ABSTRACT
The dimorphic alphaproteobacterium Prosthecomicrobium hirschii has both short-stalked and long-stalked morphotypes. Notably, these morphologies do not arise from transitions in a cell cycle. Instead, the maternal cell morphology is typically reproduced in daughter cells, which results in microcolonies of a single cell type. In this work, we further characterized the short-stalked cells and found that these cells have a Caulobacter-like life cycle in which cell division leads to the generation of two morphologically distinct daughter cells. Using a microfluidic device and total internal reflection fluorescence (TIRF) microscopy, we observed that motile short-stalked cells attach to a surface by means of a polar adhesin. Cells attached at their poles elongate and ultimately release motile daughter cells. Robust biofilm growth occurs in the microfluidic device, enabling the collection of synchronous motile cells and downstream analysis of cell growth and attachment. Analysis of a draft P. hirschii genome sequence indicates the presence of CtrA-dependent cell cycle regulation. This characterization of P. hirschii will enable future studies on the mechanisms underlying complex morphologies and polymorphic cell cycles.
IMPORTANCE Bacterial cell shape plays a critical role in regulating important behaviors, such as attachment to surfaces, motility, predation, and cellular differentiation; however, most studies on these behaviors focus on bacteria with relatively simple morphologies, such as rods and spheres. Notably, complex morphologies abound throughout the bacteria, with striking examples, such as P. hirschii, found within the stalked Alphaproteobacteria. P. hirschii is an outstanding candidate for studies of complex morphology generation and polymorphic cell cycles. Here, the cell cycle and genome of P. hirschii are characterized. This work sets the stage for future studies of the impact of complex cell shapes on bacterial behaviors.
INTRODUCTION
The Alphaproteobacteria comprise a diverse group of bacteria, including important pathogens of animals (Brucella spp. and Rickettsia spp.) and plants (Agrobacterium spp.), plant symbionts (Rhizobium spp., Sinorhizobium spp., and Mesorhizobium spp.), photosynthetic bacteria (Rhodobacter spp.), freshwater bacteria (Caulobacter spp.), and marine bacteria (Hyphomonas spp.). Despite the diverse habitats and lifestyles of these bacteria, many alphaproteobacterial species have a characteristic life cycle that culminates in asymmetric cell division (1–4). Caulobacter crescentus has served as a model bacterial system for the study of cell cycle regulation, and decades of research have provided insights into the mechanism underlying the precise cell cycle control that allows the production of two morphologically and functionally diverse daughter cells, a motile swarmer cell and an adherent stalked cell (5). A polar adhesin, termed holdfast, is found at the tip of the stalk and is required for permanent attachment to a surface (6).
Bioinformatic analysis of alphaproteobacterial genomes suggests that the core architecture of the regulatory genes that govern Caulobacter cell cycle progression is broadly conserved within at least two clades of Alphaproteobacteria, Rhizobiales and Caulobacterales (7, 8). Furthermore, genes shown to be essential for cell cycle progression in C. crescentus have also been shown to have important functions in cell cycle regulation of Agrobacterium tumefaciens (9–12), Sinorhizobium meliloti (3, 13–16), and Brucella abortus (14, 17–20). Notably, the essentiality and regulons of cell cycle regulators are varied in Alphaproteobacteria, suggesting that mechanisms of cell cycle regulation are plastic and may provide a means of adapting to a particular environment (12, 15, 21).
The ability to produce a polar polysaccharide appears to be a conserved trait among the Alphaproteobacteria. Polar polysaccharides are described for a number of species within the Caulobacterales and Rhizobiales clades (4, 6, 10, 22–26). A comparison of the genes responsible for the synthesis of the polar polysaccharide among C. crescentus and several Rhizobiales species suggests that these genes are largely conserved (see Fig. S1 and Table S1 in the supplemental material) (27). The production of polar polysaccharides is stimulated by contact with a surface in three alphaproteobacterial species, C. crescentus, Asticcacaulis biprosthecum, and A. tumefaciens, indicating that this may be a general phenomenon (28).
Asymmetric cell division and polar polysaccharide production are likely ancestral traits within the Caulobacterales and Rhizobiales clades, and yet, there are many alphaproteobacterial species for which these traits have not been described. In this work, we have characterized Prosthecomicrobium hirschii, a member of the Rhizobiales clade (see Fig. S2 in the supplemental material), which at first glance does not appear to use a regulated cell cycle to generate distinct morphotypes or produce a polar polysaccharide (29). P. hirschii cells adopt one of two morphologies: (i) numerous short stalks or (ii) 3 to 12 markedly longer stalks (Fig. 1A) (29). The short-stalked morphotype of P. hirschii has a single polar or subpolar flagellum (Fig. 1E) (29), which raises the possibility that this bacterium undergoes asymmetric cell division.
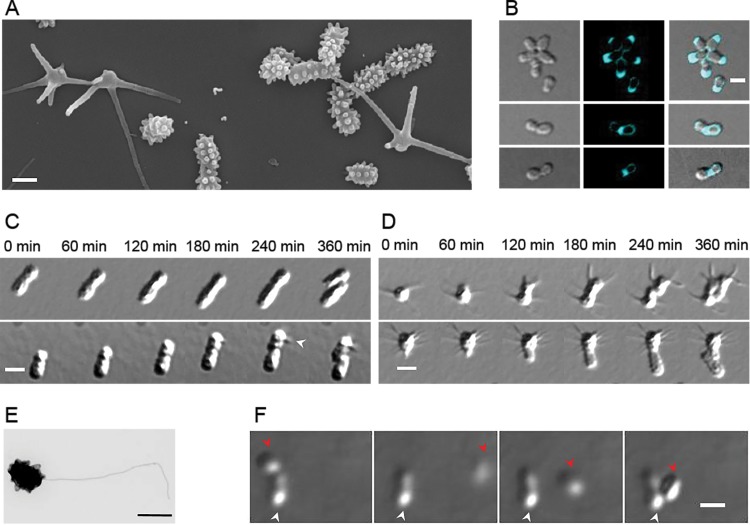
FIG 1.
P. hirschii has two distinct morphotypes. (A) Scanning electron microscope image of P. hirschii cells highlights the short- and long-stalked morphologies. The image was acquired at 60,000× magnification. Scale bar = 1 μm. (B) Fluorescent d-amino acid staining of cells reveals polar and midcell peptidoglycan synthesis. Scale bar = 2 μm. (C) Time-lapse differential interference contrast (DIC) images taken every 60 min on MMB agar pads show a short-stalked mother cell giving rise to a short-stalked daughter cell (top) and a short-stalked mother cell giving rise to a long-stalked daughter cell (bottom). The white arrowhead indicates the formation of a long stalk. Scale bar = 2 μm. (D) A long-stalked mother cell gives rise to a long-stalked daughter cell (top), and a long-stalked mother cell gives rise to a short-stalked daughter cell (bottom). Scale bar = 2 μm. (E) Transmission electron micrograph of an individual short-stalked P. hirschii cell with a single polar flagellum. Scale bar = 1 μm. (F) Montage showing a nonmotile mother cell producing a motile daughter cell. The white arrowheads indicate the stationary mother cell. The red arrowheads indicate the position of the motile daughter cell in the each image. The images shown were acquired at 20-min intervals. Scale bar = 2 μm.
Traditional microscopy techniques are integral to better understand the division of this bacterium. However, there are advantages to smaller-scale platforms that reduce medium consumption, precisely control the cell environment, and automate analysis. In addition, significant advances have been made in the development of microfluidic systems for cell biology (30–33). Of particular importance to this work, precise fluid handling on microfluidic devices provides more control over the microenvironment of the cells (34, 35). The integration of automated valves and pumps on-chip has enabled the addition of sample preparation and analysis on one device (36–38). A recent work on the development of a microfluidic device for bacterial synchronization and high-resolution analysis (39) of C. crescentus highlights the utility of these devices. Swarmer cell enrichments on the microfluidic device are comparable to standard synchronization techniques, but synchronized populations are collected on-demand over 4 days, and cells are tracked in downstream microchannels postsynchronization to observe single-cell behavior.
In this study, we carefully examined the short-stalked morphotype of P. hirschii with traditional microscopy techniques, and our observations indicate that these cells may have a Caulobacter-like cell cycle in which cell division gives rise to two functionally distinct daughter cells. Although the two cells are similar in appearance, one cell is motile and the other cell is nonmotile. Furthermore, we observed that short-stalked P. hirschii cells produce a polar adhesin. The presence of a polar adhesin renders this bacterium a suitable organism to integrate into our previously developed microfluidic platform, due to the ability of the cell to permanently adhere to the microchannel walls. With our device, we were able to synchronize P. hirschii cells and observe the behavior of newborn cells, confirming that short-stalked P. hirschii cells exhibit a Caulobacter-like cell cycle. Finally, a study of the P. hirschii genome confirmed the presence of a complex regulatory circuit consistent with the presence of a CtrA-regulated cell cycle.
MATERIALS AND METHODS
Strain and culture conditions.
P. hirschii (ATCC 27832T) was obtained from James Staley (University of Washington, Seattle, WA) and grown in minimal medium broth (MMB) (29) or peptone yeast extract (PYE) medium (6) at 26°C with shaking.
Materials.
We purchased glass slides (50 mm by 75 mm by 1.5 mm) from Corning Incorporated; poly(dimethylsiloxane) (Sylgard 184) from Dow Corning, Inc.; no. 1 Gold Seal cover glass (48 by 60 mm) from VWR International, LLC; Shipley 1813 photoresist, SU-8 2010, and Nano PG Developer from MicroChem Corp.; Alexa Fluor 488-wheat germ agglutinin lectin from Life Technologies; and all other chemicals from Sigma-Aldrich.
Scanning electron microscopy.
P. hirschii cells were grown to an optical density at 600 nm (OD600) of 0.3 in PYE medium, centrifuged, and washed in sterile water. Cell suspensions were spotted onto poly-l-lysine-coated cover glass, fixed with 3% glutaraldehyde for 5 min and 1% osmium tetroxide for 5 min, washed twice in deionized water, and dehydrated with sequential washes of 50, 70, 90, 95, and 100% ethanol. The cover glass was dried in a CPD 030 (Balzers) critical point drying apparatus with liquid CO2, mounted on a stub, sputter coated in an E5100 (Polaron Equipment) vacuum evaporator with a palladium-gold (80%–20%) target, and examined with a JEOL JSM-5800 LV scanning electron microscope at 15 kV.
Transmission electron microscopy.
Droplets of exponentially growing cultures were deposited onto a piece of Parafilm, and electron microscopy (EM) carbon-Formvar-coated copper grids (200 mesh) were floated onto the droplets for 2 min. Excess liquid was removed with filter paper, and the grids were quickly washed four times by floating onto droplets of double-distilled water, transferred to droplets of 1% uranyl acetate in water for 2 min, washed once in double-distilled water, dried, and examined with a JEOL JEM-1010 transmission electron microscope.
Light microscopy and FDAA staining.
Epifluorescence microscopy was performed with an inverted Nikon Eclipse TiE, a QImaging Rolera EM-C2 1K EMCCD camera, and the Nikon NIS-Elements imaging software. Short pulses of fluorescent d-amino acid (FDAA) labeling were completed as described previously (40). Briefly, exponentially growing cells (OD600, ∼0.3) were stained with 1 mM HADA (7-hydroxycoumarin-3-carboxylic acid-amino-d-alanine; emission maximum, 450 nm; blue) for 5 min and then washed three times in PYE to remove excess dye prior to imaging.
Initial attachment and polar adhesion detection.
Wheat germ agglutinin lectin conjugated to Alexa Fluor 488 (WGA-AF488) (Life Technologies) was used to detect polar polysaccharide in P. hirschii. In all experiments, a concentration of 0.5 μg/ml WGA-AF488 was used. Epifluorescence microscopy was performed with a Nikon Eclipse 90i equipped with a Photometrics Cascade 1K EMCCD camera and MetaMorph imaging software (Molecular Devices, LLC).
An inverted optical microscope, equipped with a total internal reflection fluorescence (TIRF) system (cell^TIRF; Olympus, Inc.) and a high-sensitivity CCD camera (C9100-13; Hamamatsu, Inc.), was used to visualize the polar holdfast in both wide-field and TIRF modes. To achieve a higher signal-to-noise ratio for images in TIRF mode, cover glass (no. 1) was cleaned in a solution of NH4OH, H2O2, and H2O (2:1:2) at 70°C for 20 min and rinsed with ultrapure water (18 MΩ · cm). The glass substrates were then sonicated in ultrapure water, rinsed again, dried in a stream of nitrogen, and heated for 10 min in a 70°C oven to remove excess water. A mixed culture of P. hirschii in MMB was dispensed onto the clean slides and placed in a humid chamber overnight to promote permanent attachment. After overnight incubation, cover glass with the attached cells was washed gently with a syringe filled with 50 μg/ml WGA-AF488 in MMB. Approximately 2 ml of the lectin-containing medium was added to the cover glass, incubated for 10 min to stain the holdfast, and rinsed with MMB to remove any unbound lectin. The holdfast-bound lectin was excited with a 491-nm laser, and images in wide-field and TIRF modes were collected to optically characterize the polar holdfast.
Biofilm formation and LIVE/DEAD staining.
Bacteria were grown to mid-log phase in MMB and diluted to an OD600 of 0.05. Three milliliters of diluted bacteria was placed in the wells of 12-well polystyrene plates containing polyvinyl chloride coverslips placed vertically in each well. The plates were incubated at 26°C for the indicated times (see Fig. 2E). After incubation, coverslips were removed and rinsed with distilled water (dH2O) to remove planktonic cells. Half a microliter of LIVE/DEAD stain mixture (LIVE/DEAD BacLight bacterial viability kit L7007; Life Technologies) was added to 500 μl of dH2O, placed on the top of the coverslip, and incubated in the dark for 15 min at room temperature. The stained coverslips were rinsed with dH2O and observed by epifluorescence microscopy.
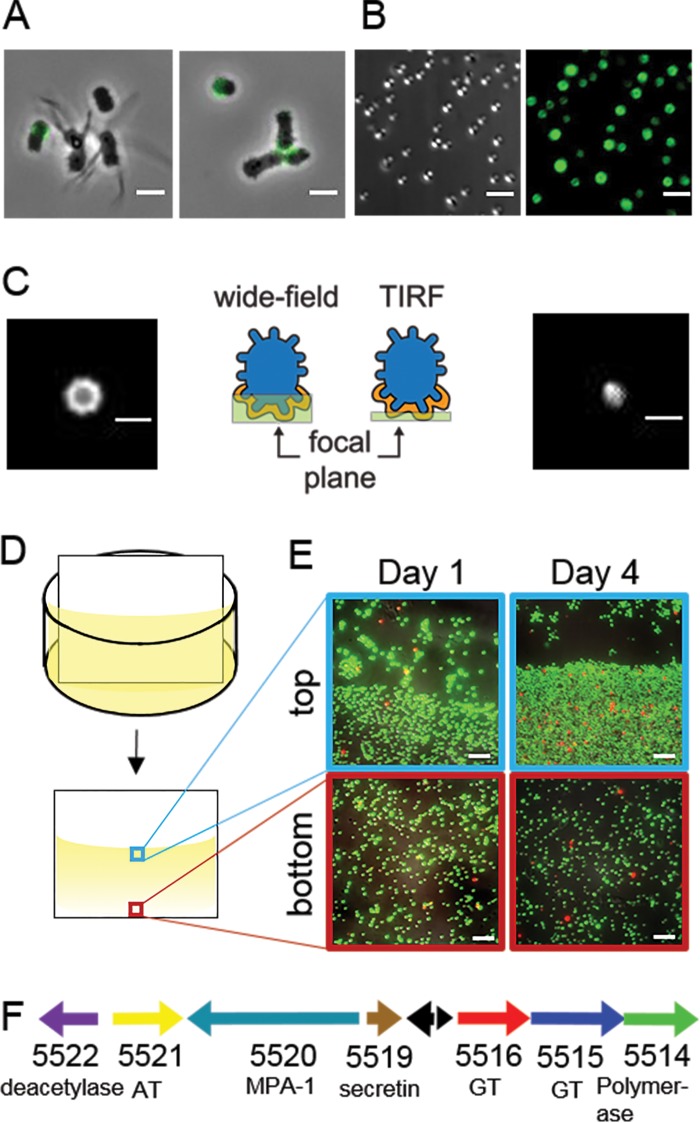
FIG 2.
Short-stalked P. hirschii cells produce a holdfast. (A) A polar polysaccharide is detected by labeling with WGA-AF488 (green) in short-stalked cells but is absent in long-stalked cells. Scale bar = 2 μm. (B) Short-stalked cells with a holdfast attach polarly to a glass surface. A DIC image (left) and fluorescence image of WGA-AF488 signal (right) of attached cells are shown. Scale bar = 2 μm. (C) Wide-field (left) and TIRF (right) images of a holdfast reveal that the holdfast is located at the surface-cell interface and forms a cap around the cell pole. Schematics indicate the focal plane imaged for the wide-field and TIRF images. Scale bar = 1 μm. (D) Schematic of the experimental setup used in the static biofilm assay in panel E. (E) Biofilm attached to polyvinyl chloride (PVC) coverslips stained with the BacLight LIVE/DEAD stain at different times of a static biofilm assay. The images represent overlays of green (live cells) and red (dead cells) collected by epifluorescence microscopy from the meniscus and bottom of the coverslip. Scale bar = 5 μm. (F) Genetic organization of the putative unipolar polysaccharide (upp) gene cluster (C1_5514 to C1_5522). The locus tag numbers and predicted functions are listed below each arrow. Black arrows indicate the positions of hypothetical proteins. AT, acetyltransferase; GT, glycosyltransferase; MPA-1, membrane-periplasmic-auxiliary protein.
Microfluidic device fabrication.
Polydimethylsiloxane (PDMS)-glass microfluidic devices were fabricated, as previously described (39), from two layers (control and fluid) of PDMS and one layer of glass. Briefly, the fluid and control layers were cast from SU-8 masters created by standard photolithography. An initial 15-μm-thick layer of SU-8 created a robust film, on top of which the fluid and control channels were created. This SU-8 layer was prebaked on a programmable hot plate (65°C for 3 min and 95°C for 1 min), exposed to UV light (270 mJ/cm2), and postbaked in a manner similar to the prebake. For the fluid control master, a 20-μm-thick SU-8 layer was spin-coated onto the initial layer and exposed to 270 mJ/cm2 through the first photomask. For the control layer master, a 40-μm-thick layer of SU-8 was spin-coated in two steps onto the substrate and exposed to 350 mJ/cm2 of UV light through a second photomask. The masters were developed for 2 min in Nano PG Developer, rinsed with 2-propanol, and dried with nitrogen.
Control layer replicas were prepared by spreading 9 ml of uncured PDMS onto the master. The PDMS was degassed under a vacuum for 30 min and cured for 2 h, at which point the pressure and vacuum access holes were punched. A fluid layer was created by spreading 3 ml of uncured PDMS across the master and degassing the PDMS under a vacuum for 10 min. The glass substrate was spun at 1,000 rpm for 35 s to achieve a 100-μm-thick layer suitable for the lifting-gate valve architecture and was partially cured for 15 min. The control and fluid layers were aligned, and the two substrates were brought together and cured for 2 h to permanently bond the layers. All PDMS replicas were cured at 65°C to minimize any shrinking in the layers that would compromise optimal alignment and valve operation. The bonded control and fluid layers were slowly peeled off the glass substrate, and fluid-access holes were punched through the PDMS.
Number 1 cover glass was cleaned in a solution of NH4OH, H2O2, and H2O (2:1:2) at 70°C for 20 min, rinsed with ultrapure water, and dried in a stream of nitrogen. The PDMS valves were covered with a mask prior to plasma cleaning to prevent irreversible adhesion that would render the valves nonfunctional. Both the bonded control and fluid layers and the glass coverslip were cleaned of plasma, the mask was removed, and the substrates were brought into direct contact with each other for an irreversible bond between the PDMS and the coverslip.
Device operation.
The fluidic device was coupled to an AssayMark system (Microfluidic Innovations, LLC). The AssayMark system interfaces with the microfluidic device through a stainless steel manifold that contains pressure (35,000 Pa) and vacuum (−89,000 Pa) connections, which align with the pressure and vacuum holes in the control layer of the device. The control software allows the user to operate the valves in manual and programmed modes. The programmed mode automates the actuation of pumps and valves over multiple days with no user input. The lifting-gate valve architecture in this device permits thin cover glass (no. 1) to be used as a cover plate for the fluid layer, and as a consequence, high-resolution imaging through the thin cover glass in both wide-field and TIRF modes is possible. Images were taken with high-numerical-aperture (NA) objectives in the analysis channel at 100× magnification.
Cell culture, seeding, and synchronization.
The microfluidic device was inoculated by pumping a mixed culture from the cell reservoir through the incubation region to the waste reservoir. After the incubation region was filled with culture, the pumps were stopped to initiate a gentle hydrostatic flow. During this time, permanent adhesion of the bacteria to the microchannel wall was observed. Fresh culture was pumped in every 10 min to replenish the population of swarmer cells. Device seeding was terminated after 1 h, and PYE medium was pumped from the incubation reservoir through the incubation region to flush away any unadhered cells.
The biofilm in the incubation region was grown over a period of 3 days. The doubling time of P. hirschii (∼6 h) is much slower than that of C. crescentus synchronized on this same platform (39). As a result, 3 days of growth was necessary to establish a biofilm large enough to produce a number of motile progeny. During this growth period, the incubation region was rinsed every 30 min to remove unadhered cells and to perfuse fresh medium over the established biofilm. To collect a synchronized population, the incubation region was flushed with medium for 15 min. The exit valve to the incubation region was closed to prevent flow, and newborn cells were released into the fresh medium for 20 min. Pumps 1 and 3 were used to route the medium containing the synchronized cell population to the analysis channel for downstream experiments. The inverted optical microscope was used to collect images from the microfluidic device experiments. WGA-AF488 was added to the medium prior to synchronization to observe the fluorescence associated with rapid cell attachment to the analysis channel surface. Video data were collected with the MetaMorph Advanced imaging software (Molecular Devices, LLC), and cell tracks were obtained from the video data with the ImageJ ParticleTracker plug-in.
P. hirschii genome sequencing, assembly, and annotation.
P. hirschii genome sequencing and assembly were completed as described previously (41). The annotation used in this analysis was completed by the University of Missouri Informatics Research Core Facility (IRCF) and is publicly available on the IRCF genomics website (http://genomics.ircf.missouri.edu/genomes/). The various scripts and files associated with the bacterial annotation pipeline are available for download from GitHub (https://github.com/sgivan/BGA). A custom genome annotation pipeline that progresses through four components was used: gene prediction, gene refinement, functional data collection (FDC), and function prediction (FP). Glimmer3 predicts initial gene boundaries in the DNA sequence of the genome assembly (42). Gene refinement proceeds through two phases. The first phase refines the gene start position based on potential upstream ribosome binding site (RBS) motifs obtained with RBSfinder (43). A consensus RBS is defined by DREME (44). The second phase of gene refinement follows FDC. During FDC, every gene identified by Glimmer3 is conceptually translated and subjected to BLASTP searches against two databases, Swiss-Prot and KEGG, and hidden Markov model searches of Pfam (45–47). Following FDC, gene start positions are confirmed or modified based on likelihood models of potential upstream and downstream start locations constructed from comparisons to similar database sequences. The data from FDC are also used to resolve significant overlaps between neighboring coding regions. Automated FP relies on a scoring matrix generated dynamically for every gene. The scoring matrix is constructed by parsing database hit descriptions to score words and phrases weighted by the quality of the database hits. Subsequently, database hits are ranked by the scoring matrix, and the highest-scoring hit is used to create a functional description of the genomic element. The distribution of scores provides a confidence estimate of the quality of the function assignment. The automated FP is an unsupervised process. A final component of the pipeline searches for conserved coding potential in intergenic regions of the genome. If identified, these open reading frames (ORFs) are subjected to FDC and FP.
Ortholog identification.
Orthologs of genes described to have an important function in cell cycle regulation or holdfast biogenesis of C. crescentus or A. tumefaciens were identified in P. hirschii, following searches for bidirectional best hits (BBH). Two query data sets composed of proteins from C. crescentus and A. tumefaciens were used for the BBH analysis. In all cases in which the protein was encoded in both query genomes, the same BBH was identified in the translated coding sequence (CDS) from the P. hirschii genome. An E value of 0.01 and sequence identity of 30% were used as thresholds in the analysis.
Genome-wide motif searches.
The P. hirschii genome was searched for the presence of conserved motifs involved in the cell cycle regulation of Alphaproteobacteria, including CtrA-binding sites (TTAAN7TTAA) and CcrM methylation sites (GANTC) (7). Our RelaxedMotifSearch (RMS) algorithm provides a fast and simple search of the patterns of short sequences of form “XYZ,” in which prefix X and suffix Z are to be matched, and Y is a variable part of the pattern of a predefined length N. The search can be done at the whole-genome scale and allows for a single-base-pair substitution in the prefix X and/or suffix Z. The input includes a specific motif(s) of interest, and the outputs are the locations in which the motif(s) is found. The RMS guarantees to find a comprehensive list of the motifs in both DNA strands. The search scans the motifs by consecutively trying to match subsequences of the genome sequence with a shift of one nucleotide. In the event the motif is found, RMS shifts |X| + |Z| + N nucleotide positions and continues the search. Searches were completed on both the total genome sequence and the predicted CDS.
Phylogenetic tree construction.
A phylogenetic tree was constructed with gyrA sequences from representative species. Sequences were aligned with MUSCLE (48). RAxML (49) reconstructed the maximum-likelihood phylogeny with the JTT amino acid substitution matrix, a four-category discrete gamma distribution, and single invariant category of sequence rate variation among sites. Analysis of 100 bootstrapped data sets determined support for clades occurring in the maximum-likelihood phylogeny.
RESULTS
Short-stalked P. hirschii cells are motile.
A mixed culture of P. hirschii cells grown in PYE or MMB contained cells of two distinct morphologies (Fig. 1A) (29). The population has cells with many short stalks that often appear to be positioned regularly around the cell surface (short-stalked cells). The population also has cells with stalks that are much longer but fewer in number (long-stalked cells). When individual cells are spotted on agarose pads and allowed to grow, two morphologies do not arise by a morphological transition in the cell cycle. Indeed, the morphologies of 367 mother cells remained fixed during 20 h of observation. This result is consistent with previous observations and presumably occurs because P. hirschii cells grow by tip elongation (29, 50). Staining of live P. hirschii cells with fluorescent d-amino acids (FDAA) (40) confirmed that new peptidoglycan is synthesized at one pole during elongation and at midcell in late-predivisional cells (Fig. 1B). When grown in PYE or MMB, the short-stalked morphology appears to be favored, and a short-stalked mother cell gave rise to a short-stalked daughter cell 99% of the time (n = 339) (Fig. 1C). In contrast, long-stalked mother cells were about equally likely to produce a short-stalked daughter cell (54% [n = 15]) or a long-stalked daughter cell (46% [n = 12]) (Fig. 1D). Time-lapse microscopy illustrated that the short- and long-stalked morphotypes reproduce at similar rates, with division times of approximately 6 h (Fig. 1C and D). Irrespective of the daughter-cell morphology, a mother cell typically produced daughters of the same morphology for many generations, resulting in the formation of microcolonies dominated by cells of a single morphology (see Fig. S3 in the supplemental material) (29).
The observation that microcolonies are composed of either long- or short-stalked cells revealed a clear functional difference in the two cell types. Microcolonies of long-stalked cells were tightly compacted due to the nonmotile nature of these cells (see Fig. S3 in the supplemental material). In contrast, microcolonies of short-stalked cells were capable of spreading on the agar (see Fig. S3 in the supplemental material), which suggests that cells of this morphology are motile by means of a polar or subpolar flagellum (Fig. 1E) (29). Notably, when a mixture of long- and short-stalked cells was observed under light microscopy, only a small fraction (∼10%) of the short-stalked cells appeared to be actively swimming, and rosettes were frequently observed (Fig. 1B, top, and 2A). Furthermore, when cells were observed in 0.5% agarose that was sufficiently wet for motile cells to swim, nonmotile mother cells were observed to give rise to motile daughter cells (Fig. 1F; see also Movie S1 in the supplemental material). These observations are consistent with the hypothesis that short-stalked P. hirschii cells may have a Caulobacter-like cell cycle during which asymmetric cell division will result in the formation of two functionally distinct daughter cells, a motile cell and a sessile cell (Fig. 1F).
Short-stalked P. hirschii cells produce a polar polysaccharide.
The ability of the cells to form rosettes is often indicative of the presence of a polar polysaccharide. Wheat germ agglutinin lectin conjugated to Alexa Fluor 488 (WGA-AF488), which binds to N-acetylglucosamine, was used to determine if the polar polysaccharide is produced in planktonic cultures of P. hirschii. Lectin bound to one pole of some short-stalked cells but was never observed to bind to long-stalked cells (Fig. 2A, left). In addition, lectin bound at the center of rosettes (Fig. 2A, right), which indicates that short-stalked P. hirschii cells are capable of producing a holdfast-like polar polysaccharide. To determine if the polar polysaccharide of P. hirschii mediates surface attachment, P. hirschii cells were incubated with cover glass for 24 h. The cover glass was washed to remove unattached cells, the polar polysaccharide of attached cells was labeled with WGA-AF488, and attached cells were visualized with epifluorescence microscopy. P. hirschii cells bound to the coverslip in an end-on configuration, and each cell attached to the surface also bound WGA-AF488 (Fig. 2B).
To ensure that the polar polysaccharide is present at the cell-surface interface, microscopy in wide-field and TIRF modes was conducted to detect WGA-AF488-labeled polar polysaccharides of cells attached to cover glass. In images collected in wide-field mode, the polar polysaccharide appeared to be a ring, which suggests that the polar polysaccharide forms a cap around the pole of the cell (Fig. 2C, left). In TIRF mode, which only visualizes fluorescence signal approximately 100 nm deep into the sample, the WGA-AF488 was detected as a single uniform spot, which suggests that the polar polysaccharide is present at the cell pole-to-surface interface and likely promotes attachment to surfaces (Fig. 2C, right).
Because P. hirschii produces a polar polysaccharide and attaches to surfaces, we hypothesized that this bacterium should be capable of robust biofilm formation. Under static conditions, we observed that P. hirschii cells readily attach to a submerged vertical coverslip (Fig. 2D), with an enrichment of cells near the liquid-air interface (Fig. 2E). After 4 days, we observed robust biofilm formation on the coverslip along the meniscus of the coverslip, and LIVE/DEAD staining indicated that the attached cells were largely viable (Fig. 2E, right). Taken together, these results suggest that P. hirschii produces a unipolar polysaccharide capable of mediating cell-to-surface and cell-to-cell attachment. This notion is supported by the presence of a nearly complete wzy-type polysaccharide gene cluster with similarity to the C. crescentus holdfast and A. tumefaciens unipolar polysaccharide (UPP) biosynthesis clusters in the P. hirschii genome (Fig. 2F; see also Fig. S1 and Table S1 in the supplemental material).
Short-stalked P. hirschii cells can be synchronized.
The presence of the P. hirschii polar adhesin and its ability to form a biofilm suggested that this bacterium could be synchronized on a microfluidic platform previously used to synchronize C. crescentus (39). Figure 3A is a schematic of the microfluidic device design. Using this device, we collected bright-field and fluorescence images of the biofilm. The bright-field image of the cells (Fig. 3C, left) suggests the biofilm develops as a monolayer on the surface of the microchannel. During the 3 days of biofilm growth, cells adhered to the microchannel wall by the polar adhesin, which was confirmed by the fluorescence of WGA-AF488 added to the medium to indicate the presence of the polar polysaccharide (Fig. 3C, middle and right). The biofilm density increased over time, and cells remained attached to the biofilm, even when the pumps perfused fresh medium over the biofilm every 30 min. Movie S2 in the supplemental material shows that the cells were adhered to the surface and remained attached during medium perfusion. In addition to attachment, the cells were confined to the incubation region by the valves (Fig. 3B), and this confinement kept the downstream analysis channel free from cells prior to analysis.
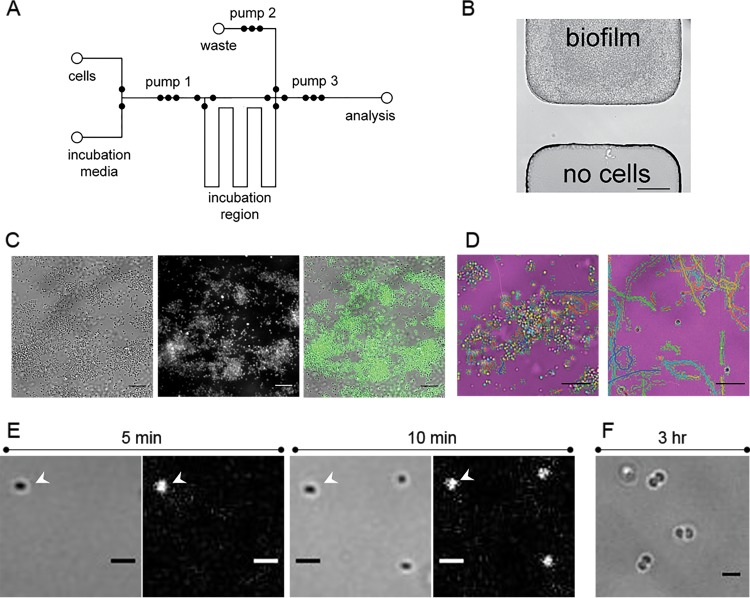
FIG 3.
Synchronization of short-stalked P. hirschii cells in the microfluidic device. (A) Fluid layer design of the microfluidic device. A biofilm is formed in the incubation region over 3 days, and the synchronized population is pumped to the analysis channel for observation. (B) Bright-field image of cell confinement in the microfluidic device. The area above the valve seat has cells grow up to, but not beyond, the valve seat. No cells were observed on the opposite side during biofilm growth. Scale bar = 50 μm. (C) Biofilm formation within the incubation region of the microfluidic device. Biofilm is labeled with WGA-AF488, which indicates the presence of the polar polysaccharide. White-light (left), fluorescent (middle), and overlay (right) images are provided. Scale bar = 5 μm. (D) Newborn cells released from the biofilm are motile. Tracks of motile cells were generated with ImageJ ParticleTracker and are overlaid on the false-colored white-light images in the incubation region (left) and the analysis channel (right). Scale bar = 5 μm. (E) Newborn cells rapidly attach to the analysis channel. White-light (left) and fluorescent (right) images of cells labeled with WGA-AF488 attached to the analysis channel 5 (left) and 10 (right) min after collection. The arrowheads indicate the position of a cell that attached within 5 min of collection. Scale bar = 1 μm. (F) Attached cells grow in the analysis channel. Three hours after collection, predivisional cells were observed attached to the analysis channel. Scale bar = 1 μm.
Using the automated operation of the microchip device by the AssayMark system, we synchronized cells, and newborn motile cells were released into the fresh medium in the incubation region for 20 min. During this time, the activity in the biofilm was monitored to ensure that cells in the biofilm were dividing. Motile cells accumulated in the biofilm region during the 20-min release time (Fig. 3D, left). After the 20-min release period, cells were pumped to the analysis channel where motile cells were highly enriched (∼75%), as evidenced by the cell tracks for the synchronized population in the analysis channel (Fig. 3D, right; see also Movie S3 in the supplemental material).
In addition to the release of newborn motile cells, we observed the rapid attachment of newborn cells to the microchannel walls (Fig. 3E; see also Fig. S4 in the supplemental material). Within 5 min after the synchrony, a cell had attached to the microchannel wall and developed a unipolar polysaccharide (Fig. 3E, left). After an additional 5 min, two more cells had attached to the microchannel in the field of view (Fig. 3E, right). Over time, the number of motile cells in the microchannel decreased, and the number of adhered cells increased. After 10 min, the proportion of cells attached to the microchannel increased from ∼30% to 55% (see Fig. S4 in the supplemental material). These data suggest that polar adhesin-mediated attachment is rapid, and our observations are consistent with the possibility that surface contact stimulates polar polysaccharide production, as described for other Alphaproteobacteria (28).
We monitored the swim behavior of the synchronized cells over the course of 3 h in the analysis channel and observed that the newly adhered cells in the microchannel continued to grow (Fig. 3F; see also Fig. S5 in the supplemental material). We observed that the cells attached to the microchannel wall were similarly sized predivisional cells, suggesting that these cells are tightly synchronized by our microfluidic platform (Fig. 3F; see also Fig. S5 in the supplemental material). The majority of cells were synchronized based on the observation that predivisional septa increased from 8.33% (n = 108) shortly after synchronization to 70.86% (n = 127) at ∼200 min postsynchrony (see Fig. S5 in the supplemental material). The observation that a synchronized motile population transitions to a sessile state is further evidence for an asymmetric Caulobacter-like cell cycle.
Conservation of cell cycle regulators and binding sites.
Many members of the Alphaproteobacteria, including C. crescentus and A. tumefaciens, have a complex life cycle resulting in the formation of two distinct cell types, a motile cell and a nonmotile sessile cell (5, 10, 27, 51). Notably, the regulatory components underlying the alphaproteobacterial cell cycle are well conserved, and transcriptional control has been hypothesized to be a conserved mechanism to govern cell cycle progression in C. crescentus, A. tumefaciens, and S. meliloti (1, 7, 16). Because short-stalked P. hirschii cells exhibit features of a similar cell cycle (Fig. 1F), the newly sequenced genome of P. hirschii (41) was searched for hallmarks of regulatory networks governing the alphaproteobacterial cell cycle.
Like other Alphaproteobacteria, the P. hirschii genome contains orthologs of proteins predicted to function in cell cycle regulation (see Table S2 in the supplemental material). The P. hirschii genome contains orthologs of the global regulators GcrA, CcrM, SciP, and CtrA. In C. crescentus, these global regulators function in regulatory modules to control progression through the cell cycle: the CtrA/MucR, GcrA/CcrM, and CtrA/SciP modules regulate sequential transcription of target genes during the G1, S, and G2 phases, respectively (8, 52). The P. hirschii genome also contains orthologs of proteins predicted to regulate CtrA function, including CckA, ChpT, DivL, and DivK. In C. crescentus, the CckA/ChpT phosphorelay is required for the phosphorylation and activation of CtrA (51). The activity of the hybrid histidine kinase CckA is modulated by the tyrosine kinase DivL and the single-domain response regulator DivK. Phosphorylated DivK can inhibit the activity of the DivL-CckA complex, resulting in the inhibition of CtrA activity. The phosphorylation state of DivK is regulated by the PdhS family of histidine kinases, which includes PleC and DivJ in C. crescentus and additional kinases in other Alphaproteobacteria (1, 10, 13, 51, 53). The P. hirschii genome encodes four PdhS proteins with similarity to the PleC, DivJ, PdhS1, and PdhS2 proteins of A. tumefaciens (see Table S2 in the supplemental material). In C. crescentus, CtrA activity is also modulated by proteolysis by the ClpPX protease, along with CpdR and RcdA. The proteolysis machinery is conserved in P. hirschii (see Table S2 in the supplemental material). The presence of genes encoding key components known to function in cell cycle regulation of Alphaproteobacteria suggests that P. hirschii may utilize temporal transcriptional control as a means to control cell cycle progression.
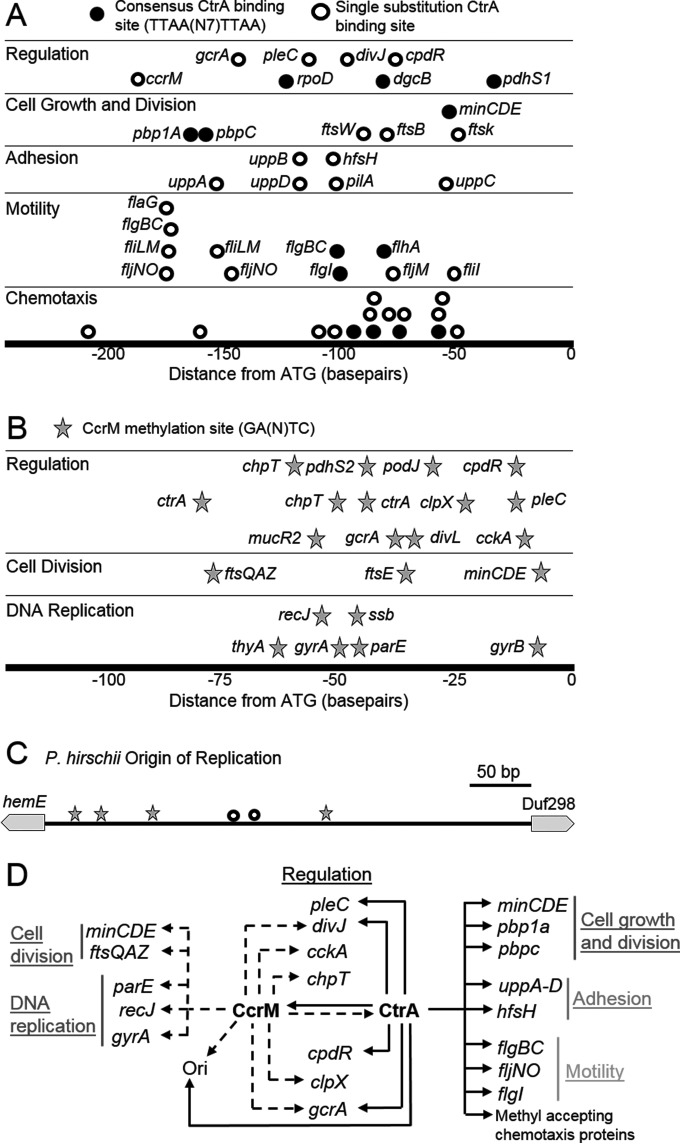
The CtrA protein sequence of P. hirschii is remarkably well conserved, with 94.2% protein sequence identity with CtrA from A. tumefaciens (see Fig. S6A in the supplemental material). Based on the high degree of CtrA protein conservation and successful complementation of C. crescentus CtrA mutants with CtrA from Rickettsia prowazekii (54), it is reasonable to hypothesize that CtrA-binding sites are highly conserved among Alphaproteobacteria (7). To determine which genes might be directly regulated by CtrA, we searched for the consensus CtrA-binding site (TTAAN7TTAA [7]) in the P. hirschii genome. Twenty exact matches to the consensus CtrA-binding site were identified, and 90% (18/20) of these sites were found in intergenic regions (Fig. 4A, closed circles). Seven of these putative CtrA-binding sites were found upstream of genes involved in flagellum biosynthesis or methyl-accepting chemotaxis proteins (Fig. 4A and D; see also Fig. S6B in the supplemental material), suggesting that CtrA may regulate the motility of P. hirschii. Three putative CtrA-binding sites were upstream of genes encoding penicillin binding proteins (PBPs) and the min genes, suggesting a role in the regulation of cell growth and division, as in S. meliloti (16). CtrA-binding sites also occupied the regions upstream of regulatory genes, including dgcB and pdhS1. Relaxing the search for CtrA-binding sites to allow one substitution in the binding site resulted in the identification of additional candidate CtrA-binding sites in the genome. Many of these putative binding sites were found upstream of genes with predicted functions in chemotaxis, motility, adhesion, cell growth and division, and regulation (Fig. 4A, open circles, and 4D; see also Fig. S6B and C in the supplemental material). In addition, putative CtrA-binding sites were identified in the origin of replication (Fig. 4C), suggesting a role for CtrA in integrating DNA replication with the cell cycle, as described for Caulobacter species (55, 56). Further support for the hypothesis that DNA replication is integrated with the P. hirschii cell cycle was provided by the presence of four consensus CcrM methylation sites (GANTC [7]) (Fig. 4C).
FIG 4.
P. hirschii genome reveals signatures of cell cycle regulation. (A) Relative distances of CtrA-binding sites to the ATG of predicted targets. The predicted targets of CtrA regulation in chemotaxis, motility, adhesion, cell growth and division, and regulation are shown. For chemotaxis, all predicted targets are methyl-accepting chemotaxis proteins. Closed circles indicated a perfect match to the TTAAN7TTAA consensus sequence. Open circles have one substitution in the predicted CtrA-binding site. (B) Relative distance of CcrM methylation sites to the ATG of predicted targets in DNA replication, cell division, and regulation. All predicted methylation sites match the GANTC consensus sequence. (C) The P. hirschii origin of replication contains CtrA-binding sites with one substitution (open circles) and multiple CcrM methylation sites (stars). (D) Schematic of genes that may be regulated via putative CtrA binding (solid arrows) or via putative CcrM methylation (dashed arrows), based on the findings in this study. Genes are grouped together according to function. Ori, origin of replication.
In C. crescentus, the methylation of DNA by CcrM plays an important role in the regulation of cell cycle progression (57–59), and methylated sites are subject to transcriptional regulation by GcrA (14). Methylation by CcrM is conserved in other Alphaproteobacteria, including B. abortus (18), A. tumefaciens (9), and S. meliloti (60). Furthermore, ccrM genes from C. crescentus and S. meliloti are able to cross-complement (60), suggesting that the methylation site may be conserved. Finally, methylation-dependent binding of GcrA from other Alphaproteobacteria has been shown in vitro, suggesting that methylation-dependent recruitment of GcrA to promoters may be conserved (14). Because orthologs of both CcrM and GcrA are present in the P. hirschii genome (see Table S2 in the supplemental material), the genome was scanned for potential methylation sites. In the P. hirschii genome, there are a total of 7,538 GANTC sites potentially methylated by CcrM. Notably, the number of possible methylation sites (7,538) is far lower than would be predicted by the expected frequency of this sequence (16,996 occurrences) in the P. hirschii genome, suggesting that these sites are underrepresented in the genome. The proportion of intergenic regions in the genome is 11.95%, and nearly a quarter (24.6%) of these methylation sites are found in intergenic regions, suggesting that these sites might be targets for transcriptional regulation. Putative methylation sites are enriched upstream of genes encoding proteins with function in DNA replication, cell division, and cell cycle regulation (Fig. 4B and D). This observation is consistent with the described role of CcrM-mediated methylation in mediating cell cycle progression, DNA metabolism, and cell division in C. crescentus (57, 58).
DISCUSSION
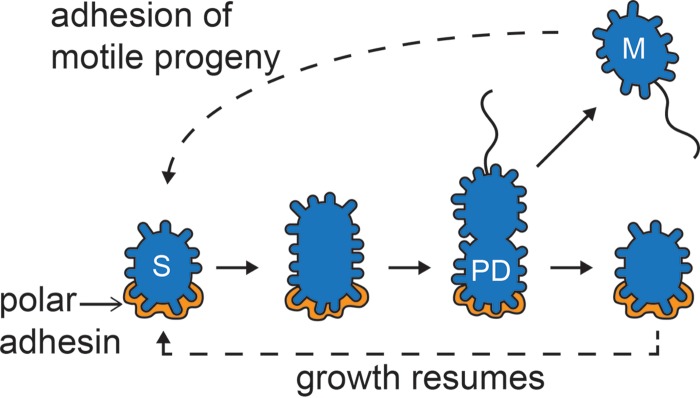
The results presented in this work suggest that the short-stalked cells of P. hirschii have a Caulobacter-like cell cycle in which cell division results in the release of a motile cell (Fig. 1F). The motile cell is capable of rapid attachment to surfaces mediated by a polar polysaccharide (Fig. 3E). Following surface attachment, the cells undergo a period of cell growth, leading to the production of a predivisional cell with a polar polysaccharide at one cell pole and a flagellum at the opposite pole. Cell division ultimately leads to the production of two functionally distinct daughter cells, a motile cell and a sessile cell (Fig. 5). Asymmetric cell division is proposed to be broadly conserved in the Caulobacterales and Rhizobiales clades of the Alphaproteobacteria (1, 7), and our results support this notion.
FIG 5.
Schematic of the short-stalked cell cycle. Sessile cells (S) are attached to a surface by means of a polar polysaccharide (gold) and are capable of elongation. Late predivisional cells (PD) produce a polar or subpolar flagellum. Cell division produces two distinct daughter cells: a motile cell (M) with a flagellum and a sessile cell (S) with a polar polysaccharide.
Rapid attachment to surfaces by means of a polar polysaccharide has been characterized in C. crescentus and other Alphaproteobacteria (28). We have observed a polar polysaccharide in P. hirschii (Fig. 2) that shares key features with the well-studied C. crescentus holdfast and A. tumefaciens UPP. First, we observed labeling of the polar polysaccharide with WGA-AF488 (Fig. 2A to C and 3C). WGA is an N-acetylglucosamine-specific lectin and is known to bind both the holdfast and the A. tumefaciens UPP, suggesting that the polar polysaccharides of C. crescentus, A. tumefaciens, and P. hirschii all contain N-acetylglucosamine. Second, we observed that newborn P. hirschii cells rapidly attach to surfaces and produce a polar polysaccharide (Fig. 3E). Both C. crescentus and A. tumefaciens have been shown to extrude a polar polysaccharide within minutes of contacting a surface (25, 28, 61), and P. hirschii cells have a similar response to surface contact. Third, we observed that the P. hirschii cells bound to surfaces within a biofilm produce the UPP (Fig. 3C). Both holdfast and UPP play essential roles in attachment and biofilm formation (25, 26, 62, 63). Finally, we have identified a gene cluster with similarity to the upp biosynthesis cluster (Fig. 2E; see also Fig. S2 in the supplemental material) in the P. hirschii genome. The P. hirschii upp gene cluster contains putative CtrA-binding sites (Fig. 4A and D; see also Fig. S6C in the supplemental material), which are suggestive of cell cycle regulation of polysaccharide biosynthesis, as has been described for holdfast (64, 65) and suggested for A. tumefaciens UPP (11). Because the putative P. hirschii UPP biosynthesis cluster is missing a flippase (see Fig. S2 in the supplemental material), we hypothesize that additional proteins involved in UPP biosynthesis and export remain to be identified. Indeed, a putative flippase at another genomic locus has been identified (see Fig. S1 and Table S1 in the supplemental material). Taken together, these observations suggest that P. hirschii is capable of producing a unipolar polysaccharide that enables rapid attachment to surfaces and likely promotes biofilm formation. Additional characterization of P. hirschii genes predicted to function in UPP biosynthesis will be necessary to understand the mechanisms underlying attachment and biofilm formation in this bacterium; however, we have been hindered by a lack of a genetic system and an inability to generate mutants in this bacterium, despite testing a number of approaches.
In this work, we provide evidence that the short-stalked cells of P. hirschii have a Caulobacter-like cell cycle and explore the potential role of global regulators in mediating cell cycle progression. Studies indicate that the P. hirschii genome contains genes that encode candidate cell cycle regulators, including CtrA and CcrM, suggesting that transcriptional control may play an important role in regulation of the P. hirschii cell cycle (Fig. 4; see also Table S2 in the supplemental material). The high degree of conservation of regulatory proteins in the Alphaproteobacteria has led to the suggestion that transcriptional control of the cell cycle is conserved among Alphaproteobacteria (1, 7). The Caulobacter paradigm of transcription regulation of cell cycle progression has recently been extended to S. meliloti, providing additional support for this notion (15). CtrA-binding sites are found upstream of cell cycle-regulated genes in both C. crescentus and S. meliloti; however, there is very little overlap between the CtrA regulons (15). In contrast, CtrA-binding sites are strongly conserved among Alphaproteobacteria closely related to S. meliloti, suggesting that the functions regulated by CtrA may be influenced by the lifestyle(s) of the bacteria (15).
The regulation of motility and flagellum biosynthesis by CtrA is posited to be an ancestral trait (21); thus, our observation of CtrA-binding sites upstream of genes involved in motility and chemotaxis is to be expected. Remarkably, in P. hirschii, the putative mechanism of CtrA regulation of motility is more similar to that of C. crescentus than to that of S. meliloti. CtrA-mediated regulation of motility is largely indirect in S. meliloti, in which CtrA is thought to regulate Rem, an important regulator of flagellar gene expression (15, 66). In contrast, CtrA directly binds to the promoter regions of the flg, fli, flm, and flj genes, representing all four tiers of the flagellar regulatory hierarchy, and many mcp genes in C. crescentus (67). Putative CtrA-binding sites are present upstream of presumptive class II flagellar genes (flgBC, fliLM, flhA, and fliI), class III flagellar genes (flgI operon), class IV flagellar genes (fljM and fljNO), and numerous chemotaxis genes in the P. hirschii genome (Fig. 4A; see also Fig. S6B in the supplemental material). These observations suggest that CtrA may have a direct role in the regulation of flagellum biosynthesis and chemotaxis in P. hirschii. In P. hirschii, CtrA-binding sites are also observed upstream of genes encoding a putative unipolar polysaccharide biosynthesis cluster (Fig. 4A; see also Fig. S6C in the supplemental material). We are tempted to speculate that because P. hirschii and C. crescentus share an environmental niche (freshwater), common mechanisms of cell cycle regulation may be beneficial. Indeed, simulations have shown that in patchy nutrient environments, such as freshwater, a developmental cell cycle with both sessile and motile cell types may confer a survival advantage (68).
Although the potential regulation of motility by CtrA in P. hirschii is similar to that of C. crescentus, this trend is not universal. We found that the predicted regulation of cell division in P. hirschii by CtrA resembles what is observed in S. meliloti, in which CtrA represses the min operon (16). Likewise, in P. hirschii, CtrA-binding sites are found upstream of the genes encoding the Min system, which regulates the placement of the septum, and upstream of ftsK, which encodes an essential cell division protein. These results support the notion that CtrA has a critical role in controlling the timing of min expression and subsequent cell division in the Rhizobiales (1, 15, 16). This finding may reflect a common mechanism for the regulation of cell division among bacteria exhibiting polar elongation (50). Additional research is necessary to test the hypothesis that P. hirschii exhibits cell cycle-dependent transcription and to understand the influence of global regulators on processes, such as cell division, biofilm formation, and motility.
Notably, the Caulobacter-like cell cycle is observed only among short-stalked P. hirschii cells. Long-stalked P. hirschii cells are nonmotile and do not produce an observable polar polysaccharide (Fig. 2A; see also Fig. S3 in the supplemental material). The long-stalked cells are capable of cell elongation and division; however, when long-stalked mother cells produce long-stalked daughter cells, the progeny are not morphologically or functionally distinct from the mother cells (Fig. 1D, top). Thus, P. hirschii exhibits polymorphic cell cycles in which two morphologically different cell types with distinct cell cycles persist in the same culture. Other Alphaproteobacteria, particularly Rhodomicrobium and Hyphomicrobium spp., are described to have polymorphic cell cycles in which one cell morphology routinely utilizes a Caulobacter-like cell cycle, which suggests that the complex elaboration of the dimorphic cell cycle may be widespread (2, 69).
The ability to synchronize short-stalked P. hirschii cells in a microfluidic device provides an opportunity to better understand the interplay among the two morphologies of P. hirschii. Under standard laboratory conditions, the short-stalked morphology is dominant and appears to be stable, with very few short-stalked cells giving rise to long-stalked daughter cells (Fig. 1C). In contrast, long-stalked cells frequently gave rise to short-stalked daughter cells (Fig. 1D, bottom). These observations raise the possibility that the different morphologies provide an advantage under certain nutrient conditions, as described for Rhodomicrobium species (2). In the microfluidic device, synchronous populations of short-stalked P. hirschii cells could be exposed to a variety of nutrient conditions in an effort to determine if a particular condition triggers short-stalked cells to produce long-stalked daughter cells. The identification of the conditions that trigger such a switch would certainly provide insights into the complex polymorphic cell cycles of P. hirschii.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by startup funds from the University of Missouri to P.J.B.B. and by grant NIH R01 GM113172 to S.C.J.
Funding Statement
Early phases of this work were initiated in the lab of Yves V. Brun (Indiana University), with support from National Institutes of Health grant GM051986 and National Science Foundation grant MCB0731950. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00896-15.
REFERENCES
- 1.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol 12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Whittenbury R, Dow CS. 1977. Morphogenesis and differentiation in Rhodomicrobium vannielii and other budding and prosthecate bacteria. Bacteriol Rev 41:754–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam H, Matroule JY, Jacobs-Wagner C. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell 5:149–159. doi: 10.1016/S1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch P. 1974. Budding bacteria. Annu Rev Microbiol 28:391–444. doi: 10.1146/annurev.mi.28.100174.002135. [DOI] [PubMed] [Google Scholar]
- 5.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panis G, Murray SR, Viollier PH. 2015. Versatility of global transcriptional regulators in alpha-proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev 39:120–133. doi: 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 9.Kahng LS, Shapiro L. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol 183:3065–3075. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heindl JE, Wang Y, Heckel BC, Mohari B, Feirer N, Fuqua C. 2014. Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front Plant Sci 5:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis PD, Brun YV. 2014. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol 93:713–735. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pini F, Frage B, Ferri L, De Nisco NJ, Mohapatra SS, Taddei L, Fioravanti A, Dewitte F, Galardini M, Brilli M, Villeret V, Bazzicalupo M, Mengoni A, Walker GC, Becker A, Biondi EG. 2013. The DivJ, CbrA and PleC system controls DivK phosphorylation and symbiosis in Sinorhizobium meliloti. Mol Microbiol 90:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fioravanti A, Fumeaux C, Mohapatra SS, Bompard C, Brilli M, Frandi A, Castric V, Villeret V, Viollier PH, Biondi EG. 2013. DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genet 9:e1003541. doi: 10.1371/journal.pgen.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. 2014. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci U S A 111:3217–3224. doi: 10.1073/pnas.1400421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pini F, De Nisco NJ, Ferri L, Penterman J, Fioravanti A, Brilli M, Mengoni A, Bazzicalupo M, Viollier PH, Walker GC, Biondi EG. 2015. Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet 11:e1005232. doi: 10.1371/journal.pgen.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- 18.Robertson GT, Reisenauer A, Wright R, Jensen RB, Jensen A, Shapiro L, Roop RM Jr. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol 182:3482–3489. doi: 10.1128/JB.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Henst C, de Barsy M, Zorreguieta A, Letesson JJ, De Bolle X. 2013. The Brucella pathogens are polarized bacteria. Microbes Infect 15:998–1004. doi: 10.1016/j.micinf.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Deghelt M, Mullier C, Sternon JF, Francis N, Laloux G, Dotreppe D, Van der Henst C, Jacobs-Wagner C, Letesson JJ, De Bolle X. 2014. G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat Commun 5:4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene SE, Brilli M, Biondi EG, Komeili A. 2012. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol 194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westmacott D, Primrose SB. 1976. Synchronous growth of Rhodopseudomonas palustris from the swarmer phase. J Gen Microbiol 94:117–125. doi: 10.1099/00221287-94-1-117. [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol 12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laus MC, Logman TJ, Lamers GE, Van Brussel AA, Carlson RW, Kijne JW. 2006. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol 59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown PJ, Hardy GG, Trimble MJ, Brun YV. 2009. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54:1–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staley JT. 1984. Prosthecomicrobium hirschii, a new species in a redefined genus. Int J Syst Bacteriol 34:304–308. doi: 10.1099/00207713-34-3-304. [DOI] [Google Scholar]
- 30.Salieb-Beugelaar GB, Simone G, Arora A, Philippi A, Manz A. 2010. Latest developments in microfluidic cell biology and analysis systems. Anal Chem 82:4848–4864. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 31.Kovarik ML, Gach PC, Ornoff DM, Wang Y, Balowski J, Farrag L, Allbritton NL. 2012. Micro total analysis systems for cell biology and biochemical assays. Anal Chem 84:516–540. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu MH, Huang SB, Lee GB. 2010. Microfluidic cell culture systems for drug research. Lab Chip 10:939–956. doi: 10.1039/b921695b. [DOI] [PubMed] [Google Scholar]
- 33.Hol FJH, Dekker C. 2014. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346:1251821. doi: 10.1126/science.1251821. [DOI] [PubMed] [Google Scholar]
- 34.Gan MZ, Su J, Wang J, Wu HK, Chen LW. 2011. A scalable microfluidic chip for bacterial suspension culture. Lab Chip 11:4087–4092. doi: 10.1039/c1lc20670b. [DOI] [PubMed] [Google Scholar]
- 35.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. 2009. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip 9:545–554. doi: 10.1039/B810571E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. 2006. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A 103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gómez-Sjöberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. 2007. Versatile, fully automated, microfluidic cell culture system. Anal Chem 79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 38.Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA. 2003. Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sensor Actuat B-Chem 89:315–323. doi: 10.1016/S0925-4005(02)00468-9. [DOI] [Google Scholar]
- 39.Madren SM, Hoffman MD, Brown PJ, Kysela DT, Brun YV, Jacobson SC. 2012. Microfluidic device for automated synchronization of bacterial cells. Anal Chem 84:8571–8578. doi: 10.1021/ac301565g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel JJ, Givan SA, Brun YV, Brown PJ. 2015. Draft genome sequence of Prosthecomicrobium hirschii ATCC 27832T. Genome Announc 3(6):e01355-15. doi: 10.1128/genomeA.01355-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123–1130. doi: 10.1093/bioinformatics/17.12.1123. [DOI] [PubMed] [Google Scholar]
- 44.Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. 2002. The Pfam protein families database. Nucleic Acids Res 30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsokos CG, Laub MT. 2012. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol 15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray SM, Panis G, Fumeaux C, Viollier PH, Howard M. 2013. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol 11:e1001749. doi: 10.1371/journal.pbio.1001749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, Letesson JJ, Jacobs-Wagner C, De Bolle X. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J 26:1444–1455. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brassinga AK, Siam R, McSween W, Winkler H, Wood D, Marczynski GT. 2002. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J Bacteriol 184:5789–5799. doi: 10.1128/JB.184.20.5789-5799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaheen SM, Ouimet MC, Marczynski GT. 2009. Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155:1215–1225. doi: 10.1099/mic.0.025528-0. [DOI] [PubMed] [Google Scholar]
- 56.Siam R, Marczynski GT. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J 19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collier J, McAdams HH, Shapiro L. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A 104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez D, Kozdon JB, McAdams HH, Shapiro L, Collier J. 2014. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res 42:3720–3735. doi: 10.1093/nar/gkt1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohapatra SS, Fioravanti A, Biondi EG. 2014. DNA methylation in Caulobacter and other alphaproteobacteria during cell cycle progression. Trends Microbiol 22:528–535. doi: 10.1016/j.tim.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 60.Wright R, Stephens C, Shapiro L. 1997. The CcrM DNA methyltransferase is widespread in the alpha subdivision of Proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol 179:5869–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffman MD, Zucker LI, Brown PJ, Kysela DT, Brun YV, Jacobson SC. 2015. Timescales and frequencies of reversible and irreversible adhesion events of single bacterial cells. Anal Chem 87:12032–12039. doi: 10.1021/acs.analchem.5b02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toh E, Kurtz HD Jr, Brun YV. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janakiraman RS, Brun YV. 1999. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J Bacteriol 181:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levi A, Jenal U. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant Microbe Interact 21:1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- 67.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A 99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kysela DT, Brown PJ, Huang KC, Brun YV. 2013. Biological consequences and advantages of asymmetric bacterial growth. Annu Rev Microbiol 67:417–435. doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyler PA, Marshall KC. 1967. Pleomorphy in stalked, budding bacteria. J Bacteriol 93:1132–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.