ABSTRACT
The death and lysis of a subpopulation in Staphylococcus aureus biofilm cells are thought to benefit the surviving population by releasing extracellular DNA, a critical component of the biofilm extracellular matrix. Although the means by which S. aureus controls cell death and lysis is not understood, studies implicate the role of the cidABC and lrgAB operons in this process. Recently, disruption of the srrAB regulatory locus was found to cause increased cell death during biofilm development, likely as a result of the sensitivity of this mutant to hypoxic growth. In the current study, we extended these findings by demonstrating that cell death in the ΔsrrAB mutant is dependent on expression of the cidABC operon. The effect of cidABC expression resulted in the generation of increased reactive oxygen species (ROS) accumulation and was independent of acetate production. Interestingly, consistently with previous studies, cidC-encoded pyruvate oxidase was found to be important for the generation of acetic acid, which initiates the cell death process. However, these studies also revealed for the first time an important role of the cidB gene in cell death, as disruption of cidB in the ΔsrrAB mutant background decreased ROS generation and cell death in a cidC-independent manner. The cidB mutation also caused decreased sensitivity to hydrogen peroxide, which suggests a complex role for this system in ROS metabolism. Overall, the results of this study provide further insight into the function of the cidABC operon in cell death and reveal its contribution to the oxidative stress response.
IMPORTANCE The manuscript focuses on cell death mechanisms in Staphylococcus aureus and provides important new insights into the genes involved in this ill-defined process. By exploring the cause of increased stationary-phase death in an S. aureus ΔsrrAB regulatory mutant, we found that the decreased viability of this mutant was a consequence of the overexpression of the cidABC operon, previously shown to be a key mediator of cell death. These investigations highlight the role of the cidB gene in the death process and the accumulation of reactive oxygen species. Overall, the results of this study are the first to demonstrate a positive role for CidB in cell death and to provide an important paradigm for understanding this process in all bacteria.
INTRODUCTION
A fundamental aspect of bacterial biofilms is the synthesis of self-produced extracellular matrix molecules that are critical for intercellular adherence and binding to surfaces (1). These molecules are diverse in nature and include specific carbohydrates, proteins, and extracellular DNA (eDNA). In Staphylococcus aureus, it has been demonstrated that eDNA is released as a consequence of the death and lysis of a subpopulation of cells in the biofilm via a process termed bacterial programmed cell death (PCD) (2, 3). The surviving cells derive benefit from the death of the subpopulation by using the eDNA as a matrix molecule (4). Although the mechanism(s) controlling cell death remains to be elucidated, this process is known to involve the products of the cidABC and lrgAB operons, which share many features in common with the regulatory components known to regulate PCD in more complex eukaryotic organisms (5–7).
The current model for the roles of the cidABC and lrgAB operons in cell death is based on our previous studies of mutants that disrupt this process (8–10). For example, we have demonstrated that disruption of the cidA gene resulted in decreased antibiotic-induced death and lysis (9), whereas disruption of the lrgAB operon had the opposite effect (8). Furthermore, CidA and LrgA were found to share sequence similarities with bacteriophage holin proteins with well-known roles in the control of death and lysis (9). Based on these findings, it was proposed that CidA and LrgA represent a bacterial holin-antiholin system, which is the foundation of bacterial PCD (11). In this model, CidA oligomerizes and forms pores in the cytoplasmic membrane, leading to membrane depolarization, activation of murein hydrolase activity, and cell lysis (7). LrgA is envisioned as an antiholin (8), opposing the activity of CidA by interfering with its ability to depolarize the membrane and cause subsequent death and lysis (9). Thus, the balance between CidA and LrgA is thought to determine whether a cell will live or die. Indeed, CidA and LrgA have been shown to associate with the cytoplasmic membrane and to oligomerize into high-molecular-weight complexes (10). More recently, cidC-encoded pyruvate oxidase (12) was shown to potentiate cell death during the stationary phase and biofilm development by promoting cytoplasmic acidification through the production of acetate (13). Thus, we hypothesize that the cid operon mediates cell death via a complex process involving the metabolic potentiation of the cell, followed by the activation of a holin-like complex associated with the cytoplasmic membrane.
The regulation of cidABC expression is known to be mediated by CidR, which is required for the induction of transcription during growth in excess glucose (9) or low oxygen (14). Recent studies have also demonstrated that cidABC expression is influenced by the SrrAB two-component system, which is known to be important for anaerobic growth and survival (15). Disruption of srrAB resulted in enhanced cell death and decreased biofilm thickness (16, 17), likely as a result of the role that this regulatory system has in adaptation to an anaerobic environment. In the current study, we demonstrated that the increased death exhibited by the ΔsrrAB mutant is a function of increased cidABC expression caused by the ΔsrrAB mutation. However, in contrast to our expectations, the increased death that was observed was not a function of increased acetate accumulation. Rather, death was associated with cidB-dependent increased reactive oxygen species (ROS) accumulation. Thus, these results are the first to demonstrate a positive role for CidB in cell death.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the bacterial strains used in this study are listed in Table 1. S. aureus strains were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI). Escherichia coli DH5α was grown in Luria-Bertani medium (Fisher Scientific). Liquid S. aureus cultures were grown in Erlenmeyer flasks at 37°C with shaking (250 rpm) in a volume that was no greater than 10% of the flask volume. All antibiotics were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the following concentrations: kanamycin, 50 μg ml−1; chloramphenicol, 10 μg ml−1; ampicillin, 100 μg ml−1; and erythromycin, 5 μg ml−1.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| S. aureus | ||
| UAMS-1 | Clinical osteomyelitis isolate, rsbU+ | 34 |
| RN4220 | Highly transformable strain; restriction deficient | 35 |
| KB1058 | UAMS-1 ΔcidC | 13 |
| KB1060 | UAMS-1 ΔcidB, markerless | This study |
| KB1065 | UAMS-1 ΔcidAΔ2–52, markerless | This study |
| KB6004 | UAMS-1 ΔsrrAB, markerless | 20 |
| KB6005 | UAMS-1 ΔsrrAB ΔcidABC, markerless | This study |
| KB6006 | UAMS-1 ΔsrrAB ΔcidA, markerless | This study |
| KB6007 | UAMS-1 ΔsrrAB ΔcidB, markerless | This study |
| KB6008 | UAMS-1 ΔsrrAB ΔcidC, markerless | This study |
| E. coli | ||
| BL21 | Expression strain, F– ompT hsdS (rB− mB−) gal dcm (DE3) | Novagen |
| DH5a | Host strain for construction of recombinant plasmids | 36 |
DNA manipulations.
Genomic DNA was isolated from S. aureus using the Wizard Plus kits from Promega, Inc. (Madison, WI). Restriction endonucleases, DNA polymerases, and the T4 DNA ligase used in this study were purchased from either New England BioLabs (Beverly, MA) or Invitrogen Life Technologies (Carlsbad, CA). Recombinant plasmids (listed in Table 2) were isolated using the Wizard Plus SV Minipreps DNA purification system (Promega Corporation, Madison, WI). PCR was performed using primers (listed in Table 3) purchased either from Integrated DNA Technologies (Coralville, IA) or from Eurofins Operon (Louisville, KY), a KOD polymerase kit (Novagen, Madison, WI), and an Applied Biosystems GeneAmp PCR system 9700 (Life Technologies Corporation, Carlsbad, CA). DNA fragments were recovered using the DNA Clean and Concentrator-5 kit (Zymo Research, Orange, CA), and recombinant DNA plasmid products were sequenced at the University of Nebraska Medical Center and analyzed using Vector NTI (Invitrogen, Carlsbad, CA).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| pAJ22 | β-Galactosidase reporter plasmid; Cmr | 25 |
| pCL52.2 | Temp-sensitive shuttle vector; Tcr Spr | 19 |
| pCM12 | E. coli/S. aureus shuttle vector with PsarA-sodARBS-gfp (superfolder); Ampr Spr | 24 |
| pCN51 | Pcad-inducible plasmid; Ampr Ermr | 22 |
| pET24b | IPTG-inducible E. coli expression plasmid | Novagen |
| pET24b-srrA | pET24b with gene encoding SrrA | This study |
| pIHW5 | Pcad-inducible lacZ reporter plasmid; Ampr Ermr | This study |
| pIHW7 | Promoterless lacZ reporter plasmid; Ampr Ermr | This study |
| pIHW10lac | cidABC reporter plasmid; Ampr Ermr | This study |
| pIHW58 | srrAB complementation plasmid; Tcr Spr | This study |
| pJB12 | Temp-sensitive UAMS-1 ΔcidB plasmid (pCL52.2) | This study |
| pJB60 | Temp-sensitive allelic-exchange plasmid with counterselection; Ampr Cmr | This study |
| pJB60-114115 | Temp-sensitive UAMS-1 ΔcidABC plasmid; Ampr Cmr | This study |
| pJB61 | Temp-sensitive allelic-exchange plasmid with counterselection; Ampr Cmr | This study |
| pJB67 | pCN51 with TIR; Ampr Ermr | This study |
| pJB94 | E. coli/S. aureus shuttle vector; Tcr Spr | This study |
| pJB97 | UAMS-1 cidB complement plasmid; Ampr Ermr | This study |
| pRN8298 | pI258 replicon; Ampr Ermr | 22 |
TIR, optimized transcription initiation region.
TABLE 3.
Primers used in this studya
| Primer | Sequence (5′ to 3′) |
|---|---|
| IW3 | gcccgggATGACCATGATTACGGATTCACTGGCCGTC |
| IW4 | gggcgcgccTTATTTTTGACACCAGACCAACTGGTAATGG |
| IW11 | ggctagCGCCATCCCTTTCTAAATATGTCTAAATTGTTAC |
| IW15 | ggatccCATATTAATAAAGCACTCATTATTTGTGATTCC |
| IW91c | gggaattcGAACAGCGTAGCCAACAATTAATTACTACTGA |
| IW92c | ggggatccACATGCTTTTCTTTACAAAAGTATTATATCAC |
| IW93c | ggggatccTAAAATTGAATATAGTTATTTCAGAACGCATG |
| IW94c | ggtctagaGTAATTGTCTTTAGTGCTAAATAAAGTTGTAA |
| IW100 | GACGCCTCATGAAGTAAAAGTGATGCGTCA |
| IW101 | ATAGTTGATATTCGCAAAAACCCTAAACCC |
| IW114 | ggctagcTGATTGAAAGGTTATCACAATTGAATTGAA |
| IW115 | ggggtcgacCCAGAACGGTGAATAGAAAATATGATGTAA |
| IW118cc | ggctagcGTGGGAGGTATGACCTGTATGTCGAACG |
| IW119ccc | gctcgagTTTAGCCGGCTCATCATTAGATTTAACCTCAAATTTATACC |
| IW138 | ggagctcACAATAACAGAAGGTCGTAATCGTCAAGTC |
| IW139 | gggatccTTATTCTGGTTTTGGTAGTTTAATAATAAA |
| IW147 | ggggatccGTGGGAGGTATGACCTGTATGTCGAACG |
| IW152 | ggggtcgacTTTAGCCGGCTCATCATTAGATTTAACCTCAAATTTATACC |
| IW154 | GCTTATGAACTTGCAATGGAG |
| IW155 | CAGTTGATACTCATGTTAAACGAC |
| IW156 | GGGCTCATCTCAAACATATATTTTG |
| IW157 | CGGAAATGCGTGATTTAGAAATG |
| IW168 | Biotin- ATAGTTATTGTAACAATTTAGACATATTTAGAAAGGGATGGCGCCATGCACAAAGTCCAA |
| IW169 | TTGGACTTTGTGCATGGCGCCATCCCTTTCTAAATATGTCTAAATTGTTACAATAACTAT |
| IW170 | ATAGTTATTGTAACAATTTAGACATATTTAGAAAGGGATGGCGCCATGCACAAAGTCCAA |
| JBGFP4 | CGAATTCTTATTTGTAGAGCTCATCCATGCCATGTG |
| JBGFP8 | CCATATGCCCGGGagcaaaggagaagaacttttcactgg |
| JBI258ORI1 | ccagatcTGGCGAATGGCGCCGTTTTATCTTCATCAC |
| JBI258ORI2 | ggtgtacaGGGCCCTCGATGATTACCAGAAGTTCTCAC |
| JBCACOMP1 | cggatCCGCATGCAAATTATCAATGATGAAGTAGATATAGGC |
| JBCDALAC3 | cgtcgacCCATGCTTGTAATGCTTTAACTAATGCTTC |
All primers were generated in this study. Lowercase letters indicate nonhomologous regions of DNA.
Chromosomal mutations were generated as previously described (18). Briefly, an in-frame deletion plasmid for cidB was generated by amplifying approximately 1,000 bp of DNA flanking the cidB gene from the UAMS-1 chromosome. Subsequent cloning of the PCR products into pCL52.2 (19) yielded a plasmid, pJB12, in which the cidB gene is replaced by an XhoI site. A similar strategy was used for cidA. Due to the presence of a cidBC promoter in the cidA open reading frame, an in-frame deletion of cidA was constructed such that the 5′ 150 bp (38%) of the cidA gene was removed, but the cidBC promoter was left intact. This construct produces a CidA protein lacking the first 50 amino acids after the start codon. The cidC mutant was published previously (12). The ΔsrrAB knockout strain was constructed as described previously (20). To create a markerless cidABC knockout strain, UAMS-1 genomic DNA was used as a template for PCR using the primers IW114 and IW115 (Table 3) to isolate the 3′ region of the cidABC operon. The PCR products were digested with SalI and NheI and were ligated into the vector pJB60 to produce the cidABC knockout plasmid pJB60-114115. Deletion of the cidABC operon in UAMS-1 was then generated by allelic replacement as described previously (21).
An srrAB complementation plasmid was generated by PCR amplification of the low-copy-number pI258 replicon from pRN8298 (22) using the primers JBI258ORI1 and JBI258ORI2. The PCR product was then ligated into the BglII and BsrGI sites of pCL52.2 (23), replacing the temperature-sensitive origin of replication to create the plasmid pJB94. The primers IW138 and IW139 were used to amplify the srrAB operon, including its promoter region, using UAMS-1 genomic DNA as a template. The PCR product was then ligated into the SmaI site of pJB94 in a blunt-end ligation reaction. The resulting plasmid, pIHW58, was confirmed to contain the proper DNA insert by DNA sequencing. A cidB complement plasmid was generated by PCR amplification of cidB from the UAMS-1 chromosome using the primers JBCACOMP1 and JBCDALAC3. The PCR product was digested with SphI and was ligated into the same site of pCN51 to produce the plasmid pJB97.
The lacZ reporter plasmid to monitor cidABC expression was created by PCR amplification of the gfp gene of pCM12 (24) using primers JBGFP4 and JBGFP8. The resulting PCR was cloned into the NdeI and EcoRI sites of pJB51, a derivative of pCN51 containing an optimized ribosome binding site (RBS), to yield the plasmid pJB66. An internal EcoRI digestion was performed, which removed the gfp gene, leaving the desired construct, pJB67. The lacZ gene in pAJ22 (25) was amplified by PCR using the primers IW3 and IW4 and was cloned into the SmaI and AscI restriction sites of pJB67 to produce the plasmid pIHW5. The cadmium-inducible promoter of pIHW5 was removed by digestion with SphI and PstI and treatment with the Klenow fragment and was self-ligated to produce the lacZ vector plasmid pIHW7. The cidABC promoter was PCR amplified from the UAMS-1 genome using the primers IW11 and IW15 and was cloned into the BamHI- and NheI-cut sites to yield the final reporter product, pIHW10lac.
β-Galactosidase assays.
β-Galactosidase assays were performed based on protocols previously described (26). Briefly, 1.0 ml of cell cultures was centrifuged, and the cell pellet was resuspended in 1 ml Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) and was disrupted using a FastPrep FP120 (MP Biomedicals, Santa Ana, CA). Cellular debris was pelleted by centrifugation, and 700 μl of the supernatant was transferred to a 1.5-ml microcentrifuge tube. After the addition of 140 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) (4 mg ml−1), the samples were incubated at 37°C until they turned slightly yellow (under an optical density at 420 nm [OD420] of 1.0). To stop the reactions, 500 μl of 1 M sodium carbonate was added, and the OD420 was measured. Protein concentrations were determined by performing Bradford assays using the protein assay dye solution (Bio-Rad, Hercules, CA). Miller units were calculated using protein concentration instead of the OD600 (27).
Expression of recombinant SrrA.
To produce purified SrrA for analysis, the srrA gene was PCR amplified using the primers IW147 and IW152 and UAMS-1 genomic DNA as a template. The PCR product was ligated into the NheI and XhoI sites of pET24b, and the DNA sequence was confirmed by sequencing the cloned product. The resulting plasmid (pET24b-srrA) was transformed into BL21 (Novagen, Madison, WI) for protein expression.
To express and purify SrrA, the BL21(pET24b-srrA) strain was grown in 1 liter of LB medium containing kanamycin at 37°C until the culture reached mid-exponential phase. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM, and srrA was expressed at 30°C overnight. The cells were collected by centrifugation and were resuspended in 50 ml lysis buffer (100 mM phosphate and 300 mM NaCl) containing phenylmethylsulfonyl fluoride (PMSF) (1.0 mM) to inhibit protease activity. Cells were lysed via serial passage through an EmulsiFlex-C3. The protein was then purified using HisPur Cobalt purification kit columns (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. The protein was then desalted via ultrafiltration using an exchange buffer (100 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM EDTA, and 0.1 mM dithiothreitol [DTT]). The purified protein was stored at −20°C in an exchange buffer containing 40% glycerol.
Protein-DNA interactions.
The binding of purified proteins to target DNA was demonstrated by electrophoretic mobility shift assays (EMSAs) using a LightShift chemiluminescence kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. The target DNA used for the SrrA EMSAs was made by annealing the 60-bp primers IW168 and IW169. The binding reaction mixture for each sample contained 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 1 μg of salmon sperm DNA, 5 mM MgCl2, 0.05% NP-40, 10 mM EDTA, and 5 fmol of labeled DNA in a total volume of 20 μl. Competitor DNA was added in 200-fold excess according to the manufacturer's instructions. After 30 min of incubation at room temperature, the protein-DNA mixtures were separated in a 6% Tris-borate-EDTA (TBE) gel in 0.5× TBE buffer at 85 V. The DNA was then transferred from the gel to a nylon membrane and was cross-linked to the nylon membrane using a UV Stratalinker 1800 (Stratagene) cross-linker instrument. The labeled DNA fragments were then imaged using an SRX-101a imager (Konica Minolta, Wayne, NJ).
Flow cytometry.
Flow cytometry was performed as previously described (13). Briefly, a BD LSR II flow cytometer (Becton and Dickinson, San Jose, CA) was used to perform analyses using 1- and 3-day-old stationary-phase cultures of S. aureus. Cell samples were washed twice and diluted to a final concentration of 107 cells/ml in phosphate-buffered saline (PBS) and were then stained for 30 min with 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) (5 mM) and 3′-(p-hydroxyphenyl)fluorescein (HPF) (15 mM). Fluorescence-activated cell sorter (FACS) analyses were performed at a flow rate of 1,000 cells/s. A total of 10,000 events were collected for each sample. Bacteria were discriminated from the background using a combination of forward-scattered light and side-scattered light. Samples were excited at 488 nm using an argon laser, and HPF emission was detected at 530 ± 30 nm (with a 505-nm long-pass mirror), whereas CTC emission was detected at 695 ± 40 nm (with a 685-nm long-pass mirror). Raw data were analyzed using FlowJo software.
Metabolite analyses.
For these analyses, bacterial growth was allowed to proceed at 37°C and 250 rpm in flasks containing TSB (35 mM glucose) in a 1:10 flask-to-volume ratio. Metabolite excretion profiles were determined from culture supernatants that were harvested at 1 and 3 days of incubation. Acetate was measured using commercial kits (R-Biopharm, Marshall, MI) according to the manufacturer's instructions.
RESULTS
Disruption of srrAB enhances weak acid-dependent death.
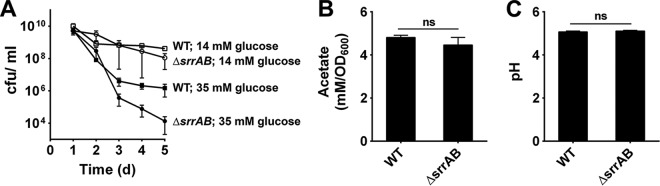
Previous studies have demonstrated a pronounced cell death phenotype associated with the ΔsrrAB mutation (16, 17). Given that transcription profiling experiments suggested that the SrrAB regulatory system has a negative effect on cidABC transcription (17) and that medium acidification due to the accumulation of acidic fermentative metabolites can influence cell fate (13), we initially tested the hypothesis that increased cidC-dependent acetate production was responsible for the decreased survival of an S. aureus ΔsrrAB mutant relative to that of the wild-type strain. Thus, we monitored the survival of the wild-type strain and the ΔsrrAB mutant in stationary phase following growth in tryptic soy broth (TSB) supplemented with excess glucose (35 mM) as previously described (13). Indeed, the ΔsrrAB mutant exhibited an increased rate of cell death in the presence of 35 mM glucose compared to that in the wild-type strain, but it demonstrated similar survival in the stationary phase when grown in the presence of 14 mM glucose (Fig. 1A), indicating that acidification is necessary for cell death to occur. Additionally, consistent with a role for acetate in modulating the rate of cell death associated with the ΔsrrAB mutant during stationary phase, cell viability of the wild-type and mutant strains was dramatically increased when grown in glucose-supplemented medium buffered to a pH of 7.4 with 50 mM morpholinepropanesulfonic acid (MOPS) (see Fig. S1A in the supplemental material). This is most likely due to the inability of acetate (pKa = 4.8) to permeate cells and acidify the cytoplasm under relatively neutral conditions (13). Much to our surprise, however, measurements of acetate production and culture pH revealed that the ΔsrrAB mutant produced nearly identical amounts of acetate compared to that of the wild-type strain (Fig. 1B) and exhibited similar culture pH values (Fig. 1C) when grown in the presence of 35 mM glucose. Thus, in contrast to our initial hypothesis, these data indicate that the decreased viability of the ΔsrrAB mutant during the stationary phase is not due to the increased production and release of acetate.
FIG 1.
Acidic conditions adversely affect the survival of the ΔsrrAB mutant in stationary phase. (A) S. aureus UAMS-1 (wild type [WT]) and the isogenic ΔsrrAB mutant cell viabilities (mean ± standard error of the mean [SEM]) were monitored every 24 h over a period of 5 days (d) in TSB plus 14 mM glucose or TSB plus 35 mM glucose. Cultures were grown at 37°C under aerobic conditions. (B) Acetate levels that accumulated in S. aureus UAMS-1 (WT) and the isogenic ΔsrrAB mutant culture supernatants were measured after 24 h of growth in TSB plus 35 mM glucose using a commercially available kit (R-Biopharm). (C) pH values of the culture supernatants from panel B. ns, differences were not significant.
Decreased ΔsrrAB mutant viability is dependent on enhanced cidABC transcription.
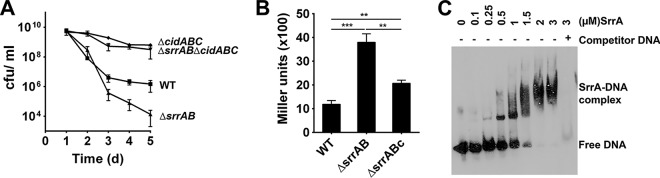
Although the increased death of the ΔsrrAB mutant in stationary phase did not correlate with acetate excretion, cidABC expression was still involved in this process. As shown in Fig. 2A, survival of the ΔsrrAB mutant during stationary phase was dramatically improved by disruption of the cidABC operon to levels similar to that associated with cells containing the ΔcidABC mutation alone, as well as to those observed previously for an S. aureus strain containing the ΔcidC deletion alone (13). To confirm that disruption of srrAB results in increased cidABC transcription, we engineered a reporter construct, pIHW10lac, that contained the cidABC promoter fused to a lacZ reporter. The wild-type and the ΔsrrAB mutant strains containing the reporter plasmid were grown in medium containing 35 mM glucose and were assayed for β-galactosidase activity. As shown in Fig. 2B, ΔsrrAB disruption resulted in an approximately 4-fold increase in cidABC expression compared to that in the wild-type strain, which is consistent with previously published results (17). Complementation of srrAB in trans under the control of its native promoter restored cidABC promoter activity to wild-type levels (Fig. 2B). To determine whether SrrA directly binds the cidABC promoter region, we performed electrophoretic mobility shift assays (EMSAs) (Fig. 2C). C-terminal His-tag-labeled SrrA was affinity purified and was incubated with 60-bp biotin-labeled DNA fragments of the cidABC promoter region, including the putative SrrA binding site (see Fig. S1B in the supplemental material). Analysis of the EMSAs clearly showed that SrrA bound the target DNA in a dose-dependent manner. Furthermore, the addition of a 200-fold excess of unlabeled specific competitor DNA effectively blocked the formation of higher-order SrrA-DNA complexes, indicating that the binding of SrrA to the target DNA was specific. Taken together, these data demonstrate that SrrA acts as a direct repressor of cidABC transcription and that the decreased viability of the ΔsrrAB mutant in stationary phase is a function of enhanced cidABC expression but not a result of increased production of CidC-mediated acetate excretion into the culture medium.
FIG 2.
The ΔsrrAB phenotype requires srrAB-dependent cidABC expression. (A) S. aureus UAMS-1 (WT) and isogenic ΔsrrAB, ΔcidABC, and ΔsrrAB ΔcidABC mutant cell viabilities (mean ± SEM) were monitored every 24 h over a period of 5 days in TSB plus 35 mM glucose. Cultures were grown at 37°C under aerobic conditions. (B) S. aureus cells containing a cidABC promoter fused to lacZ were grown to postexponential phase and assayed for β-galactosidase activity. (C) Electrophoretic mobility shift assays (EMSAs) were performed using increasing amounts of purified SrrA protein and biotin-labeled cidABC promoter DNA as a target. Reaction mixtures were incubated for 30 min at room temperature and separated in a 6% TBE polyacrylamide gel.
The ΔsrrAB mutant generates increased levels of reactive oxygen species.
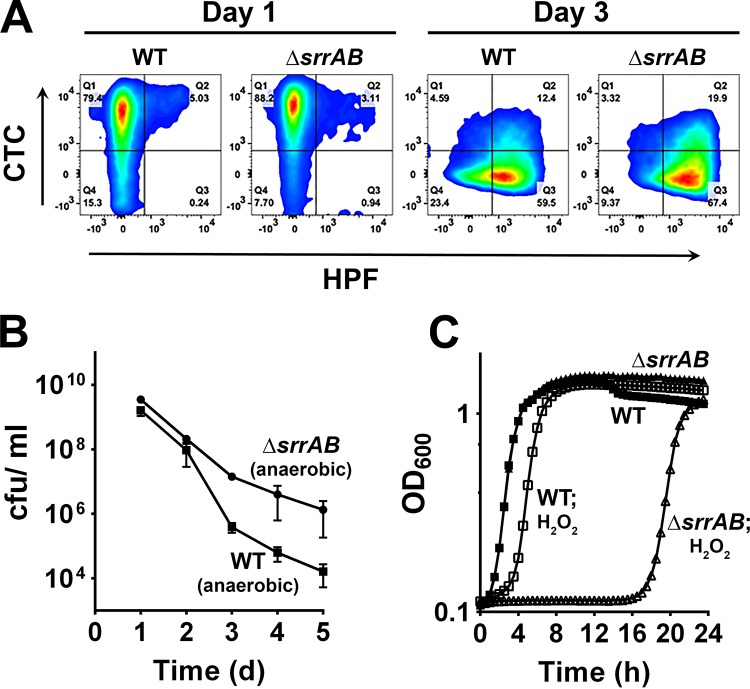
Given that death induced by weak acids is associated with the inhibition of respiration and the production of reactive oxygen species (ROS) in S. aureus (13), we also tested the hypothesis that the decreased survival of the ΔsrrAB mutant in stationary phase was due to increased sensitivity to ROS. The wild-type strain and the ΔsrrAB mutant were grown in TSB supplemented with 35 mM glucose and costained with 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) and 3′-(p-hydroxyphenyl)fluorescein (HPF) at 24 h and 72 h, which have previously been used to distinguish between respiring bacterial populations and those generating deleterious hydroxyl radicals (28, 29). As expected under these conditions, the wild-type strain and ΔsrrAB mutant exhibited respiring populations at 24 h (Fig. 3A). However, these populations declined by 72 h and were replaced by a ROS-generating population (HPF positive) (Fig. 3A). Interestingly, the mutant produced 15% more HPF-positive cells at 72 h of growth than the wild-type strain (Fig. 3A). These observations suggest that increased cell death observed in the ΔsrrAB mutant may be a consequence of ROS accumulation. Evidence supporting this hypothesis was obtained by shifting the ΔsrrAB mutant to anaerobic conditions following 24 h of aerobic growth in TSB supplemented with 35 mM glucose (Fig. 3B). This strategy allowed the cells to remain under weak acid stress but to be devoid of the ROS observed at 72 h of growth due to the anoxic growth conditions. Consistent with a significant role for ROS in catalyzing cell death of the ΔsrrAB mutant, we observed that a shift to anaerobiosis improved the survival of the ΔsrrAB mutant, even beyond that observed for the wild-type strain. Finally, to test the relative sensitivity of the ΔsrrAB mutant to oxidative stress, we performed growth experiments in the presence and absence of hydrogen peroxide. As shown in Fig. 3C, analysis of cell growth (OD600) revealed that the ΔsrrAB mutant was more sensitive to hydrogen peroxide challenge (5 mM) than the wild-type strain, as indicated by the greatly increased lag phase for the mutant. Together, these data suggest a role for SrrAB in negatively regulating cell death under acidic conditions by decreasing the generation of ROS while at the same time increasing resistance to these toxic molecules, possibly by modulating antioxidant activities.
FIG 3.
Effect of the ΔsrrAB mutation on ROS production and survival. (A) S. aureus UAMS-1 (WT) and isogenic ΔsrrAB mutant cells were collected at 1 and 3 days of growth in TSB plus 35 mM glucose at 37°C under aerobic conditions, stained with CTC and HPF, and then analyzed by flow cytometry. (B) Aerobically grown cells of UAMS-1 and the ΔsrrAB mutant (TSB and 35 mM glucose) were shifted to an anaerobic chamber after 24 h of growth and monitored for cell viability every 24 h over a period of 5 days in TSB plus 35 mM glucose. (C) The growth of S. aureus UAMS-1 (WT) and ΔsrrAB mutant cultures containing 5 mM H2O2 was monitored for 24 h at 37°C in a Tecan Infinite 200 spectrophotometer under maximum aeration.
Inactivation of cidB rescues the ΔsrrAB mutant from stationary-phase death.
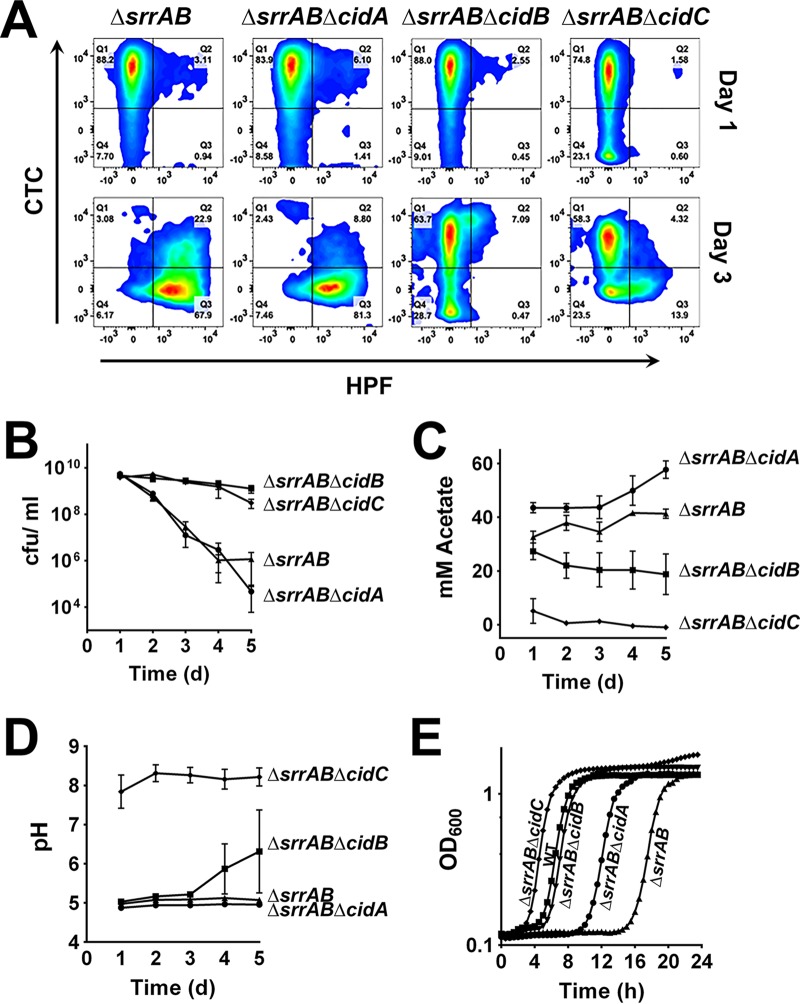
Although a role for the SrrAB regulon in an adaptive response to anaerobiosis has been demonstrated, its ability to modulate ROS sensitivity has not been reported. Since knocking out cidABC increased stationary-phase survival (Fig. 2A) and since CidA is an integral membrane protein that has holin-like properties proposed to be involved in the control of cell death, we reasoned that its expression in the context of the ΔsrrAB mutant background may affect stationary-phase survival. To test this hypothesis, in-frame and isogenic deletion mutants (ΔsrrAB ΔcidA, ΔsrrAB ΔcidB, and ΔsrrAB ΔcidC) were generated and monitored for their survival relative to the ΔsrrAB mutant. Consistently with our previous studies of a cidC mutant, a mutant in which ΔsrrAB and ΔcidC were disrupted (ΔsrrAB ΔcidC) exhibited a decrease in ROS generation and an increased population of respiring cells at day 3 (Fig. 4A), resulting in increased stationary-phase survival (Fig. 4B). As shown in Fig. 4C, these phenotypes were likely a result of the reduced acetate excreted by this strain. Interestingly, despite generating higher acetate levels than the ΔsrrAB ΔcidC mutant (Fig. 4C), as well as a lower pH (Fig. 4D), the ΔsrrAB ΔcidB mutant phenocopied the ΔsrrAB ΔcidC mutant in terms of survival (Fig. 4B) and in the presence of a healthy respiring population and reduced generation of ROS relative to those of the wild-type strain (Fig. 4A). A polar effect of ΔcidB mutation on the cidC allele was ruled out, as transcomplementation of cidB in the ΔsrrAB ΔcidB mutant restored the rates of cell death (see Fig. S1B in the supplemental material) to levels observed in the ΔsrrAB mutant. Consistent with its decreased ROS production and like the ΔsrrAB ΔcidC mutant, the ΔsrrAB ΔcidB mutant was more resistant to oxidative stress upon hydrogen peroxide challenge (Fig. 4E), a phenotype that may also be complemented in the latter strain by expressing cidB in trans (see Fig. S1 in the supplemental material). In contrast, although the cidA gene is coexpressed with cidB and cidC, the ΔsrrAB ΔcidA mutant exhibited phenotypes similar to those of the ΔsrrAB mutant with respect to ROS production, acetate generation, and survival in stationary phase (Fig. 4A to D). Taken together, these data suggest that overexpression of CidB in the ΔsrrAB mutant induces cell death by enhancing ROS production and increasing sensitivity to oxidative stress.
FIG 4.
Role of the cidABC genes on ROS production and survival. (A) S. aureus ΔsrrAB, ΔsrrAB ΔcidA, ΔsrrAB ΔcidB, and ΔsrrAB ΔcidC mutants were collected at 1 and 3 days of growth in TSB plus 35 mM glucose at 37°C under aerobic conditions, stained with CTC and HPF, and then analyzed by flow cytometry. (B) Cell viabilities (mean ± SEM) were monitored every 24 h over a period of 5 days in TSB plus 35 mM glucose. (C) Acetate concentrations in culture supernatants were measured every 24 h over a period of 5 days using a commercially available kit (R-Biopharm). (D) pHs of the culture supernatants. (E) Growth of the S. aureus mutants in the presence of 5 mM H2O2 was monitored for 24 h at 37°C in a Tecan Infinite 200 spectrophotometer under maximum aeration.
DISCUSSION
The results generated in this study demonstrate that the SrrAB two-component system functions to directly repress expression of the cidABC operon under conditions of excess glucose (Fig. 2B and C), resulting in the suppression of cell death. The decreased stationary-phase survival of the srrAB mutant is not due to changes in the levels of acetate or changes in the pH (Fig. 1B and C) but instead to the result of the increased sensitivity of this strain to acetate-dependent ROS (Fig. 3). Importantly, this study revealed for the first time the critical role of cidB in this process.
Previous studies performed by our laboratory have reported that S. aureus cidC-encoded pyruvate oxidase plays a critical role in cell death during stationary phase under excess glucose conditions (12) by producing acetate and promoting the acidification of the growth medium (13). Thus, given that cidABC transcription was enhanced in a ΔsrrAB mutant (Fig. 2B) (17), we initially hypothesized that the decreased survival of these cells was due to increased acetate, which would in turn lead to cytoplasmic acidification and ROS accumulation. However, the results of the current study revealed that this phenotype was clearly not associated with increased acetate accumulation or decreased pH of the culture medium (Fig. 1B and C). Instead, experiments where the cultures were switched to anaerobic conditions (Fig. 3B) suggest that the decreased stationary-phase survival of the ΔsrrAB mutant is due to the increased sensitivity to the acetate-dependent generation of ROS.
Acetate potentiates cell death via a complicated process (13). When the pH of the environment surrounding the cell is higher than the pKa of acetate (∼4.8), most of the acetate is unprotonated and has a negative charge (13), allowing the cell membrane to act as a barrier to entry. As the extracellular pH lowers and approaches the pKa of acetate, there is an increased percentage of acetate that is protonated to acetic acid. Now neutrally charged, the acetic acid can diffuse across the membrane into the bacterial cytoplasm. Once across the membrane, the protons disassociate from acetic acid, leading to a decrease in the intracellular pH and, through an unknown mechanism, an increase in the accumulation of ROS. Over time, this leads to increasing amounts of cellular damage and cell death. It has been suggested that acetate may contribute to a bottleneck in electron transport by reducing the functionality of the respiratory chain and catalyzing the reduction of oxygen, resulting in the production of ROS (13), but it is not clear how acetate initiates ROS production at the molecular level. Interestingly, the disruption of cidB improved stationary-phase survival (Fig. 4B) and sensitivity to H2O2 (Fig. 4E), implicating cidB in these processes. Importantly, any involvement of catalase or superoxide dismutase seems unlikely, as there was no alteration in the expression of the genes encoding these enzymes in the ΔsrrAB mutant (data not shown). Thus, the ΔsrrAB mutant background may be an important genetic context in which the role of CidB manifests itself as a direct mediator of cell death in response to ROS.
Although the model for CidAB and LrgAB function until now has focused primarily on the CidA and LrgA proteins as a holin and antiholin, respectively, the results presented here, as well as recent data generated by our laboratory, cast this system in a different light. Although an early report suggested that the cidB gene can affect the expression/activity of a 25- to 30-kDa murein hydrolase (30), little is known about CidB and LrgB other than that they are predicted to be ∼25-kDa membrane-associated proteins (9, 30). The results of the current studies indicate that in the context of the ΔsrrAB mutation, disruption of cidB has a dramatic effect on stationary-phase survival (Fig. 4B) but only a modest effect on acetate secretion (Fig. 4C) and culture pH (Fig. 4D). Importantly, ongoing studies in our laboratory indicate that CidB plays a positive role in the regulation of CidC activity and acetate secretion (our unpublished data). Combined with the studies presented here, these results suggest that CidB may have a more impactful role in the cell death pathway and acetate metabolism than had been previously recognized.
In addition to revealing the effect of the cidB mutation on H2O2 sensitivity, this study revealed that the disruption of cidA also decreased the sensitivity of the ΔsrrAB mutant to H2O2 (Fig. 4E). In contrast to these findings, disruption of the Streptococcus mutans ΔlrgAB and ΔlytSR operons was previously shown to cause increased sensitivity to H2O2 (17). Indeed, our investigations of the S. aureus ΔlrgAB and ΔlytSR operons revealed a similar increase in sensitivity to this ROS (data not shown). With respect to the H2O2 phenotype, it is interesting to note that SrrAB and LrgAB have a negative effect on cidABC; SrrAB represses cidABC expression at the transcriptional level, and LrgA has been proposed to inhibit CidA activity as an antiholin (11). Although we did not observe altered ROS sensitivity resulting from cidA or cidB mutations in the wild-type background (data not shown), this is likely due to the fact that cidABC expression is repressed by the SrrAB regulatory system in this genetic context. Overall, these results are consistent with the model in which the cid and lrg operons play opposing roles in cell death.
Although we noted that cidABC expression is increased nearly 4-fold in the ΔsrrAB mutant background (Fig. 2B), a corresponding increase in extracellular acetate levels was not observed (Fig. 1B). One explanation for this is that under these conditions, an alternative pathway, the pta-ackA pathway (31), produces most of the acetate in the ΔsrrAB mutant. Alternatively, while we have focused on cidABC transcription in this study, the cid operon also produces a cidBC transcript, whose expression is regulated by sigma factor B independently of CidR (32). Since the cidBC transcript is highly elevated when there is excess glucose (32), it is possible that the increase in cidABC transcription does not result in a significant increase in CidC protein levels and, as a result, acetate production. The elevated cidBC transcript may also be the reason that there is a large difference in the level of ROS detected in the ΔsrrAB ΔcidA mutant compared to that detected in the ΔsrrAB ΔcidB and ΔsrrAB ΔcidC mutants (Fig. 4A). These possibilities are under investigation.
It is also interesting to note that while the ΔsrrAB mutant has previously been shown to exhibit poor growth under anaerobic conditions (15), we have demonstrated that it survives better in stationary phase under these conditions than the wild-type strain (Fig. 3B). These data suggest that the ΔsrrAB mutant is necessary for anaerobic growth but conversely may be detrimental to survival within an anaerobic environment. It has been suggested that the decreased expression of genes involved in anaerobic metabolism may be responsible for the altered growth and survival of the ΔsrrAB mutant under these conditions (16). For example, the discrepancy between growth and survival may be explained by the dysregulation of the nrdDG and nar operons (17). The nrdDG operon encodes ribonucleoside-triphosphate reductase, which is essential for anaerobic growth (33); thus, the decreased expression of nrdDG in a ΔsrrAB mutant (17) would lead to poor growth under low-oxygen conditions. On the other hand, the nar genes (nitrate reductase) are repressed by SrrAB (17), so increased expression may lead to greater consumption of nitrate, a more active electron transport chain, and more energy for the cell, potentially allowing for better survival during stationary phase under anaerobic conditions.
In conclusion, our results indicate that the disruption of the srrAB regulatory operon in S. aureus results in reduced stationary-phase viability due to the increased production and sensitivity to ROS. The ROS are formed in the presence of acetic acid produced by CidC, encoded by the cidABC operon, which is repressed by SrrAB. Furthermore, we demonstrate that the increased cell death observed in the ΔsrrAB mutant is dependent on cidB. These results are the first to demonstrate a role for cidB in cell death and provide greater insight into the functions of the cidABC-encoded proteins and into the transcriptional control of this cell death regulatory system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victoria Smith of the UNMC Cell Analysis Facility for technical assistance with flow cytometry, as well as the High-Throughput DNA Sequencing and Genotyping Core Facility for their technical assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00954-15.
REFERENCES
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. [DOI] [PubMed] [Google Scholar]
- 4.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice KC, Bayles KW. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol 50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- 6.Bayles KW. 2014. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice KC, Bayles KW. 2008. Molecular control of bacterial cell death and lysis. Microbiol Mol Biol Rev 72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol 182:1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice KC, Firek BA, Nelson JB, Yang SJ, Patton TG, Bayles KW. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol 185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjit DK, Endres JL, Bayles KW. 2011. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol 193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 12.Patton TG, Rice KC, Foster MK, Bayles KW. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol Microbiol 56:1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 13.Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, Ahn JS, Jawa RS, Zimmerman MC, Bayles KW. 2014. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog 10:e1004205. doi: 10.1371/journal.ppat.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moormeier DE, Bose JL, Horswill AR, Bayles KW. 2014. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Zhu T, Han H, Liu H, Xu T, Francois P, Fischer A, Bai L, Gotz F, Qu D. 2015. Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J Bacteriol 197:459–476. doi: 10.1128/JB.02231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sau S, Sun J, Lee CY. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol 179:1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis AM, Matzdorf SS, Endres JL, Windham IH, Bayles KW, Rice KC. 2015. Examination of the Staphylococcus aureus nitric oxide reductase (saNOR) reveals its contribution to modulating intracellular NO levels and cellular respiration. Mol Microbiol 96:651–669. doi: 10.1111/mmi.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fey PD. (ed). 2014. Methods in molecular biology, vol. 1106. Staphylococcus epidermidis: methods and protocols. Springer-Verlag, New York, NY. [Google Scholar]
- 22.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin WS, Cunneen T, Lee CY. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol 176:7005–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. 2010. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res 28:55–61. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill AJ, Miller K, Oliva B, Chopra I. 2004. Comparison of assays for detection of agents causing membrane damage in Staphylococcus aureus. J Antimicrob Chemother 54:1127–1129. doi: 10.1093/jac/dkh476. [DOI] [PubMed] [Google Scholar]
- 26.Lehman MK, Bose JL, Sharma-Kuinkel BK, Moormeier DE, Endres JL, Sadykov MR, Biswas I, Bayles KW. 2014. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol Microbiol 95:723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haskell RE, Hughes SM, Chiorini JA, Alisky JM, Davidson BL. 2003. Viral-mediated delivery of the late-infantile neuronal ceroid lipofuscinosis gene, TPP-I to the mouse central nervous system. Gene Ther 10:34–42. doi: 10.1038/sj.gt.3301843. [DOI] [PubMed] [Google Scholar]
- 28.Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. 2003. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem 278:3170–3175. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- 29.Smith J, McFeters G. 1997. Mechanisms of INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride), and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli K-12. J Microbiol Methods 29:161–175. doi: 10.1016/S0167-7012(97)00036-5. [DOI] [PubMed] [Google Scholar]
- 30.Brunskill EW, Bayles KW. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol 178:5810–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadykov MR, Thomas VC, Marshall DD, Wenstrom CJ, Moormeier DE, Widhelm TJ, Nuxoll AS, Powers R, Bayles KW. 2013. Inactivation of the Pta-AckA pathway causes cell death in Staphylococcus aureus. J Bacteriol 195:3035–3044. doi: 10.1128/JB.00042-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice KC, Patton T, Yang SJ, Dumoulin A, Bischoff M, Bayles KW. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J Bacteriol 186:3029–3037. doi: 10.1128/JB.186.10.3029-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masalha M, Borovok I, Schreiber R, Aharonowitz Y, Cohen G. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J Bacteriol 183:7260–7272. doi: 10.1128/JB.183.24.7260-7272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun 63:3373–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 36.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.