Abstract
Histone methylation regulates DNA repair. However, the mechanisms that underlie the regulation of histone methylation during this repair remain to be further defined. Here, we show that ionizing radiation (IR) induces DNA-PK-dependent phosphorylation of nuclear fumarase at T236, which leads to an interaction between fumarase and the histone variant H2A.Z at DNA double-strand break (DSB) regions. Locally generated fumarate inhibits KDM2B histone demethylase activity, resulting in enhanced dimethylation of histone H3 K36; in turn, this increases the accumulation of the Ku70-containing DNA-PK at DSB regions for non-homologous end joining (NHEJ) DNA repair and cell survival. These findings reveal a feedback mechanism that underlies DNA-PK regulation by chromatin-associated fumarase and an instrumental function of fumarase in regulating histone H3 methylation and DNA repair.
Keywords: fumarase, fumarate, DNA-PK, histone H2AZ, KDM2B, histone H3, methylation, phosphorylation, NHEJ
INTRODUCTION
DNA double-strand breaks (DSBs) are one of the most deleterious forms of DNA damage1, 2. Higher eukaryotic cells primarily repair DSBs by one of two genetically separable pathways, homologous recombination (HR) or nonhomologous end joining (NHEJ)3, 4. HR operates in dividing cells in late S/G2 phase, as it requires homologous sister chromatid sequences as the template for mediating faithful repair3. By contrast, NHEJ repairs broken ends, with little or no need for sequence homology; it can function in both dividing and non-dividing cells and can occur in all cell cycle stages. In NHEJ, DSBs are recognized by the DNA-dependent protein kinase (DNA-PK) holoenzyme, which consists of the DNA end-binding heterodimer Ku70–Ku80 and the catalytic subunit DNA-PKcs. This binding results in activation of DNA-PKcs and the subsequent recruitment and activation of end-processing enzymes, polymerases, and DNA ligase IV1, 5.
DSB repair requires chromatin and nucleosome remodeling, which creates open, relaxed chromatin domains6. The histone-variant H2A.Z is exchanged onto nucleosomes at DSBs, which promotes specific patterns of histone modification and reorganization of the chromatin architecture. H2A.Z exchange at DSBs is required for the acetylation and ubiquitination of histones, loading the BRCA1 complex during HR DNA repair, and loading Ku70/Ku80 for NHEJ DNA repair7. Among the posttranslational modifications of histone, histone methylation is instrumental for DNA repair. Histone methylation mediates localization of 53BP1 to DSBs during HR repair8, 9. During NHEJ, IR was found to primarily induce dimethylated K36 of histone H3 (H3K36me2); this process was regulated by recruiting the DNA repair protein Metnase (also known as SETMAR), a SET histone methylase domain-containing protein, to DSBs. H3K36me2 near DSBs can be counteracted by expression of the KDM2 histone demethylase. H3K36 dimethylation improved the association of early DNA repair components, including Ku70, with DSBs10–12. Although H2A.Z exchange and histone H3 methylation play a vital role in NHEJ, how these events are regulated is unknown.
Metabolic enzymes can possess non-metabolic functions in response to extracellular stimuli, as reported by our research group and others13–17. Fumarase (FH) catalyzes the reversible hydration and dehydration of fumarate to malate in the tricarboxylic-acid cycle to facilitate a transition step in the production of energy in the form of NADH. In the cytosol, FH metabolizes fumarate, which is a byproduct of the urea cycle and amino acid catabolism18. FH translocates from the cytosol into the nucleus upon DNA damage. FH knockdown increases the sensitivity of cells to DNA damage, which can be complemented by high concentrations of fumarate but not by the expression of a catalytically inactive FH mutant. These findings indicate that the enzymatic activity of FH and its product, fumarate, are critical elements of the DNA damage response and that FH deficiency promotes tumorigenesis due to impairment of DNA repair15. However, the mechanism that underlies fumarase-dependent DNA damage repair remains largely elusive.
In this study, we found that ionizing radiation (IR) induced binding of FH to DSB regions in a manner that was dependent on DNA-PK-regulated phosphorylation of FH and the binding of FH to H2A.Z. Fumarate generated by DSB region-associated FH inhibited KDM2B, leading to enhanced H3K36me2 and promoted DNA repair.
RESULTS
H2A.Z-regulated recruitment of FH to DSB regions promotes the accumulation of Ku70 at DSB regions
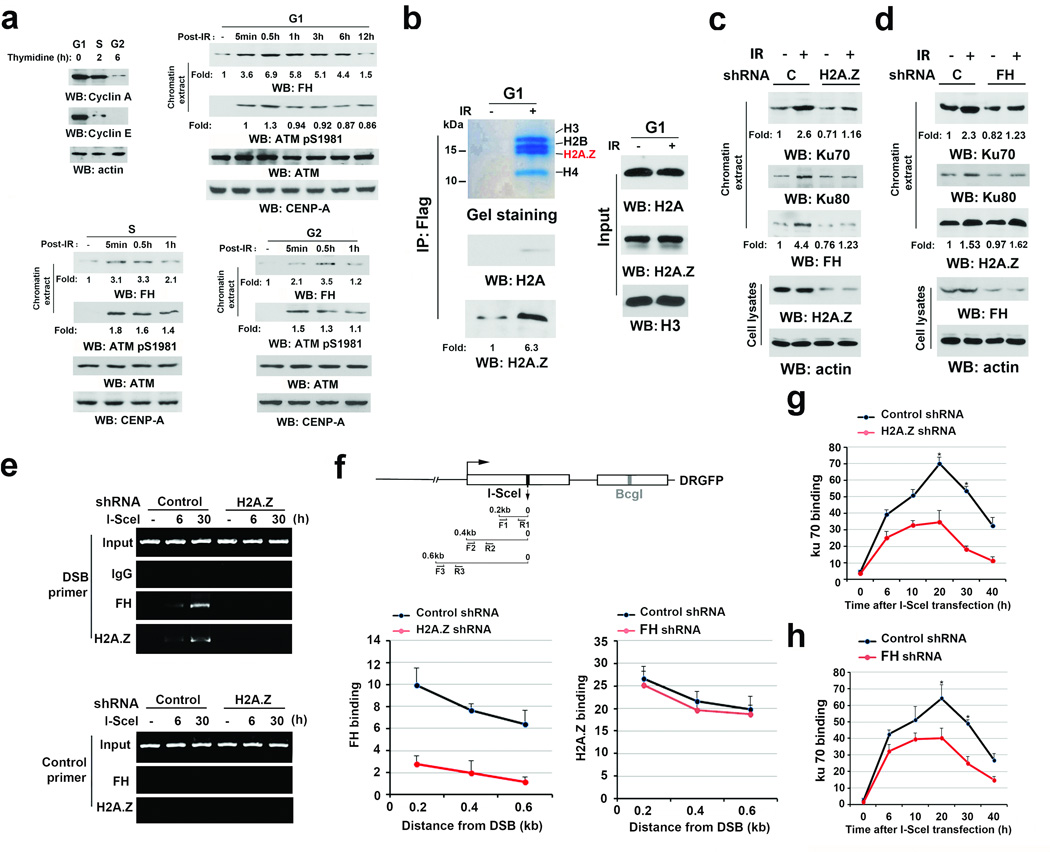
To determine whether FH is involved in IR-induced DSB repair in a cell cycle-dependent manner, we synchronized U2OS human osteosarcoma cells at G1 phase using a double-thymidine block or released the cells into S or G2 phase by removing thymidine for 2 or 6 h, respectively. The high expression of cyclin A in G1 and S phases and cyclin E in G1 phase reflected the status of different phases of cell cycle (Fig. 1a, left upper panel), which were also validated by flow cytometry analyses and by the detection of histone H3 S10 phosphorylation, a marker of late G2 phase and cell mitosis (Supplementary Fig. 1a)16. Immunoblotting analyses showed that IR, which resulted in ATM S1981 phosphorylation, enhanced the binding of FH to chromatin in G1 phase (Fig. 1a, right upper panel) and to a less extent at S or G2 phase (Fig. 1a, bottom panel). Given that HR occurs in late S/G2 phase and that NHEJ occurs throughout the whole cell cycle, these results suggest that chromatin-associated FH plays a role in DSB repair by NHEJ during G1 phase as well as other phases of the cell cycle. Similar results were also observed using GSC11 human primary glioblastoma cells (Supplementary Fig. 1b).
Figure 1. H2A.Z-regulated recruitment of FH to DSB regions promotes the accumulation of Ku70 at DSB regions.
a–d, Immunoblotting and e–h, ChIP analyses were performed using the indicated antibodies. a–e, Data represent one out of 3 experiments.
a, U2OS cells synchronized by thymidine double block (2 mM) underwent no release (upper right panel, G1 phase) or release for 2 h (bottom left panel, S phase) or 6 h (bottom right panel, G2 phase). These cells were then exposed to IR (10 Gy) and harvested at the indicated time after IR. Chromatin extracts were prepared. CENP-A was used as a control for chromatin-associated proteins.
b, Thymidine double block-synchronized U2OS cells expressing Flag-FH were exposed to IR (10 Gy) and harvested 1 h after IR. Chromatin extracts subjected to immunoprecipitation with an anti-FH antibody were analyzed by Coomassie brilliant blue staining and immunoblotting analyses.
c, d, Thymidine double block-synchronized U2OS cells, with or without H2A.Z shRNA (c) or FH shRNA (d) expression, were exposed to IR (10 Gy) and harvested 1 h after IR. Chromatin extracts and total cell lysates were prepared.
e, f, g, h, U2OS cells expressing the DR-GFP reporter, with or without H2A.Z shRNA, were transfected with a vector expressing I-SceI. ChIP analyses with the indicated antibodies were performed at the indicated time points (e, g, h) or 30 h (f) after I-SceI transfection. The indicated primers covering a range of distances from the cutting open site (f) or F1/R1 (e, g, h) primers were used for the PCR. Control primers were selected against a specific region of chromosome 12. The y-axis stands for the value of the I-SceI-induced fold increase of specific protein binding (the IP value was normalized to the input). The data represent the mean ± SD (n=3 independent experiments). * stands for P < 0.01 between the cells expressing control shRNA and the cells expressing the indicated shRNA.
We stably expressed Flag-FH in U2OS cells. Immunoprecipitation of chromatin-associated proteins with an anti-Flag antibody followed by mass spectrometry (Supplementary Fig. 1c) and immunoblotting analyses (Fig. 1b) showed that IR induced a dramatic increase in FH’s association with histones, and more histone variant H2A.Z than conventional H2A was detected in the immunoprecipitates. H2A.Z depletion in U2OS cells largely reduced the IR-induced chromatin association of FH, Ku70, and Ku80 (Fig. 1c). In contrast, FH depletion reduced the amount of chromatin-associated Ku70 and Ku80 but not H2A.Z (Fig. 1d).
To support this finding, we expressed I-SceI restriction enzyme to create DSBs in U2OS cells that contained an integrated DR-GFP gene with a unique I-SceI cutting site19, 20. A chromatin immunoprecipitation (ChIP) assay with antibodies against H2A.Z and FH showed that both H2A.Z and FH bound to the DNA adjacent to the I-SceI cutting site (Fig. 1e, 1f). In addition, H2A.Z depletion blocked the recruitment of FH to DSB regions (Fig. 1e; 1f, bottom left panel), while FH depletion had no significant effect on H2A.Z recruitment to DSBs (Fig. 1f, bottom right panel). Time-course studies showed that H2A.Z (Fig. 1g) and FH (Fig. 1h) depletion moderately affected the initial binding of Ku70 but led to dampened enrichment of Ku70 at the I-SceI cutting site. These results suggest that H2A.Z promotes the binding of FH to DSB regions, which is required for the DNA-PK complex accumulation at DNA damage areas.
DNA-PK phosphorylates FH at T236
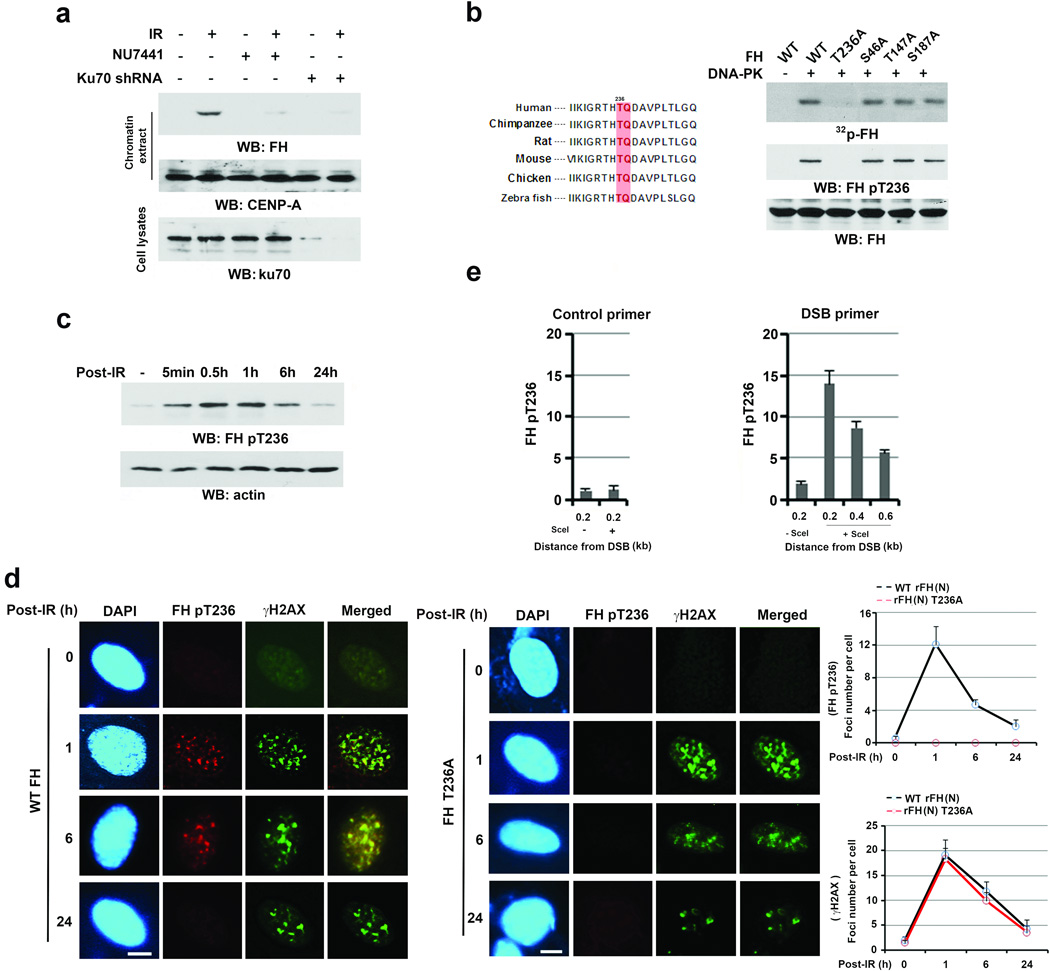
To determine the relationship between FH and the Ku70/Ku80-containing DNA-PK complex, we irradiated U2OS cells after DNA-PK inhibitor NU7441 treatment or Ku70 depletion. Fig. 2a shows that Ku70 depletion and DNA-PK inhibition blocked the IR-induced FH’s binding to chromatin, indicating an essential role of DNA-PK activity in this binding. In line with this result, an in vitro protein phosphorylation assay showed that purified, activated DNA-PK phosphorylated purified recombinant FH; this phosphorylation was only detected by immunoblotting analyses with an anti-phospho-threonine antibody (Supplementary Fig. 2a). We next mutated Scansite analysis-identified potential DNA-PK-phosphorylated residues and found that only the mutation of evolutionally conserved T236 abolished DNA-PK-mediated FH phosphorylation, as demonstrated by autoradiography and immunoblotting analysis with a specific FH pT236 antibody (Fig. 2b, Supplementary Fig. 2b). FH T236 phosphorylation was also detected in IR-treated U2OS cells (Fig. 2c) and blocked by pretreating the cells with NU7441 DNA-PK inhibitor but not KU55933 ATM inhibitor (Supplementary Fig. 2c). These results indicate that DNA-PK phosphorylates FH at T236 in vitro and in vivo.
Figure 2. DNA-PK phosphorylates FH at T236 and subsequently promotes Ku70 accumulation at DSB regions and NHEJ.
a–c, e, Immunoblotting analyses were performed with the indicated antibodies. Data represent one out of 3 experiments.
a, U2OS cells with or without Ku70 shRNA expression were pretreated with or without NU7441 (1 µM) before being exposed to IR (10 Gy) and harvested 1 h after IR. Chromatin extracts and cell lysates were prepared.
b, In vitro phosphorylation analyses were performed by mixing the purified DNA-PK complex with the indicated bacterially purified His-FH proteins in the presence of [γ-32P]ATP. T236/Q237 of FH, a DNA-PK phosphorylation motif, is evolutionally conserved in the indicated species.
c, Thymidine double block-synchronized U2OS cells (G1 phase) were exposed to IR (10 Gy) and harvested at the indicated time points after IR.
d, U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins were irradiated with IR (3 Gy). Immunofluorescence analyses with the indicated antibodies were performed at the indicated time points after IR. The number of foci in 30 cells were counted and quantified. Bar, 5 µm. The data represent the mean ± SD (n=3 independent experiments).
e, U2OS cells expressing the DR-GFP reporter were transfected with a vector expressing I-SceI. ChIP analyses with an anti-FH pT236 antibody were performed 30 h after I-SceI transfection. The primers described in Fig. 1f were used for the PCR. Control primers were selected against a specific region of chromosome 12. The y-axis stands for the value of the I-SceI-induced fold increase of binding of FH pT236 to the specific regions in DSBs (the IP value was normalized to the input). The data represent the mean ± SD (n=3 independent experiments).
Cell fractionation analyses showed that nuclear FH was much less in amount than cytosolic FH, and IR enhanced both nuclear and chromatin-associated FH (Supplementary Fig. 2d, left panel). Immunoblotting analyses of the nuclear extract with an anti-FH antibody after immunodepletion with an anti-FH pT236 antibody showed that about 23% of nuclear FH was phosphorylated (Supplementary Fig. 2d, right panel), and most phosphorylated FH was associated with chromatin upon IR (Supplementary Fig. 2d, left panel). In line with these findings, IR enhanced the co-localization of γH2AX with FH pT236 (Fig. 2d), and FH T236A expression did not affect IR-induced γH2AX or ATM pS1981 (Fig. 2d, Supplementary Fig.2e and 2f-top panel). These results suggest that DNA-PK-mediated FH phosphorylation is involved in NHEJ and does not affect ATM-regulated γH2AX or HR-dependent DNA repair; they also suggest that IR induces the recruitment of FH specifically to the DNA damage regions. The latter conclusion was supported by ChIP analyses, which showed that phosphorylated FH T236 was primarily located in the region within 0.5 kb of the I-SceI cutting site in U2OS cells (Fig. 2e). Consistent with a previous report15, FH depletion did not affect the initial stage of γH2AX but affected the duration of γH2AX upon IR (Supplementary Fig. 2f, bottom panel), suggesting that FH participates in DNA repair at a late stage in an FH pT236-independent manner.
DNA-PK-phosphorylated FH promotes the DNA-PK complex accumulation at DSB regions and NHEJ
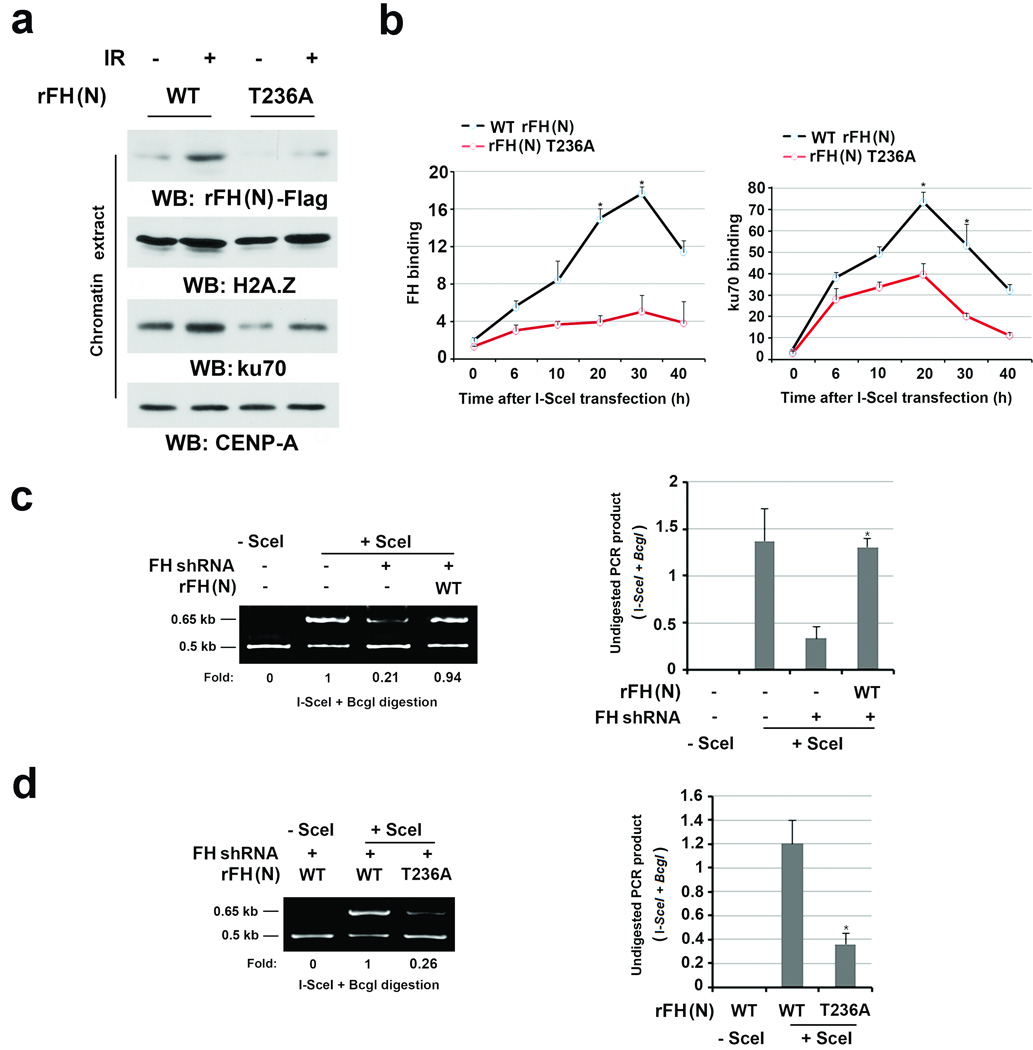
To determine whether IR-induced FH pT236 phosphorylation is dependent on mitochondrial localization of FH, we depleted endogenous FH and reconstituted the expression of RNAi-resistant (r) FH (N), in which the 21 N-terminal amino acids containing the mitochondrial localization sequence were deleted21, 22, rFH (N) T236A, or full-length rFH (FL) (Supplementary Fig. 3a). Immunofluorescence analysis confirmed that rFH (N) and rFH (N) T236A were not localized in mitochondria (data not shown). rFH (N) showed efficient binding to chromatin (Supplementary Fig. 3b) and phosphorylation at T236 compared to its full-length counterpart upon IR (Supplementary Fig. 3c), indicating that FH T236 phosphorylation is independent of its mitochondrial localization. In contrast, FH T236A failed to associate with chromatin (Fig. 3a) and bind to the I-SceI cutting site (Fig. 3b, left panel) and reduced Ku70 accumulation at DSB regions (Fig. 3b, right panel). These results indicate that the DNA-PK complex and FH regulate each other and that DNA-PK-dependent FH phosphorylation promotes the accumulation of FH and the DNA-PK complex at DSB regions.
Figure 3. DNA-PK-phosphorylated FH promotes the DNA-PK complex accumulation at DSB regions and NHEJ.
a, Thymidine double block-synchronized U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins were exposed to IR (10 Gy) and harvested 1 h after IR. Chromatin extracts or total cell lysates were prepared.
b, DR-GFP-expressed U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins were transfected with a vector with or without expressing I-SceI. ChIP analyses with antibodies that recognize FH (left panel) or Ku70 (right panel) and F1/R1 primers for the PCR were performed at the indicated time points after I-SceI transfection. The y-axis stands for the value of the I-SceI-induced fold increase of specific protein binding (the IP value was normalized to the input). The data represent the mean ± SD (n=3 independent experiments).
c, d, DR-GFP-expressed U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins were transfected with a vector with or without expressing I-SceI. PCR analyses for NHEJ were performed 42 h after transfection. A representative image of PCR products digested by I-SceI and BcgI is shown (left panel). The data represent the mean ± SD (n=3 independent experiments, right panel). * stands for P < 0.05 between the FH-depleted cells, with or without WT rFH (N) expression (c), and between the cells expressing WT rFH (N) and rFH (N) T236A (d).
To determine the role of FH T236 phosphorylation in DNA repair, we used a PCR strategy and a DR-GFP system to analyze NHEJ repair and HR, respectively (Supplementary Fig. 3d)19, 20. The NHEJ-regulated repair of I-SceI-induced DSBs in the GFP locus resulted in a 0.65-Kb PCR fragment that was resistant to both I-SceI and BcgI digestion (Supplementary Fig. 3e). As expected, depletion of Ku70 (Supplementary Fig. 3e) and FH (Fig. 3c) largely dampened NHEJ-regulated repair, and this defect induced by FH depletion was rescued by reconstituted expression of rFH (N) but not FH (N) T236A (Fig.3c and 3d) while FH depletion and reconstituted expression of rFH (N) and FH (N) T236A had no effect on the efficiency of DSB production by I-Sce1 (Supplementary Fig. 3f, 3g). In contrast, reconstituted expression of FH (N) and FH (N) T236A did not affect the HR-regulated repair of the I-SceI-induced DSB, which was represented by HR-restored GFP expression (Supplementary Fig. 3h). These results indicate that DNA-PK-regulated FH pT236 plays an instrumental role in NHEJ- but not HR-mediated DNA repair.
The binding of FH pT236 to H2A.Z promotes the DNA-PK complex accumulation at DSB regions and NHEJ
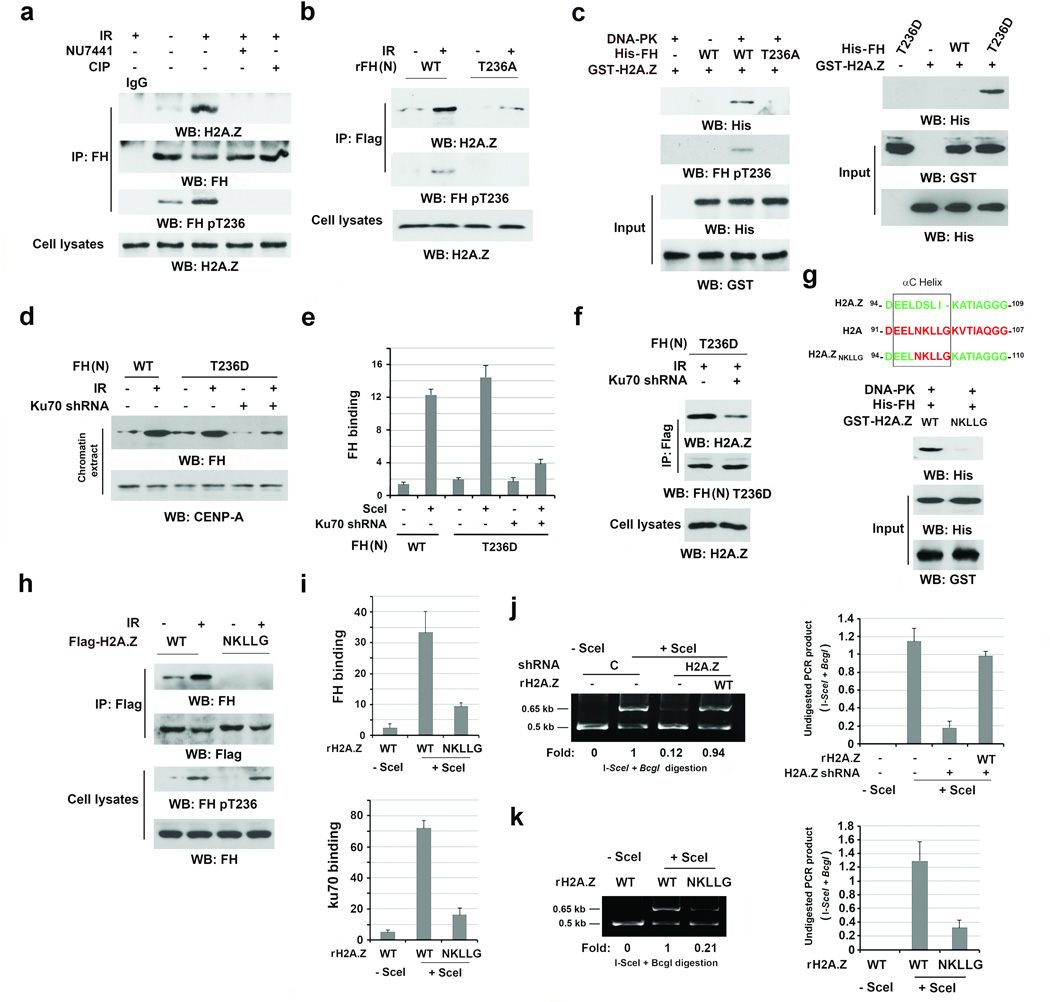
H2A.Z mediates the association between FH and chromatin upon DNA damage (Fig.1c and 1f, bottom left panel). To determine the relationship between H2A.Z and FH T236 phosphorylation, we pretreated U2OS cells with NU7441, treated immunoprecipitated FH with calf intestinal alkaline phosphatase (CIP), or expressed FH (N) T236A. We demonstrated that DNA-PK inhibition, CIP treatment (Fig. 4a), and FH (N) T236A expression (Fig. 4b) blocked IR-induced interaction between FH and H2A.Z. Of note, purified WT His-FH failed to bind to purified H2A.Z (Fig. 4c). However, the inclusion of DNA-PK enabled WT His-FH but not His-FH T236A to bind to H2A.Z (Fig. 4c, left panel). These results indicate that DNA-PK-mediated FH phosphorylation promotes the binding of FH to H2A.Z. Consistent with this finding, purified phosphorylation-mimic FH T236D mutant directly bound to H2A.Z in vitro (Fig. 4c, right panel). In response to DNA damage, FH T236D largely increased its association with chromatin (Fig. 4d) and accumulation at DSB regions (Fig. 4e) and this increase was blocked by Ku70 depletion. In addition, IR-induced binding of FH T236D to H2A.Z was also dramatically impaired by Ku70 depletion (Fig. 4f). These results suggest that the DNA-PK complex, in addition to recruiting and phosphorylating FH at DSB regions, alters the local chromatin structure and makes H2A.Z accessible for interaction with phosphorylated FH.
Figure 4. The binding of FH pT236 to H2A.Z promotes the DNA-PK complex accumulation at DSB regions and NHEJ.
a–d, f–h, Immunoprecipitation and Immunoblotting were performed with the indicated antibodies. a, b, d, f, h, U2OS cells were harvested 1 h after IR (10 Gy). Data represent one out of 3 experiments. e, i, j, k, The data represent the mean ± SD (n=3 independent experiments).
a, Thymidine double block-synchronized U2OS cells were pretreated with or without NU7441 (1 µM) before IR for 30 min. The immunoprecipitates were treated with or without CIP (10 units).
b, U2OS cells with depleted FH and reconstituted expression of the indicated FH proteins were exposed to IR.
c, Immobilized, purified GST-H2A.Z protein was mixed with the indicated purified His-FH proteins with or without DNA-PK. GST pull-down analyses were performed.
d, U2OS cells with or without Ku70 depletion expressed the indicated Flag-FH protein and were treated with or without IR. Chromatin extracts were prepared.
e, DR-GFP-expressing U2OS cells with depleted Ku70 expressed the indicated Flag-FH proteins with or without I-SceI. ChIP analyses were performed with an anti-Flag antibody. The y-axis stands for the value of the I-SceI-induced fold increase of FH binding.
f, U2OS cells with depleted FH and reconstituted expression of Flag-FH T236D protein contained with or without Ku70 depletion and were treated with IR.
g, The schematic diagram shows the replacement of the αC Helix sequence in H2A.Z with H2A (top panel). Purified and DNA-PK-phosphorylated His-FH protein was mixed with the indicated purified GST-H2A.Z proteins. A GST pull-down assay was performed (bottom panel).
h, Thymidine double block-synchronized U2OS cells with depleted H2A.Z and reconstituted expression of the indicated H2A.Z proteins were exposed to IR.
i, j, k, DR-GFP-expressing U2OS cells with or without I-SceI expression were depleted endogenous H2A.Z and reconstituted the indicated H2A.Z protein expression. ChIP analyses were performed with antibodies that recognize FH (i, top panel) or Ku70 (i, bottom panel). An NHEJ analysis was performed (j, k).
The M6 cassette that encompasses amino acids 89–100 of H2A.Z specifically interacts with other proteins23. The main differences between the M6 cassettes of H2A.Z and H2A, which had limited binding to FH (Fig. 1b), reside in the small αC-helix23. Replacing DSLI in the αC-helix of H2A.Z with the corresponding NKLLG in H2A showed that the purified H2A.Z (NKLLG) mutant lost its binding to purified FH in vitro (Fig. 4g). In addition, Flag-H2A.Z (NKLLG) mutant failed to bind to FH in endogenous H2A.Z-depleted U2OS cells upon IR (Fig. 4h, Supplementary Fig. 4a). In addition, expression of the H2A.Z (NKLLG) mutant, which accumulated at DSB regions like its WT counterpart (Supplementary Fig. 4b), largely reduced its binding of FH (Fig. 4i, top panel; Supplementary Fig. 4b) and Ku70 (Fig. 4i, bottom panel) at DSB regions. Analyses of NHEJ showed that H2A.Z depletion (Fig. 4j) or reconstituted expression of rH2A.Z (NKLLG) (Fig. 4k), which did not affect the efficiency of DSB production by I-Sce1 (Supplementary Fig. 4c), inhibited DSB repair. These results indicate that the interaction between H2A.Z and FH pT236 is critical for the binding of FH and the DNA-PK complex at DSB regions and for NHEJ-mediated DNA repair.
Fumarate produced by chromatin-associated FH promotes NHEJ
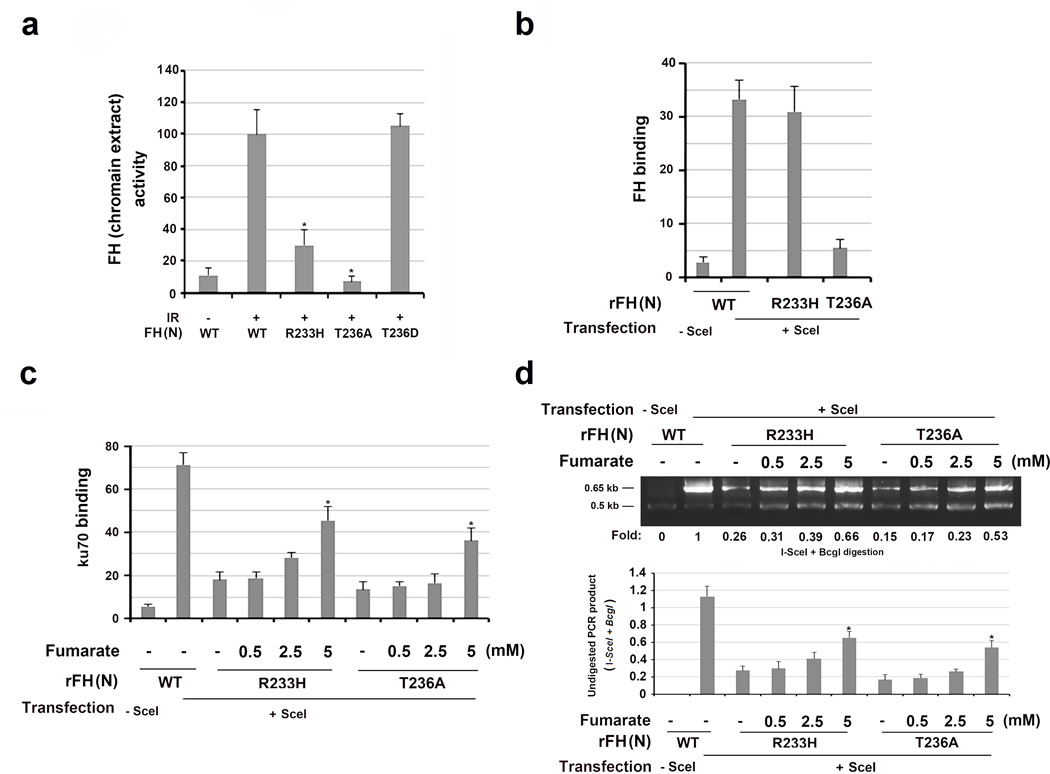
To determine whether FH activity is involved in NHEJ repair, we examined the chromatin-associated FH activity in converting malate to fumarate. As shown in Fig. 5a, IR enhanced comparable chromatin-associated activity between FLAG-tagged WT FH (N) and FH (N) T236D mutant, which was in line with the increased enrichment of FH in chromatin (Fig.1a and 4d). In contrast, the catalytically inactive FH (N) R233H mutant, which was phosphorylated and recruited to chromatin (Supplementary Fig. 5a, left panel), exhibited little activity, and FH (N) T236A, which had similar activity to its WT counterpart and FH (N) T236D in vitro (Supplementary Fig. 5b), did not bind to chromatin (Fig. 3a) or exhibit chromatin-associated activities (Fig. 5a). FH (N) R233H, which retained its ability to co-immunoprecipitate with H2A.Z (Supplementary Fig. 5a, right panel) and bind it in vitro (Supplementary Fig. 5c), was able to translocate to I-SceI-cutting regions (Fig. 5b); however, it acted like FH T236A, reducing Ku70 accumulation at the DSB regions (Fig. 5c) and blocking the repair of I-SceI-induced DSB by NHEJ (Fig. 5d). These results indicate that FH activity is required for DSB repair by NHEJ.
Figure 5. Fumarate produced by chromatin-associated FH promotes NHEJ.
The data represent the mean ± SD (n=3 independent experiments). c, d, * stands for P < 0.05 between the indicated samples and the WT counterparts without adding exogenous monoethyl-fumarate.
a, Chromatin extracts of thymidine double block-synchronized U2OS cells were collected 1 h after IR. Relative FH activity was measured. a, * stands for P < 0.05 between the indicated samples and the WT counterpart.
b, DR-GFP-expressing U2OS cells with depleted FH and reconstituted expression of the indicated FH proteins were harvested 30 h after transfection with a vector expressing I-SceI. ChIP analyses with an anti-FH antibody were performed.
c, DR-GFP-expressing U2OS cells with depleted FH and reconstituted expression of the indicated FH proteins were incubated with the indicated concentration of monoethyl-fumarate for 20 h after I-SceI transfection. ChIP analyses with an anti-Ku70 antibody were performed.
d, DR-GFP-expressed U2OS cells with depleted FH and reconstituted expression of the indicated FH proteins were incubated with the indicated concentration of monoethyl-fumarate for 20 h after I-SceI transfection. An NHEJ analysis was performed. A representative image of the PCR products digested by I-SceI and BcgI is presented.
FH depletion-dampened DNA repair can be partially rescued by high concentrations of fumarate15, indicating the important role of fumarate in this process. Enzymatically active FH T236A failed to mediate DSB repair, suggesting that local accumulated fumarate in DNA damage regions is critical for DNA repair. This assumption was supported by the addition of various amounts of exogenous monoethyl-fumarate into the cell medium, which resulted in accordingly increased concentrations of intracellular fumarate (up to 15 fold) (Supplementary Fig. 5d). Fig. 5c shows a fumarate dosage-dependently partial rescue of Ku70 accumulation at DSB regions and restoration of NHEJ repair (Fig. 5d) in U2OS cells expressing FH R233H or FH T236A mutants. In contrast, the addition of exogenous malate in a high dosage, which cannot be converted into fumarate in the DNA damage regions by FH R233H or FH T236A, did not exert any effect on Ku70 binding at DSB regions (Supplementary Fig. 5e) or NHEJ repair (Supplementary Fig. 5f). Intriguingly, reconstituted expression of inactive FH (N) R233H, but not of its WT counterpart or enzyme activity-intact FH (N) T236A, in endogenous FH-depleted cells increased total cellular fumarate production, as did FH depletion21 (Supplementary Fig. 5g). Compared to FH (N) T236A expression, expression of FH (N) R233H resulted in less dampened Ku70 accumulation at DSB regions (Fig. 5c) and DNA repair (Fig. 5d) due to increased cellular fumarate levels. However, this increased cellular level of fumarate had limited effects on the local concentration of fumarate at DSB regions. This observation is consistent with the finding that the incubation of a large amount of exogenous fumarate only partially affected Ku70 accumulation at DSB regions (Fig. 5c) and NHEJ repair (Fig. 5d). These results strongly suggest that local fumarate production at DSB regions by chromatin-associated FH play a critical role in NHEJ repair.
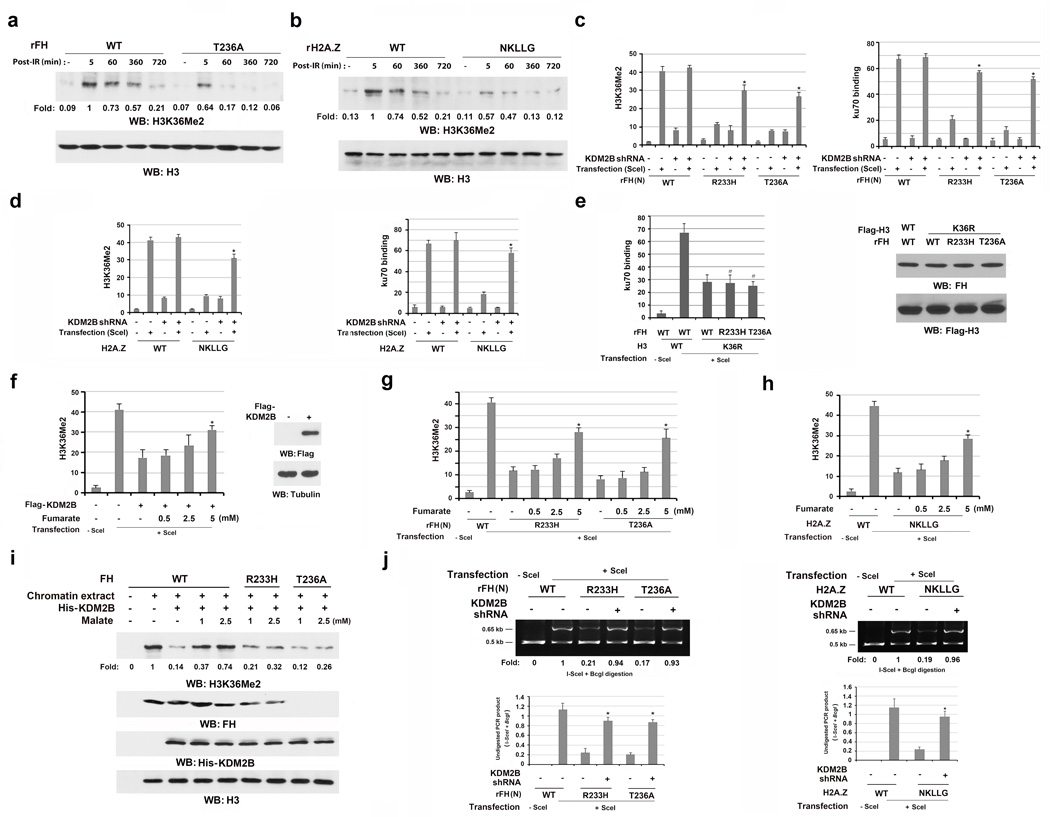
Fumarate produced by chromatin-associated FH inhibits KDM2B-mediated H3K36Me2 demethylation
Fumarate can inhibit α-ketoglutarate (αKG)-dependent dioxygenases, altering histone methylation24. H3K36 dimethylation improves the association of Ku70 at DSBs10. As expected, H3K36Me2, but not H3K9me2, H3K9me3, or H3K27me2, were dramatically increased at the DSBs induced by I-SceI expression (Supplementary Fig. 6a). In addition, H3K36me2 primarily accumulated in the region within 0.5 kb of the I-SceI cutting site (Supplementary Fig. 6b), and expression of the H3K36R mutant largely compromised the recruitment of Ku70 to I-SceI-cutting sites (Supplementary Fig. 6c). To determine whether chromatin-associated FH regulates H3K36Me2 and the subsequent Ku70 accumulation at DSB regions, we performed immunoblotting analyses and showed that reconstituted expression (Supplementary Fig. 6d) of rFH (N) T236A (Fig. 6a) or H2A.Z (NKLLG) (Fig. 6b) largely inhibited IR-induced chromation-associated H3K36Me2. Consistent with these findings, reconstituted expression of rFH T236A, rFH R233H (Fig. 6c, left panel), and H2A.Z (NKLLG) (Fig. 6d, left panel) notably suppressed H3K36Me2 at the DSB regions. In addition, rFH T236A or rFH R233H expression did not further reduce H3K36R-suppressed Ku70 accumulation at DSB regions (Fig. 6e). These results indicate that chromatin-associated FH regulates H3K36Me2 and the subsequent accumulation of the DNA-PK complex at DSB regions.
Figure 6. Fumarate produced by chromatin-associated FH inhibits KDM2B-mediated H3K36Me2 demethylation.
a, b, e, f, i, Immunoblotting were performed with the indicated antibodies. Data represent one out of 3 experiments. c–h, j, The data represent the mean ± SD (n=3 independent experiments). c, d, f, g, h * stands for P < 0.05 between the indicated samples and the counterparts without adding monoethyl-fumarate.
a, b, Thymidine double block-synchronized GSC11 cells with depleted FH and reconstituted expression of the indicated FH proteins were exposed to IR (10 Gy) and harvested 1 h after IR. Chromatin extracts were prepared at the indicated time points after IR.
c, d, g, h, j, DR-GFP-expressing U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins (c, g, j left panel) or with depleted endogenous H2A.Z and reconstituted expression of the indicated H2A.Z proteins (d, h, j right panel) expressed or did not express KDM2B shRNA and/or I-SceI (c, d, j). (c, d) ChIP analyses were performed with antibodies for H3K36Me2 and Ku70. (g, h) The cells were incubated with or without the indicated concentration of monoethyl-fumarate for 20 h after I-SceI transfection. ChIP analyses were performed with an anti-H3K36Me2 antibody. (j) An NHEJ analysis was performed 42 h after I-SceI transfection. A representative image of PCR products digested by I-SceI and BcgI is shown. * stands for P < 0.01 between the indicated samples and the counterparts without KDM2B shRNA.
e, U2OS cells with depleted FH and reconstituted expression of the indicated FH proteins were expressed with the indicated H3 proteins. ChIP analyses with an anti-Ku70 antibody were performed 30 h after I-SceI transfection. # stands for no statistical significance between the indicated samples and the WT counterparts.
f, DR-GFP-expressing U2OS cells with or without expression of Flag-KDM2B were incubated with or without the indicated concentration of monoethyl-fumarate for 20 h after I-SceI transfection. ChIP analyses were performed with an anti-H3K36Me2 antibody.
i, GSC11 cells with depleted endogenous KDM2B and FH and reconstituted expression of the indicated FH proteins were exposed to IR. Chromatin extracts were incubated with the indicated concentration of malate for 30 min, followed by incubation with KDM2B.
KDM2B preferentially demethylates H3K36Me212. KDM2B depletion (Supplementary Fig. 6e) rescued rFH (N) R233H-, rFH (N) T236A-, H2A.Z (NKLLG)-suppressed H3K36Me2 at DSB regions (Fig.6c and 6d, left panel) and Ku70 accumulation at DSB regions (Fig.6c and 6d, right panel). Notably, expression of these mutants did not affect the localization of KDM2B at the DSB regions (Supplementary Fig. 6f). These results strongly suggest that chromatin-associated FH regulates H3K36Me2 by inhibiting KDM2B.
To determine the involvement of fumarate in KDM2B-mediated H3K36Me2 demethylation, we overexpressed KDM2B and added exogenous monoethyl-fumarate to the cell medium. Fig. 6f shows that ectopic expression of KDM2B reduced H3K36Me2 levels at the DSB regions; this suppression was counteracted by exogenous fumarate in a dosage-dependent manner. In addition, exogenous fumarate partially rescued the suppression of H3K36Me2 at the DSB regions induced by the expression of FH T236A, FH R233H (Fig. 6g), or H2A.Z (NKLLG) (Fig. 6h). Furthermore, mixing purified KDM2B with chromatin extracted from WT FH-expressing GSC11 cells demethylated IR-induced H3K36Me2 (Fig. 6i). However, this demethylation was inhibited by adding malate in a dosage-dependent manner (Fig. 6i), and this demethylation inhibition was abrogated by inclusion of αKG (Supplementary Fig. 6g). In sharp contrast, malate did not affect H3K36Me2 levels in cells expressing rFH R233H that lost its ability to convert malate to fumarate (Fig. 6i).
We next determined the effect of FH-regulated H3K36Me2 on NHEJ-mediated DSB repair. As shown in Fig. 6j, KDM2B depletion in U2OS rescued impaired DSB repair in cells expressing rFH T236A, rFH R233H (left panel), and H2A.Z (NKLLG) (right panel). These results indicate that FH-produced fumarate promotes NHEJ-dependent DNA repair by inhibiting KDM2B-mediated demethylation at DSB regions.
Fumarate produced by chromatin-associated FH promotes cell survival
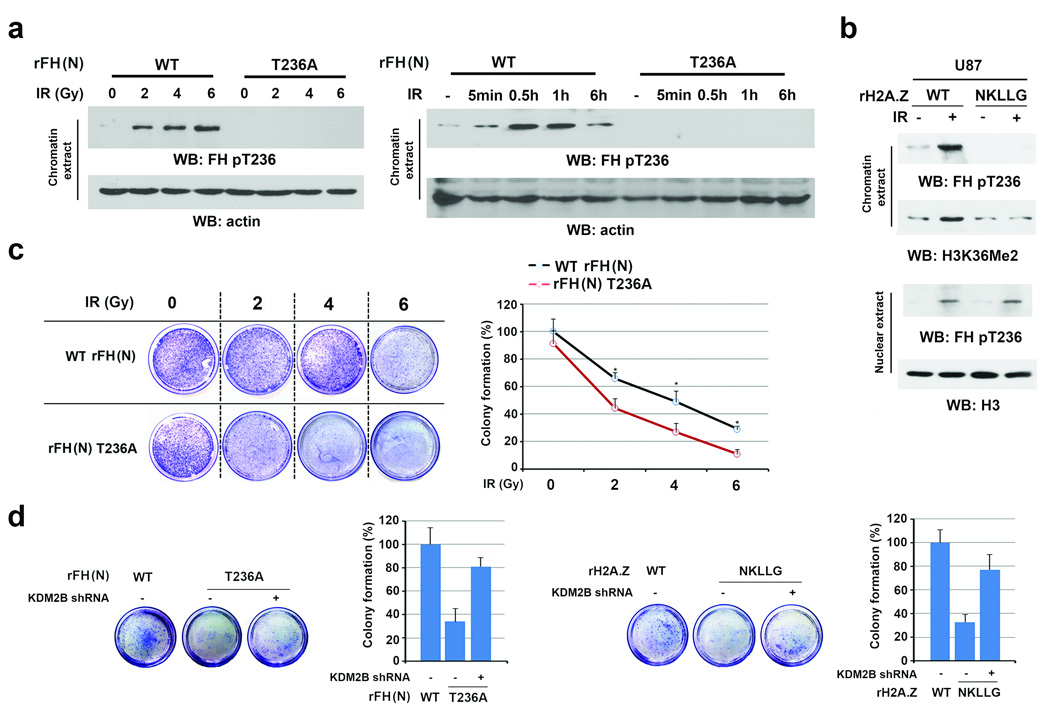
We found that IR-induced and FH phosphorylation-regulated H3K36Me2 occurred in both U2OS and GSC11 cells. To determine whether this finding also applied to other cell lines, we deleted FH or H2A.Z and reconstituted the expression of rFH (N) T236A (Supplementary Fig. 7a), H2A.Z (NKLLG) (Supplementary Fig. 7b), or their WT counterparts in U87 GBM cells, HeLa cervical cancer cells, and A549 non-small lung cancer cells. In contrast to the WT protein expression, rFH T236A (Fig. 7a, Supplementary Fig. 7c) and H2A.Z (NKLLG) (Fig. 7b, Supplementary Fig. 7d) expression largely reduced IR-enhanced chromatin-associated FH pT236 and H3K36Me2. These results indicate that FH phosphorylation-regulated H3K36Me2 occurs in different types of cancer cells.
Figure 7. Fumarate produced by chromatin-associated FH promotes cell survival.
a, b, Immunoblotting analyses were performed with the indicated antibodies. Data represent one out of 3 experiments. c, d, Cell viability was examined 12 h after IR by using clonogenic assay. The data represent the mean ± SD (n=3 independent experiments).
a, Thymidine double block-synchronized U87 cells with depleted endogenous FH and reconstituted expression of FH WT or T236A were exposed to IR as the indicated doses and harvested at 1 h after IR (left panel) or were exposed to IR (10 Gy) and harvested at the indicated time points (right panel). Chromatin extracts were prepared.
b, Thymidine double block-synchronized U87 cells with depleted endogenous H2A.Z and reconstituted expression of the indicated H2A.Z proteins were exposed to IR (10 Gy) and harvested at 1 h after IR. Nuclear fractions and chromatin extracts were prepared.
c, U2OS cells with depleted endogenous FH and reconstituted expression of FH proteins were exposed to IR as the indicated doses. * stands for P < 0.05.
d, U2OS cells with depleted endogenous FH and reconstituted expression of the indicated FH proteins (left panel) or with depleted endogenous H2A.Z and reconstituted expression of the indicated H2A.Z proteins (right panel) were transfected with or without a vector expressing KDM2B shRNA and exposed to IR (6 Gy).
Successful DNA repair prohibits DNA damage-induced cell death. Clonogenic assay showed that IR enhanced the death rate of U2OS cells expressing rFH T236A (Fig. 7c) or H2A.Z (NKLLG) (Fig. 7d, right panel) compared to that of cells with expression of their WT counterparts; this enhanced cell death by the mutant protein expression was blocked by KDM2B depletion (Fig. 7d, Supplementary Fig. 7e). Similar results were obtained by using the trypan blue analyses of different types of cancer cells (Supplementary Fig. 7f, 7g). These results indicate that FH phosphorylation regulates NHEJ by antagonizing H3 demethylation and promotes cell survival in response to IR.
DISCUSSION
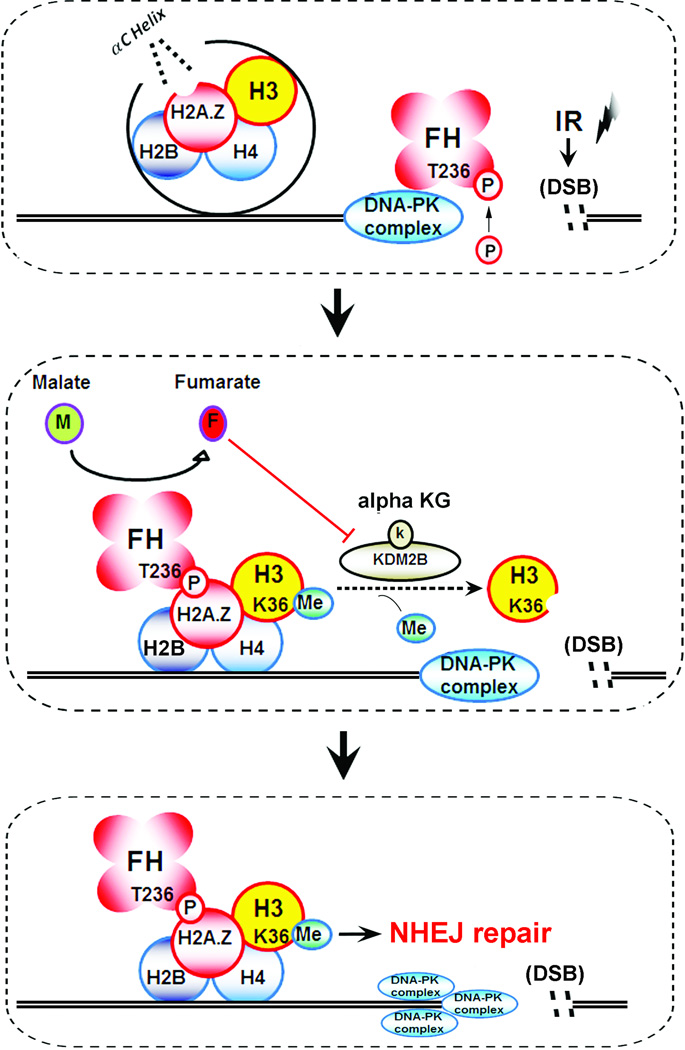
Metabolic enzymes execute their metabolic functions in cytosol and mitochondria. However, in response to extracellular stimuli, these enzymes, which include PKM2, FH, and MATIIα (a methionine adenosyltransferase isozyme), possess functions that are not directly linked to metabolism regulation when they alter their subcellular localization12–17, 25, 26. In this report, we demonstrated that IR results in the binding of FH to H2A.Z at DSB regions in a manner that is dependent on DNA-PK-mediated FH T236 phosphorylation. FH pT236, which dissociated from DNA-PK, interacted with the adjacent αC-helix of H2A.Z, leading to DSB region-enriched FH and the subsequent fumarate production in this region. The locally accumulated FH inhibited KDM2B activity at DSB regions and enhanced H3K36Me2, which in turn promoted the accumulation of the DNA-PK complex at DSB regions for subsequent NHEJ-mediated DNA repair and cell survival (Fig. 8). The results of this study underscore the instrumental role of chromatin-associated FH in IR-induced DSB repair.
Figure 8. A schematic model: DNA-PK–mediated phosphorylation of fumarase promotes DNA repair by local fumarate-inhibited histone H3 demethylation.
IR induces DNA-PK-mediated phosphorylation of FH T236, leading to the binding of FH to H2A.Z adjacent to DSBs. Local FH-produced fumarate inhibits KDM2B, which results in enhanced H3K36Me2 and accumulation of DNA-PK at DSBs for NHEJ.
HR, which requires complete histone eviction adjacent to the DSBs, regulates DNA repair during S-G2 phase3. In our study, FH increased its association with chromatin after IR. In addition, DNA-PK, which is active at the early stage of the NHEJ process, phosphorylates FH T236. Importantly, FH T236A expression, which had little effect on HR-regulated DSB repair, blocked H3K36Me2 at DSB regions and impaired the function of NHEJ. These results revealed that FH T236 phosphorylation plays an important role in NHEJ-mediated DNA repair in a cell cycle-dependent manner. Since FH is also involved in regulating γH2AX15, which was not affected by FH T236A expression (data not shown), these findings suggest that FH participates in HR repair independently of the DNA-PK-mediated FH phosphorylation.
FH T236 is rapidly phosphorylated by DNA-PK at the early phase of NHEJ in response to IR. FH depletion or FH T236A expression had less effect in the early phase than in the late stage on the binding of the DNA-PK complex to the DNA open end. These results suggest that FH T236 prevents the dissociation of DNA-PK from chromatin and are in line with the previous observation that H3K36me2 in DSB regions decreased the rates of dissociation of DNA-PK from chromatin10. These findings imply that accumulated local fumarate production by FH and enhanced H3K36me2 at DNA damage regions stabilizes the repair components of NHEJ at the DSBs. Our studies reveal a feedback mechanism for regulating DNA-PK and thus NHEJ-mediated DSB repair through DNA-PK-mediated FH T236 phosphorylation, fumarate-dependent H3K36me2, and the subsequent maintenance of the DNA-PK complex at the DSB regions for recruiting other DNA repair components.
Chromatin-localized metabolite biosynthesis plays an important role in nucleosome and chromatin modification. For instance, protein acetyltransferases require acetyl-CoA as a donor of acetyl groups in histone acetylation, and poly (ADP-ribose) polymerases require oxidized nicotinamide adenine dinucleotide as a donor of ADP-ribose units in poly(ADP-ribosyl)ation reactions27. The demonstration of the critical roles of chromatin-localized FH and FH-produced fumarate in regulating histone H3 methylation and DNA repair provides instrumental insight into how subcellular compartment- or locally localized metabolic enzymes and their products regulate cellular activities that are not directly linked to metabolism.
MATERIALS AND METHODS
Cell culture and synchronization
U2OS cells were maintained in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS). U87, HeLa, and A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum (HyClone, Logan, UT). Human primary GSC11 cells were maintained in DMEM/F-12 50/50 supplemented with B27, EGF (10 ng/ml), and basic fibroblast growth factor (10 ng/ml).
Cells (30%-40% in confluence) were washed twice with phosphate-buffered saline (PBS), treated with 2 mM thymidine for 17 h, washed twice with PBS again, released in complete medium containing 10 µM deoxycytidine for 9 h, treated with 2 mM thymidine for 17 h, and released in complete medium with 10 µM deoxycytidine before being assayed.
After the double thymidine block, cells were washed twice with PBS and released in complete medium for 2 h (S phase) or 6 h (G2 phase).
Materials
Rabbit polyclonal antibodies that recognize FH (#32975, 1:1000), and histone H3 pS10 (#12084, 1:1000) were obtained from Signalway Antibody (College Park, MD). Antibodies that recognize cyclin A (ab87359, 1:1000), cyclin E (ab7959, 1:1000), FH (ab113963, 1:1000), H2A.Z (ab18262, 1:1000 for immunoblotting, 1:400 for immunofluorescence), H2A (ab18255, 1:1000), CENP-A (ab13939, 1:1000), H3 (ab1791, 1:1000), H3K36Me2 (ab9049, 1:1000), and KDM2B (ab108276, 1:1000) were purchased from Abcam (Boston, MA). Rabbit polyclonal antibodies against Ku70 (#4588, 1:1000) were from Cell Signaling Technology (Beverly, MA). Thymidine, monoethyl-fumarate, micrococcal nuclease, KU55933, and mouse monoclonal antibodies for tubulin (T9026, 1:10,000), Flag (F3165, 1:10,000), GST (G1160, 1:10,000), and His (SAB4301134, 1:10,000) were purchased from Sigma (St. Louis, MO). I-SceI and BcgI restriction enzymes were purchased from New England Biolabs (Ipswich, MA). Hygromycin, puromycin, DNase-free RNase A, and propidium iodide were purchased from EMD Biosciences (San Diego, CA). NU7441 was purchased from Selleckchem (Houston, TX). HyFect transfection reagent was from Denville Scientific (Metuchen, NJ). GelCode blue stain reagent was obtained from Pierce (Rockford, IL).
Rabbit polyclonal FH pT236 antibody was made by Signalway Antibody (College Park, MD). A peptide containing FH pT236 was injected into rabbits. The rabbit serum was collected and purified using an affinity column with non-phosphorylated FH T236 peptide to exclude the antibodies for non-phosphorylated FH, followed by an affinity column with phosphorylated FH T236 peptide to bind to and purify the FH pT236 antibody. The FH pT236 antibody was then eluted and concentrated.
Transfection
Cells were plated at a density of 4 × 105/60-mm dish 18 h prior to transfection. The transfection was performed using HyFect reagent (Denville Scientific) according to the vendor’s instructions.
Analyses of NHEJ and HR repair at the DR-GFP locus and the efficiency of I-SceI-dependent cleavage at the DR-GFP locus
A schematic of NHEJ and HR repair at the DR-GFP locus is presented in Supplementary Figure 3d. The expression of I-SceI induces a DSB in the SceGFP (defective in GFP expression). PCR products were amplified using the indicated primers against sequences on the flank of the I-SceI-induced DSB site before being subjected to enzyme digestion with I-SceI and BcgI. The product from NHEJ caused a loss of the I-SceI restriction enzyme site, while HR repair caused I-SceI to be replaced with the BcgI cutting sequence. The product (0.65-Kb PCR fragment) that is resistant to both I-SceI and BcgI digestion represents DNA repair by NHEJ (0.5-Kb PCR fragment created by both I-SceI and BcgI digestion reflected a comparable amount of I-Sce1-uncut SceGFP or Sce1-uncut SceGFP with HR repair in cells). NHEJ repair efficiency is quantitated by the percentage of DNA in PCR products that is resistant to I-SceI and BcgI digestion. HR repair restores the expression of GFP by replacing the I-SceI-damaged SceGFP with an iGFP sequence. Flow cytometry analyses of GFP expression indicate the efficiency of HR repair.
A schematic of the efficiency of I-SceI-dependent cleavage at the DR-GFP locus is presented in Supplementary Figure 3f. DR-GFP-expressing U2OS cells with FH depletion were transfected with a vector expressing I-SceI, which induces a DSB in SceGFP. After transfection, the cells were treated with NU7441 (1 µM) and KU55933 (10 µM) for 12 h to inhibit DNA repair and eliminate the effect of DNA repair on the evaluation of I-SceI-cut efficiency. The cells were then harvested, and the PCR products were amplified using the indicated primers against sequences on the flank of the I-SceI-induced DSB site. The efficiency of DSB production by I-SceI was measured by the amount of 0.65 Kb PCR products (I-Sce1-uncut SceGFP) in cells. The amount of I-Sce1-uncut SceGFP gives an indirect value of I-Sce1-uncut SceGFP. GAPDH was used as an internal control for the PCR product.
Mass spectrometry analysis
Flag-FH-associated proteins from the immunoprecipitation assay were acetone-precipitated in vitro at −20°C overnight and re-suspended in 50 mM ammonium bicarbonate buffer containing Rapigest (Waters Corp, Milford, MA). The sample was heated to 95°C for 10 minutes and allowed to cool down before 100 ng of sequencing-grade modified trypsin (Promega, Madison, WI) was added. The digestion proceeded overnight at 37°C and was analyzed by LC-MS/MS using an Obitrap-Elite mass spectrometer (Thermo Fisher Scientific, Waltham, MA).
Proteins were identified by comparing the fragment spectra against those in the SwissProt protein database (EBI) using Mascot v.2.3 (Matrix Science, London, UK) and Sequest v.1.20 via Proteome Discoverer v.1.3 (Thermo Fisher Scientific) software.
Immunoprecipitation and immunoblotting analysis
Proteins were extracted from cultured cells using a modified buffer, followed by immunoprecipitation and immunoblotting with the corresponding antibodies, as described previously28.
Chromatin extract isolation
Cells (2 × 106) were washed with PBS and resuspended in 200 µl of solution A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, 10 mM NaF, 1 mM Na2VO3, and protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and the cells were incubated for 5 min on ice, followed by low-speed centrifugation (4 min at 1300 × g at 4°C) to separate the cytoplasmic proteins from the nuclei. The isolated nuclei were then lysed in 200 µl of solution B (3 mM ethylenediamine tetraacetic acid, 0.2 mM EGTA, 1 mM dithiothreitol, and protease inhibitors). Insoluble chromatin was collected by centrifugation (4 min at 1700 × g at 4°C), washed once with solution B, and centrifuged at high speed (10,000 × g) for 1 min. The chromatin pellet was resuspended in Laemmli buffer and sonicated for 15 s. For immunoprecipitation and GST pull-down assays, the chromatin pellet was subjected to sonication and suspended with micrococcal nuclease (MNase) buffer (20 mM Tris-HCl, 1 mM CaCl2, 2 mM MgCl2, 0.1 mM KCl, 0.1% Triton X-100, 0.3 mM sucrose, 1 mM DTT, 1 mM benzamidine, 0.2 mM PMSF, 1 mM STZ [pH 7.9], and 0.5 µg/ml ethidium bromide). The samples were incubated with 50 units of micrococcal nuclease for 30 min at room temperature. The reaction was stopped by adding 5 mM EGTA and 5 mM EDTA. After centrifugation at 3,000 g for 30 min at 4°C, the supernatant were passed through a WGA agarose column (Vector). The flow-through fraction was mixed with α-Flag M2 agarose beads and rotated for 12 h at 4°C. Finally, the chromatin extract was washed three times with buffer (20 mM Tris-HCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1 mM KCl, 0.05% NP-40, 10% glycerol, 1 mM DTT, 1 mM benzamidine, 0.2 mM PMSF, and 1 mM STZ [pH 7.9]).
ChIP assay
A ChIP assay was performed using an Upstate Biotechnology kit, as described previously29. Quantitative real-time PCR was used to measure the amount of bound DNA, and the value of enrichment was calculated according to the relative amount of input and the ratio to IgG. The specific primers used in PCR for the sequences that were 0.2 Kb, 0.4 Kb, and 0.6 Kb distant from I-SceI-induced DSBs were 5’-GATCAGGCAGAGCAGGAACC-3’ (forward) and 5’-GAACAGCTCCTCGCCCTTGC-3’ (reverse), 5’-TTATTGTGCTGTCTCATCATT-3’ (forward) and 5’-GTGCTGCATGCTTCTTCGGCA-3’ (reverse), and 5’-TCCATCTCCAGCCTCGGGGCT-3’ (forward) and 5’-AGGCTCTAGAGCCGCCGGTCA-3’ (reverse), respectively. Control primers selected against a specific region of chromosome 12 were 5´-ATGGTTGCCACTGGGGATCT-3´ (sense) and 5´-TGCCAAAGCCTAGGGGAAGA-3´(antisense). Control primers were selected against a specific region of chromosome 12.
Cell viability assay
Cells (2 × 106) were plated and exposed to IR (10 Gy). Cell viability was measured 12 h after IR using the trypan blue staining assay.
Clonogenic cell survival assay
The indicated cells (5 × 103) were plated in 100-mm dishes and treated with various doses of IR (0, 2, 4, and 6 Gy). Cells were then re-cultured for 10 days and stained with 0.5% crystal violet in methanol to determine colony formation efficiency. The cluster of staining cells was considered a colony at more than 50 cells.
DNA constructs and mutagenesis
The Flag-tag of FH in the pcDNA6 vector is located at the C-terminus of FH. Flag-FH (N) with deletion of 21 N-terminal amino acids was constructed by PCR reactions. pcDNA 3.1/hygro (+) FH T236A and H2A.Z (NKLLG) were created using the QuikChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA). pGIPZ human FH shRNA was generated using the GGAATTTAGTGGTTATGTT oligonucleotide. pGIPZ human H2A.Z shRNA was generated with the oligonucleotides TCTAGGACGACCAGTCATG and ATACTCTAACAGCTGTCCA. pGIPZ human KDM2B shRNA was generated with the oligonucleotide GCTCCACCTCAGTTACTGTAA. Two pGIPZ human Ku70 shRNAs were generated with the oligonucleotides GACACAGGTGGAGAATATA and TCACTGTACCTACACTGAA. The pGIPZ controls were generated with control oligonucleotide GCTTCTAACACCGGAGGTCTT or GCCCGAAAGGGTTCCAGCTTA.
DNA-PK kinase assay
Purified DNA-PK complexes were purchased from Promega (Madison, WI). This complex is composed of an approximately 460-kDa catalytic subunit and heterodimeric DNA-binding subunits, including Ku80 and Ku70. Kinase reactions were performed as described previously30. In brief, the purified DNA-PK complex was incubated with FH (100 ng) in kinase buffer with 50 mM HEPES (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 0.2 mM EGTA, 10 mM MgCl2, 0.1 M KCl, 80 µg/ml bovine serum albumin, 10 µg/ml linear double-stranded DNA, and 40 Ci/ml [γ-32P]ATP in 50 µl at 25°C for 1 h. DNA-PK was added last. The reaction mixtures were incubated at 30°C for 10 minutes and terminated by adding sodium dodecyl sulfate–polyacrylamide gel electrophoresis loading buffer and heating to 100°C.
NHEJ repair analysis and PCR analysis of I-SceI-induced DSBs
The genomic DNA of cells with or without I-SceI expression was extracted. PCR was performed as previously described using primers that flanked the DSB site and PCR products were digested with I-SceI and BcgI19. After gel electrophoresis of PCR products, the intensity of the enzyme-resistant and enzyme-cleaved fragments was quantified using NIH IMAGE software. To assay the cutting efficiency of I-SceI in cells, genomic DNA was extracted and adjusted to equal concentrations. Semi-quantitative PCR was carried out using the same primers for NHEJ analysis (30 cycles). The sequences of primers were as follows: DRGFP-F, 5-CTGCTAACCATGTTCATGCC-3 and DRGFP-R, 5-AAGTCGTGCTGCTTCATGTG-3. β-actin primers (Applied Biosystems) were used as an internal control (25 cycles). The cycle number was determined to generate PCR products in the range of linear amplification.
HR repair analysis
U2OS cells containing a single copy of the HR repair reporter substrate DR-GFP were used19. GFP-expressing plasmid (pEGFP-C1) was used for transfection efficiency control. Seventy-two hours after transfection of a vector expressing DR-GFP, cells were re-seeded and transfected with mock or pCBASce plasmid. Forty-eight hours later, a flow cytometry analysis was performed to detect GFP-positive cells using a FACSCalibur apparatus with Cellquest software (Becton Dickinson, San Jose, CA) at the MD Anderson Flow Cytometry Facility. To induce chromatin relaxation, cells were incubated for 16 h in sodium butyrate (5 mM) or trichostatin A (200 ng/ml) before being analyzed by flow cytometry.
Flow cytometry analysis
Cells (1×106) were fixed in 70% ethanol on ice for 3 h, spun down, and incubated for 1 h at 37°C in PBS with DNase-free RNase A (100 µg/ml) and propidium iodide (50 µg/ml). Cells were then analyzed by fluorescence-activated cell sorting.
Recombinant protein purification
WT and mutant His-FH and GST-H2A.Z were expressed in bacteria and purified, as described previously29.
Enzyme activity assay
To measure FH activity, we collected chromatin extract from cells 1 h after IR (10 Gy). Immunoprecipitated Flag-FH proteins were eluted using a Flag peptide. Eluted proteins were added to an enzyme assay buffer (50 mM malate and 10 mM potassium phosphate at pH 7.3), and the absorbance at OD240 was recorded.
A KDM2B demethylase assay was conducted in the presence of 300 ng of KDM2B and 1 mg of chromatin extract in 20 ml of buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 50 mM [NH4]2Fe[SO4]2, 100 mM α-ketoglutarate, 2 mM Vc, and 10 mM PMSF) for 3 h.
Fumarate measurement
Relative fumarate concentrations in tumor cells were measured using Fumarate Detection Kit purchased from Abcam (Boston, MA).
Statistical analysis
Statistical analysis was conducted with the two-tailed unpaired Student’s t-test. All data represent the mean ± SD of three independent experiments.
Supplementary Material
Acknowledgments
We thank David Hawke, Bih-Fang Pan, and Mohammad B. Hossain at MD Anderson for their technical assistance and Ann Sutton for her critical reading of this manuscript.
This work was supported by National Cancer Institute grants 2R01CA109035 (Z.L.) and 1R0CA169603 (Z.L.), National Institute of Neurological Disorders and Stroke grant 1R01NS089754 (Z.L.), the MD Anderson Cancer Center Support Grant CA016672, James S. McDonnell Foundation 21st Century Science Initiative in Brain Cancer Research Award 220020318 (Z.L.), 2P50CA127001 (Brain Cancer SPORE), a Sister Institution Network Fund from The University of Texas MD Anderson Cancer Center (Z.L.), and the Odyssey Fellowship from MD Anderson (J.Y.). Z.L. is a Ruby E. Rutherford Distinguished Professor.
Footnotes
CONTRIBUTIONS
This study was conceived by Z.L.; Z.L. and Y.J. designed the study; Y.J., X.Q., J.S., Y.W., X.L., R.L., and Y.X. performed experiments; Q.C., G.P., and S.L. provided reagents and conceptual advice; Z.L. wrote the paper with comments from all authors
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohgaki T, Bohgaki M, Hakem R. DNA double-strand break signaling and human disorders. Genome integrity. 2010;1:15. doi: 10.1186/2041-9414-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annual review of biochemistry. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. The mechanism of human nonhomologous DNA end joining. The Journal of biological chemistry. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 5.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & development. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 6.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 10.Fnu S, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 12.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nature cell biology. 2012;14:457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yogev O, et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS biology. 2010;8:e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, et al. PKM2 Regulates Chromosome Segregation and Mitosis Progression of Tumor Cells. Molecular cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, et al. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nature communications. 2014;5:5566. doi: 10.1038/ncomms6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature. 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 19.Peng G, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nature cell biology. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakanishi K, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Flaherty L, et al. Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Human molecular genetics. 2010;19:3844–3851. doi: 10.1093/hmg/ddq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam J, et al. A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell reports. 2013;3:1440–1448. doi: 10.1016/j.celrep.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obri A, et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 24.Xiao M, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z. PKM2 functions as a histone kinase. Cell Cycle. 2012;11:4101–4102. doi: 10.4161/cc.22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z. Nonmetabolic functions of pyruvate kinase isoform M2 in controlling cell cycle progression and tumorigenesis. Chinese journal of cancer. 2012;31:5–7. doi: 10.5732/cjc.011.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson BA, Kraus WL. Small molecules, big effects: a role for chromatin-localized metabolite biosynthesis in gene regulation. Molecular cell. 2011;41:497–499. doi: 10.1016/j.molcel.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 28.Lu Z, et al. Activation of protein kinase C triggers its ubiquitination and degradation. Molecular and cellular biology. 1998;18:839–845. doi: 10.1128/mcb.18.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia Y, et al. c-Jun downregulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Molecular cell. 2007;25:219–232. doi: 10.1016/j.molcel.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang D, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. The Journal of biological chemistry. 2007;282:11221–11229. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.