC8 phospholipid, 45 mM HEPES (pH 7.8), and 1.0 μCi n-[3H]butanol in a liposome form. Samples were incubated for 20 min at 30°C with continuous shaking. Addition of 0.3 ml ice-cold chloroform-methanol (1:2) stopped the reactions. Lipids were then isolated and resolved by thin-layer chromatography. The amount of [3H]phosphatidylbutanol-di-16:0 ([3H]Pbut) that comigrated with PBut standards was measured by scintillation spectrometry. mTOR and S6K activities were measured as indicated by our lab in reference 17.
Immunoprecipitation/Western blotting.
Cells were transfected with several expression plasmids, as described in the figure legends, for 2 days. Cells were solicited in special lysis buffer (SLB; consisting of 5 mM HEPES, 1 mM leupeptin, 768 nM aprotinin, 100 μM sodium orthovanadate, and 0.4% Triton X-100). After sonication, aliquots of the supernatant were resolved by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, followed by immunoblot analysis with anti-PLD1, anti-PLD2, or antiactin antibodies, and visualized using enhanced chemiluminescence (ECL) reagents.
Gene expression measurement by qRT-PCR.
Total RNA was isolated from cells with the RNeasy minikit according to the manufacturer's protocol (Qiagen, Valencia, CA). RNA concentrations were determined using a NanoDrop, and samples were normalized to 50 ng/μl RNA. Reverse transcription was performed with 210 ng RNA, 210 ng random hexamers, 500 μM deoxynucleotide triphosphates (dNTPs), 84 units RNaseOUT, and 210 units of Superscript II reverse transcriptase, and the mixture was incubated at 42°C for 55 min. Quantitative real-time reverse transcription-PCR (qRT-PCR) assays were run with 100 ng total input RNA, 2 μl of PLD1 gene expression assay mixture (6-carboxyfluorescein [FAM] labeled), or 2 μl of the PLD2 gene expression assay mixture (FAM labeled) multiplexed with the housekeeping gene (β-actin) (FAM labeled), with the final concentrations being 200 pmol and 400 pmol for the primers and probe, respectively. Primers and fluorescent probes were synthesized by ThermoFisher Scientific. qRT-PCR conditions for the Stratagene Cycler were 95°C for 3 min and then 50 cycles of the next 3 steps, i.e., 30 s at 95°C, 1 min at 60°C, and then 1 min at 72°C. The cycle threshold (CT) values were arbitrarily chosen from the linear part of the PCR amplification curve, where an increase in fluorescence can be detected at >10 standard errors of the means (SEM) above the background signal. ΔCT was calculated as follows: ΔCT = avg PLD CT − avg housekeeping CT (where avg is average), and the gene fold increase expression was calculated as 2−ΔΔCT = 2−(experimental condition ΔCT − control ΔCT).
Quantitative reverse transcription-PCR for microRNAs.
Cells that were transfected with the PLD miExpress Precursor miRNA expression clones were used for RNA lysates 48 h posttransfection using the TaqMan miroRNA Cells-to-CT kit according to the manufacturer's protocol (catalog no. 4391848; Life Technologies). RNA concentrations were determined using a NanoDrop, and samples were normalized to ∼66 ng/μl RNA. Reverse transcription was performed in a 15-μl reaction volume with 1 μg of RNA, 1.5 μl 10T buffer, 1 mM dNTPs, 3.8 units of RNase inhibitor, and 1 μl of Multiscribe reverse transcriptase, and the mixture was incubated in one cycle at 16°C for 30 min, 42°C for 30 min, and then 85°C for 5 min. Quantitative PCRs were run in a 20-μl reaction volume using 10 μl TaqMan master mix, ∼88 ng of total input RNA, 1 μl of the relevant microRNA gene expression assay (FAM labeled) multiplexed with the housekeeping gene (U6). TaqMan miRNA primers and fluorescent probes were from Life Technologies. Quantitative PCR conditions for the Stratagene Cycler were 95°C for 10 min and then 40 cycles of the next 3 steps, i.e., 15 s at 95°C , 1 min at 55°C, and then 30 s at 72°C. The CT values were chosen from the linear part of the PCR amplification curve, where an increase in fluorescence can be detected at >10 standard errors (SE) above the background signal. ΔCT was calculated as follows: ΔCT = avg PLD CT − avg housekeeping CT. The gene expression fold change was calculated as 2−ΔΔCT = 2−(experimental condition ΔCT − control ΔCT).
Immunofluorescence microscopy.
Cells were fixed onto coverslips using 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature, and then incubated in 10% FCS–0.1% Triton X-100 in PBS for up to 4 h at room temperature. For detection of miRNA overexpression, miRNA plasmids were constructed with a green fluorescent protein (GFP) gene. For the detection of endogenous PLD2, cells were incubated with a goat anti-PLD2 (N-20 antibody) IgG primary antibody overnight at 4°C, followed by incubation with a donkey anti-goat tetramethyl rhodamine isothiocyanate (TRITC)-conjugated IgG secondary antibody for 1 h at room temperature. Nuclei were stained using a 1:2,000 dilution of 4,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min at room temperature. Coverslips were mounted onto glass microscope slides using VectaShield mounting medium, and cells were visualized using a Nikon 50 Eclipse epifluorescence microscope.
Luciferase reporter assay.
Luciferase reporter assays were performed using the LightSwitch luciferase assay kit from Active Motif (catalog no. 32031). The reporter vectors contain a 3′ UTR region and a downstream RenSP luciferase region. Cells were cotransfected as previously described with either a miRNA mimic or plasmid and a LightSwitch 3′ UTR positive control vector or PLD2 3′ UTR vector in a 96-well plate for 36 h. LightSwitch luciferase assay reagent was added to the wells, and the signal was read on a luminometer. The signal knockdown from the control was normalized to a negative-control mimic.
Cell invasion assay.
MDA-MB-231 human breast cancer cells were serum starved for 2 h and resuspended at a concentration of 1.5 × 106 cells/ml in chemotaxis buffer (DMEM–0.5% bovine serum albumin). Next, ∼3 × 105 cells were applied to the upper chambers of 8 μm PET Matrigels (24-well format) with a 6.5-mm-diameter membrane, and cells were allowed to invade for 6 h at 37°C in a humidified 5% CO2 cell culture incubator. The final concentration of chemoattractant used was 0 or 30 nM EGF in 500 μl of chemotaxis buffer placed in the lower wells of 24-well plates. Cells were scraped from the Matrigel insert and then stained for 1 h with hematoxylin. Six separate fields of cells were counted for each invasion assay, and results were expressed in terms of total number of invading cells ± SEM.
Statistical analysis.
The data presented in the figures as bars are means and SEM (standard deviation/n1/2, where n is the sample size). Experiments were performed in duplicates for at least 3 independent experiments. The difference between means was assessed by the single-factor analysis of variance (ANOVA) test, calculated using SigmaPlot version 10 (Systat Software Inc., San Jose, CA). A probability (P) of <0.05 indicates a significant difference.
RESULTS
PLD gene, protein, and activity are enhanced in cancer cells under conditions of cellular stress, such as hypoxia, starvation, or overconfluence.
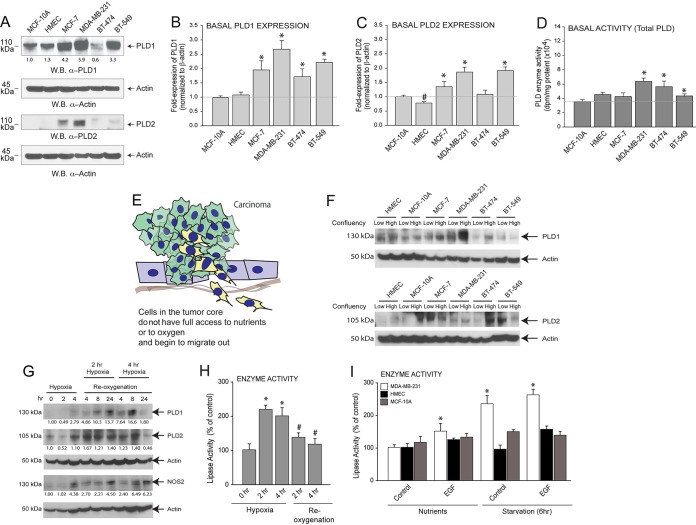
Endogenous expressions of both isoforms of PLD (PLD1 and PLD2) were elevated in MCF-7 (low-invasive luminal) and MDA-MB-231 or BT-549 (high-invasive, epidermal growth factor receptor [EGFR]-high) cancer cells compared to normal MCF-10A cells or HMEC (Fig. 1A). Quantitative analysis of basal PLD2 protein expression in noncancerous and breast cancer cell lines is presented underneath the appropriate Western blots. Additionally, relative endogenous PLD1 and PLD2 gene expression levels were increased in the four cancer cell lines tested (MCF-7, MDA-MB-231, BT-474, and BT-549) compared to the two normal breast cell lines (MCF-10A and HMEC) (Fig. 1B and C), with MDA-MB-231 and BT-549 having the highest (>2.5-fold over controls) basal levels. In Fig. 1D, basal lipase activity seems to be elevated in MDA-MB-231 and BT-474 cell lines compared to HMEC, although not in the MCF-7 cell line and only marginally in the BT-549 cell line. Taken together, these data indicate that high-invasive breast cancer cells have higher levels of PLD gene and protein expression, which was represented by increased PLD activity compared to that of the respective controls.
FIG 1.
PLD protein and activity are enhanced in cancer cells under conditions of cellular stress, such as hypoxia, starvation, or overconfluence. (A) Protein expression of PLD1 and PLD2 in several cell lines identified by Western blotting (W.B.). β-Actin was used as a control. Quantitative analysis (density of bands) of basal PLD2 protein expression in noncancerous and breast cancer cell lines is presented underneath the appropriate Western blots. (B, C) Basal PLD1 and PLD2 gene expression, respectively, in several cell lines as determined by qRT-PCR and normalized to β-actin. (D) Basal PLD enzyme activity in several cell lines. (E) Cartoon depicting core cancer cells leaving a primary tumor (after EMT) as conditions inside the core encounter hypoxia, starvation, and overconfluence. (F) Expression of PLD in several cell lines cultured at low (∼40%) confluency or maximal (100%) confluency. (G) Expression of PLD under chemically induced hypoxia (iNOS) or under hypoxia-plus-reoxygenation conditions in MCF-7 cells. (H) Same as for panel G, with cell lysates used for measurement of PLD enzymatic activity. (I) Either normal (HMEC and MCF-10A) or malignant (MDA-MB-231) epithelial breast cancer cells were subjected to normal serum in the medium (“nutrients”) and to nutrient deprivation/starvation (“starvation” for 6 h) and then incubated with 3 nM EGF. Data presented in the figures as bars are means plus standard errors of the means (SEM). Experiments were performed in technical triplicates (for qPCR assays) for 5 independent experiments. The difference between means was assessed by the single-factor analysis of variance (ANOVA) test, calculated using SigmaPlot version 10 (Systat Software Inc., San Jose, CA). A probability (P) of <0.05 indicates a significant difference. Asterisks denote statistically significant (P < 0.05) ANOVA increases between samples and controls. Pound signs denote statistically significant (P < 0.05) ANOVA decreases between samples and controls.
To understand the reason for this high level of PLD in highly invasive cells, we have hypothesized that cells in the tumor core do not have full access to nutrients and begin to migrate out. Since this would be more evident in post-epithelium-to-mesenchymal transition (EMT) in cells like MDA-MB-231 and BT-549 (Fig. 1E), PLD could act as a “stress-response signal” that could change with the stress conditions encountered in the tumor core. Dividing cells and cancer cells have highly anabolic metabolisms in order to generate the molecules needed to increase cell mass and successfully divide. It has been proposed that phosphatidic acid is an indicator of nutrient sufficiency due to its role in activating mTOR and the role of PLD as a stress response protein and further that cancer cell proliferation is PLD dependent (18, 19). We tested the effect of cell density in both PLD1 and PLD2 levels, defining states of low (∼40% to 50%) and high (100%) confluence in a wide array of cell lines (Fig. 1F). We have also tested hypoxia in low-invasive cells (MCF-7) (Fig. 1G and H) and nutrient starvation (Fig. 1I) in PLD1 and PLD2 protein levels. In Fig. 1I, we included the results of incubation of cells with EGFR because the growth factor is an activator of PLD, as our laboratory has demonstrated earlier (20), and the data confirm previous results indicating that the increase is more profound in starved cells, as expected with cells (quiescent, i.e., in the G0 phase of the cell cycle) that are treated with a growth factor (21).
In most cases, PLD protein and enzyme levels were elevated under the stress conditions induced to the cancer cells. The exceptions are illustrated in Fig. 1F, where PLD1 expression in BT-549 cells and PLD2 expression in MCF7, MDA-MB-231, and BT-549 cells are not elevated at high cell density. Interestingly, reoxygenation after 4 h of hypoxia returned PLD levels to basal conditions (Fig. 1G and H). Since we observed the most robust changes as a result of nutrient starvation, we chose to pursue nutrient starvation to study.
Serum starvation alters the cellular profile of PLD in breast cancer cells.
Of the three phenomena indicated in Fig. 1 (overconfluence, hypoxia, and starvation), we chose to examine starvation in greater detail and searched for the molecular mechanism underlying the need for higher PLD expression and activity. Knowing that nutrient starvation of cancer cells is important to the expression of certain transcription factors and key signaling proteins (22) that ultimately contribute to cancer cell survival, proliferation, and migration (23, 24), we tested PLD gene and protein expression and enzymatic activity in several human breast cancer cell lines and normal cells following serum starvation as a function of time.
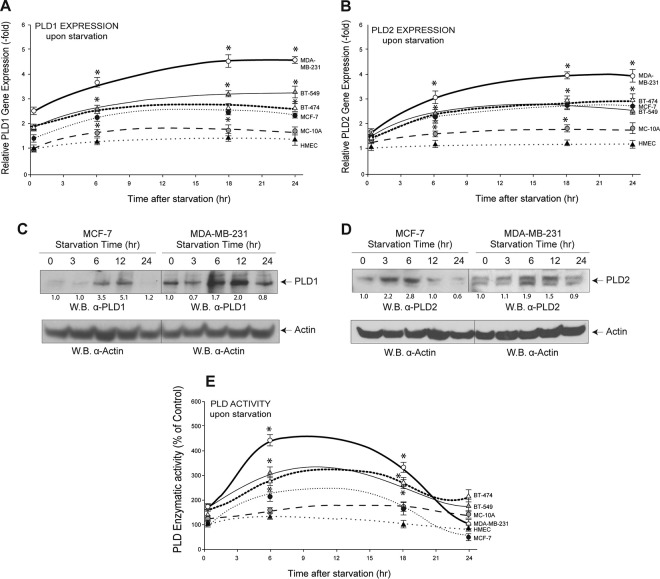
We observed an increase in relative PLD gene expression (Fig. 2A and B) that after 6 h remained relatively constant in most cell lines, with PLD1 expression and MDA-MB-231 cells showing a steady increase. Moving to protein expression, starvation resulted in an initial steady increase in protein expression (Fig. 2C and D), which returned to basal levels (or below) after prolonged (24-h) starvation. Enzyme activity followed that pattern as well (Fig. 2E), showing biphasic changes. We posited that the reason for a steady increase in PLD gene and protein expression yet a decrease in activity at prolonged (>12-h) activity could be due to posttranslational changes in the proteins or in the way they were synthesized in the ribosome. For the latter, we chose to study translational regulation by microRNAs.
FIG 2.
Cells experience a large change in PLD gene and protein expression in response to starvation. (A, B) qRT-PCR was performed to show PLD1 and PLD2 gene expression, respectively, in several cell lines after increasing the duration of starvation. β-Actin was used as a control. (C, D) Western blot analysis shows changes in PLD1 and PLD2 protein expression, respectively, with increasing duration of nutrient starvation in MCF-7 and MDA-MB-231 cells. β-Actin was used as a control. The density of the Western blot bands is included at the bottom of pertinent panels to provide an accurate judging of the protein levels. (E) PLD enzymatic activity was measured in many cell lines after increasing the duration of cell culture starvation. Statistics and symbols are as indicated in the legend to Fig. 1.
Endogenous gene expression of PLD-targeted miRNAs varies between normal and cancerous breast cell lines.
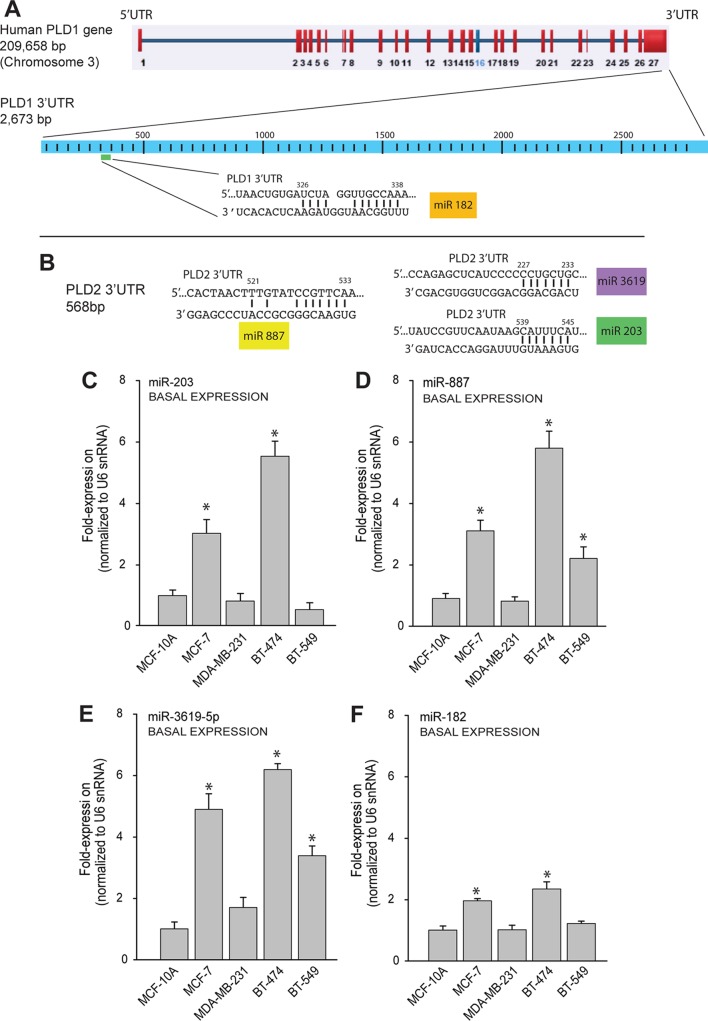
Using the TargetScan Human and EMBL-EBI databases, we identified four miRNAs that were predicted to associate with the 3′ UTR of PLD1 (miRNA 182 [miR-182]) (Fig. 3A) or PLD2 (miR-203, miR-887, and miR-3619-5p) (Fig. 3B). Next, we sought to determine the endogenous expression levels of these PLD-targeted miRNAs in the same normal and cancerous breast cell lines as those that were used in the experiment illustrated in Fig. 1. As seen in Fig. 3C to F, the relative levels of expression of miR-203, miR-887, miR-3619-5p, and miR-182 were significantly increased (>4-fold) in the low-invasive cancer cells (MCF-7 and BT-474) compared to highly invasive cells (MDA-MB-231 and BT-549). These data could provide the answer as to why PLD protein expression is different in MCF-7 versus MDA-MB-231 cells. An elevated level of basal PLD-targeting PLD miRNAs could maintain low levels of PLD in MCF-7 cells in comparison to MDA-MB-231 cells. These fundamental differences warranted the subsequent study of the underlying mechanism of PLD response to starvation.
FIG 3.
Basal expression of microRNAs in cancer cells. (A, B) Repertoire of microRNAs predicted to bind PLD1 (A) or PLD2 (B) 3′ untranslated regions (3′ UTR) by TargetScan Human and EMBL-EBI. (C to F) MicroRNA profiles under basal conditions of breast tissue cells, cancerous and noncancerous. qRT-PCR was performed to measure endogenous levels of several miRNAs, i.e., miR-203 (C), miR-887 (D), miR-3619-5p (E), and miR-182 (F), in the indicated cell lines. U6 snRNA was used as a control. qRT-PCR, quantitative reverse transcription-PCR; snRNA, small nuclear RNA. Statistics and symbols are as indicated in the legend to Fig. 1.
Starvation affects endogenous gene expression of miRNAs in breast cancer cells.
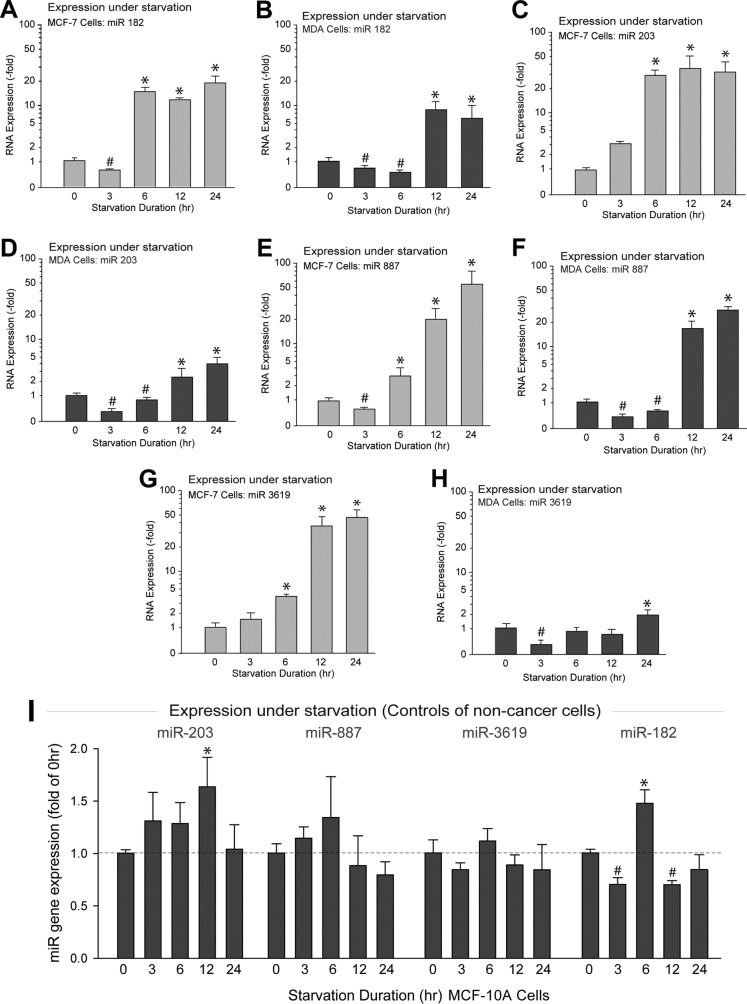
As shown in Fig. 4A to H, the expression of all of the PLD-specific miRNAs tested here varied upon starvation, following a biphasic pattern. Initially (between 0 and 3 to 6 h), miRNAs were underexpressed, whereas at prolonged starvation (12 to 24 h), miRNAs were overexpressed. The largest increases in miRNA expression for the MDA-MB-231 cells were the PLD2-targeted miR-887 and the PLD1-targeted miR-182. We measured miRNA levels in MCF10A cells for every time point measured for MCF7 and MDA-MB-231 cells (Fig. 4I). Minor changes in expression were found but at no point compared to the large quantitative changes observed in the cancer cells (that can reach in some cases 50-fold); therefore, Fig. 4I, showing results for noncancer cells, serves as a good negative control for the cancerous cell lines in other panels of Fig. 4. All in all, the data in Fig. 4 indicate that one of the fundamental and significant differences between cancer and noncancer cells is that the former are able to modulate their response to environmental cues (in this case starvation) by increasing or decreasing microRNAs.
FIG 4.
PLD-targeting microRNA expression is increased in breast cancer cells in response to long-term starvation. (A to H) Cells were deprived of serum for the indicated lengths of time. qRT-PCR was performed to measure endogenous levels of miRNAs, i.e., miR-182 (A, B), miR-203 (C, D), miR-887 (E, F), and miR-3619-5p (G, H) in MCF-7 cells and MDA-MB-231 cells, respectively. (I) Endogenous gene expression of PLD-targeting miRNAs under nonstarved and starved (24 h) conditions in noncancer cells, MCF-10. Error bars are for results from three independent experiments performed in duplicate; n = 5 for measurement of miR-3619-5p at 24 h of starvation. * and #, statistically significant (P < 0.05) ANOVA increases or decreases, respectively, between samples and controls.
mTOR and S6K involvement in PLD-modulated starvation.
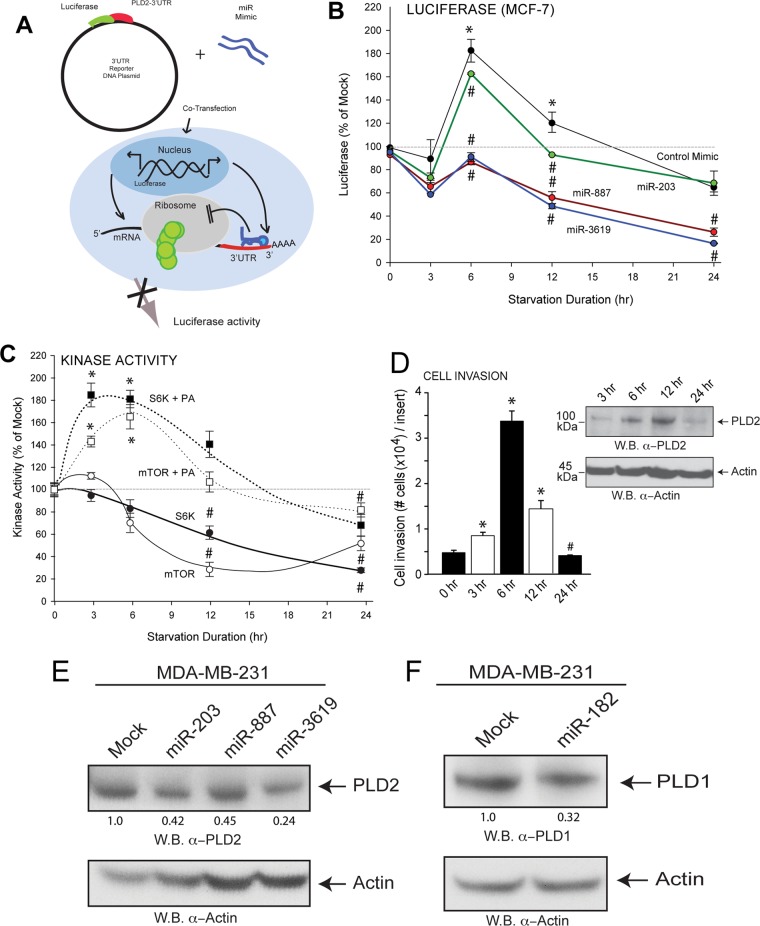
We used luciferase reporter assays (schematic in Fig. 5A) to study PLD2 translation in the presence of miRNA and under starvation conditions. We observed an initial increase, followed by decreased PLD2 translation (measured by decreased luciferase signal) as a product of prolonged nutrient starvation (Fig. 5B). With the addition of miRNA, we observed a greater decrease in PLD2 translation after prolonged starvation, suggesting an additive effect of endogenous and exogenous miRNA (Fig. 5B). We confirmed the decreased expression of PLD2 and PLD1 protein in MDA-MB-231 cells after transfection with miRNA mimics by Western blotting as seen in Fig. 5E and F, respectively.
FIG 5.
Biphasic pattern of PLD2 protein expression in starvation. (A) Luciferase reporter and scheme of cotransfections with miRNAs. (B) Luciferase reporter assay shows decreased PLD2 protein production in the presence of miR-203 with increasing duration of starvation. (C) Measurements of mTOR and S6K activities in MDA-MD-231 starved cells at different lengths of time in the presence or absence of PA. (D) Cell invasion assays of MDA-MD-231 cells in Matrigels in response to 3 nM EGF after the indicated starvation lengths of time. The inset shows a Western blot with the same samples, probed with anti-PLD2 antibodies. (E, F) Western blot analysis of PLD2 and PLD1 protein expression in MDA-MB-231 cells after transfection with miRNA mimics. The density of the Western blot bands is included at the bottom of pertinent panels to provide an accurate judging of the protein levels. Protein expression is relative to β-actin expression. Statistics and symbols are as indicated in the legend to Fig. 1.
The initial increase caught our attention, and we hypothesize that the increase in PLD activity could be related to other well-known biological phenomena in which PLD is implicated, namely, regulation of mTOR, as it is linked to nutrient response. An activated mTOR pathway (including ribosomal S6K) results in increased cell growth and proliferation. However, after prolonged starvation, both mTOR and S6K basal activities decreased with time (Fig. 5C). mTOR and S6K activities were elevated in the presence of PA, which is consistent with the fact that PA activates both mTOR and S6K (17, 25). We predict that as a highly sensitive stress response protein, PLD protein mass initially increases during nutrient starvation, facilitating cell migration. After prolonged starvation, mTOR, the major inhibitor of autophagy, plays a less important role and contributes to its lack of activity in the absence of PA. We could reason at this point that the initial increase in PLD would serve in the cell as a stress response that would allow functions other than nutrient related, such as cell migration that could be encountered initially in cells in the core of a tumor lacking nutrients, and a second phase when mTOR and S6K were no longer activated by PA (as PLD is low). Under these conditions, cell migration would cease, whereas nutrient conservation and autophagy could begin. In support of this dual response during migration, results in Fig. 5D show different responses of MDA-MB-231 cells to cell invasion through Matrigel. mTOR and S6K enzymatic activities are elevated during starvation, as they apply to PA (17, 25, 26).
A PA-dependent negative-feedback regulation of PLD protein expression mediated by miRNAs.
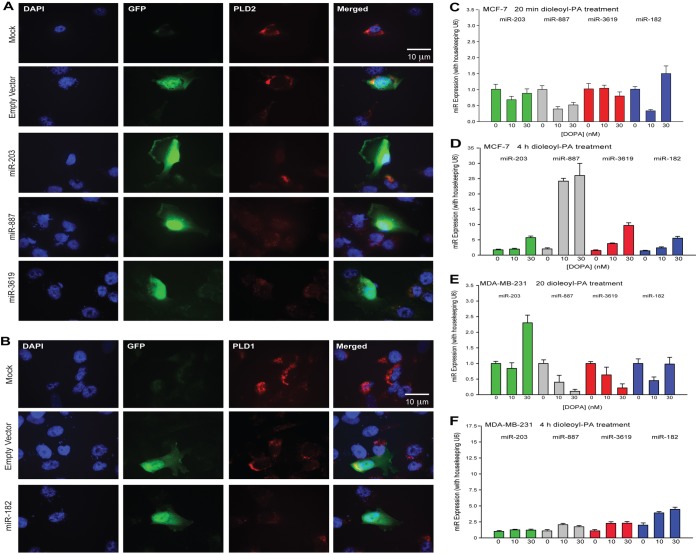
As a necessary control, the data shown in Fig. 6A and B confirm that miRNAs impeded protein (PLD) expression. MDA-MB-231 breast cancer cells transfected with GFP-tagged miRNA-containing plasmid (GFP panels) showed a decrease in endogenous PLD2 or PLD1 protein expression (TRITC panels) by immunofluorescence (Fig. 6A and B). Based on the results of this study so far and the biphasic nature of PLD protein expression during starvation, we posited that a mechanism of negative feedback exists in which the enzymatic product of PLD, PA, increases expression of miRNA with posttranslational regulation of PLD. We sought to reproduce the effects of nutrient starvation on miRNA by incubating cancer cells with PA. Prolonged incubation (4 h) resulted in an increase in miRNA gene expression (Fig. 6D and F) compared to a slight decrease in gene expression after only 20 min of incubation (Fig. 6C and E).
FIG 6.
PLD2 protein expression decreases in the presence of miRNA in MDA-MB-231 breast cancer cells, and miRNA gene expression increases in response to PA. (A, B) Immunofluorescence of cells transfected with plasmids containing an miRNA gene and a GFP gene. miRNA gene expression was represented by GFP expression. The empty plasmid backbone contained the GFP gene and no miRNA gene. PLD2 and their targeting miR-203, miR-887, and miR-3619-5p (A) or PLD1 and its targeting miR-182 (B) were represented with TRITC. (C to F) Gene expression of miR-203, miR-887, miR-3619-5p, and miR-182 after incubation with increasing amounts of DOPA (1,2-dioleoyl-sn-glycero-3-phosphate) for 20 min or 4 h in MCF-7 or MDA-MB-231 cells. Note that the two y axis scales in panels D and F are different since expression in MCF-7 is lower than in MDA-MB-231 cells.
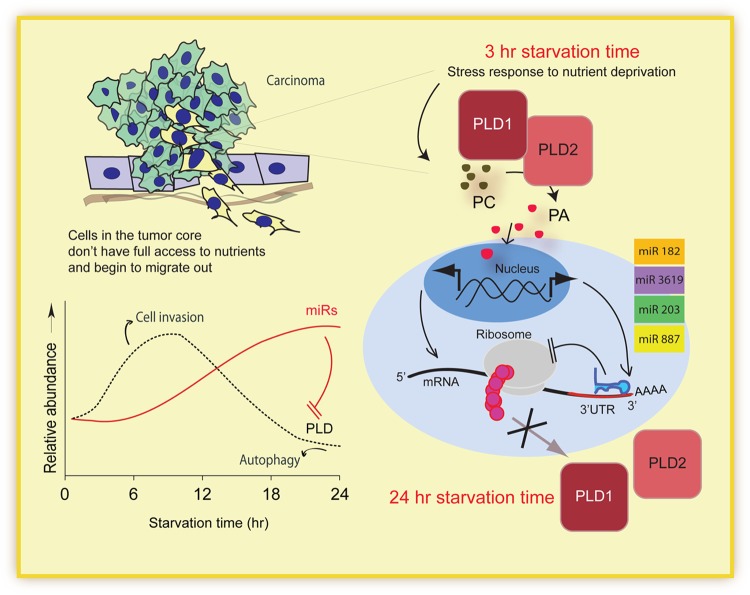
In tumors, cells in the core often lack full access to nutrients. Starvation induces a cellular stress response in these cells, leading to many cellular changes, including elevated protein expression of PLD. For its established roles in signaling pathways and cell migration, we propose that the observed increase in PLD protein aids cell invasion. With prolonged starvation (24 h), PLD protein levels decreased to below basal level. Additionally, we found an increase in gene expression of the miRNAs targeting PLD. The increase in miRNA gene expression correlated with decreased PLD protein expression. We, therefore, propose a model in which increased miRNAs gene expression is mediated by PA, the enzymatic product of PLD (Fig. 7).
FIG 7.
Model depicting the proposed effect of starvation on microRNA in cancer cells. Initially, when placed under starvation conditions, cancer cells increase protein expression of the stress response protein PLD. The enzymatic product of PLD, PA, subsequently serves to signal miRNA gene expression, thereby regulating PLD protein translation in a negative-feedback pathway. The model compiles the repertoire of miRNAs that suppress PLD translation in response to an elevated PA level. The elevated PLD level results in a rise of the cellular PA level, therefore inducing expression of miRNAs that suppress PLD translation. PLD gene, protein, and activity are enhanced specifically in cancer cells under conditions of cellular stress frequently occurring at the tumor core, such as hypoxia, starvation, or overconfluence. This elevation of PLD activity results in an increased expression of multiple miRNAs in low-invasive tumor cells leading to feedback inhibition of PLD expression. However, high-invasive tumor cells lose the ability to induce these miRNAs, resulting in an uncontrolled PA level and the subsequent inhibition of apoptosis through the mTOR pathway.
DISCUSSION
It is well known that mTOR is a strong inhibitor of apoptosis. One of the intracellular activators of mTOR is PA. The activity of the enzyme that synthesizes PA, PLD, is increased in cancer cells subjected to serum deprivation, making PLD a “survival” or “stress response” protein (13). However, the mechanism for this increase during starvation has remained unsolved. We found that this is because the expression of the PLD protein is upregulated through a novel mechanism involving a specific set of miRNAs that we have uncovered here for the first time, which are miR-203, miR-887, miR-3619-5p, and miR-182. PLD rises and decreases as indicated in Fig. 7; miRNA expression steadily rises with time of starvation, and miRNAs are responsible for the decrease of PLD expression. The interaction between PLD/PA and the microRNAs is biphasic and depends on a feedback loop that is absent in noncancer cells. It should be pointed out that the in vitro conditions presented in this study were made to mimic conditions that are likely to be present in the in vivo situation, which boiled down to three specific microenvironment situations, i.e., starvation, high cellularity (high cell density), and hypoxia, and these should be the first steps for future translational studies in vivo.
We report for the first time a new mechanism of regulating nutrient starvation by microRNAs targeting PLD. PLD plays an important role in the metastasis of breast cancer cells (13), and several protein-protein interactions relevant to cell migration have been discovered. The aim of the current study was to identify and further characterize microRNAs, which bound with imperfect complementarity to the 3′ UTR of PLD mRNA, leading to repression of its translation. Additionally, we have also determined the effect, or biological function, of nutrient starvation on miRNA-mediated posttranscriptional regulation of the signaling enzyme PLD.
We found that several miRNAs do interact with the 3′ UTR of either PLD1 or PLD2 isoforms. It is known that several miRNAs can regulate the expression of a particular protein, and we present here a specific set of miRNAs that affect PLD translation. In combination with current software, we determined that miR-182 was predicted to bind to PLD1 and that miR-203, miR-887, and miR-3619-5p were predicted to bind to PLD2. We aimed to determine the regulatory role of these miRNAs on PLD protein expression in breast cells, both noncancerous and cancerous.
There have been numerous studies describing the dysregulation of miRNAs in cancer tissue and cell culture compared to noncancerous samples (27–30). Most often, miRNAs are repressed in cancer, as we have found with our PLD regulating miRNAs. However, there have been examples of oncogenic miRNAs that are overexpressed in cancer, such as miR-155 and miR-21, where overexpression was sufficient to induce lymphomagenesis in mice (31, 32). miRNAs have vast roles in cell function, are pivotal intersections of communication, and thus have been implicated in many areas of cancer disease, including initiation (31, 32), proliferation, metastasis, drug resistance (33), and biomarkers (29, 30). This highlights the crucial role of miRNAs in cancer development and progression. We proposed in this study that miRNA regulation of PLD protein expression is a vital component of the nutrient depletion stress response in breast tumor cells.
As shown by the Foster and Frohman groups, starvation and lack of nutrients in cancer cells initiate a survival program (15, 16, 34–36). Cells under hypoxic conditions and that also lack nutrients have elevated PLD activity. We found that the endogenous levels of PLD expression and phospholipase activity in luminal MCF-7 and claudin-low MBA-MB-231 breast cancer cells followed a biphasic pattern with a maximal expression at ∼6 to 12 h after starvation and a decrease below control levels at ∼24 h. After 12 to 24 h of starvation, microRNAs were elevated in MDA-MB-231 cells and MCF-7 cells ∼20- to 40-fold, respectively, and after 24 h of starvation, PLD2 protein mass was minimal. We observed that miRNA expression levels changed prior to significant decreases in PLD2 protein mass. We determined if this response was the result of modulation by miRNAs and have characterized the regulatory pattern of miRNAs acting upon PLD during starvation. The four PLD-specific miRNAs (miR-203, miR-182, miR-3619-5p, and miR-887) played a key role in decreasing PLD protein mass. Still, all four of these miRNAs that had elevated expression (miR-203, miR-887, miR-3619, and miR-182) could, in combination, be responsible for influencing PLD2 protein mass levels.
These data are consistent with the current model for phosphatidic acid (PA) activation of mTOR. PA, the enzymatic product of PLD, binds and activates mTOR (37). The activated mTOR pathway results in increased cell growth and proliferation (16). As a highly sensitive stress response protein, the PLD protein mass initially increased during nutrient starvation, facilitating cell migration. Additionally, this increase in PA results in mTOR-induced cell growth. After prolonged starvation, cells can no longer afford the cost of increased mTOR activity and cell growth associated with increased PLD and PLD-mediated cell migration. Therefore, we propose a negative-feedback mechanism wherein cells undergoing prolonged starvation increase gene expression of PLD-regulating microRNAs, resulting in the downregulation of PLD protein mass.
Starvation induces autophagy, which is associated with protein degradation (38, 39). It is still possible that PLD2 protein degradation could be mediated by an additional mechanism in parallel with miRNAs. For example, autophagy is coupled with a reduction in histone H4 lysine 16 acetylation (H4K16ac) via downregulation of hMOF, a histone acetyltransferase (40). If hMOF is degraded (leading to a decrease in H4K16ac at the PLD2 promoter), then this could influence the levels of PLD2 protein mass.
p53 mutation status is one of the molecular differences between MCF-7 (wild-type p53) and MDA-MB-231 (p53 replacement mutation at codon 280; this mutation is located in the DNA binding domain of p53, which causes altered transcriptional activity). miR-203 expression is p53 dependent (41), and miR-182 expression is regulated by p53. In cardiac fibroblasts, serum starvation resulted in an elevated expression of p53 (42). Upon starvation, levels of p53 might be altered differently in MCF-7 cells and MB-231 cells. These altered p53 levels might influence miRNAs and also PLD2 levels.
Loss or gain of function of specific miRNAs probably contributes to breast epithelial cellular transformation and tumorigenesis (43). A model of starvation effects on PLD in cancer cells by the David Foster group indicates that cells in the core of a tumor mass become hypoxic and leave the tumor, seeking a new environment; these cells resemble mesenchymal cells, and the expression of PLD is augmented (19). It could very well be that the miRNAs that we have documented here are downregulated under starvation conditions.
The present study was conducted with four breast cancer cell lines, which are powerful experimental tools from which to derive information for potential clinical benefit. Luminal A and B subtypes are amenable to hormone therapy, and the HER2 group is a potential candidate for trastuzumab therapy (44). Basal tumors are difficult to treat, are biologically aggressive, and often have a poor prognosis. The claudin-low subtype is unique by the additional downregulation of claudin-3 and claudinin-4, low expression of the proliferation marker Ki67, enrichment for markers associated with the epithelial-to-mesenchymal transition, and expression of features associated with mammary cancer stem cells (45).
We provide for the first time direct evidence for a feedback loop, where PLD induction upon starvation leads to PA, which induces expression of miRNAs, which in turn inhibits PLD2 translation. The physiological relevance for breast cancer cells is that PA can activate cell invasion and then, due to the negative feedback, can deprive mTOR and S6K of their natural activator, prevent inhibition of apoptosis, and survive nutrient deprivation, which normal cells cannot do. A better understanding of PLD regulation in these cells could also lead to the design of better treatment options.
ACKNOWLEDGMENTS
We thank Karen M. Henkels for excellent editorial assistance in the preparation of the manuscript and K. Dougherty for data on kinase activities.
Funding Statement
The following grants to J.G.-C. have supported this work: HL056653-14 from the National Institutes of Health (NIH) and 13GRNT17230097 from the American Heart Association.
REFERENCES
- 1.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. 2012. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol 41:1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM, Calin GA. 2005. miRNAs, cancer, and stem cell division. Cell 122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki S, Tomari Y. 2009. Argonaute-mediated translational repression (and activation). Fly (Austin) 3:204–206. [PubMed] [Google Scholar]
- 5.Fukaya T, Iwakawa HO, Tomari Y. 2014. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol Cell 56:67–78. doi: 10.1016/j.molcel.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Aleckovic M, Kang Y. 2015. Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta 1855:24–42. doi: 10.1016/j.bbcan.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue J, Niu J, Wu J, Wu ZH. 2014. MicroRNAs in cancer therapeutic response: friend and foe. World J Clin Oncol 5:730–743. doi: 10.5306/wjco.v5.i4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki HI, Katsura A, Matsuyama H, Miyazono K. 2014. MicroRNA regulons in tumor microenvironment. Oncogene 34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Liu J, Wang G. 2014. The role of microRNAs in human breast cancer progression. Tumour Biol 35:6235–6244. doi: 10.1007/s13277-014-2202-8. [DOI] [PubMed] [Google Scholar]
- 10.Shah MY, Calin GA. 2014. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip Rev RNA 5:537–548. doi: 10.1002/wrna.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho JH, Hong SK, Kim EY, Park SY, Park CH, Kim JM, Kwon OJ, Kwon SJ, Lee KS, Han JS. 2008. Overexpression of phospholipase D suppresses taxotere-induced cell death in stomach cancer cells. Biochim Biophys Acta 1783:912–923. doi: 10.1016/j.bbamcr.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Hui L, Abbas T, Pielak RM, Joseph T, Bargonetti J, Foster DA. 2004. Phospholipase D elevates the level of MDM2 and suppresses DNA damage-induced increases in p53. Mol Cell Biol 24:5677–5686. doi: 10.1128/MCB.24.13.5677-5686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Cambronero J. 2014. Phosphatidic acid, phospholipase D and tumorigenesis. Adv Biol Regul 54:197–206. doi: 10.1016/j.jbior.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Zheng Y, Foster DA. 2003. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene 22:3937–3942. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Rodrik V, Foster DA. 2005. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene 24:672–679. doi: 10.1038/sj.onc.1208099. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, Foster DA. 2006. Phospholipase D couples survival and migration signals in stress response of human cancer cells. J Biol Chem 281:15862–15868. doi: 10.1074/jbc.M600660200. [DOI] [PubMed] [Google Scholar]
- 17.Lehman N, Ledford B, Di Fulvio M, Frondorf K, McPhail LC, Gomez-Cambronero J. 2007. Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR. FASEB J 21:1075–1087. doi: 10.1096/fj.06-6652com. [DOI] [PubMed] [Google Scholar]
- 18.Foster DA. 2013. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol Metab 24:272–278. doi: 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, Foster DA. 2011. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1). J Biol Chem 286:25477–25486. doi: 10.1074/jbc.M111.249631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkels KM, Peng HJ, Frondorf K, Gomez-Cambronero J. 2010. A comprehensive model that explains the regulation of PLD2 activity by phosphorylation-dephosphorylation. Mol Cell Biol 30:2251–2263. doi: 10.1128/MCB.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Q, Kantonen S, Gomez-Cambronero J. 2013. Serum deprivation confers the MDA-MB-231 breast cancer line with an EGFR/JAK3/PLD2 system that maximizes cancer cell invasion. J Mol Biol 425:755–766. doi: 10.1016/j.jmb.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz-Pinedo C, El Mjiyad N, Ricci JE. 2012. Cancer metabolism: current perspectives and future directions. Cell Death Dis 3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. 2006. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 24.Pucer A, Brglez V, Payre C, Pungercar J, Lambeau G, Petan T. 2013. Group X secreted phospholipase A(2) induces lipid droplet formation and prolongs breast cancer cell survival. Mol Cancer 12:111. doi: 10.1186/1476-4598-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 26.Tabatabaian F, Dougherty K, Di Fulvio M, Gomez-Cambronero J. 2010. Mammalian target of rapamycin (mTOR) and S6 kinase down-regulate phospholipase D2 basal expression and function. J Biol Chem 285:18991–19001. doi: 10.1074/jbc.M110.111542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acunzo M, Romano G, Wernicke D, Croce CM. 2015. MicroRNA and cancer—a brief overview. Adv Biol Regul 57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Tanic M, Yanowski K, Andres E, Gomez-Lopez G, Socorro MR, Pisano DG, Martinez-Delgado B, Benitez J. 2015. miRNA expression profiling of formalin-fixed paraffin-embedded (FFPE) hereditary breast tumors. Genomics Data 3:75–79. doi: 10.1016/j.gdata.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertoli G, Cava C, Castiglioni I. 2015. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi RU, Miyazaki H, Ochiya T. 2015. The roles of microRNAs in breast cancer. Cancers 7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A 103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina PP, Nolde M, Slack FJ. 2010. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 33.Egeland NG, Lunde S, Jonsdottir K, Lende TH, Cronin-Fenton D, Gilje B, Janssen EA, Soiland H. 2015. The role of microRNAs as predictors of response to tamoxifen treatment in breast cancer patients. Int J Mol Sci 16:24243–24275. doi: 10.3390/ijms161024243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodrik V, Gomes E, Hui L, Rockwell P, Foster DA. 2006. Myc stabilization in response to estrogen and phospholipase D in MCF-7 breast cancer cells. FEBS Lett 580:5647–5652. doi: 10.1016/j.febslet.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA. 2006. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 25:7305–7310. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- 36.Dall'Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, Yu WH, Robinson KS, Yeku O, Small SA, Duff K, Frohman MA, Wenk MR, Yamamoto A, Di Paolo G. 2010. The phospholipase D1 pathway modulates macroautophagy. Nat Commun 1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. 2009. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Fang Y. 2002. A novel pathway regulating the mammalian target of rapamycin (mTOR) signaling. Biochem Pharmacol 64:1071–1077. doi: 10.1016/S0006-2952(02)01263-7. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N. 2007. Autophagy: process and function. Genes Dev 21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 40.Dunn WA., Jr 1994. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 41.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna DJ, McDade SS, Patel D, McCance DJ. 2010. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol 84:10644–10652. doi: 10.1128/JVI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ubil E, Duan J, Pillai IC, Rosa-Garrido M, Wu Y, Bargiacchi F, Lu Y, Stanbouly S, Huang J, Rojas M, Vondriska TM, Stefani E, Deb A. 2014. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 514:585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG. 2010. microRNA, cell cycle, and human breast cancer. Am J Pathol 176:1058–1064. doi: 10.2353/ajpath.2010.090664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holliday DL, Speirs V. 2011. Choosing the right cell line for breast cancer research. Breast Cancer Res 13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]