Abstract
Background:
MicroRNA-206 (miR-206) and connexin 43 (Cx43) are related with the distant metastasis of breast cancer. It remains unclear whether the regulatory effect of miR-206 on Cx43 is involved in metastasis of breast cancer.
Methods:
Using quantitative real-time polymerase chain reaction and Western blot, the expressions of miR-206 and Cx43 were determined in breast cancer tissues, hepatic and pulmonary metastasis (PM), and cell lines (MCF-10A, MCF-7, and MDA-MB-231). MCF-7/MDA-M-231 cells were transfected with lentivirus-shRNA vectors to enhance/inhibit miR-206, and then Cx43 expression was observed. Cell counting kit-8 assay and Transwell method were used to detect their changes in proliferation, migration, and invasion activity. The mutant plasmids of Cx43-3’ untranslated region (3’UTR) at position 478–484 and position 1609–1615 were constructed. Luciferase reporter assay was performed to observe the effects of miR-206 on luciferase expression of different mutant plasmids and to confirm the potential binding sites of Cx43.
Results:
Cx43 protein expression in hepatic and PM was significantly higher than that in the primary tumor, while no significant difference was showed in messenger RNA (mRNA) expression. MiR-206 mRNA expression in hepatic and PM was significantly lower than that in the primary tumor. Cx43 mRNA and protein levels, as well as cell proliferation, migration, and invasion capabilities, were all significantly improved in MDA-MB-231 cells after reducing miR-206 expression but decreased in MCF-7 cells after elevating miR-206 expression, which demonstrated a significantly negative correlation between miR-206 and Cx43 expression (P = 0.03). MiR-206 can drastically decrease Cx43 expression of MCF-7 cells but exerts no effects on Cx43 expression in 293 cells transfected with the Cx43 coding region but the lack of Cx43-3’UTR, suggesting that Cx43-3’UTR may be the key in Cx43 regulated by miR-206. Luciferase expression showed that the inhibition efficiency was reduced by 46.80% in position 478–484 mutant, 16.72% in position 1609–1615 mutant; the inhibition was totally disappeared in double mutant (P = 0.02).
Conclusions:
MiR-206 can regulate the expression of Cx43, the cytobiological activity, and the metastasis of breast cancer through binding to the two binding sites in Cx43-3’UTR: position 478–484 and position 1609–1615.
Keywords: Breast Cancer, Connexin 43, Metastasis, MicroRNA-206
INTRODUCTION
Breast cancer is the most common malignant tumor in women and the second leading cause of cancer death among women worldwide.[1,2] Despite well-organized screening and early treatment programs that have been effective in preventing breast cancer, many breast tumors are not eradicated completely due to acquired resistance or relapse following an initial response, thus resulting in metastatic disease at later stages that leads to patient death.[3] Gap junction is a membrane structure between adjacent cells, which composed of connexin (Cx) and mediates the mutual, direct electric or chemical signal communication. There are 21 members in Cx family of humans gene spectrum, which are named according to relative molecular mass (Mr), for example, Cx43 refers to a Cx with Mr = 43,000. Human breast tissues mainly express Cx26 and Cx43.[4] It is observed that breast cancer metastasis is accompanied by increasing Cx43 expression and intercellular exchange disorders.[5,6,7] The expression of Cx43 protein is regulated through two mechanisms: transcriptional and posttranscriptional. There are lots of regulatory factors in transcriptional level. Messenger RNA (mRNA) must be translated into protein to exert its function. The posttranscriptional regulation mechanism of Cx43 has not been clarified yet. MicroRNA (miRNA) is one of the largest groups of posttranscriptional regulatory factors,[8] whose 2–8 bases at 5’ end can bind to 3’ untranslated region (3’UTR) of target gene so as to regress gene translation and reduce protein expression.[9] miRNA is involved in a series of biological processes including cell cycle, growth, apoptosis, differentiation, and stress reaction.[10] The first discovered miRNA associated with breast cancer is miR-206. A significant decline of miR-206 is noticed in breast cancer tissues (CTs), and low miR-206 level is closely associated with advanced clinical stage and shorter overall survival.[11] In our previous study, we compared Cx43 protein and miR-206 mRNA expression in the resected primary lesion of human breast cancer and paired metastatic axillary lymph node, and found that in metastatic axillary lymph node, miR-206 expression was significantly reduced while Cx43 protein expression was significantly increased, and a negative correlation was observed between them.[11] However, whether the posttranscriptional regulation of Cx43 by miR-206 is involved in the distal metastasis of breast cancer is still controversial. In this study, we observed miR-206 and Cx43 expression in primary tumor, hepatic, and pulmonary metastatic lesions of breast cancer, then explored the effect of miR-206 on Cx43 expression and biological characteristics of human breast cancer cells (MCF-7 and MDA-MB-231). At last, we validated the possible binding sites in Cx43-3’UTR by which miR-206 regulates the expression of Cx43. The aim of this research was to provide references to the further mechanic study of breast cancer metastasis and develop new therapeutic targets in the treatment of breast cancer.
METHODS
Tissue collection
From October 1, 2012, to July 1, 2013, we enrolled 40 cases of invasive ductal carcinoma, 10 hepatic metastases, and 10 pulmonary metastases (PMs) of breast cancer which were pathologically diagnosed in the Breast Disease Center of Southwest Hospital affiliated to the Third Military Medical University and the Department of Breast, Thyroid and Pancreas Surgery of the Second Affiliated Hospital of Chongqing Medical University. All the patients were aged from 32 to 67 years (median 53 years), females, with tumor diameter of 1.5–4.7 cm (2.8 cm on average). They did not receive any neoadjuvant chemotherapy or endocrine therapy preoperatively. Two kinds of specimens were sterilely taken during modified radical mastectomy: normal tissues (NTs, at 3 cm from the tumor margin) and CTs. 2–3 pieces of specimens from hepatic metastasis (HMs) and PMs were obtained from metastatic patients during computed tomography-guided biopsy. The specimens for quantitative real-time polymerase chain reaction (qRT-PCR) were quickly immersed into RNA inhibitor solution overnight at 4°C and removed into the protective liquid, preserved at −80°C.
This study was approved by the Ethics Committee of Southwest Hospital affiliated to the Third Military Medical University and Second Affiliated Hospital of Chongqing Medical University. All patient-derived tissues were obtained with written informed consent.
Cell lines and culture conditions
Human breast cancer cell lines MCF-7 and MDA-MB-231 were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences. The normal breast cell line MCF-10A was preserved in our laboratory. The cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Hyclone, USA) containing 10% fetal bovine serum (Gibco, USA) in 5% CO2 incubator at 37°C, digested with 0.25% trypsin and passaged in vitro. Cells used in the experiment were in the logarithmic growth phase.
MicroRNA-206 and connexin 43 expressions in tissues and cells
Messenger RNA expression of microRNA-206 and connexin 43 by quantitative real-time polymerase chain reaction
Total RNA was extracted from tissues or cells with TRIzol reagent (Invitrogen, USA). For qPCR, RNA was reversely transcribed to cDNA from 1 mg of total RNA using a Reverse Transcription Kit (Takara, Japan). qRT-PCR was conducted with SYBR® premix Ex Taq™ II (perfect real-time) (Takara). All protocols were performed according to the manufacturer's instructions. Results were normalized to the expression of U6/glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and then expressed as a relative expression compared with related control samples. The primer sequences were listed in Table 1.
Table 1.
Primer sequence and reaction condition of miR-206 and Cx43 by qRT-PCR
| Primer | Sequence (5’-3’) | Reaction condition of qRT-PCR |
|---|---|---|
| Cx43 | F: GGTGGAGAGGGAGGGGATAAGAG R: CCACCCGCTCATTCACATACAC |
95°C 1: 30→(95°C, 5 s→62.5°C, 30 s) × 40→65°C, 1 s |
| GAPDH | F: AGAGGCAGGGATGATGTTCTG R: GACTCATGACCACAGTCCATGC |
95°C 1: 30→(95°C 5 s→62.5°C 30 s) × 40→65°C 1 s |
| miR-206 Reverse-transcribed primer: GTCGTATCCAGTGCAGGGTCCG AGGTATTCGCACTGGATACGACCCACACA |
F: GGCCGCTGGAATGTAAGGAAG R: GTGCAGGGTCCGAGGT |
95°C 1: 30→(95°C, 5 s→60°C, 30 s) × 40→65°C, 1 s |
| U6 Reverse-transcribed primer: AAAATATGGA ACGCTTCACGAATTTG |
F: CTCGCTTCGGCAGCACATATACT R: GTAACGCTTCACGAATTTGCGTGTC |
95°C 1: 30 →(95°C 5 s→60°C 30 s) × 40→65°C 1 s |
Cx43: Connexin 43; qRT-PCR: Quantitative real-time polymerase chain reaction; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; miR-206: MicroRNA-206.
Protein expression of connexin 43 by Western blotting assay
Total proteins were extracted with cell lysis solution, and the concentration was determined by Bradford method. About 50 μg protein was separated in 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to PCDF membrane, and incubated with rabbit anti-human Cx43 antibody (1:1000, Santa, USA) at 37°C for 4 h, with anti-rabbit secondary antibody (1:2500, Sigma, USA) at 37°C for 2 h. Protein bands were quantified from film by densitometry using Adobe Photoshop V7.01 imaging system.
Altering microRNA-206 level in breast cancer cell lines
The cells in logarithmic growth phase were seeded in 6-well plates at the density of 105/well and grown for 12 h, then switched to fresh DMEM medium. In order to improve the expression of miR-206, 20 μl 6 × 108 TU/ml (multiplicity of infection [MOI] = 20) LV-hsa-miR-206 (1569-11) virus (lentivirus-shRNA vectors, Genechem, China) was added into MCF-7 cells (low expression of miR-206). 10 μl 6 × 108 TU/ml (MOI = 10) LV-hsa-miR-206-inhibition (lentivirus-shRNA vectors, Genechem, China) was added into MDA-MB-231 (with high expression of miR-206) to inhibit the expression of miR-206. After 12 h, the cells were switched to fresh medium and harvested after 96 h for subsequent experiments.
Connexin 43 expressions after altering microRNA-206
Cx43 mRNA and protein expression of MCF-7/MDA-MB-231 transfected by LV-hsa-miR-206 (1569-11) virus/LV-hsa-miR-206-inhibition were checked by qRT-PCR and Western blot as previously described.
Cytobiological changes after changing microRNA-206
Cell proliferation assay by cell counting kit-8
The proliferation of MCF-7/MDA-MB-231 transfected by LV-hsa-miR-206 (1569-11) virus/LV-hsa-miR-206-inhibition was checked by cell counting kit-8 (CCK-8) (Yiyuan, China). According to the instruction, five wells in each group were for the experiment, another five wells were to add DMEM medium and CCK-8 solution as a control. At 12, 24, 48, 72, and 96 h after cell seeding, CCK-8 cell proliferation experiment was performed. The optical density at 450 nm wavelength (D[450]) was determined by Thermo Varioskan Multifunction Reader (Thermo Scientific, USA). Cell proliferation curve was drawn with Time as X-axis, D (450) as Y-axis.
Transwell migration assay
Serum-free DMEM containing 0.1% bovine serum albumin was used to suspend and dilute the cells to a concentration of 1 × 105/ml. About 200 μl of cell suspension was sucked to the upper chamber of Transwell (PC membrane with 8.0 μm pore size, BD, USA), high-glucose DMEM containing 10% fasting blood sugar (FBS) to the lower chamber. The cells were cultured in a 37°C incubator at 5% CO2 for 12 h. The upper chamber was removed, air-dried, fixed with paraformaldehyde for 15 min, rinsed with phosphate-buffered saline (PBS) for 2 min, and stained with 0.1% crystal violet. Under a microscope (200×), randomly selected 5 view fields to count cells and averaged the numbers.
Transwell invasion assay
Before the experiment, precooled serum-free DMEM was mixed with matrigel (BD, USA) at the ratio of 2:1, and then 30 μl of mixture per well was drawn into Transwell upper chamber and solidified at 37°C for 1 h. Add 200 μl suspension at the cell concentration of 1 × 105/ml into the upper chamber, high-glucose DMEM medium containing 10% FBS into the lower chamber. The cells were cultured in a 37°C incubator at 5% CO2 for 12 h. The upper chamber was removed, air-dried, fixed with paraformaldehyde for 15 min, rinsed with PBS for 2 min, and stained with 0.1% crystal violet. Under a microscope (×200), randomly selected 5 view fields to count cells and averaged the numbers.
Studying on binding sites of connexin 43 for microRNA-206
MicroRNA-206 may regulate connexin 43 protein expression via connexin 43-3’ untranslated region
Human keratinocytes cell line HaCaT cells in the logarithmic phase were lysated using RNAiso Reagent. Total cellular RNA was extracted. According to the instructions of PrimeScript 1st strand cDNA Synthesis Kit (TaKaRa, Japan), transverse transcription was performed to prepare the corresponding cDNA, which was used as a template for PCR amplification of the coding region of Cx43. The primer sequence: Cx43 F: 5’-CCCAAGCTTATGGGTGACTGGAGCGCCTT-3’ (designed Hind III restriction enzyme cutting sites were underlined), Cx43 R: 5’-CTAGTCTAGACTAGATCTCCAGGTCATCAG-3’ (designed Xba I restriction enzyme cutting sites were underlined). The 1.0% agarose gel electrophoresis was conducted to observe the amplified fragments 293 and MCF-7 cells were seeded in a 6-well plate 24 h earlier. According to the instructions of Lipofectamine 2000 (Life Tech, USA), pcDNA3.1-Cx43 eukaryotic expression plasmid (2.5 μg/well) and 60 nmol of pre-miR negative control or pre-miR has-miR-206 (Ambion, USA) were premixed, and then the mixture was used to transfect 293 cells. The MCF-7 cells were only transfected by 60 nmol of pre-miR negative control or pre-miR has-miR-206. After 6 h, changed the medium and cultured for another 48 h, and then harvested the protein. The protein was separated by 12% SDS-PAGE gel electrophoresis, transferred to a membrane, incubated with antibodies (Cx43: 1:250; GAPDH: 1:1000), and finally visualized.
The target straps (1149 bp) became purer and more abundant as the annealing temperature increased. We set the annealing temperature at 65°C, when the abundance of target strap was the highest. Plasmid pcDNA3.1 and purified PCR amplifying products of Cx43 underwent Hind III/Xba I double enzyme digestion. The products were connected after the secondary purification and preserved at 16°C overnight. The ligation product was dealt with Escherichia coli DH5α, then painted in Amp+ plate and cultured overnight. Monoclonal bacteria were selected and identified by PCR. The plasmid with correct sequence was named as pcDNA3.1-Cx43.
MicroRNA-206 can regulate connexin 43 by binding specific sites of connexin 43-3’ untranslated region
Using EZNA Tissue DNA Kit (Omega, USA), genomic DNA was extracted from human keratinocyte cell line HaCaT cells in the logarithmic growth phase, and stored at −20°C after determining the concentration. Primers were designed based on the sequence of Cx43-3’UTR in GeneBank as follows: C43-3’UTR F: 5’-GGACTAGTATACAGGCTTGAAAGCATCA-3’ (designed Spe I restriction enzyme cutting sites were underlined); R: 5’-CGACGCGTTATGTT TATACTAAATTAAAA-3’ (designed Mul I restriction enzyme cutting sites were underlined). With human genomic DNA as template for amplification, firstly Cx43 3’UTR (1751 bp) was amplified by gradient PCR, then 1.0% agarose gel electrophoresis was performed. It was found that the amplification effect was optimal when the annealing temperature was 50°C. The fragment size was nearly equal to the real value. The purified PCR amplification product of Cx43 3’UTR, and plasmid pmiR-Luc underwent Hind III/Xba I double enzyme digestion. The products were connected after the secondary purification and preserved at 160C overnight. The ligation product was dealt with E. coli DH5α, then painted in Amp+ plate and cultured overnight. Monoclonal bacteria were selected and identified by PCR. The plasmid with correct sequence was named as pmiR-Luc-Cx43-3’UTR. The amplification effect was optimal when the annealing temperature was 50°C. The fragment size was nearly equal to the real value. Two bioinformatic predictive softwares TargetScan and PicTar (http://www.targetscan.org/; http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi) were used to screen the 3’UTR of Cx43–GJA1 (located on human chromosome 6q22-q23). There are two possible binding sites in Cx43-3’UTR which are located in sites 478–484 (ACAUUC) and 1609–1615 (ACAUUC) and complementary to the 6 bases at position 58–63 of miR-206, i.e., TGTAAG. In order to clarify whether both of them should bind to miR-206 and regulate the cytobiological activity, we designed the plasmid mutated at the two possible binding sites in Cx43-3’UTR. In the median sites of Cx43-3’UTR: 480–483 and 1611–1614, AUUC was mutated to UAAG. So the primer sequences were designed as follows: for the first binding site (478–484, BS1) F: 5’-ACTAAGCATTGTTAAAATTTGCACTT-3’, BS1 R: 5’-TTTAGCAGGGACTTAAGGACAATCCT-3’. For the second binding site (1609–1615, BS2) F: 5’-ACTAAGCATGTTAAACTACGGTCATG-3’, BS2 R: 5’-CAAACAAATTAGTAATTTTCATTGTA-3’. According to the instruction of KOD-Plus-Mutagenesis Kit (Toyobo, Japan), the clones were sent for sequence detection. The plasmid with correct mutation was reserved based on the sequencing results.
Cells transfection was performed using Lipofectamine 2000 (Life Tech, USA) according to the instructions of the manufacturer. The 6 purified luciferase reporter plasmids (pmiR-Luc-con, wild-type Cx43-3’UTR, Cx43-3’UTR BS-1 mut, Cx43-3’UTR BS-2 mut, Cx43-3’UTR BS-1, 2 mut, and pmiR-β-gal) were diluted to the same concentration. The former five plasmids (150 ng/well) were mixed with pre-miR negative control or pre-miR has-miR-206 (50 nmol, Ambion, USA), respectively. pmiR-β-gal (200 ng/well) was added as a control for transfection efficiency in each group. 293 cells were co-transfected by the three plasmids for 6 h and cultured for another 24 h after changed the medium. The luciferase and β-galactosidase expression were assayed using luciferase detection kit and β-galactosidase detection kit (Beyotime, China). The ratio of Luc/β-gal was expressed as a mean ± standard deviation (SD). With the value of control group (pmiR-Luc-con) as the correction, luciferase expression was calculated (×100%).
Statistical analysis
All experiments were done in duplicate and repeated three times. Using SPSS 17.0 software package (SPSS Inc., USA), the data were expressed as a mean ± standard deviation (SD). Statistical significance between groups was assessed by one-way analysis of variances, followed by Tukey's multiple comparison tests. The values between two groups were compared with t-test. The simple correlation analytical method was used to analyze the dependability. A P value of < 0.05 was considered to be statistically significant.
RESULTS
MicroRNA-206 and connexin 43 expressions in tissues and cells
Messenger RNA expression
As shown in Table 2, miR-206 mRNA in CTs, HMs, and PMs was significantly lower than that in NTs (P = 0.02), while miR-206 mRNA in HMs and PMs was lower than that in CTs (P = 0.015). Cx43 mRNA in CTs, HMs, PMs was significantly lower than that in NTs (P = 0.023). There was no statistical difference between primary tissues and metastatic tissues (HMs, PMs) (P > 0. 05) [Table 2].
Table 2.
mRNA expression of miR-206 and Cx43 in breast cancer tissue
| Tissue | miR-206 | Cx43 | ||
|---|---|---|---|---|
| Ct value (mean ± SD) | 2−ΔΔCt | Ct value (mean ± SD) | 2−ΔΔCt | |
| NTs | 32.510 ± 1.420 | 1 | 29.490 ± 0.790 | 1 |
| CTs | 19.550 ± 0.940 | 0.045* | 16.820 ± 1.400 | 0.039‡ |
| HMs | 15.030 ± 0.830 | 0.022*† | 18.900 ± 0.890 | 0.042‡ |
| PMs | 16.470 ± 0.910 | 0.029*† | 17.370 ± 0.930 | 0.032‡ |
*P<0.05, compared with miR-206 NTs subgroup; †P<0.05, compared with miR-206 CTs subgroup; ‡P<0.05, compared with Cx43 NTs subgroup. miR-206 mRNA in CTs, HMs and PMs was significantly lower than that in NTs (P<0.05), miR-206 mRNA in HMs and PMs was lower than that in CTs (P<0.05). NTs: Normal tissues; CTs: Cancer tissues; HMs: Hepatic metastasis; PMs: Pulmonary metastases; SD: Standard deviation; Cx43: Connexin 43; miR-206: MicroRNA-206; mRNA: Messenger RNA; SD: Standard deviation.
As shown in Figure 1a and 1b, when miR-206 expression in MCF-7 cells was set at 1, miR-206 expression in MCF-10A (normal breast cell line) was 639.130 ± 13.040, significantly higher than that in breast cancer cell lines MCF-7 (1) and MDA-MB-231 (15.540 ± 0.840), while that in MDA-MB-231 was higher than that in MCF-7. Cx43 mRNA in MDA-MB-231 was significantly lower than that in MCF-7 (7.190 ± 0.640) (P = 0.031), [Figure 2c].
Figure 1.
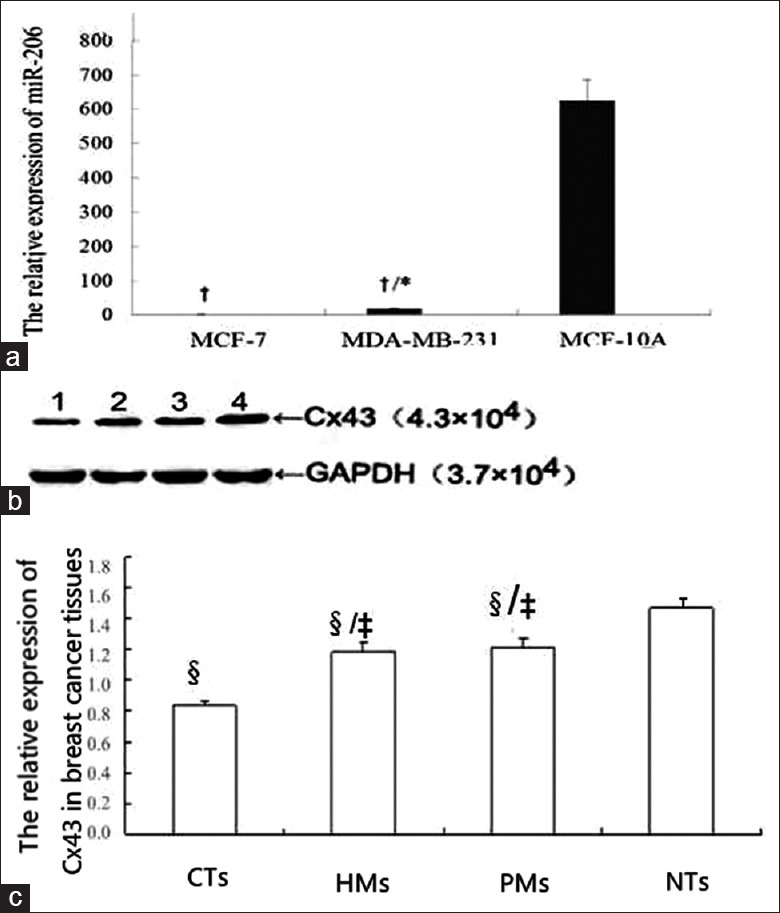
The expression of MiR-206 and connexin 43 in breast tissues and cells. (a): *P < 0.05, compared with MCF-7; †P < 0.05, compared with MCF-10A. When miR-206 expression in MCF-7 cells was set at 1, miR-206 expression in normal breast cell line MCF-10A was 639.13±13.04, significantly higher than that in MCF-7 and MDA-MB-231 (15.54±0.84), while miR-206 expression in MDA-MB-231 was higher than that in MCF-7. (b): Western blotting image; (c): Statistic graph of graph b. Lane 1: breast cancer tissues; lane 2: hepatic metastasis; lane 3: pulmonary metastasis; lane 4: normal breast tissues; NTs: normal breast tissues; CTs: breast cancer tissues; HMs: hepatic metastasis; PMs: Pulmonary metastasis; ‡P < 0. 05, compared with CTs group; §P < 0. 05, compared with NTs group; Cx43 protein expression in CTs, HMs and PMs was significantly lower than that in NTs (P < 0. 05). Cx43 protein expression in HMs and PMs was significantly higher than that in CTs (P < 0. 05).
Figure 2.
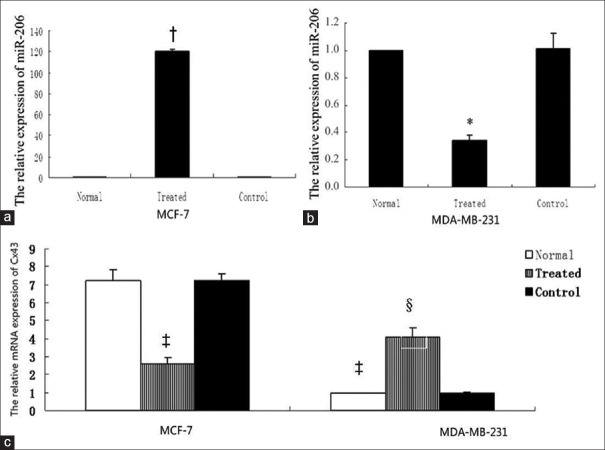
Connexin 43 expression after changing microRNA-206. (a) MCF-7 cells. When microRNA-206 expression in nontransfected cells was set at 1, microRNA-206 in MCF-7 cells was significantly increased to 120.331 ± 1.801 after transfected LV-has-microRNA-206 (1569-11) (P < 0.05), while no statistically significant difference was observed in normal and negative control group (P > 0.05). (b) MDA-MB-231 cells. MicroRNA-206 in MDA-MB-231 cells after LV-has-microRNA-206-inhibition transfection was decreased from 1 to 0.342 ± 0.037 (P < 0.05), while no statistically significant difference was observed in normal and negative control group (P > 0.05). *P < 0.05, compared with normal MDA-MB-231 cells; †P <0.05, compared with normal MCF-7 cells. (c) Connexin 43 mRNA in MDA-MB-231 was significantly lower than that in MCF-7 (7.190 ± 0.640). After MCF-7 cells were transfected with LV-has-microRNA-206 (1569-11), connexin 43 mRNA was decreased from 7.190 ± 0.640 to 2.580 ± 0.340 (P < 0.05), while no significant change in the negative control group (P > 0.05). After MDA-MB-231 cells transfected with LV-has-miR-206-inhibition, connexin 43 mRNA was increased from 1.012±0.035 to 4.256±0.095 (P < 0.05), while no significant change in the negative control group (P > 0.05). ‡P < 0.05, compared with normal MCF-7 cells, §P < 0.05, compared with normal MDA-MB-231.
Connexin 43 protein expression
Cx43 protein expression in CTs, HMs, and PMs was significantly lower than that in NTs (P < 0. 05) [Figure 1c]. Cx43 protein expression in HMs and PMs was significantly higher than that in CTs (P < 0. 05) [Figure 1c], Cx43 protein level in MCF-7 cells was significantly higher than that in MDA-MB-231 cells (P < 0. 05) [Figure 3].
Figure 3.
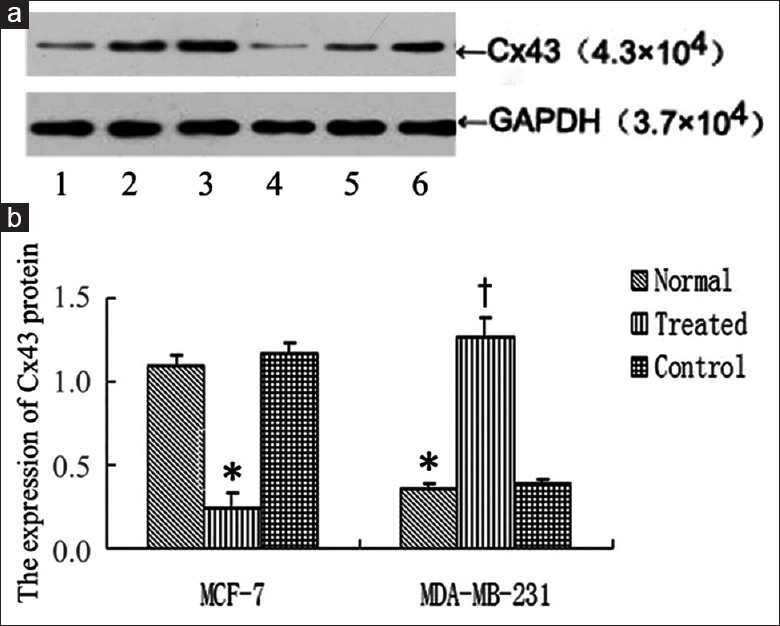
Connexin 43 protein expression after interfering microRNA-206 expression. (a) Western blotting image; (b) Statistic graph of graph a; Lane 1: MDA-MB-231; lane 2: MCF-7; lane 3: Transfected MDA-MB-231 group; lane 4: Transfected MCF-7 group; lane 5: MDA-MB-231 negative control group; lane 6: MCF-7 negative control group. *P < 0.05, compared with normal MCF-7 cells; †P < 0.05, compared with normal MDA-MB-231 cells. Connexin 43 protein level in MCF-7 cells was significantly higher than that in MDA-MB-231 cells. In MCF-7 cells, connexin 43 protein expression in transfection group was significantly lower than that in normal group and negative control group (P < 0.05). In MDA-MB-231 cells, connexin 43 protein expression in transfection group was significantly higher than that in normal group and negative control group (P < 0.05).
MicroRNA-206 expression after lentivirus transfection
Lentiviral transfection efficiency was >80% in all groups. The change of miR-206 expression after lentivirus transfection was shown in Figure 2, when miR-206 expression in nontransfected cells was set at 1, miR-206 in MCF-7 cells transfected by LV-hsa-miR-206 (1569-11) was significantly increased to 120.331 ± 1.801 (P = 0.042), while no statistically significant difference was observed in normal group and negative control group (P > 0.05). miR-206 in MDA-MB-231 cells transfected by LV-hsa-miR-206-inhibition was decreased from 1 to 0.342 ± 0.037 (P = 0.022), while no statistically significant difference was observed in normal group and negative control group (P > 0.05).
Connexin 43 expressions after changing microRNA-206
Lentiviral transfection efficiency was >80% in MCF-7. Lentiviral transfection efficiency was >80% in MDA-MB-231.
In mRNA level, qRT-PCR was used to detect the Cx43 mRNA after altering miR-206 expression. As shown in Figure 2c after MCF-7 cells were transfected with LV-hsa-mir-206 (1569–11), Cx43 mRNA was decreased from 7.190 ± 0.640 to 2.580 ± 0.340 (P = 0.018 < 0.05), while no significant change in the negative control group (P > 0.05). In MDA-MB-231 cells, Cx43 mRNA was increased from 1 to 4.090 ± 0.500 (P = 0.033), no significant change was shown in the negative control group (P > 0.05). In MDA-MB-231 cells, Cx43 mRNA was increased from reference value 1 to 4.090 ± 0.500 (P = 0.011), no significant change was showed in the negative control group (P > 0.05).
In protein level, as shown in Figure 3, Cx43 protein expression in MCF-7 cells was significantly higher than that in MDA-MB-231 cells (P = 0.021) [Figure 3]. In MCF-7 cells, Cx43 protein expression in transfection group was significantly lower than that in normal and negative control group (P = 0.024). In MDA-MB-231 cells, Cx43 protein expression in transfection group was significantly higher than that in normal and negative control group (P = 0.011), [Figure 3].
In correlation analysis of Cx43 and miR-206, the correlation coefficients were as follow: r = −0.591, P = 0.043 in mRNA level; r = −0.724, P = 0.008 in protein level, which showed that miR-206 expression was negatively correlated with Cx43 mRNA and protein expression.
Changes of biological activity after altering microRNA-206
Proliferation capability
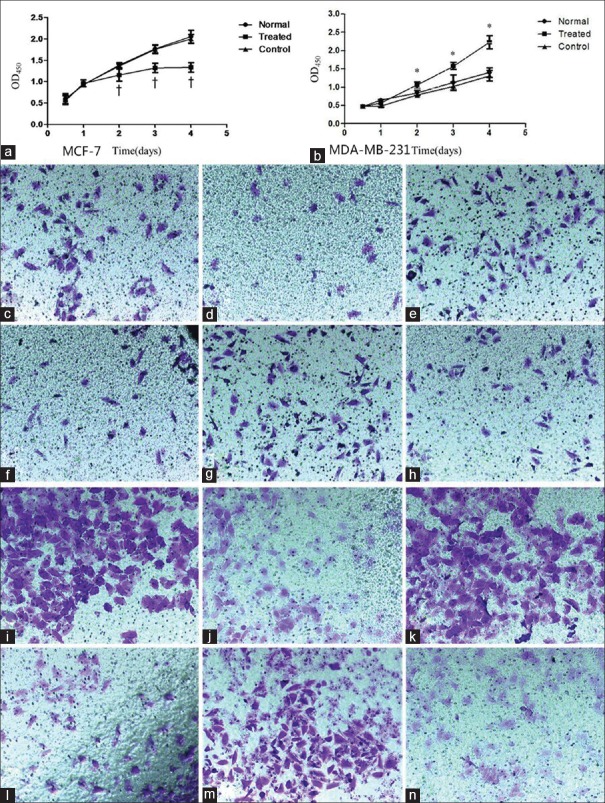
As shown in Figure 5 compared with untransfected cells, 2 days after LV-hsa-mir-206 (1569-11) transfection, an obvious decline in proliferation capability was noticed in MCF-7 cells, while a remarkable elevation was observed in MDA-MB-231 cells (P < 0.05) [Figure 5].
Figure 5.
Changes of biological activity after altering microRNA-206 expression. (a-b): Cell proliferation after altering miR-206 expression. a: MCFc-7 cells †P< 0.05, as compared with the corresponding time point subgroup in normal MCF-7 cells. b: MDA-MB-231 cells *P < 0.05, as compared with the corresponding time point subgroup in normal MDA-MB-231 cells. (c-h): Cell migration after altering miR-206 expression (crystal violet staining, original magnification ×200). c: Normal MCF-7 cells. d: Transfected MCF-7 cells. e: Negative control MCF-7 cells. f: Normal MDA-MB-231 cells. d: Transfected MDA-MB-231 cells. e: Negative control MDA-MB-231 cells. (i-n): Cell invasion after altering miR-206 expression (crystal violet staining, original magnification×200). i : Normal MCF-7 cells. j: Transfected MCF-7 cells. k: Negative control MCF-7 cells. l: Normal MDA-MB-231 cells. m: Transfected MDA-MB-231 cells. n: Negative control MDA-MB-231 cells.
Cell migration
Transwell migration assay showed that the cell count in transfected MCF-7 group was 19.600 ± 3.040 in each field of view, significantly lower than 43.000 ± 7.410 in normal MCF-7 group, suggesting that the migration capability was significantly reduced (P < 0.05). The cell count in the transfected MDA-MB-231 group was 81.800 ± 6.610 in each field of view, significantly higher than 61.200 ± 5.260 in the normal MDA-MB-231 group, suggesting that migration capability was significantly enhanced (P < 0.05) [Figure 5c and 5h].
Cell invasion
As Figure 5i–5n shown, the number of cells passing through Transwell chamber in the transfected MCF-7 group was 48.200 ± 10.010, significantly lower than 88.400 ± 7.230 in the normal MCF-7 group (P < 0.05), indicating that the cells’ invasion activity was decreased after enhancing miR-206 expression. The number of cells passing through Transwell chamber in the transfected MDA-MB-231 group was 123.000 ± 4.690, significantly higher than 99.000 ± 6.240 in the normal MDA-MB-231 group (P < 0.05), indicating that the cells’ invasion activity was increased after reducing miR-206 expression.
Possible binding sites of connexin 43 for microRNA-206
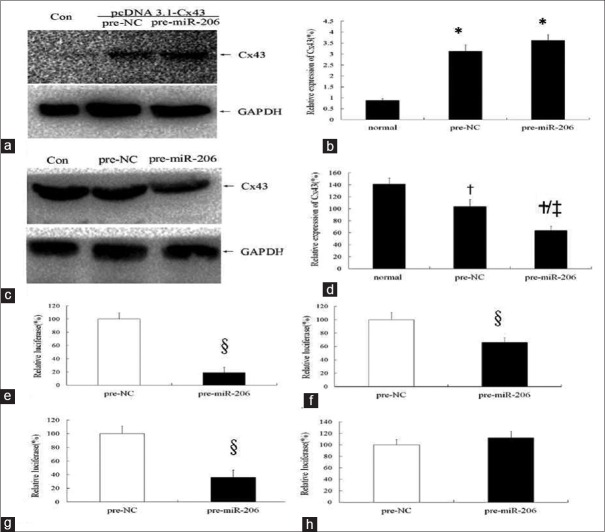
As shown in Figure 4a, Cx43 was not expressed in 293 cells. Cx43 expression became strongly positive (P < 0.05) in 293 cells transfected by eukaryotic expression plasmid pcDNA3.1-Cx43 (containing the Cx43 coding region and lack of Cx43-3’UTR), while Cx43 remained the same after transfected by miR-206. In MCF-7 cells (bearing Cx43 3’UTR) [Figure 4b], Cx43 was highly expressed but significantly decreased after transfected by miR-206 (P < 0.05).
Figure 4.
The possible binding sites of connexin 43 for microRNA-206. (a-b) MicroRNA-206 showed no regulatory effects on connexin 43 of 293 cells lack of 3’UTR. a: Western blotting image; b: Statistic graph of graph a. *P < 0.05, compared with the normal group; pre-NC: Negative control group. Con: Normal group; Pre-NC: Negative control group; Pre-microRNA-206: MicroRNA-206 transfected group. c: Western blotting image; d: Statistic graph of graph c. †P < 0.05, compared with normal group; ‡P < 0.05, compared with negative control group; Con: normal group; Pre-NC: Negative control group; Pre-microRNA-206: MicroRNA-206 transfected group. (e-h) Results of luciferase activity assay. e: wild type of Cx43-3’UTR; f: mutated at the first binding sites in Cx43-3’UTR (position 480-483); g: mutated at the second binding sites in Cx43-3’UTR (position 1611-1614); h: mutated at the first and second binding sites in Cx43-3’UTR (position 480-483 and 1611-1614); pre-NC: negative control group; pre-miR-206: transfected by miR-206 group. 3’UTR: 3’ untranslated region. §ìP < 0.05, compared with pre-NC group. For the wild-type Cx43-3’UTR luciferase reporter plasmid, miR-206 can inhibit the luciferase expression (e, P <0.05). With position 480-483 mutation or 1611-1613 mutation, luciferase expression increased to 65.89% or 35.81%, respectively (f-g, P <0.05). The inhibition disappeared with mutations at positions 480-483 and 1611-1613 (h, P > 0.05).
MiR-206 can regulate Cx43 by binding to specific sites of Cx43 3’UTR [Figure 4].
As shown in Figure 4c–4f, for the wild-type Cx43-3’UTR luciferase reporter plasmid, miR-206 can inhibit the luciferase expression, which decreased to 19.090% of the value in the control group [Figure 4c] (P < 0.05). With position 480–483 mutant, luciferase expression was increased to 65.890%, an increment of 46.800% [Figure 4b], (P < 0.05). With position 1611–1613 mutant, luciferase expression was increased to 35.81%, an increment of 16.72% [Figure 4c], (P < 0.05); the inhibition was disappeared with double mutant at positions 480–483 and 1611–1613 [Figure 4d].
DISCUSSION
MiRNAs are the largest group of posttranscriptional regulatory factor.[12] A variety of miRNA plays a role in regulating cell proliferation, angiogenesis, invasion, and metastasis of breast cancer via targeting genes.[13] For example, miR-125 can inhibit metastasis of breast cancer by ErBb2, miR-145 by RTNK.[12,14] miR-206 gene, located on human chromosome 6p12.2, is the first discovered miRNA associated with breast cancer. Breast CTs show a significant decline of miR-206 and the low miR-206 level is closely correlated with higher clinical stage and shorter overall survival in breast cancer patients.[11] The patients with high miR-206 expression have a remarkable delay of distant metastases and longer metastasis-free survival compared with those with low miR-206 expression.[15] miR-206 expression is obviously reduced in ERα -positive human breast CT, and transfecting miR-206 into human breast cancer cells MDA-MB-231 can inhibit the cells’ growth.[16] It has been acknowledged that as a posttranscriptional regulatory factor, miR-206 can regulate the progression and metastasis of breast cancer, but its target gene is not clear yet. In our previous study, metastatic axillary lymph node showed decreased expression of miR-206 and increased expression of Cx43 protein. According to the results of TargetScan and PicTar, Cx43 has two possible binding sites for miR-206: sites 478–484 and 1609–1615. Cx43 may be one of the target genes of miR-206.[11] This study focused on whether Cx43 is the target gene of miR-206, if so, what are their potential binding sites.
First, we researched the Cx43 expression in primary lesion, HMs, and PM of breast cancer and found that Cx43 protein expression was significantly lower than that in normal breast tissue, and the value of HMs and PM was significantly higher than that in primary breast cancer, whereas mRNA expression presented no significant difference, which was consistent with the results of Kanczuga-Koda et al.[17] Besides the mechanic coupling, gap junction can mediate intercellular transfer of electrical and chemical signals by electrical coupling and chemical coupling, allowing the passage of electrical signals and small molecules or ions with milliroentgen <1000 or the diameter <1.0 nm, including small molecule metabolites, hydrogen, second messengers (Ca2+, inositol triphosphate, cyclic nucleotides), etc.[18] When it is aroused by external or internal stimuli, the body can adapt to this change by altering Cx expression. The decreased Cx43 level which is positively correlated with the permeability of gap junction[19] causes the reduction or loss of intercellular gap junction exchange, leading to the increased cell heterogeneity, which is typical of malignant cancer cells.[20] On the contrary, Cx43 protein expression in hepatic and PMs of breast cancer is significantly higher than that in the primary tumor. Because in distant metastasis, malignant cancer cells need to interact with the surrounding stromal cells and vascular endothelial cells. The body upregulates gap junctions such as Cx43 to increase the information exchange and prepares for the metastasis.[7] Our further study showed that miR-206 mRNA expression in the primary breast tumor was significantly lower than that in NT, so was Cx43. The main role of miR-206 is to suppress gene translation and reduce the protein expression through its 2–8 bases at 5’ end binded to the 3’UTR of the target gene.[9] It indicated that the role of miR-206 in regulating Cx43 expression is not obvious in happen of breast cancer. In the metastatic lesions of liver and lung, miR-206 mRNA expression was significantly lower than that in the primary tumor and Cx43 protein expression was significantly higher than that in the primary tumor, thus miR-206 mRNA expression showed the opposite tendency compare with Cx43 protein expression in hepatic and PM, indicating that miR-206 may play a role in regulating Cx43 expression during breast cancer metastasis.
We chosed normal breast cell line MCF-10A, breast cancer cell lines MCF-7 and MDA-MB-231 as our study object, and observed Cx43 expression and cytobiological changes by interfering miR-206 expression. MDA-MB-231 is a metastatic breast cancer cell line with ERα (-), PR (-), HER-2(-),[21] commonly used in triple negative breast cancer study.[22,23] MCF-7 is a nonmetastatic breast cancer cell line with ERα (+), HER-2 (+).[21] Our study showed that miR-206 expression in MDA-MB-231 or MCF-7 was significantly lower than that in normal breast cells, miR-206 expression in MDA-MB-231 was significantly higher than that in MCF-7, mRNA and protein levels of Cx43 in MDA-MB-231 were lower than those in MCF-7. We constructed lentivirus LV-hsa-miR-206-inhibition and LV-hsa-miR-206 (1569–11), and transfected them into MDA-MB-231 and MCF-7 cells, respectively. Transfected by lentivirus LV-hsa-miR-206-inhibition, miR-206 expression was decreased from 1 to 0.342 ± 0.037 in MDA-MB-231. With lentivirus LV-hsa-miR-206 (1569–11), miR-206 expression was increased from 1 to 120.331 ± 1.801 in MCF-7. Cx43 mRNA and protein, as well as biological behaviors of cells, showed a concomitant change with miR-206, i.e., in MDA-MB-231 cells, miR-206 expression decreased, Cx43 mRNA and protein expression, the proliferation, migration, and invasion capability of cells significantly increased; in MCF-7 cells, miR-206 expression increased, Cx43 expression, the proliferation, migration and invasion capability of cells significantly decreased. Correlation analysis showed a significantly negative correlation between miR-206 and Cx43. We analyzed that under normal conditions, 2–8 bases at 5’ end of miR-206 bind to the target sites of Cx43 mRNA so as to inhibit Cx43 translation and induce degradation, thereby suppressing the metastasis of cancer cells. With the down-regulation of miR-206 mRNA, posttranscriptional regulation of Cx43 mRNA could not be effectively inhibited, so Cx43 expression was upregulated, as well as cell proliferation, migration, and invasion. Cx43 can mediate the adhesion of cancer cells with vascular endothelial cells,[24] change cytoskeleton,[25,26] increase cell movement,[27] thus promoting distal metastasis of breast cancer. These evidences suggest that miR-206 gene may regulate proliferation, invasion and migration of breast cancer cells through negative regulation of Cx43, so as to play a regulatory role in metastasis of breast cancer.
MiRNAs are a new class of small noncoding RNAs that posttranscriptionally regulate the expression of target mRNA transcripts. Basic research has proved that miRNAs play a regulatory role mainly in two aspects: affecting target mRNA cleavage or inhibiting gene translation. It is believed that if miRNAs are perfectly complementary to the noncoding area at 3’ end of mRNA, miRNAs direct specific mRNA cleavage and degradation; if there is less complementarity, miRNAs direct the inhibition of translation, i.e., they only affect protein expression without changing mRNA stability.[28] In this study, changes of miR-206 expression affect both mRNA and protein levels of Cx43 in cell lines, indicating that both of regulation mechanisms exist in the cell lines. However, no Cx43 mRNA change was found in breast cancer specimens, suggesting that the actual regulation of Cx43 protein may be achieved through the latter mechanism.
To further clarify the interaction between miR-206 and Cx43, we constructed eukaryotic expression plasmid pcDNA3.1-Cx43 (containing the coding region of Cx43 and lack of Cx43-3’UTR) and transfected it into 293 cells (not expressing Cx43) to highly express Cx43. Then, we transfected pre-miR-206 into normal MCF-7 cells and 293 cells transfected by pcDNA3.1-Cx43 to observe the Cx43 expression. The results demonstrated that miR-206 can significantly decrease Cx43 expression in MCF-7 cells but take no effect on 293 cells. The difference between 293 cell transfected by pcDNA3.1-Cx43 and normal MCF-7 is a lack of Cx43-3’UTR in 293 cells. It suggested that Cx43-3’UTR may play a key role in miR-206 regulating on Cx43. The bioinformatics analysis further showed that Cx43-3’UTR contains two potential sites binding to miR-206:478–484 and 1609–1615. Therefore, based on pMIR-REPORT Luciferase, using serial PCR clone and mutation techniques, we constructed 4 plasmids (wild type of Cx43-3’UTR, Cx43-3’UTR BS-1 mut, Cx43-3’UTR BS-2 mut, and Cx43-3’UTR BS-1, 2 mut) to determine the influence of mutants of different binding sites on luciferase expression. The results showed that the inhibition rate was reduced by 46.80% with position 478–484 mutant, 16.72% with position 1609–1615 mutant, completely disappeared with double mutant at the positions 478–484 and 1609–1615. These results indicated that the negative regulation of Cx43 by miR-206 is realized via two specific sites of Cx43-3’UTR, in which position 478–484 contributes more than position 1609–1615 does.
In summary, we have shown that miR-206 is dramatically down-regulated in breast CT compared with normal breast tissues and lower expression of miR-206 in breast cancer metastatic tissues is associated with higher expression of Cx43 and a more advanced activity of breast cancer cell. Moreover, up-regulation of miR-206 suppresses breast cancer cell proliferation and metastasis by blocking the expression of Cx43. MiR-206 may take effect through positions 478–484 and 1609–1615 of Cx43-3’UTR, in which position 478–484 is more responsible than position 1609–1615. Our experimental data may provide a strategy for targeting miR-206/Cx43 interaction in a novel therapeutic application to treat breast cancer patients.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Li-Min Chen
REFERENCES
- 1.Coughlin SS, Ekwueme DU. Breast cancer as a global health concern. Cancer Epidemiol. 2009;33:315–8. doi: 10.1016/j.canep.2009.10.003. doi: 10.1016/j.canep. 2009.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. doi: 10.1158/1055-9965. [DOI] [PubMed] [Google Scholar]
- 3.Lin SX, Chen J, Mazumdar M, Poirier D, Wang C, Azzi A, et al. Molecular therapy of breast cancer: Progress and future directions. Nat Rev Endocrinol. 2010;6:485–93. doi: 10.1038/nrendo.2010.92. doi: 10.1038/nrendo.2010.92. [DOI] [PubMed] [Google Scholar]
- 4.Conklin CM, Bechberger JF, MacFabe D, Guthrie N, Kurowska EM, Naus CC. Genistein and quercetin increase connexin43 and suppress growth of breast cancer cells. Carcinogenesis. 2007;28:93–100. doi: 10.1093/carcin/bgl106. doi: 10.1093/carcin/bgl106. [DOI] [PubMed] [Google Scholar]
- 5.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–87. doi: 10.1158/1078-0432.CCR-09-1532. doi: 10.1158/1078-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elzarrad MK, Haroon A, Willecke K, Dobrowolski R, Gillespie MN, Al-Mehdi AB. Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med. 2008;6:20. doi: 10.1186/1741-7015-6-20. doi: 10.1186/1741-7015-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanczuga-Koda L, Sulkowski S, Lenczewski A, Koda M, Wincewicz A, Baltaziak M, et al. Increased expression of connexins 26 and 43 in lymph node metastases of breast cancer. J Clin Pathol. 2006;59:429–33. doi: 10.1136/jcp.2005.029272. doi: 10.1136/jcp.2005.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petri A, Lindow M, Kauppinen S. MicroRNA silencing in primates: Towards development of novel therapeutics. Cancer Res. 2009;69:393–5. doi: 10.1158/0008-5472.CAN-08-2749. doi: 10.1158/0008-5472. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–48. doi: 10.1038/onc.2009.406. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 10.Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–87. doi: 10.1038/sj.onc.1209912. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Hong F, Yu Z. Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res. 2013;41:596–602. doi: 10.1177/0300060513485856. doi: 10.1177/0300060513485856. [DOI] [PubMed] [Google Scholar]
- 12.Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG. MicroRNA, cell cycle, and human breast cancer. Am J Pathol. 2010;176:1058–64. doi: 10.2353/ajpath.2010.090664. doi: 10.2353/ajpath.2010.090664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–58. doi: 10.1016/S1470-2045(12)70073-6. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 15.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–8. doi: 10.1158/0008-5472.CAN-08-0180. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 17.Kanczuga-Koda L, Sulkowski S, Tomaszewski J, Koda M, Sulkowska M, Przystupa W, et al. Connexins 26 and 43 correlate with Bak, but not with Bcl-2 protein in breast cancer. Oncol Rep. 2005;14:325–9. doi: 10.3892/or.14.2.325. [PubMed] [Google Scholar]
- 18.Parthasarathi K, Ichimura H, Monma E, Lindert J, Quadri S, Issekutz A, et al. Connexin 43 mediates spread of Ca2+ dependent proinflammatory responses in lung capillaries. J Clin Invest. 2006;116:2193–200. doi: 10.1172/JCI26605. doi: 10.1172/JCI26605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg GS, Moreno AP, Lampe PD. Gap junctions between cells expressing connexin 43 or 32 show inverse permselectivity to adenosine and ATP. J Biol Chem. 2002;277:36725–30. doi: 10.1074/jbc.M109797200. doi: 10.1074/jbc.M109797200. [DOI] [PubMed] [Google Scholar]
- 20.Oktem G, Bilir A, Ayla S, Yavasoglu A, Goksel G, Saydam G, et al. Role of intercellular communications in breast cancer multicellular tumor spheroids after chemotherapy. Oncol Res. 2006;16:225–33. doi: 10.3727/000000006783981071. doi: 10.3727/000000006783981071. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Chen D, Wang J, Yin Y, Gao Q, Zhang Y. Downregulation of the transcription factor, FoxD3, is associated with lymph node metastases in invasive ductal carcinomas of the breast. Int J Clin Exp Pathol. 2014;7:670–6. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30:4437–46. doi: 10.1038/onc.2011.145. doi: 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 23.Stratford AL, Reipas K, Hu K, Fotovati A, Brough R, Frankum J, et al. Targeting p90 ribosomal S6 kinase eliminates tumor-initiating cells by inactivating Y-box binding protein-1 in triple-negative breast cancers. Stem Cells. 2012;30:1338–48. doi: 10.1002/stem.1128. doi: 10.1002/stem.1128. [DOI] [PubMed] [Google Scholar]
- 24.Cotrina ML, Lin JH, Nedergaard M. Adhesive properties of connexin hemichannels. Glia. 2008;56:1791–8. doi: 10.1002/glia.20728. doi: 10.1016/j.yjmcc.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, Francis R, Wei CJ, Linask KL, Lo CW. Connexin 43-mediated modulation of polarized cell movement and the directional migration of cardiac neural crest cells. Development. 2006;133:3629–39. doi: 10.1242/dev.02543. doi: 10.1242/dev.02543. [DOI] [PubMed] [Google Scholar]
- 26.Homkajorn B, Sims NR, Muyderman H. Connexin 43 regulates astrocytic migration and proliferation in response to injury. Neurosci Lett. 2010;486:197–201. doi: 10.1016/j.neulet.2010.09.051. doi: 10.1016/j.neulet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 27.Behrens J, Kameritsch P, Wallner S, Pohl U, Pogoda K. The carboxyl tail of Cx43 augments p38 mediated cell migration in a gap junction-independent manner. Eur J Cell Biol. 2010;89:828–38. doi: 10.1016/j.ejcb.2010.06.003. doi: 10.1016/j.ejcb.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]