Abstract
Amino acids have a dual role in cellular metabolism, as they are both the building blocks for protein synthesis and intermediate metabolites which fuel other biosynthetic reactions. Recent work has demonstrated that deregulation of both arms of amino acid management are common alterations seen in cancer. Among the most highly consumed nutrients by cancer cells are the amino acids glutamine and serine, and the biosynthetic pathways that metabolize them are required in various cancer subtypes and the object of current efforts to target cancer metabolism. Also altered in cancer are components of the machinery which sense amino acid sufficiency, nucleated by the mechanistic target of rapamycin (mTOR), a key regulator of cell growth via modulation of key processes including protein synthesis and autophagy. The precise ways in which altered amino acid management supports cellular transformation remain mostly elusive, and a fuller mechanistic understanding of these processes will be important for efforts to exploit such alterations for cancer therapy.
Keywords: Amino acids, Metabolism, Cancer, Serine, Glutamine, mTOR
1. Introduction
Proliferative cells alter their metabolism to support the biosynthetic reactions required for accumulation of biomass; indeed, alterations in tumor cell metabolism are recognized as a hallmark of cancer [1–3]. Robust macromolecular biosynthesis is required to support a proliferative cell metabolism [4], and proper sensing of the diverse nutrients required to support such biosynthesis is important to orchestrate these complex events [5]. Proliferative metabolism is supported by cellular programs which ensure that there is sufficient nutrient uptake and energy generation, management of redox potential, appropriate activation of autophagy to recycle macromolecules and damaged organelles, and elimination of toxic byproducts [1]. As such, alterations in these processes have been described over the past decades with increasing levels of sophistication, and strategies which target the altered metabolism of cancer are emerging [6].
In particular, it is becoming more apparent that proper amino acid management is critical to support proliferative metabolism via alterations in pathways that support their biosynthesis and sensing (Fig. 1). Moreover, alterations in the recycling of amino acids by autophagy and their scavenging from the environment by micropinocytosis of serum proteins are observed in proliferative cells, and may support the transformed state by providing amino acids during periods of starvation [7–10]. Many amino acids cannot be synthesized by the cell, and therefore their uptake is essential for protein biosynthesis and cell viability [11]. Among these essential amino acids, there is evidence that two – leucine and arginine – are sensed by the cell to determine if there is sufficient material available for protein biosynthesis [12–14]. This sensing likely occurs within or near the surface of the lysosome, and permits the activation of the mechanistic target of rapamycin complex I (mTORC1) [5]. Amino acid insufficiency provides a dominant signal to turn off the mTORC1 pathway over other inputs, such as insulin signaling, placing amino acids as key regulators of cell growth via mTORC1 [14]. Several amino acids can also be readily synthesized by the cell, and among them, serine and glutamine are consumed greatly in excess of that required for protein biosynthesis for downstream reactions providing one carbon units, TCA cycle intermediates, fatty acids, membrane lipids, and other amino acids for biosynthetic reactions [15,16]. As such, recent work has identified genetic alterations in cancer of both the machinery which senses amino acid sufficiency as well as those pathways which utilize amino acids as intermediate metabolites (Table 1). Here, we will first describe cancer-specific alterations in the use of amino acids as intermediate metabolites, followed by a discussion of the role of the mTORC1 pathway in amino acid sensing, including alteration of mTORC1 pathway components and functional outputs in cancer.
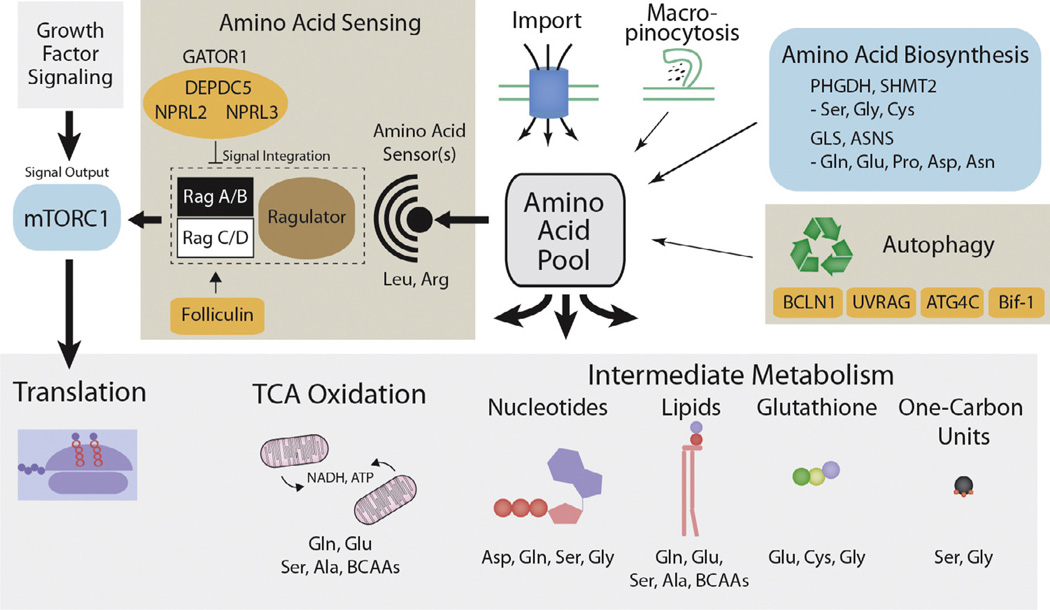
Fig. 1.
Amino acid sensing and biosynthesis are altered in cancer. Amino acid import and biosynthesis, as well as the processes of autophagy and macropinocytosis contribute to the pool of amino acids available to the cell for macromolecular biosynthesis. Amino acid biosynthetic pathways are activated in subsets of cancer and drive the production of specific amino acids and their utilization as intermediate metabolites for the production of important biomolecules such as nucleotides, lipids, glutathione, and one-carbon units. Amino acids are also oxidized in the TCA cycle as an alternative to glucose for the production of ATP and NADH. The specific amino acids which most directly contribute to these biomolecules and processes are listed. Unknown amino acids sensor(s) assess the availability of specific amino acids, likely including leucine and arginine. The Ras-like small GTPase Rag complex, modulated by the action of its GEF, Ragulator, and GAPs, tumor suppressive complexes GATOR1 and folliculin, integrate this amino acid signal and effect a change in localization of the mTORC1 complex, leading to its activation. The mTORC1 complex can then activate pathways promoting cell growth, including protein biosynthesis. Activating mutations in the mTORC1 core component mTOR, are recurrently observed in cancer. Processes exhibiting activation in cancer are colored green, tumor suppressive genes and complexes are colored orange.
Table 1.
Alteration of amino acid management pathways in cancer.
| Amino acid metabolism | |||
| Gene | Function | Cancer alteration/relevance | References |
| GLS | Rate limiting step in glutaminolysis | Increased translational efficiency downstream of MYC, miR-23a/b | [58] |
| PHGDH | Rate limiting step in serine biosynthesis | Genomic amplification and over-expression | [23,24] |
| SHMT2 | Diversion of serine into mitochondrial one-carbon metabolism | Over-expression, drives hypoxia resistance | [23,35,36] |
| GLDC | Key component of the glycine cleavage complex | Prevents toxic glycine accumulation, drives broad metabolic changes | [36,42] |
| ASNS | Asparagine biosynthesis | Increased expression in glioblastoma | [65] |
| Amino acid sensing | |||
| Gene | Function | Cancer alteration/relevance | References |
| mTOR | Protein kinase, controls translation in response to nutrient sufficiency signals | Activating point mutations, amino acid starvation fails to inactivate mTORC1 | [120,155] |
| FLCN | mTORC1 positive regulator, GTPase activating protein for RAG C/D | Loss of function mutations in Birt–Hogg–Dubé syndrome | [105–107] |
| DEPDC5 | GATOR1 component, negative regulator of mTORC1, GTPase activating protein for RAG A/B | Deletion of 22q12.2, amino acid starvation fails to inactivate mTORC1 | [116,117] |
| NPRL2 | GATOR1 component, negative regulator of mTORC1, GTPase activating protein for RAG A/B | Deletion of 3p21.3, amino acid starvation fails to inactivate mTORC1 | [116,118,119] |
2. Amino acids as intermediate metabolites
2.1. Serine and glycine metabolism in cancer
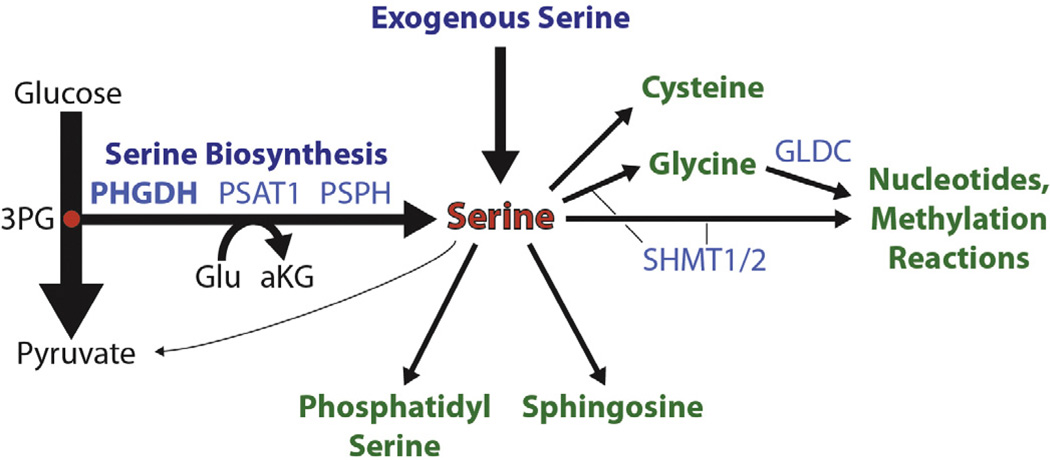
Serine is unique among the amino acids in that it is not only rapidly taken up by the transformed cell and heavily utilized as an intermediate metabolite, but is also produced biosynthetically by a subset of tumors [17]. Serine biosynthesis occurs in a three step pathway off of glycolysis (Fig. 2), beginning with the glycolytic intermediate 3-phosphoglycerate, which can be converted to 3-hydroxypyruvate by the action of the enzyme phosphoglycerate dehydrogenase (PHGDH), using the cofactor NAD(P)H [18,19]. 3-Hydroxypyruvate then participates in a transamination reaction with glutamate catalyzed by the enzyme phosphoserine aminotransferase (PSAT1), producing phosphoserine and alpha-ketoglutarate [17]. Phosphoserine can then be dephosporylated by the final enzyme in the pathway, phosphoserine phosphatase (PSPH), to produce serine [17]. Subsequent to its production, serine can be converted to glycine by the action of serine hydroxymethyltransferase either in the cytoplasm (via SHMT1) or upon transport into the mitochondrion (via SHMT2), whereupon serine contributes to the one-carbon pool via production of 5,10-methylene–tetrahydrofolate (5,10-MeTHF) [20]. Mitochondrially localized glycine can be further consumed by the glycine cleavage system, a multiprotein complex which includes the enzyme glycine decarboxylase (GLDC), producing another molecule of 5,10-MeTHF [21]. However, the significance of the compartmentalization of these reactions is just beginning to be appreciated (see below). The one carbon pool is managed principally by the methylene–tetrahydrofolate dehydrogenases (MTHFD1, 1L and 2) and supports a variety of cellular methylation reactions through the production of S-adenosyl methionine (SAM), a key methyl-donor in the cell, as well as nucleotide biosynthesis through several reactions including via the trifunctional enzyme GART, which utilizes 10-formyl-THF and glycine in purine biosynthesis, and the enzyme thymidylate synthase (TYMS), which utilizes 5,10-MeTHF in dTMP biosynthesis [22]. Serine also contributes to the production of glutathione (via glycine), cysteine, phosphatidylserine, and sphingosine [16]. In the liver, serine can be deaminated to pyruvate for subsequent gluconeogenesis via the action of serine dehydratase (SDS); however, this enzyme is universally silenced in other tissues, including transformed liver cells [17]. Thus, serine produced biosynthetically or taken up from the environment can contribute to all major classes of macromolecules (Fig. 2).
Fig. 2.
Serine biosynthesis and utilization. Serine can be imported into the cell or biosynthetically produced from the glycolytic intermediate 3-phosphoglycerate (3PG) in a three step pathway. Flux through serine biosynthesis additionally drives the production of cytosolic alpha-ketoglutarate (aKG). Serine is catabolized to generate phosphatidylserine, sphingosine, cysteine, glycine or pyruvate, the latter conversion being restricted to non-transformed liver cells. The conversion of serine to glycine or the subsequent catabolism of glycine in the mitochondria provides one-carbon units for nucleotide biosynthesis and the methylation reactions of the cell. Key enzymes are shown in light blue.
Activation of serine biosynthesis has been reported in a subset of tumors from a growing number of cancer types, including melanoma, ER-negative negative breast cancer, non-small cell lung cancer, and ovarian cancer [23–26]. Globally, PHGDH lies in a locus whose amplification is selected for during tumorigenesis in about 16% of cancers, arguing that PHGDH expression could be driving tumorigenesis in these cases [23,24]. Importantly, PHGDH expression is rate limiting for serine biosynthesis, as assessed in breast cancer cell lines in vitro [23]. Moreover, PHGDH expression and catalytic activity are required for the optimal proliferation of cell lines that over-express it, despite the presence of serine in the environment [23,24,26,27]. In ER-negative breast cancer, PHGDH protein levels are upregulated in ~70% of cases, including those without genomic amplification, arguing for additional epigenetic mechanisms for its activation in breast cancer [23]. PHGDH over-expression in breast cancer is correlated to the expression of several enzymes in the serine biosynthetic and downstream pathways including PSAT1, PSPH, SHMT2, and MTHFD1, 1L and 2 [23]. These data suggest that serine biosynthesis is coordinately regulated, and that PHGDH is a key enzyme in this regulation. In support of this narrative, now decades-old experiments have demonstrated that PHGDH and SHMT1/2 activity correlate with the incorporation of labeled serine into nucleotides [28], observations supported by recent work in activated T cells [29], and SHMT1 has been observed to undergo sumoylation and translocation into the nucleus during S-phase to support the synthesis of dTTP [30], indicating a cell cycle dependent role for serine biosynthesis. Serine biosynthetic enzymes are also upregulated upon serine or glutamine starvation, a response that is mediated by the direct activity of the transcription factor ATF4 [31,32].
While serine is an important intermediate metabolite, the precise reason for activation of serine biosynthesis and high PHGDH levels in specific cancers has remained elusive. Indeed, some cancer cell lines have little to no measurable serine biosynthetic flux, arguing that the metabolic advantage provided by PHGDH over-expression can be compensated for by the activation of other pathways, or is not required in all contexts [23,24]. Interestingly, suppression of serine biosynthesis by PHGDH inhibition does not result in an appreciable effect on the intracellular levels of serine, as serine transporters rapidly equilibrate intracellular and extracellular serine [23]. However, serine availability from the environment may not be sufficient to compensate for biosynthetic demand in the context of a nutrient-poor tumor with low serine biosynthesis. Indeed, elevated intracellular serine levels cause feedback activation of the M2 isoform of pyruvate kinase (PKM2), thereby balancing glycolytic flux into serine biosynthesis when intracellular serine levels fall [32,33], and allowing cells to survive inhibition of oxidative phosphorylation [34]. The presence of this feedback mechanism suggests that serine levels can be limiting in a physiologic context. Supporting the importance of this feedback mechanism, activation of SHMT2 simultaneously drives flux of serine into one carbon metabolism while suppressing pyruvate kinase flux, thereby suppressing oxygen consumption and permitting cancer cell survival in hypoxia [35,36]. Moreover, feeding mice a diet in which glycine and serine are withheld reduces circulating serine and glycine levels by ~50%, and negatively impacts tumor growth in cell lines dependent upon extracellular serine and glycine for growth [37]. As such, in nutrient poor conditions, the simultaneous serine and glycine demand for nucleotide, glutathione, and protein biosynthesis becomes overwhelming in the absence of p53 dependent growth controls [37]. These data suggest that serine can become limiting in physiological contexts with broad consequences on cellular metabolism, mediated to some extent by PKM2 regulation.
Despite the presence of abundant serine in an in vitro environment, PHGDH catalytic activity is required for the viability of cell lines with pathway activation, suggesting that a requirement for serine biosynthetic flux itself exists in this context. Explaining this apparent contradiction has remained elusive, although several hypotheses have been proposed. Serine produced biosynthetically might be utilized in a different way than serine imported into the cell via channeling into some other compartment or being coupled to some key downstream reaction. Alternatively, a metabolite upstream of serine may be regulating metabolism or cell growth in some way. For example, phosphoserine levels are almost entirely dependent upon serine biosynthetic flux [23,24]. Finally, another product of the pathway may be supporting cellular metabolism. Indeed, levels of the TCA cycle intermediate alpha-ketoglutarate, produced by the serine biosynthesis pathway in equimolar amounts to serine, are reduced upon inhibition of serine biosynthesis using a variety of experimental methods [23], although this change has not been observed in all cell systems [24]. Additional work disentangling these possibilities will be needed to understand why serine biosynthesis is elevated in specific types of cancer and how it supports cellular metabolism in serine replete and limiting conditions.
Investigation of how reactions downstream of serine contribute to proliferative metabolism has focused on the contribution of serine to glycine production and consequently to one carbon metabolism (Fig. 3). When compared to other metabolites in cell culture medium, consumption versus production of glycine by cancer cells best correlates with proliferative activity, suggesting that high metabolic flux into one carbon metabolism is either important for robust cell proliferation, or a consequence of it [38]. Somewhat counter-intuitive to this observation, increasing the medium concentration of glycine above 1 mM (approximately 5-fold above plasma levels) decreases cell proliferation, especially when coupled with serine depletion [39]. This anti-proliferative effect is a result of high glycine levels leading to a SHMT1/2-dependent depletion of 5,10-MeTHF as excess glycine is converted to serine, and an unexpected failure of cytoplasmic glycine to contribute to one carbon metabolism, particularly in the absence of serine, in those cell lines analyzed [39–41].
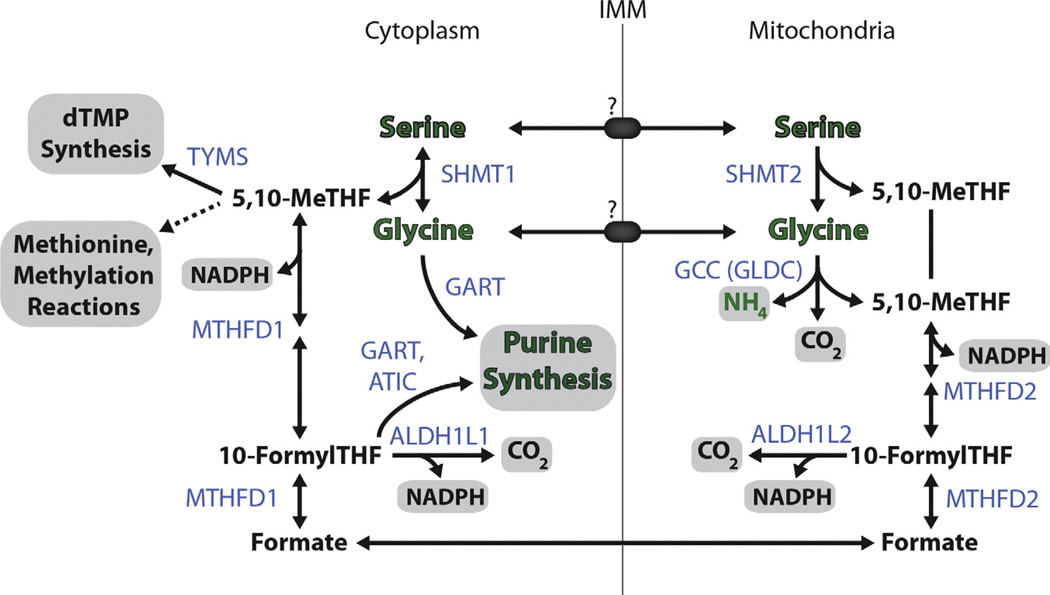
Fig. 3.
Cytosolic and Mitochondrial Compartmentalization of One-Carbon Metabolism. Serine and glycine contribute to one-carbon metabolism in largely parallel cytoplasmic and mitochondrial pathways. Serine hydroxymethyltransferate (SHMT1/2) catalyzes the contribution of the serine beta carbon into the one carbon pool by production of 5,10-methyl tetrahydrofolate (MeTHF). Cytoplasmic 5,10-MeTHF can then contribute to dTMP synthesis or to most major methylation reactions of the cell via production of S-Adenosyl methionine. In the mitochondria, glycine can be further cleaved to form another molecule of 5,10-MeTHF whilst in the cytoplasm, glycine instead can contribute en masse to purine biosynthesis. One-carbon units are managed by the methylenetetrahydrofolate dehydrogenases (MTHFD1/2), which can produce appropriate THF derivatives for nucleotide biosynthesis in the cytoplasm or for the production of NADPH in either compartment. Serine and glycine are thought to move across the inner mitochondrial membrane (IMM) via unknown transporters, whilst THF derivatives and NADPH are not thought to translocate between these two compartments. Green text denotes the fate of serine derived nitrogen, gray boxes indicate major endpoint metabolites produced from serine. Key enzymes are shown in light blue.
Glycine can be catabolized by the mitochondrial glycine cleavage complex, a key component of which, GLDC, is required for the viability of lung cancer stem cells [42]. Surprisingly, the GLDC requirement in cancer is dependent upon the expression of the upstream enzyme SHMT2, which produces mitochondrial glycine from serine [36]. In SHMT2 replete cells, GLDC suppression results in an accumulation of mitochondrial glycine, which may be detrimental to cells as a result of its diversion into minor pathways of glycine metabolism, producing toxic metabolites such as aminoacetone and methylglyoxyl [36]. This toxic accumulation of glycine and its metabolites in the mitochondria upon GLDC inhibition contrasts the mechanism of toxicity seen by the aforementioned extracellular serine/glycine imbalance. Indeed these mechanisms of glycine toxicity appear to be distinct, but both are important depending on the physiological and genetic context.
In addition to moderating the level of mitochondrial glycine, the glycine cleavage complex and downstream one carbon metabolic enzyme MTHFD2 can participate in the complete catabolism of mitochondrial glycine, producing NADPH in the process [41]. NADPH is a key currency in cellular redox management via reduction of glutathione, and an important cofactor in biosynthesis, particularly of fatty acids [41]. Studies using enzymatic sensors for cytoplasmic versus mitochondrial NADPH production have demonstrated that NADPH produced downstream of serine almost exclusively occurs in the mitochondria, with the corresponding cytoplasmic reactions running in the reverse direction [40]. Indeed, individuals with mutations in MTHFD1, which manages the cytoplasmic folate pool, have defects in formate flux into dTMP and methionine synthesis [43]. However, cytoplasmic NADPH production from serine has been observed in other cellular systems, indicating that the way in which serine contributes to one carbon metabolism is cell type or context dependent [41]. Together with the observations that SHMT2 expression correlates with in vivo tumorigenicity [36], these data implicate mitochondrial serine metabolism as playing an important role in transformation. Interestingly, the contribution of amino acids to one carbon metabolism may be different in humans compared to most other organisms, as a key enzyme in this process, threonine dehydratase (Tdh), has been mutated during the course of human evolution and rendered nonfunctional [44]. Threonine is a major contributor to one carbon metabolism in mouse ES cells, which are threonine auxotrophs and require abundant threonine to maintain proper histone methylation [45,46]. However, the significance of Tdh and the ability of threonine to contribute to one carbon metabolism in murine cancers has not been explored, and expression of Tdh seems to be tightly restricted to mouse ES cells. Future work on one carbon metabolism in cancer will likely focus on the control of flux through cytoplasmic versus mitochondrial one carbon metabolism, its coordination with serine metabolism, and the relevance of these pathways to transformation. A fuller understanding of one carbon metabolism would be greatly assisted by better technologies to measure mitochondrial versus cytoplasmic metabolite pools to better understand the compartmentalization of these reactions.
2.2. TCA-adjacent amino acids: glutamine, glutamate, asparagine, and aspartate
Glutamine is the most abundant amino acid in both cell culture medium and blood. In vitro experiments have identified glutamine as the most highly consumed amino acid [47,48], and regulation of the glutamine transporters SLC1A5 and SLC38A2 have been implicated in breast and lung tumorigenesis [49,50]. While glutamine can be generated biosynthetically, in vitro most cancer cell lines engage exclusively in conversion of glutamine to glutamate, by contribution of nitrogen from the glutamine side-chain amine either to de novo nucleotide biosynthesis pathways (in five steps, catalyzed by PPAT, PFAS, CTPS1/2, CAD1/2, and GMPS) to asparagine synthesis (via ASNS), or through release of the amine as free ammonia by the action of mitochondrial glutaminases (GLS and GLS2, Fig. 4). Glutamate can be subsequently converted to alpha-ketoglutarate by the action of several enzymes, which, mirroring glutamine conversion, either transfer the alpha amine from glutamate for use in alanine, aspartate, and serine biosynthetic reactions (GPT/GPT2, GOT1/2, and the aforementioned PSAT1) or release the amine as free ammonia by the action of mitochondrially localized glutamate dehydrogenases (GLUD1 and, to a not-appreciated extent in cancer, GLUD2). Glutamate can also be converted to pro-line through a two-step, NADPH-consuming process [41]. Because each step in the conversion of glutamine to alpha-ketoglutarate can either retain nitrogen for use in biosynthetic reactions or contribute to the release of nitrogen as ammonia into the environment, these reactions play an important role in macromolecular nitrogen assimilation. Under hypoxia [51] or upon inhibition of mitochondrial oxidative phosphorylation [52,53], glutamine can contribute reductively to the biosynthesis of fatty acids by a process termed reductive carboxylation, in which alpha-ketoglutarate is converted first to citrate and then to acetyl-CoA, a key substrate for fatty acid generation, via the action of IDH1/2 and ATP-citrate lyase. However, the significance of this pathway to generating net flux into fatty acid biosynthesis in hypoxia is controversial [54]. Flux of glutamine oxidatively into the TCA cycle is termed glutamine anaplerosis, and can drive a substantial proportion of cellular ATP generation [55]. Glutamine anaplerosis can be fueled by oncogene activation, particularly MYC [56,57], via modulation of mir-23a/b [58]. Indeed, cells over-expressing MYC have a greater dependence upon exogenous glutamine due to an enhancement of glutamine anaplerotic flux [56,57], depend on expression of glutaminase for viability [58], and respond to glutaminase inhibitors [59–61]. Furthermore, inhibition of glutaminolysis via suppression of GLUD1 increases the dependence of cells upon glycolysis and AKT signaling to maintain metabolic balance and viability [48]. These observations have inspired the production of additional glutaminase inhibitors for anti-cancer therapy [62].
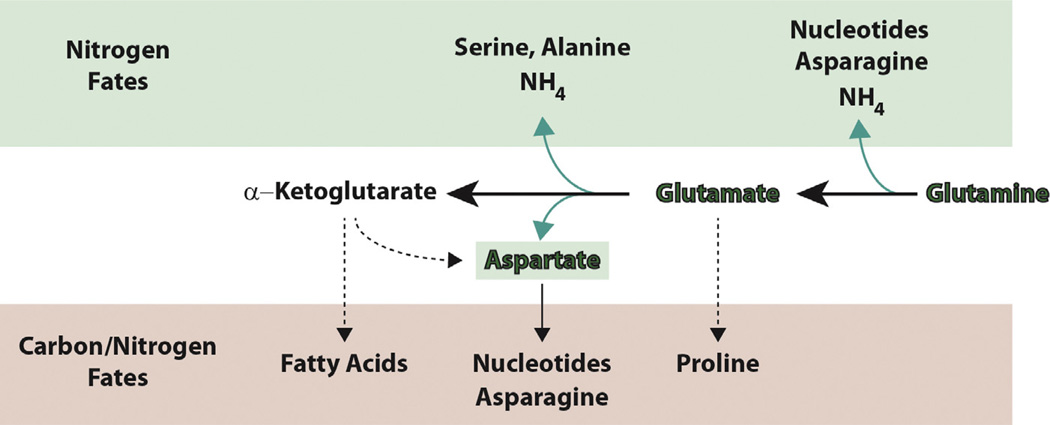
Fig. 4.
Major fates of glutamine. Glutamine and its major metabolite, glutamate, are used as nitrogen donors to contribute to the production of amino acids, via transamination reactions, or to nucleotide biosynthesis (green box). The amino groups of glutamine and glutamate can also be hydrolyzed to form ammonia, thereby balancing nitrogen assimilation and release. Glutamate and alpha-ketoglutarate produced by these deamination reactions can be further utilized for biosynthesis (pink box). Glutamate can be converted to proline or, via the TCA cycle, to aspartate, which is incorporated en masse in nucleotide biosynthesis or converted to asparagine. Alpha-ketoglutarate can be metabolized via various routes to acetyl CoA for use in de novo fatty acid biosynthesis.
The biosynthesis of the amino acid aspartate, and by extension asparagine which is synthesized from it, is tightly linked to glutamine/glutamate and TCA cycle metabolism. Asparagine is moderately abundant in the circulation, and unlike all other amino acids asparagine is only used for protein biosynthesis, never as an intermediary metabolite [63]. It is likely for these reasons that asparagine biosynthesis is dispensable for extra-neuronal organismal development [64]. However, asparagine synthetase (ASNS) levels correlate with tumor aggressiveness, potentially through a mechanism in which asparagine suppresses apoptosis [65]. In contrast, aspartate levels in the circulation are very low and its plasma membrane permeability poor, potentially rendering its biosynthesis essential. Aspartate biosynthesis involves coordination of mitochondrial and cytoplasmic compartmentalized enzymes and mitochondrial transporters, members of the malate–aspartate shuttle [66]. Key to aspartate biosynthesis is the action of GOT2, which produces this amino acid in the mitochondria using TCA cycle intermediate oxaloacetate and glutamate derived nitrogen. Mitochondrial aspartate can then be transported to the cytoplasm where ASNS synthesizes asparagine utilizing nitrogen from glutamine, in an irreversible reaction [67]. Thus, while the nitrogen from aspartate and asparagine derives from glutamine, the carbon required for biosynthesis of these amino acids can be derived from either glucose or glutamine. This decision can be impacted by oncogene activation as mutant KRAS expression results in favoring aspartate production almost entirely from glutamine [68]. Another key fate of aspartate is its assimilation en masse into nucleotide precursors in the de novo biosynthesis of pyrimidines (via CAD1/2) and contribution of nitrogen in the biosynthesis of purines (via PAICS or ADSS).
The demand for aspartate as an intermediate metabolite may partly underlie the anti-tumor efficacy of l-asparaginase, which is widely used in the treatment of ALL, and acts by depleting asparagine (and to a lesser extent, glutamine) from the circulation by converting it to aspartate [69,70]. ASNS-null or low cell lines and tumors are auxotrophic for asparagine and therefore sensitive to l-asparaginase [71], but ASNS expression is not entirely predictive of drug sensitivity, particularly in clinical samples [72]. Interestingly, recent work has shown that asparagine depletion by l-asparaginase increases the rate of glutamine depletion from the media [73], indicating that l-asparaginase treatment places an increased demand on glutamine anaplerosis and TCA cycle function to provide sufficient precursors for asparagine biosynthesis. Therefore, the regulation of asparagine biosynthesis and its interaction with glutamine and TCA cycle metabolism will likely be important for understanding asparagine auxotrophy and l-asparaginase sensitivity. Such an investigation may be complicated by the absence of asparagine from common cell culture media such as DMEM, to which many cancer cell lines have adapted.
3. Regulation of growth by mTORC1
As described above, tumors exhibit increased biosynthesis of amino acids and upregulate their utilization as intermediate metabolites. Recent work to elucidate the specific mechanisms by which amino acid levels are sensed has additionally uncovered mutations in the mTOR complex 1 (mTORC1) pathway, nucleated by the mTOR kinase, resulting in inappropriate amino acid sufficiency signals and thus increased mTORC1 activity (Table 1). As a central regulator of growth, the mTORC1 pathway integrates nutrient sufficiency and growth factor signals to regulate important processes like translation, lipid and nucleotide biosynthesis, and autophagy [74–76]. These cancer promoting mutations affecting the amino acid sensing arm of the mTORC1 pathway are distinct from mutations which have long been appreciated to impact growth factor signaling upstream of mTORC1, mediated by the phosphoinositide-3-kinase (PI3K) and AKT pathways [77,78]. We will first briefly describe how activation of growth factor signaling pathways upstream of mTORC1 promote tumorigenesis before reviewing recent work to elucidate the role in cancer of altered amino acid sensing.
3.1. Pathways altered in cancer upstream of mTORC1
The growth factor signaling input of the mTORC1 pathway is one of the most frequently mutated in cancer, and is comprised of well-established oncogenes of the Ras, PI3K, and AKT family [79,80]. Tumor suppressors associated with the mTORC1 pathway are mutated in both sporadic cancers and familial tumor-prone syndromes [81]. These tumor suppressors are downstream of growth factor receptor tyrosine kinases (RTKs), themselves subject to oncogenic amplification [82], and modulate the subsequent activation of the Ras/PI3K/AKT signal transduction pathways [83]. Neurofibromin-1 (NF1) modulates the activation of Ras as a GTPase activating protein (GAP) [84]. Germline mutations in NF1 cause the tumor syndrome neurofibromatosis [85], and somatic mutations have also been broadly identified in sporadic cancers [86]. Both Ras and RTKs activate the catalytic subunit p110α of the phosphatidylinositol-3 lipid kinase (PI3K) [87], which often harbors activating mutations or is amplified in cancer [77]. The most common mechanism of activating this pathway, however, is loss of the PTEN lipid phosphatase, which performs the reverse reaction of PI3K. PTEN frequently undergoes somatic mutation [88], and germline mutations in PTEN cause familial tumors in Cowden syndrome [89]. Growth factor signals are conveyed to the tuberous sclerosis complex (composed of TSC1 and TSC2) by AKT, a major effector of PI3K signaling [90], and ATP levels are reported by AMP-activated protein kinase AMPK [91]. In order to signal low ATP levels, AMPK requires activation by LKB1/STK11 [92,93], which frequently undergoes somatic mutation in lung adenocarcinomas [94] and germline mutation in the Peutz–Jeghers familial cancer syndrome [95]. TSC1/2 integrates growth factor signals and ATP levels to regulate mTORC1 activation via the Rheb GTPase [96]. Germline mutations in TSC1/2 are found in TSC familial cancer syndromes, although rarely in non-TSC cancers [97–99].
3.2. Amino acid sensing by mTORC1
Although cancer-relevant genes involved in growth factor signaling are well established, emerging components of the amino acid signaling machinery are already implicated in cancer. Growth factor signaling and ATP levels regulate TSC1/2 GAP activity toward Rheb [96], which in turn stimulates mTORC1 kinase activity. Rheb represents the first half of a lysosome-based coincidence detector that is comprised of GTPases and controls activation of mTORC1. The other half signals amino acid levels via the Rag GTPases, which bind mTORC1 and promote its lysosomal localization via an amino acid regulated interaction with the lysosomal bound Ragulator complex [100,101]. Thus, the pathway ensures that appropriate growth conditions are met by independently regulating mTORC1 lysosomal localization and kinase activation via the Rag and Rheb GTPases [102].
The Rag GTPases are obligate heterodimers comprised of RagA or RagB bound to RagC or RagD [103]. As is the case for all GTPases, the nucleotide state of the Rags determines their functional output, which is to bind mTORC1 and recruit it to the lysosome in the presence of amino acids [100]. In the case of the Rag heterodimers, amino acid stimulation results in Rag A/B bound to GTP, whereas amino acid starvation results in Rag C/D bound to GTP [100,104]. Both known GTPase activating proteins (GAPs) for the Rag het-erodimer (FLCN and the GATOR1 complex) are tumor suppressive. Inactivating mutations in FLCN, the GAP for RagC/D, cause the Birt–Hogg–Dubé (BHD) tumor syndrome characterized by benign tumors of the hair follicles and, in some cases, kidney tumors [105–107]. Suppression of FLCN in cell based systems results in mTORC1 inactivation, consistent with its role as a positive regulator of the mTOR pathway [108–112]. However, kidney tumors derived from BHD patients have hyperactive mTORC1 signaling [113–115]. As such, how loss of FLCN activity leads to BHD, and whether it is related to altered mTORC1 signaling, is still being debated. DEPDC5 and NPRL2, components of GATOR1, the GAP for RagA/B, are mutated in glioblastoma and ovarian cancer [116–119]. Cancer cells with mutated GATOR1 components have hyperactivated mTORC1 signaling and are resistant to amino acid starvation [116]. These cells are hypersensitive to rapamycin, thus mutations in GATOR1 components may help identify tumors that will respond to mTORC1 inhibition [116].
Interestingly, a significant number of recurrent mutations have been identified in MTOR itself at a prevalence of 5–10% in several cancer subtypes [86,120]. A subset of these mutations are activating and confer resistance to the effects of amino acid and glucose starvation on mTORC1 signaling [120]. While activating MTOR mutations can disrupt the interaction between mTOR and its inhibitor DEPTOR [120], the mechanisms by which these mutations liberate mTORC1 activity from amino acid dependence or promote transformation are unclear. Nevertheless, cell lines with activating mutations in MTOR are also hypersensitive to mTORC1 inhibition by rapamycin [120]. Interestingly, a recent report described an excellent anti-tumor response to inhibition of mTORC1 in an anaplastic thyroid carcinoma patient, which recurred upon acquisition of a mutation in MTOR conferring resistance to allosteric mTORC1 inhibitors, but maintaining sensitivity to ATP-competitive kinase inhibitors [121]. Future work will be required to determine how best to treat patients whose tumors harbor specific MTOR mutations, and to understand precisely how deregulation of amino acid or growth factor sensing promotes tumorigenesis. These investigations may reveal other upstream inputs of the amino acid signaling arm of mTORC1 which are deregulated in cancer.
It is now well appreciated that the lysosome is a signal integration hub for the mTORC1 pathway [102]. The convergence of so many mTORC1 pathway components to this compartment raises the possibility that amino acids can be sensed within or at the surface of the lysosome. Indeed, emerging evidence suggests that amino acids within the lysosomal lumen can be sensed in an inside-out fashion for mTORC1 pathway activation [122]. While mTORC1 dependent amino acid sensing mechanisms may be present in other compartments, recent work supports the hypothesis that at least two parallel signals emerge from within the lysosome. The vacuolar ATPase (v-ATPase) is required for the inside-out signaling of amino acids by mTORC1 via its interaction with Ragulator [122]. However, whether the v-ATPase directly senses amino acids is still unclear. The most promising candidate for a direct amino acid sensor is SLC38A9, a lysosomal amino acid transporter required for signaling arginine sufficiency [13,123]. While it is still unknown whether the v-ATPase and SLC38A9 contribute to oncogenesis, given their importance in amino acid signaling it would not be surprising to find alterations in these components of the mTORC1 pathway in cancer, as well as additional direct amino acid sensors and pathway components predicted to exist.
While the precise machinery required to sense and respond to tumor-relevant amino acid levels is being uncovered, it is interesting to speculate why mutations in the amino acid sensing arm of mTORC1 signaling occur. Indeed, it is reasonable to expect that active mTORC1 signaling in tumor cells outgrowing their nutrient supply would be deleterious. So how could constitutive amino acid sufficiency signals be pro-tumorigenic? Before angiogenesis catches up with tumorigenesis – early in the disease process – constitutive mTORC1 signaling could promote the survival of cancer cells during this stressful period of nutrient limitation. The cells that emerge could be poised to rapidly proliferate once a nutrient supply becomes adequate. Alternatively, activation of the amino acid sensing arm may prime mTORC1 for activation by the growth factor signaling arm. Indeed, given the spectrum of cancers in which mTORC1 pathway mutations have been found, the answers to these questions are probably context-dependent.
3.3. Outputs of mTORC1 altered in cancer
Given the emerging importance of amino acid sensing and biosynthesis in tumor progression, it is not surprising that processes regulated by mTORC1 which affect amino acid pools, such as translation and autophagy, have been implicated in cancer. S6 kinase 1 (S6K1) and eIF4E-binding protein (4E-BP1) are well known mTORC1 substrates that regulate translation. Phosphorylation of 4E-BP1 by mTORC1 relieves its inhibition of the eIF4E translation initiation factor, allowing translation to proceed [124]. The 4E-BP1–eIF4E axis is emerging as the major downstream effector of mTORC1 in cancer [125]. eIF4E is amplified or overexpressed in various sporadic cancers and increased 4E-BP1 phosphorylation correlates with poor survival outcomes [126,127]. Expression of phosphorylation defective 4E-BP1 reduces tumor progression in KRAS and PI3K driven tumors, suggesting eIF4E plays an important role in maintaining cancer growth [128,129]. Conversely, loss of 4E-BP1 and 4E-BP2 increased tumorigenesis in p53 null mice [130]. How 4E-BP1–eIF4E deregulation promotes oncogenesis is unclear, but it is thought to increase translation of oncogenic proteins supporting cell survival [125,131] and cell-cycle progression [132].
Autophagy, or ‘self-eating’, is a stress-induced survival mechanism that degrades and recycles damaged or superfluous proteins and organelles by fusion of autophagic vessels with the lysosome. As amino acid levels are also influenced by autophagy especially during metabolic stress [133], the mTORC1 pathway is a primary autophagy regulator, limiting the process during abundant nutrient and growth factor signaling [134]. Although regulation of amino acid levels by autophagy has not been shown to impact tumorigenesis specifically, autophagy overall plays an interesting dual role as an oncogenic and tumor suppressive process [7]. On one hand, autophagy can be activated during early tumor formation before adequate blood supply is established to buffer metabolic stress [135], and contributes to chemotherapy resistance [136,137]. In contrast, monoallelic loss of BECN1, an essential autophagy component, is frequently observed in human breast, ovarian, and prostate cancer, and is sufficient to cause spontaneous tumor development in mice [138–141]. Loss of other autophagy genes such as UVRAG, ATG4C, and SH3GLB1 (Bif-1) also increases susceptibility to tumorigenesis [142–144]. How autophagy suppresses tumor formation is still under investigation, but as an important contributor to cellular housekeeping, autophagy clears damaged proteins, damaged organelles such as mitochondria, and reduces reactive oxygen species, which can promote oncogenesis [145]. Indeed, deletion of the key autophagy component Atg7 in the context of a Kras-driven lung tumor model results in the massive accumulation of damaged mitochondria and limits tumorigenesis, indicating that activation of mitophagy to maintain mitochondrial quality is a key function of autophagy in Kras-driven cancer [146]. Consistent with this observation, maintenance of mitochondrial health by autophagy promotes tumors driven by mutation of the Ras-pathway member BRAF [147,148]. Autophagy is also thought to prevent necrotic death in apoptotic deficient tumor cells, thereby limiting local inflammation, which can increase tumor growth [135]. Finally, autophagy-deficient cells are prone to chromosome instability and DNA damage [149]. Thus, targeting autophagy in cancer may require context-specific and disease-stage specific modulation. Inhibition of autophagy has been shown to limit tumor growth in mouse models of pancreatic cancer [150–152], and the autophagy inhibitor hydroxychloroquine has shown promise in combination with chemotherapy in clinical trials [153,154]. The development of new and more potent inhibitors of autophagy will likely be critical for evaluating the plausibility of targeting this process in future anti-cancer therapy.
4. Concluding remarks
Proper amino acid management is critical for cell and organismal viability, particularly under conditions of metabolic stress. Amino acids such as serine and glutamine are not only utilized for protein biosynthesis, but also for the biosynthesis of all other major biomolecule classes. Key to our further understanding of amino acid management will be a clearer picture of how the levels of individual amino acids are sensed by the cell as well as how the process of transformation alters amino acid catabolic pathways in ways that are critical for cancer cell survival. We are finding new ways in which autophagy supports transformation, including through the recycling of damaged mitochondria, leading to novel anti-cancer strategies and a resurgence in interest in mitochondrial health and function. Moreover, the field is revisiting the importance of macropinocytosis, a process for scavenging amino acids from the environment, as part of the arsenal of cancer cells to maintain amino acid pools. For example, recent work has shown that this Ras-activated process, like autophagy, can support cell survival during periods of amino acid starvation, and contribute to the overall biomass of the cell [8,9]. Future work will be required to uncover how this process is activated and its significance to oncogenesis. Finally, the identification of an expanding number of proteins involved in amino acid management via mTORC1 will likely continue to reveal additional tumor-relevant genes and expand our understanding of these ancient pathways controlling cell growth.
Acknowledgments
RP is funded by NIH grant R00CA168940 and Susan G. Komen for the Cure.
References
- 1.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2(10):881–898. doi: 10.1158/2159-8290.CD-12-0345. PMCID: 3491070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. PMCID: 3311998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Heiden MG. Exploiting tumor metabolism: challenges for clinical translation. J Clin Investig. 2013;123(9):3648–3651. doi: 10.1172/JCI72391. PMCID: 3754281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E. The role for autophagy in cancer. J Clin Investig. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. PMCID: 3810415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75(3):544–553. doi: 10.1158/0008-5472.CAN-14-2211. PMCID: 4316379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Nat Acad Sci U S A. 2013;110(22):8882–8887. doi: 10.1073/pnas.1307237110. PMCID: 3670379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122(3168):501–504. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 12.Ban H, Shigemitsu K, Yamatsuji T, Haisa M, Nakajo T, Takaoka M, Nobuhisa T, Gunduz M, Tanaka N, Naomoto Y. Arginine and leucine regulate p70 S6 kinase and 4E-BP1 in intestinal epithelial cells. Int J Mol Med. 2004;13(4):537–543. [PubMed] [Google Scholar]
- 13.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, Straub C, Kim C, Park J, Sabatini BL, Sabatini DM. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–194. doi: 10.1126/science.1257132. PMCID: 4295826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 15.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat-tissues. Adv Enzyme Regul. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- 18.Achouri Y, Rider MH, VanSchaftingen E, Robbi M. Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem J. 1997;323:365–370. doi: 10.1042/bj3230365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh DA, Sallach HJ. Purification and properties of chicken liver D-3-phosphoglycerate dehydrogenase. Biochemistry. 1965;4(6):1076–1085. doi: 10.1021/bi00882a015. [DOI] [PubMed] [Google Scholar]
- 20.Stover P, Schirch V. Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. J Biol Chem. 1990;265(24):14227–14233. [PubMed] [Google Scholar]
- 21.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad B: Phys. 2008;84(7):246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibbetts AS, Appling DR. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 23.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. PMCID: 3353325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. PMCID: 3677549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Wu J, Cai J, He Z, Yuan J, Zhu X, Li Y, Li M, Guan H. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. J International Cancer. 2015;136(4):E39–E50. doi: 10.1002/ijc.29150. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Wei H, Liu X, Wang N, Qi Y, Zhang Y, Zhang S. Downregulation of phosphoglycerate dehydrogenase inhibits proliferation and enhances cisplatin sensitivity in cervical adenocarcinoma cells by regulating Bcl-2 and caspase-3. Cancer Biol Ther. 2015:541–548. doi: 10.1080/15384047.2015.1017690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattaini KR, Brignole EJ, Kini M, Davidson SM, Fiske BP, Drennan CL, Vander Heiden MG. An epitope tag alters phosphoglycerate dehydrogenase structure and impairs ability to support cell proliferation. Cancer Metab. 2015;3:5. doi: 10.1186/s40170-015-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J. 1987;245(2):609–612. doi: 10.1042/bj2450609. PMCID: 1148165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jun do Y, Taub D, Chrest FJ, Kim YH. Requirement of the expression of 3-phosphoglycerate dehydrogenase for traversing S phase in murine T lymphocytes following immobilized anti-CD3 activation. Cellular Immunol. 2014;287(2):78–85. doi: 10.1016/j.cellimm.2013.12.003. PMCID: 4169173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282(24):17623–17631. doi: 10.1074/jbc.M702526200. [DOI] [PubMed] [Google Scholar]
- 31.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282(23):16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 32.Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Nat Acad Sci U S A. 2012;109(18):6904–6909. doi: 10.1073/pnas.1204176109. PMCID: 3345000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O’Reilly M, Gottlieb E. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. PMCID: 3894725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M, Zhao YH, Topisirovic I, St-Pierre J, Pollak M. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res. 2014;74(24):7521–7533. doi: 10.1158/0008-5472.CAN-14-2643-T. [DOI] [PubMed] [Google Scholar]
- 35.Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, Qing G, Liu Z, Simon MC, Rabinowitz JD, Thompson CB. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4(12):1406–1417. doi: 10.1158/2159-8290.CD-14-0250. PMCID: 4258153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, Birsoy K, Freinkman E, Kami K, Possemato R, Chudnovsky Y, Pacold ME, Chen WW, Cantor JC, Shelton LM, Gui DY, Kwon M, Ramkissoon SH, Ligon KR, Kang SW, Snuderl M, Vander Heider MG, Sabatini DM. SHMT2 drives glioma cell survival in the tumor microenvironment but imposes a dependence on glycine clearance. Nature. 2015;520(7547):363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595. PMCID: 3526189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7(4):1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 40.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55(2):253–263. doi: 10.1016/j.molcel.2014.05.008. PMCID: 4106038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. doi: 10.1038/nature13236. PMCID: 4104482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Pang YH, Ang HS, Mitchell W, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148(1–2):259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 43.Field MS, Kamynina E, Watkins D, Rosenblatt DS, Stover PJ. Human mutations in methylenetetrahydrofolate dehydrogenase 1 impair nuclear de novo thymidylate biosynthesis. Proc Nat Acad Sci U S A. 2015;112(2):400–405. doi: 10.1073/pnas.1414555112. PMCID: 4299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar AJ. The human l-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. PMCID: 131051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325(5939):435–439. doi: 10.1126/science.1173288. PMCID: 4373593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. doi: 10.1126/science.1226603. PMCID: 3652341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Nat Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. PMCID: 2148292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. PMCID: 2764330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeon YJ, Khelifa S, Ratnikov B, Scott DA, Feng Y, Parisi F, Ruller C, Lau E, Kim H, Brill LM, Jiang T, Rimm DL, Cardiff RD, Mills GB, Smith JW, Osterman AL, Kluger Y, Ronai ZA. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 2015;27(3):354–369. doi: 10.1016/j.ccell.2015.02.006. PMCID: 4356903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassanein M, Qian J, Hoeksema MD, Wang J, Jacobovitz M, Ji X, Harris FT, Harris BK, Boyd KL, Chen H, Eisenberg R, Massion PP. Targeting SLC1A5-mediated glutamine dependence in non-small cell lung cancer. Int J Cancer. 2015 doi: 10.1002/ijc.29535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. PMCID: 3710581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang YF, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fendt SM, Bell EL, Keibler MA, Davidson SM, Wirth GJ, Fiske B, Mayers JR, Schwab M, Bellinger G, Csibi A, Patnaik A, Blouin MJ, Cantley LC, Guarente L, Blenis J, Pollak MN, Olumi AF, Vander Heiden MG, Stephanopoulos G. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer Res. 2013;73(14):4429–4438. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan J, Kamphorst JJ, Rabinowitz JD, Shlomi T. Fatty acid labeling from glutamine in hypoxia can be explained by isotope exchange without net reductive isocitrate dehydrogenase (IDH) flux. J Biol Chem. 2013;288(43):31363–31369. doi: 10.1074/jbc.M113.502740. PMCID: 3829450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. PMCID: 3882799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Nat Acad Sci U S A. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. PMCID: 2596212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178(1):93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. PMCID: 2729443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. PMCID: 3345194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. PMCID: 3078749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, Sun L, Song L, Yan B, Slusher BS, Zhuo J, Ooi LL, Lee CG, Mancuso A, McCallion AS, Le A, Milone MC, Rayport S, Felsher DW, Dang CV. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Investig. 2015;125(6):2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, Mackinnon AL, Parlati F, Rodriguez ML, Shwonek PJ, Sjogren EB, Stanton TF, Wang T, Yang J, Zhao F, Bennett MK. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 63.Ubuka T, Meister A. Studies on the utilization of asparagine by mouse leukemia cells. J Nat Cancer Inst. 1971;46(2):291–298. [PubMed] [Google Scholar]
- 64.Ruzzo EK, Capo-Chichi JM, Ben-Zeev B, Chitayat D, Mao H, Pappas AL, Hitomi Y, Lu YF, Yao X, Hamdan FF, Pelak K, Reznik-Wolf H, Bar-Joseph I, Oz-Levi D, Lev D, Lerman-Sagie T, Leshinsky-Silver E, Anikster Y, Ben-Asher E, Olender T, Colleaux L, Decarie JC, Blaser S, Banwell B, Joshi RB, He XP, Patry L, Silver RJ, Dobrzeniecka S, Islam MS, Hasnat A, Samuels ME, Aryal DK, Rodriguiz RM, Jiang YH, Wetsel WC, McNamara JO, Rouleau GA, Silver DL, Lancet D, Pras E, Mitchell GA, Michaud JL, Goldstein DB. Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron. 2013;80(2):429–441. doi: 10.1016/j.neuron.2013.08.013. PMCID: 3820368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Fan J, Venneti S, Cross JR, Takagi T, Bhinder B, Djaballah H, Kanai M, Cheng EH, Judkins AR, Pawel B, Baggs J, Cherry S, Rabinowitz JD, Thompson CB. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56(2):205–218. doi: 10.1016/j.molcel.2014.08.018. PMCID: 4224619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Safer B, Smith CM, Williamson JR. Control of the transport of reducing equivalents across the mitochondrial membrane in perfused rat heart. J Mol Cell Cardiol. 1971;2(2):111–124. doi: 10.1016/0022-2828(71)90065-4. [DOI] [PubMed] [Google Scholar]
- 67.Milman HA, Cooney DA. Partial purification and properties of l-asparagine synthetase from mouse pancreas. Biochem J. 1979;181(1):51–59. doi: 10.1042/bj1810051. PMCID: 1161124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, Kang Y, Fleming JB, Bardeesy N, Asara JM, Haigis MC, DePinho RA, Cantley LC, Kimmelman AC. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. PMCID: 3656466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Broome JD. Evidence that l-asparaginase activity of guinea pig serum is responsible for its antilymphoma effects. Nature. 1961;191(479):1114-&. doi: 10.1084/jem.118.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clavell LA, Gelber RD, Cohen HJ, Hitchcockbryan S, Cassady JR, Tarbell NJ, Blattner SR, Tantravahi R, Leavitt P, Sallan SE. 4-Agent induction and intensive asparaginase therapy for treatment of childhood acute lymphoblastic-leukemia. New Engl J Med. 1986;315(11):657–663. doi: 10.1056/NEJM198609113151101. [DOI] [PubMed] [Google Scholar]
- 71.Prager MD, Bachynsk N. Asparagine synthetase in asparaginase resistant and susceptible mouse lymphomas. Biochem Biophys Res Commun. 1968;31(1):43–47. doi: 10.1016/0006-291x(68)90028-4. [DOI] [PubMed] [Google Scholar]
- 72.Fine BM, Kaspers GJL, Ho M, Loonen AH, Boxer LM. A genome-wide view of the in vitro response to l-asparaginase in acute lymphoblastic leukemia. Cancer Res. 2005;65(1):291–299. [PubMed] [Google Scholar]
- 73.Purwaha P, Lorenzi PL, Silva LP, Hawke DH, Weinstein JN. Targeted metabolomic analysis of amino acid response to l-asparaginase in adherent cells. Metabolomics. 2014;10(5):909–919. doi: 10.1007/s11306-014-0634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339(6125):1323–1328. doi: 10.1126/science.1228792. PMCID: 3753690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339(6125):1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 77.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 79.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204. PMCID: 3994981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. PMCID: 3632399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(Suppl. 2):S43–S51. doi: 10.1038/onc.2009.352. PMCID: 3752670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gschwind A, Fischer OM, Ullrich A. Timeline – the discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4(5):361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 83.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 84.Xu GF, Oconnell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, Weiss R. The neurofibromatosis type-1 gene encodes a protein related to Gap. Cell. 1990;62(3):599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 85.Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type-1 gene in a neurofibrosarcoma supports a tumor suppressor gene hypothesis. Nat Genet. 1993;3(2):122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- 86.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta S, Ramjaun AR, Haiko P, Wang YH, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of Ras to phosphoinositide 3-kinase p110 alpha is required for Ras-driven tumorigenesis in mice. Cell. 2007;129(5):957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 88.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133(3):403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 89.Liaw D, Marsh DJ, Li J, Dahia PLM, Wang SI, Zheng ZM, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16(1):64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 90.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 91.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Nat Acad Sci U S A. 2004;101(10):3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LGD, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13(22):2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 94.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62(13):3659–3662. [PubMed] [Google Scholar]
- 95.Avizienyte E, Roth S, Loukola A, Hemminki A, Lothe RA, Stenwig AE, Fossa SD, Salovaara R, Aaltonen LA. Somatic mutations in LKB1 are rare in sporadic colorectal and testicular tumors. Cancer Res. 1998;58(10):2087–2090. [PubMed] [Google Scholar]
- 96.Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.vanSlegtenhorst M, deHoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, vandenOuweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JBM, Ward S, Green AJ, Yates JRW, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277(5327):805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 98.Kandt RS, Haines JL, Smith M, Northrup H, Gardner RJM, Short MP, Dumars K, Roach ES, Steingold S, Wall S, Blanton SH, Flodman P, Kwiatkowski DJ, Jewell A, Weber JL, Roses AD, Pericakvance MA. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney-disease. Nat Genet. 1992;2(1):37–41. doi: 10.1038/ng0992-37. [DOI] [PubMed] [Google Scholar]
- 99.Nellist M, Janssen B, Brookcarter PT, Hesselingjanssen ALW, Maheshwar MM, Verhoef S, Vandenouweland AMW, Lindhout D, Eussen B, Cordeiro I, Santos H, Halley DJJ, Sampson JR, Ward CJ, Peral B, Thomas S, Hughes J, Harris PC, Roelfsema JH, Saris JJ, Spruit L, Peters DJM, Dauwerse JG, Breuning MH. Identification and characterization of the tuberous sclerosis gene on chromosome-16. Cell. 1993;75(7):1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 100.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. PMCID: 3024592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. PMCID: 2475333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24(7):400–406. doi: 10.1016/j.tcb.2014.03.003. PMCID: 4074565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276(10):7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 104.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. PMCID: 3517996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Birt AR, Hogg GR, Dube WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113(12):1674–1677. [PubMed] [Google Scholar]
- 106.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, Sharma N, Walther M, Munroe D, Hill R, Maher E, Greenberg C, Lerman MI, Linehan WM, Zbar B, Schmidt LS. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt–Hogg–Dube syndrome. Cancer Cell. 2002;2(2):157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 108.Bastola P, Stratton Y, Kellner E, Mikhaylova O, Yi Y, Sartor MA, Medvedovic M, Biesiada J, Meller J, Czyzyk-Krzeska MF. Folliculin contributes to VHL tumor suppressing activity in renal cancer through regulation of autophagy. PLOS ONE. 2013;8(7):e70030. doi: 10.1371/journal.pone.0070030. PMCID: 3726479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hudon V, Sabourin S, Dydensborg AB, Kottis V, Ghazi A, Paquet M, Crosby K, Pomerleau V, Uetani N, Pause A. Renal tumour suppressor function of the Birt–Hogg–Dube syndrome gene product folliculin. J Med Genet. 2010;47(3):182–189. doi: 10.1136/jmg.2009.072009. [DOI] [PubMed] [Google Scholar]
- 110.Takagi Y, Kobayashi T, Shiono M, Wang L, Piao X, Sun G, Zhang D, Abe M, Hagiwara Y, Takahashi K, Hino O. Interaction of folliculin (Birt–Hogg–Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27(40):5339–5347. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- 111.Hartman TR, Nicolas E, Klein-Szanto A, Al-Saleem T, Cash TP, Simon MC, Henske EP. The role of the Birt–Hogg–Dube protein in mTOR activation and renal tumorigenesis. Oncogene. 2009;28(13):1594–1604. doi: 10.1038/onc.2009.14. PMCID: 2664853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. PMCID: 3787382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, Yao M, Bernardo M, Ileva L, Choyke P, Warren MB, Zbar B, Linehan WM, Schmidt LS. Kidney-targeted Birt–Hogg–Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Nat Cancer Inst. 2008;100(2):140–154. doi: 10.1093/jnci/djm288. PMCID: 2704336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen J, Futami K, Petillo D, Peng J, Wang P, Knol J, Li Y, Khoo SK, Huang D, Qian CN, Zhao P, Dykema K, Zhang R, Cao B, Yang XJ, Furge K, Williams BO, Teh BT. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia. PLoS ONE. 2008;3(10):e3581. doi: 10.1371/journal.pone.0003581. PMCID: 2570491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong SB, Yamaguchi TP, Schmidt LS, Linehan WM. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Nat Acad Sci U S A. 2009;106(44):18722–18727. doi: 10.1073/pnas.0908853106. PMCID: 2765925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins VP. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosome Cancer. 2005;43(2):181–193. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 118.Li F, Wang F, Haraldson K, Protopopov A, Duh FM, Geil L, Kuzmin I, Minna JD, Stanbridge E, Braga E, Kashuba VI, Klein G, Lerman MI, Zabarovsky ER. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Res. 2004;64(18):6438–6443. doi: 10.1158/0008-5472.CAN-03-3869. [DOI] [PubMed] [Google Scholar]
- 119.Lerman MI, Minna JD. Chromosome ILC. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res. 2000;60(21):6116–6133. [PubMed] [Google Scholar]
- 120.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4(5):554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA, Guo Y, Swanson SJ, Ruan DT, Hanna GJ, Haddad RI, Getz G, Kwiatkowski DJ, Carter SL, Sabatini DM, Janne PA, Garraway LA, Lorch JH. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med. 2014;371(15):1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. PMCID: 3211112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. PMCID: 4376665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. PMCID: 3610329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP–eIF4E. Cancer cell. 2010;17(3):249–261. doi: 10.1016/j.ccr.2010.01.021. PMCID: 2901095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bjornsti MA, Houghton PJ. Lost in translation: dysregulation of cap-dependent translation and cancer. Cancer Cell. 2004;5(6):519–523. doi: 10.1016/j.ccr.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 127.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, Baselga J, Ramon, Cajal S. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 128.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, Solit DB, Rosen N. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. PMCID: 3286650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16(20):4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Petroulakis E, Parsyan A, Dowling RJ, LeBacquer O, Martineau Y, Bidinosti M, Larsson O, Alain T, Rong L, Mamane Y, Paquet M, Furic L, Topisirovic I, Shahbazian D, Livingstone M, Costa-Mattioli M, Teodoro JG, Sonenberg N. p53-Dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell. 2009;16(5):439–446. doi: 10.1016/j.ccr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 131.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW. Dissecting eIF4E action in tumorigenesis. Genes Devel. 2007;21(24):3232–3237. doi: 10.1101/gad.1604407. PMCID: 2113024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328(5982):1172–1176. doi: 10.1126/science.1187532. PMCID: 2893390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mizushima N. Autophagy: process and function. Genes Devel. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 134.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. PMCID: 3127250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. PMCID: 2857533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Investig. 2007;117(2):326–336. doi: 10.1172/JCI28833. PMCID: 1765515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. PMCID: 1896119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 139.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Investig. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. PMCID: 297002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Nat Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. PMCID: 299911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res: MCR. 2014;12(4):485–490. doi: 10.1158/1541-7786.MCR-13-0614. PMCID: 3989371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. PMCID: 2254521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282(25):18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 144.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8(7):688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 145.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. PMCID: 2802318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, Snyder E, Santanam U, Dipaola RS, Jacks T, Rabinowitz JD, White E. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Devel. 2013;27(13):1447–1461. doi: 10.1101/gad.219642.113. PMCID: 3713426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strohecker AM, White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10(2):384–385. doi: 10.4161/auto.27320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov. 2015;5(4):410–423. doi: 10.1158/2159-8290.CD-14-1473. PMCID: 4390491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Devel. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. PMCID: 1877749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Devel. 2011;25(7):717–729. doi: 10.1101/gad.2016111. PMCID: 3070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang A, Kimmelman AC. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy. 2014;10(9):1683–1684. doi: 10.4161/auto.29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–913. doi: 10.1158/2159-8290.CD-14-0362. PMCID: 4125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17(4):654–666. doi: 10.1158/1078-0432.CCR-10-2634. PMCID: 3075808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, Fuchs CS, McCleary NJ, Meyerhardt JA, Ng K, Schrag D, Sikora AL, Spicer BA, Killion L, Mamon H, Kimmelman AC. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19(6):637–638. doi: 10.1634/theoncologist.2014-0086. PMCID: 4041680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29(18):2746–2752. doi: 10.1038/onc.2010.28. PMCID: 2953941. [DOI] [PMC free article] [PubMed] [Google Scholar]