Abstract
Healthy cells utilize intricate systems to monitor their environment and mount robust responses in the event of cellular stress. Whether stress arises from external insults or defects due to mutation and disease, cells must be able to respond precisely to mount the appropriate defenses. Multi-faceted stress responses are generally coupled with arrest of growth and cell-cycle progression, which both limits the transmission of damaged materials and serves to reallocate limited cellular resources toward defense. Therefore, stress defense versus rapid growth represent competing interests in the cell. How eukaryotic cells set the balance between defense versus proliferation, and in particular knowledge of the regulatory networks that control this decision, are poorly understood. In this perspective, we expand upon our recent work inferring the stress-activated signaling network in budding yeast, which captures pathways controlling stress defense and regulators of growth and cell-cycle progression. We highlight similarities between the yeast and mammalian stress responses and explore how stress-activated signaling networks in yeast can inform on signaling defects in human cancers.
Keywords: Stress response, Growth control, Transcription, Signal transduction
Introduction
All cells must allocate resources to balance conflicting physiological demands. From free-living microbes to multicellular animals, actively growing cells funnel resources to biosynthesis needed for division. A key consumer of cellular resources is translation, since production of ribosomes and translation itself require substantial amounts of energy (Warner 1999; Thomas 2000). The drive for proliferation often comes at the cost of stress tolerance (Fig. 1). For example, rapidly growing yeast cells are the most sensitive to environmental insults (Zakrzewska et al. 2011), whereas slow-growing clones are extremely resistant to adversity (Elliott and Futcher 1993; Lu et al. 2009; Levy et al. 2012). The same relationship exists in higher organisms, most notably in rapidly dividing cancer cells that are often the most susceptible to chemotherapy drugs and treatments (Jones and Thompson 2009). Understanding how cells allocate cellular resources toward proliferation versus stress defense is therefore critically important for understanding stress defense and, in turn, human disease.
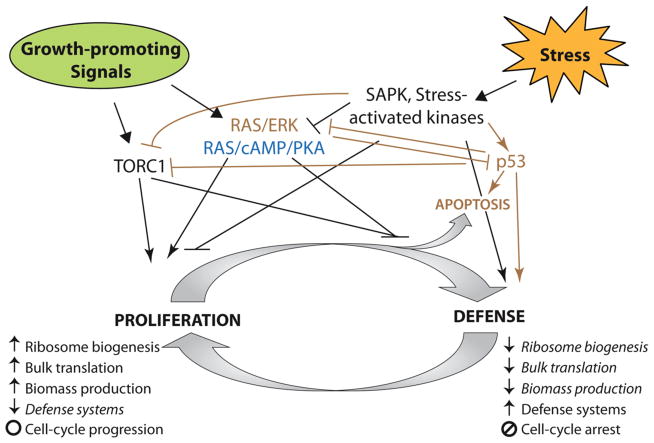
Fig. 1.
Proliferation and stress defense compete for limited resources in the cell. A simplified view of the signaling processes that occur in yeast and/or mammalian cells responding to growth-promoting signals versus cellular stress signals. Signaling molecules relevant in yeast are shown in blue and those unique to mammalian cells are shown in brown. See text for details
The balance between cellular growth and stress defense can be modulated in response to stressful situations, and thus the mobilization of defense strategies is often coordinated with arrest of growth and cell-cycle progression (Lopez-Maury et al. 2008). At the same time, defense responses include orchestrated changes to transcription, translation, and post-translational protein modifications that mediate changes in protein abundance, localization, and function. Although much is known about the signaling pathways that control individual physiological responses to stress, how signaling is integrated into a single cellular network that coordinates a multi-faceted response is only beginning to emerge. To address this question, we recently developed an experimental and computational pipeline in budding yeast Saccharomyces cerevisiae. The approach integrates stress-activated transcriptome alterations, phospho-proteomic changes, and gene-fitness contributions with large-scale protein interaction data, to implicate a single network coordinating the response to, in this case, salt stress (Chasman et al. 2014). The resulting network identified known and novel salt-regulated signaling proteins, uncovered previously unrecognized cross-connections between signaling pathways, and implicated important decision points in the growth-versus-defense decision. Importantly, the network is enriched with statistical significance for genes whose human orthologs cause cancer when mutated in somatic tissues.
In this perspective, we first provide a broad overview of signaling pathways that control proliferation versus stress defense in yeast and mammalian cells. We then highlight opportunities for using the stress-activated signaling network in yeast to understand the growth-versus-defense decision points in eukaryotic cells.
The environmental stress response: a common yeast response to diverse stresses
Actively growing microbial cells maintain high translational capacity to support division, in part by promoting ribosome biogenesis. Supporting high translational capacity therefore consumes a significant amount of the cell’s resources. Under optimal conditions, cells produce ~2000 ribosomes per minute (Warner 1999). The high demand for ribosomes requires active transcription of the rDNA locus; nearly 60 % of cellular transcription is devoted to rRNA production by RNA polymerase (Pol) I. Furthermore, over half the activity of Pol II transcription occurs at genes encoding ribosomal proteins (RPs), while Pol III is dedicated to transcribing 5S rRNA and tRNAs (Warner 1999; Rudra and Warner 2004). A significant fraction of Pol II is also dedicated to transcribing genes encoding ribosome assembly factors (commonly called the RiBi regulon) (Jorgensen et al. 2004). In addition to consuming transcriptional capacity, transcripts emerging from RP and RiBi genes are highly translated and thus monopolize a large fraction of the cell’s ribosomes (90 % of which are actively involved in translation during rapid growth) (Arava et al. 2003; von der Haar 2008). Thus, significant resources support ribosome biogenesis and translational capacity in cells actively growing under optimal conditions.
Upon a sudden shift to stressful conditions, cells must rapidly adjust resource allocation to mount cellular defense strategies (Fig. 1). Much has been gleaned from studying transcriptome changes in response to stress. Stressed yeast cells activate condition-specific transcript changes at specialized genes that specifically address the particular stress condition. Concurrently, cells activate the environmental stress response (ESR) (Gasch et al. 2000; Causton et al. 2001), a common response to diverse types of stress. The ESR includes ~300 induced ESR (iESR) genes that are broadly involved in stress defense and ~600 repressed (rESR) genes that encode RPs and RiBi factors (Gasch et al. 2000; Gasch 2002). The expression patterns of the iESR and rESR genes are strikingly anti-correlated across diverse types of stress, indicating that they are likely regulated by the same upstream signaling systems (Gasch 2002).
The ESR is postulated to relate to stress defense, given the functions of the encoded proteins. But surprisingly, activation of the ESR—and of stress-activated transcript changes in general—is not required to survive the initial stress treatment, at least in yeast (Berry and Gasch 2008; Westfall et al. 2008; Mitchell et al. 2009; Berry et al. 2011). Instead, transcript changes are critical for surviving subsequent stressful insults, through a phenomenon known as acquired stress resistance (Berry and Gasch 2008). Induction of iESR transcripts correlates with increased abundance of the encoded proteins, which most likely serve to defend against stress (Berry and Gasch 2008; Lee et al. 2011; Vogel et al. 2011). However, the role of the rESR transcript changes has remained somewhat elusive. Previous observations found that the abundance of RP and RiBi transcripts correlates with growth rate upon nutrient restriction (Jorgensen et al. 2002, 2004; Regenberg et al. 2006; Castrillo et al. 2007; Brauer et al. 2008), leading to the suggestion that rESR gene repression underlies growth and translational arrest during stress. However, we later showed that stress-dependent repression of RP and RiBi genes is not required for translational arrest or growth reduction, at least during salt stress (Lee et al. 2011). Instead, we proposed that rESR transcript reduction serves to release engaged ribosomes, thereby redirecting limited translational capacity to newly made mRNAs (Lee et al. 2011). More recent work from our lab also implies that reduced transcription of rESR genes is required to reallocate RNA Pol II to iESR and other defense genes (Chasman et al. 2014). Thus, the anti-correlation between rESR and iESR gene expression is at least partly explained by competition of these gene modules for limited resources.
Regulation of the growth-versus-defense decision in yeast
Two central signaling pathways regulating ribosome biogenesis are the target of rapamycin (TOR) and the RAS/cAMP/Protein Kinase A (PKA) signaling pathways, which respond to nitrogen and carbon availability, respectively (Neuman-Silberberg et al. 1995; Wang et al. 2004; Xiao and Grove 2009; Broach 2012). Both pathways are largely conserved from yeast to mammals. The TOR complex 1 (TORC1) directly binds the rDNA locus under optimal conditions (Li et al. 2006) and promotes rDNA transcription, by promoting activity of the rDNA transcription factor Rrn3 and, indirectly, by inhibiting the Pol III repressor Maf1 (Claypool et al. 2004; Lee et al. 2009; Philippi et al. 2010). Maf1 is also directly regulated by PKA, which suppresses the inhibitory activity of Maf1 during rapid growth (Moir et al. 2006; Willis and Moir 2007). Both TORC1 and PKA pathways have been implicated in regulating the RP genes and RiBi regulon, by modulating transcriptional activators (Fhl1/Crf1/Ifh1, Rap1, Sfp1) and repressors (Dot6/Tod6) (Klein and Struhl 1994; Jorgensen et al. 2004; Marion et al. 2004; Martin et al. 2004; Lippman and Broach 2009; Huber et al. 2011). The TOR and RAS/PKA pathways are clearly interconnected, even though the precise relationships in regulating ribosomal biogenesis are still controversial. Although some studies have suggested PKA as a downstream effector of the TOR pathway, possibly via the TORC1 downstream target Sch9 (Martin et al. 2004; Schmelzle et al. 2004; Soulard et al. 2010), other evidence has indicated that PKA and TOR are two parallel pathways activating RP gene expression (Zurita-Martinez and Cardenas 2005; Ramachandran and Herman 2011).
Along with promoting biogenesis of ribosomes and translational machinery, PKA and TOR act in parallel to antagonize ESR activation, in part by suppressing Msn2 and Msn4, the so-called ‘general stress’ transcription factors (Görner et al. 1998; Smith et al. 1998; Beck and Hall 1999). Thus, under stressful conditions TOR and PKA pathways must be suppressed—although the mechanisms remain poorly understood—while stress-activated signaling pathways are mobilized. For example, the MAP kinase Hog1 (orthologous to human kinase p38) is activated by osmotic shock and related stresses to coordinate myriad responses, including induction and repression of ~2000 transcripts (including iESR and rESR genes) (O’Rourke and Herskowitz 2004; Chasman et al. 2014), reallocation of RNA Pol II distribution (Cook and O’Shea 2012; Nadal-Ribelles et al. 2012; Chasman et al. 2014), alterations in translational capacity (Teige et al. 2001; Uesono and Toh-E 2002; Nagiec and Dohlman 2012), altered metabolism including osmolyte production (Albertyn et al. 1994; Proft and Struhl 2004), and transient cell-cycle arrest (Bellí et al. 2001; Escoté et al. 2004; Clotet et al. 2006; Yaakov et al. 2009). Alternate signaling pathways are specifically activated by other stresses (such as PKC in the case of cell wall stress (Heinisch et al. 1999), ATM/ATR in response to DNA damage (Abraham 2001), and the AMPK ortholog Snf1 during starvation (Conrad et al. 2014)). How activation of these stress-regulated pathways is coordinated with suppression of the TOR and PKA pathways remains unclear. Nonetheless, coordination and integration of signaling pathways is likely to be key for precise allocation of limited resources and adaptation to a new environment.
Stress responsive signaling in mammalian cells
There are many similarities in the molecular responses to stress in yeast and mammalian systems, despite the added complexities of multi-celled organisms. Studying cellular stress responses in the organismal context is particularly challenging, and thus much of the knowledge comes from culturing immortalized lines or primary cells responding to external stresses. A variety of environmental stresses have been studied extensively in mammalian systems, with the greatest focus on stresses linked to infection and disease, including DNA damaging agents, oxidants, elevated temperature, chemotherapy and other drugs, and hypoxic conditions that mimic the inter-tumor environment. Like yeast, cells must decide between maintaining proliferation or mediating cell-cycle arrest and stress defense. In addition, cells in a multicellular context have a third option in apoptosis, to clear cells that simply cannot recover from the insult. Thus, during adversity, mammalian cells face a multi-pronged decision point directing cells toward proliferation, defense, or death. Although there are many specific aspects of stress responses that are critically influenced by tissue type, developmental phase, and stress identity, several themes can be generalized from research in this area.
As in yeast, high ribosome biogenesis and translational activity are maintained in actively growing mammalian cells, promoted via signaling through mTORC1 and RAS to the ERK MAP kinase (Hannan et al. 2011). MTORC1 supports ribosome biogenesis through rRNA production, by stimulating transcription of rDNA genes as well as genes encoding rRNA processing factors (Mayer et al. 2004; Mayer and Grummt 2006; Kantidakis et al. 2010; Chauvin et al. 2014). mTORC signaling also promotes ribosome production by favoring translation of so-called 5-TOP transcripts that encode RPs and translation factors (Meyuhas and Dreazen 2009). mTOR also broadly enables cap-dependent translation by suppressing the eIF4e inhibitory binding protein 4E-BP1 (Gingras et al. 1998) and by activating the S6K kinase (orthologous to yeast Sch9), which further stimulates translation initiation and elongation (Magnuson et al. 2012; Roux and Topisirovic 2012). Mitogen-activated RAS/ERK signaling also provides a dual boost to translational activity: ERK signaling stabilizes the growth-enhancing transcription factor Myc, which induces Pol I, II, and III-dependent transcription of ribosome components, and phosphorylates rDNA transcription factors Tif1-A (orthologous to Rrn3 in yeast) and UBF to stimulate rDNA transcription (Hannan et al. 2011; Kusnadi et al. 2015). At the same time, ERK phosphorylation activates downstream kinases that promote cap-dependent translation initiation and elongation (Roux and Blenis 2004; Roux and Topisirovic 2012). As in yeast, there are multiple points of cross-talk between the RAS and mTORC pathways that are thought to produce precisely tuned growth behavior (Mendoza et al. 2011).
In response to stressful situations, many cell types mount a response that shares hallmarks with the yeast ESR. Murray et al. (2004) was one of the first to compare and contrast stress-activated transcriptome changes in primary and immortalized human cells: at least under the conditions and time frames studied, there were relatively few commonly induced or repressed genes upon diverse stress treatments (that included heat, oxidants, ER stress, and crowding). However, more recent studies have identified common responses to different stresses within a given cell type, albeit with smaller magnitude transcript changes than seen in yeast. Nayak et al. (2014) leveraged statistical power in a large study of B cell responses to ER stress and ionizing radiation, finding substantial overlap in response to the two stresses. Several studies interrogating p53 activity (see below) identified a common response that persists across several cell lines and conditions: induced genes are related to stress defense and regulation of cell cycle or apoptosis and repressed genes are linked to rDNA transcription, ribosome biogenesis, translation and cell cycle/apoptotic factors that work antagonistically to induced genes (Cairns and White 1998; Budde and Grummt 1999; Zhai and Comai 2000; Wei et al. 2006; Menendez et al. 2009; Nikulenkov et al. 2012; Schlereth et al. 2013). The coordinated induction of defense genes with repression of rDNA/protein synthesis genes is reminiscent of the coordinated expression changes of the induced and repressed gene modules of the yeast ESR.
Beyond the level of gene expression, other aspects of the stress response are conserved from yeast to humans. Upon stressful insults, transcription of rDNA is generally sharply decreased and coupled to an overall drop in cap-dependent translation (Spriggs et al. 2010; Liu and Qian 2014; Kusnadi et al. 2015). These responses are mediated by abrogated signaling through the mTOR/S6K and RAS/ERK pathways, and via activation of the stress-activated protein kinases (SAPKs) JNK and p38. p38 isoforms, including the broadly expressed p38α and β along with more tissue-specific γ and δ isoforms, vary in their responsiveness according to tissue type and developmental stage, but they generally respond to diverse types of cellular stress, cytokines and cell–cell contact as well as mitogens and cellular development (Zarubin and Han 2005; Ashwell 2006). The broad responsiveness of p38 isoforms distinguishes them from the yeast ortholog, Hog1, which responds primarily to osmotic and related stresses (Saito and Posas 2012). Both JNK and p38 are activated by a number of upstream kinases, some that have overlapping affinity for both SAPKs and others with specificity for only one of the kinases (Roux and Blenis 2004). Both SAPKs in turn activate a slew of downstream kinases and a host of transcription factors that mediate many aspects of the stress response.
Among the most famous of the SAPK targets is the transcription factor p53, which lies at the crux of the growth/defense/apoptosis decision (Beckerman and Prives 2010). Recent transcriptomic experiments have identified at least 280 common p53 targets activated by diverse stresses and in multiple tissues, with perhaps hundreds of additional targets identified by studies looking at different tissue and stress types (Mirza et al. 2003; Wei et al. 2006; Menendez et al. 2009; Huarte et al. 2010; Nikulenkov et al. 2012; Gambino et al. 2013; Schlereth et al. 2013; Leveille et al. 2015). Upon activation and translocation from the cytosol to nucleus, p53 can function both as an inducer and as a repressor (working directly and indirectly via its transcriptional targets); the genes regulated by p53 can vary in a given tissue depending on severity of the stress (see below) (Vousden and Lu 2002; Menendez et al. 2009; Levav-Cohen et al. 2014). Commonly induced genes include those linked to stress defense, regulators of cell-cycle arrest, and pro- or anti-apoptotic factors (Wei et al. 2006; Menendez et al. 2009; Nikulenkov et al. 2012; Schlereth et al. 2013), while repressed genes include genes that promote cell-cycle progression and proliferation and genes inked to ribosome biogenesis and translation. In fact, p53 can repress activity of all three RNA polymerases, and in the case of Pol I and Pol III, it does so by interfering with proper assembly of general transcriptional machinery (Cairns and White 1998; Budde and Grummt 1999; Zhai and Comai 2000; Becker-man and Prives 2010). Interestingly, the severity and duration of p53 activation are thought to influence whether cells arrest the cell cycle or sacrifice themselves through apoptosis (Vousden and Lu 2002). p53 binding elements upstream of pro-arrest and anti-apoptosis genes have higher affinity for p53 and lower dependence on cooperative binding, while promoters of pro-apoptosis factors are more likely to harbor degenerate p53 elements that require cooperative tetrameric association (Schlereth et al. 2013). This has led to the model that weak p53 activation (producing transient or incomplete nuclear localization) may promote cell survival, while strong or prolonged activation directs cells toward death (Beckerman and Prives 2010).
Signaling crosstalk and the role of the growth-defense-death decision in human cancers
The correct balance between growth versus stress defense or apoptosis is fundamental for proper organismal function. Improper balance leading to unchecked growth is thought to be a critical driver in diseases such as cancer (Jones and Thompson 2009). Regulators promoting RAS/ERK and mTORC signaling harbor gain-of-function mutations in many human cancers (Shaw and Cantley 2006; Fernández-Medarde and Santos 2011), underscoring the importance of these pathways in promoting growth. Furthermore, many cancerous cells show aberrantly high ribosome production and altered translation regulation (consistent with the oncogene status of eIF4e (Mamane et al. 2004)). Thus, elevated flux toward growth and cell division is a driving force in cancer emergence.
But mutations in stress-activated regulators can also contribute to cancer. For example, 5–10 % of diverse cancers harbor mutations in the SAPK-activating kinase MEK4, which is also associated with poor prognosis in several cancer types (Taylor et al. 2008), while a striking 50 % of human cancers have inactivating alleles of p53 (Vousden and Lu 2002). Several stress-activated signaling pathways play dual roles in suppressing cancer, by triggering apoptosis and by inhibiting signaling through the growth-promoting pathways. Thus, their mutation both prevents cells death and results in unchecked proliferation signaling. For example, p53 suppresses tumorogenesis not only by activating arrest or apoptosis, but also by suppressing mTORC and ERK via up-regulating expression of their inhibitors, at least in certain tissues (Matthew et al. 2009; Feng and Levine 2010; Hasty et al. 2013; Drosten et al. 2014; Akeno et al. 2015). There are other examples of stress-activated pathways directly repressing proliferation signals: upon nutrient limitation in mouse embryonic fibroblasts, stress-activated AMPK and p38β suppress mTORC activity through at least three independent routes (Zheng et al. 2011). Thus, it is clear that stress-activated pathways work at several levels to shift the balance away from proliferation and toward a productive stress response.
Stress-activated networks in yeast: a model for understanding design principles
The design principles of stress-activated signal integration remain poorly understood in both yeast and higher mammals. One strategy to investigate signaling organization is through systems-biology approaches to infer signaling networks. Several recent studies have conducted computational inference of the stress-activated signaling networks, most commonly based on transcriptome data (Friedman 2004; Schadt et al. 2005; Gat-Viks and Shamir 2007; Gitter et al. 2013; Wu et al. 2013). In our recent work, we integrated disparate high-throughput yeast datasets using an integer linear programming approach to infer the salt-activated signaling network (Chasman et al. 2014). The resulting network of ~400 proteins captured known and novel salt-activated pathways as well as key regulators in the growth-promoting TORC1, RAS, and cAMP/PKA pathways, which are suppressed upon salt treatment. It also uncovered previously unrecognized cross-connections between what are generally studied as discrete pathways. We defined pathways based on the literature and scored the number of cross-pathway connections between them. Among those with the greatest connections to other pathways were the TORC1, RAS, and cAMP/PKA pathways. The consequences of this inter-pathway connectivity remain to be tested, but we hypothesize that it reflects an intricate level of control exerted by stress-activated pathways on growth-promoting signaling.
Many of the regulatory connections in the yeast salt-responsive signaling network are orthologous to known signaling connections in mammals. But even more striking is the link to disease: the salt-activated signaling network we inferred is significantly enriched (p = 8e–4) for proteins whose human orthologous cause cancer when mutated in somatic tissues, according to the latest release of the COSMIC database (Forbes et al. 2015). Using a stringent method of identifying orthologs (Deluca et al. 2006), we found 11 of 49 orthologs from the COSMIC database in the salt-activated network: nearly half were chromatin regulators or transcription factors (including the ortholog of Sfp1 which regulates RP genes), two were involved in RNA metabolism, and one was the Hog1 activator Pbs2 (which is assigned orthologous to the ERK-activating MKK2 kinase). Remarkably, the network is also enriched nearly 3.5-fold for orthologs of p53 interacting proteins captured in the Biogrid database (p = 1e–5 (Chatr-Aryamontri et al. 2015))—even though budding yeast lacks a p53 ortholog. The group of yeast genes orthologous to p53 interactors is enriched for kinases (p = 1.5e–7) and cell-cycle regulators (p = 4e–5). These results indicate that the salt-activated signaling network in yeast shares key features with cancer-related signaling in humans, suggesting that the networks represent modern-day renditions of an ancient signaling system.
Much remains to be dissected about how diverse signals are integrated into a single signaling network, and how cells set the balance between growth-versus-stress defense. It is in this light that yeast research can contribute fundamental insights. Yeast provides an excellent test bed for systems-biology approaches to this question, since detailed follow-up studies can test predictions from network science. Despite the differences in complexity between yeast and mammalian systems, we believe that continued exploration of stress-activated yeast signaling networks in the context of disease-causing orthologs could provide a new perspective on such signaling decisions. Understanding how these fundamental decision are made and disseminated in cells will expand our understanding of stress biology and foster our eventual ability to modulate it.
Acknowledgments
We apologize to the authors of many important research studies that we were unable to cite due to space constraints. We thank M. MacGilvray for useful comments on the manuscript. This work was supported by NIH R01 GM083989 to A. P. G.
Footnotes
Communicated by M. Kupiec.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Akeno N, Miller AL, Ma X, Wikenheiser-Brokamp KA. p53 suppresses carcinoma progression by inhibiting mTOR pathway activation. Oncogene. 2015;34:589–599. doi: 10.1038/onc.2013.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6:532–540. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellí G, Garí E, Aldea M, Herrero E. Osmotic stress causes a G1 cell cycle delay and downregulation of Cln3/Cdc28 activity in Saccharomyces cerevisiae. Mol Microbiol. 2001;39:1022–1035. doi: 10.1046/j.1365-2958.2001.02297.x. [DOI] [PubMed] [Google Scholar]
- Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet. 2011;7:e1002353. doi: 10.1371/journal.pgen.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DC, Cornell MJ, Petty J, Hakes L, Wardleworth L, Rash B, Brown M, Dunn WB, Broadhurst D, O’Donoghue K, Hester SS, Dunkley TP, Hart SR, Swainston N, Li P, Gaskell SJ, Paton NW, Lilley KS, Kell DB, Oliver SG. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D, Ho Y-H, Berry DB, Nemec CM, MacGilvray ME, Hose J, Merrill AE, Lee MV, Will JL, Coon JJ, Ansari AZ, Craven M, Gasch AP. Pathway connectivity and signaling coordination in the yeast stress-activated signaling network. Mol Syst Biol. 2014;19:759. doi: 10.15252/msb.20145120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, Pende M. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene. 2014;33:474–483. doi: 10.1038/onc.2012.606. [DOI] [PubMed] [Google Scholar]
- Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KE, O’Shea EK. Hog1 controls global reallocation of RNA Pol II upon osmotic shock in Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:1129–1136. doi: 10.1534/g3.112.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca TF, Wu IH, Pu J, Monaghan T, Peshkin L, Singh S, Wall DP. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- Drosten M, Sum EY, Lechuga CG, Simón-Carrasco L, Jacob HK, García-Medina R, Huang S, Beijersbergen RL, Bernards R, Barbacid M. Loss of p53 induces cell proliferation via Ras-independent activation of the Raf/Mek/Erk signaling pathway. Proc Natl Acad Sci USA. 2014;111:15155–15160. doi: 10.1073/pnas.1417549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Futcher B. Stress resistance of yeast cells is largely independent of cell cycle phase. Yeast. 1993;9:33–42. doi: 10.1002/yea.320090105. [DOI] [PubMed] [Google Scholar]
- Escoté X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Gambino V, De Michele G, Venezia O, Migliaccio P, Dall’Olio V, Bernard L, Minardi SP, Della Fazia MA, Bartoli D, Servillo G, Alcalay M, Luzi L, Giorgio M, Scrable H, Pelicci PG, Migliaccio E. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell. 2013;12:435–445. doi: 10.1111/acel.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP. The environmental stress response: a common yeast response to environmental stresses. Springer-Verlag; Heidelberg: 2002. [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat-Viks I, Shamir R. Refinement and expansion of signaling pathways: the osmotic response network in yeast. Genome Res. 2007;17:358–367. doi: 10.1101/gr.5750507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitter A, Carmi M, Barkai N, Bar-Joseph Z. Linking the signaling cascades and dynamic regulatory networks controlling stress responses. Genome Res. 2013;23:265–276. doi: 10.1101/gr.138628.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB. Signaling to the ribosome in cancer—It is more than just mTORC1. IUBMB Life. 2011;63:79–85. doi: 10.1002/iub.428. [DOI] [PubMed] [Google Scholar]
- Hasty P, Sharp ZD, Curiel TJ, Campisi J. mTORC1 and p53: clash of the gods? Cell Cycle. 2013;12:20–25. doi: 10.4161/cc.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large inter-genic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30:3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene. 2015;556:27–34. doi: 10.1016/j.gene.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem. 2009;8:12604–12608. doi: 10.1074/jbc.C900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;19:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levav-Cohen Y, Goldberg Z, Tan KH, Alsheich-Bartok O, Zuckerman V, Haupt S, Haupt Y. The p53-Mdm2 loop: a critical juncture of stress response. Subcell Biochem. 2014;85:161–186. doi: 10.1007/978-94-017-9211-0_9. [DOI] [PubMed] [Google Scholar]
- Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia AR, Wijchers PJ, Geeven G, den Boer ML, Kalluri R, de Laat W, Esteller M, Agami R. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- Lippman SI, Broach JR. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci USA. 2009;106:19928–19933. doi: 10.1073/pnas.0907027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Qian SB. Translational reprogramming in cellular stress response. Wiley Interdiscip Rev RNA. 2014;5:301–315. doi: 10.1002/wrna.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maury L, Marguerat S, Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- Lu C, Brauer MJ, Botstein D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell. 2009;20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6 K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E–from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O’Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Matthew EM, Hart LS, Astrinidis A, Navaraj A, Dolloff NG, Dicker DT, Henske EP, El-Deiry WS. The p53 target Plk2 interacts with TSC proteins impacting mTOR signaling, tumor growth and chemosensitivity under hypoxic conditions. Cell Cycle. 2009;8:4168–4175. doi: 10.4161/cc.8.24.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3 K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Dreazen A. Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog Mol Biol Transl Sci. 2009;90:109–153. doi: 10.1016/S1877-1173(09)90003-5. [DOI] [PubMed] [Google Scholar]
- Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier P, Greene JR, Liu S. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Ribelles M, Conde N, Flores O, González-Vallinas J, Eyras E, Orozco M, de Nadal E, Posas F. Hog1 bypasses stress-mediated down-regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol. 2012;13:R106. doi: 10.1186/gb-2012-13-11-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MJ, Dohlman HG. Checkpoints in a yeast differentiation pathway coordinate signaling during hyperosmotic stress. PLoS Genet. 2012;8:e1002437. doi: 10.1371/journal.pgen.1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak RR, Bernal WE, Lee JW, Kearns MJ, Cheung VG. Stress-induced changes in gene interactions in human cells. Nucleic Acids Res. 2014;42:1757–1771. doi: 10.1093/nar/gkt999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Bhattacharya S, Broach JR. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol Biol Cell. 1995;15:3187–3196. doi: 10.1128/mcb.15.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, Selivanova G. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi A, Steinbauer R, Reiter A, Fath S, Leger-Silvestre I, Milkereit P, Griesenbeck J, Tschochner H. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 2010;38:5315–5326. doi: 10.1093/nar/gkq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. doi: 10.1016/j.cell.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Herman PK. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics. 2011;187:441–454. doi: 10.1534/genetics.110.123372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther O, Fausbøll A, Akesson M, Bro C, Hansen LK, Brunak S, Nielsen J. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol. 2012;4:a012252. doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T. Characterization of the p53 cistrome–DNA binding cooperativity dissects p53’s tumor suppressor functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Biol Cell. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schütz F, Jenö P, Hall MN. The rapamycin-sensitive phosphoproteome reveals that TOR controls PKA toward some but not all substrates. Mol Biol Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Taylor JL, Szmulewitz RZ, Lotan T, Hickson J, Griend DV, Yamada SD, Macleod K, Rinker-Schaeffer CW. New paradigms for the function of JNKK1/MKK4 in controlling growth of disseminated cancer cells. Cancer Lett. 2008;272:12–22. doi: 10.1016/j.canlet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci U S A. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nat Cell Biol. 2000;2:E71–E72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- Uesono Y, Toh-E A. Transient inhibition of translation initiation by osmotic stress. J Biol Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Vogel C, Silva GM, Marcotte EM. Protein expression regulation under oxidative stress. Mol Cell Proteomics. 2011;10(M111):009217. doi: 10.1074/mcp.M111.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol. 2008;2:87. doi: 10.1186/1752-0509-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Güldal CG, Zhang X, Tavazoie S, Broach JR. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:E128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Patterson JC, Chen RE, Thorner J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA. 2008;105:12212–12217. doi: 10.1073/pnas.0805797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Grove A. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov G, Duch A, García-Rubio M, Clotet J, Jimenez J, Aguilera A, Posas F. The stress-activated protein kinase Hog1 mediates S phase delay in response to osmostress. Mol Biol Cell. 2009;20:3572–3582. doi: 10.1091/mbc.E09-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska A, van Eikenhorst G, Burggraaff JE, Vis DJ, Hoefsloot H, Delneri D, Oliver SG, Brul S, Smits GJ. Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol Biol Cell. 2011;22:4435–4446. doi: 10.1091/mbc.E10-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang YH, Wu XN, Wu SQ, Lu BJ, Dong MQ, Zhang H, Sun P, Lin SC, Guan KL, Han J. Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol. 2011;13:263–272. doi: 10.1038/ncb2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase a: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]