Abstract
The view of the lysosome as the terminal end of cellular catabolic pathways has been challenged by recent studies showing a central role of this organelle in the control of cell function. Here we show that a lysosomal Ca2+ signaling mechanism controls the activities of the phosphatase calcineurin and of its substrate TFEB, a master transcriptional regulator of lysosomal biogenesis and autophagy. Lysosomal Ca2+ release via mucolipin 1 (MCOLN1) activates calcineurin, which binds and de-phosphorylates TFEB, thus promoting its nuclear translocation. Genetic and pharmacological inhibition of calcineurin suppressed TFEB activity during starvation and physical exercise, while calcineurin overexpression and constitutive activation had the opposite effect. Induction of autophagy and lysosomal biogenesis via TFEB required MCOLN1-mediated calcineurin activation, linking lysosomal calcium signaling to both calcineurin regulation and autophagy induction. Thus, the lysosome reveals itself as a hub for the signaling pathways that regulate cellular homeostasis.
Lysosomes are membrane-bound organelles present in all cell types. Their role in degradation and recycling processes has been extensively characterized1–3. The lysosomal lumen acidic pH with the presence of a broad variety of hydrolases able to degrade an ample spectrum of substrates, make these organelles extraordinary machineries for the recycling of cellular waste. Extracellular substrates reach the lysosome mainly via the endocytic and phagocytic pathways, while intracellular substrates are the delivered to the lysosome by the autophagic pathway via the fusion of autophagosomes with lysosomes4, 5. Thus, lysosomes are the “terminal end” of most cellular catabolic pathways. The role of the lysosomes in degradation and recycling processes has always been considered as a cellular “housekeeping” function and little attention has been paid to the regulation of these processes and to the possible influence of environmental cues, such as starvation and physical exercise.
The discovery that the Transcription Factor EB (TFEB) is a master regulator of lysosomal and autophagic function and of energy metabolism6–8 suggested that environmental cues may control lysosomal function via the induction of a broad transcriptional program. TFEB activity is regulated by phosphorylation9–13, which keeps TFEB inactive in the cytoplasm, while dephosphorylated TFEB travels to the nucleus to activate transcriptional target genes. TFEB phosphorylation is mediated by mTORC1, a major kinase complex that positively regulates cell growth and negatively regulates autophagy. Interestingly, it is known mTORC1 exerts its activity on the lysosomal surface and is positively regulated by lysosomal nutrients14, 15. The regulation of TFEB by lysosomal mTORC1 and the shuttling of TFEB to the nucleus revealed a lysosome-to-nucleus signaling mechanism9. Thus, these studies identify the lysosome as a signaling hub that controls cellular homeostasis via both post-translational and transcriptional mechanisms14–17.
Another aspect of lysosomal function underestimated in the past is the ability of lysosomes to store Ca2+ and to participate to calcium signaling processes. Several calcium channels reside on the lysosomal membrane. Recent studies have investigated the role of these lysosomal calcium channels in fundamental cellular processes and their involvement in disease mechanisms18. In addition, the recent discoveries of calcium microdomains, which mediate local calcium signals from several intracellular compartments (e.g. mitochondria)19, further suggest the involvement of the lysosome in intracellular calcium signaling.
In the present study, while searching for a phosphatase that de-phosphorylates TFEB, we discovered another example of lysosomal signaling. We revealed a calcium signaling mechanism that starts at the lysosome and controls autophagy via calcineurin-mediated induction of TFEB.
Calcineurin modulates TFEB subcellular localization
Previous studies demonstrated that mTOR-mediated phosphorylation of TFEB serine residues Ser142 and Ser211 promotes the interaction of TFEB with the 14-3-3 protein and results in a cytoplasmic localization. Conversely, conditions that lead to mTOR inhibition, such as starvation and lysosomal stress, promote TFEB nuclear translocation and transcriptional activation of lysosomal and autophagic genes6, 7, 9, 14, 15, 17. While the role of the kinases that mediate TFEB phosphorylation has been defined by previous studies9–13, the phosphatase(s) involved in its de-phosphorylation have remained elusive. To identify the phosphatase(s) that de-phosphorylate(s) TFEB we performed a High Content (HC) screening of a phosphatase siRNA library using a cellular assay based on cytoplasm-to-nucleus shuttling of TFEB during starvation9. We tested the effects of the specific inhibition of each of 231 phosphatases or putative phosphatases on TFEB subcellular localization. The most significant hit identified by the primary screening was the calcineurin catalytic subunit isoform beta (PPP3CB; Gene ID:5532)20, thus we focused subsequent studies exclusively on this phosphatase.
Fig. 1a shows that inhibition of PPP3CB suppressed starvation-induced nuclear translocation of TFEB. The ability of PPP3CB to inhibit TFEB nuclear translocation was further confirmed by using a set of 3 different PPP3CB siRNAs, as well as the pool of 3, in the HeLaTFEB-GFP cell line (Fig. 1b). In addition, the calcineurin inhibitors Cyclosporin A and FK506, both jointly and individually, reduced TFEB nuclear translocation during starvation (Fig. 1c). Importantly, overexpression of ΔCaN, a constitutively active form of calcineurin, induced TFEB nuclear translocation in normally fed cells, suggesting that calcineurin activity was not only necessary but sufficient to induce TFEB nuclear translocation (Fig. 1d,e). Consistent with its effect on TFEB nuclear translocation, ΔCaN induced the expression of TFEB transcriptional target genes in a TFEB-dependent manner (Fig. 1f). Both starvation and calcineurin overexpression induced nuclear translocation as well as de-phosphorylation of the Nuclear Factor of Activated T cells (NFAT), which is known to depend on calcineurin activity21 (Supplementary Fig. 1). In vivo studies showed that transfection by electroporation of both calcineurin subunit ΔCaN and its essential regulatory subunit PPP3R1 (CnB)22 in adult muscles from wild type mice significantly induced TFEB nuclear translocation (Fig. 1g). In addition, exercise, a physiological stimulus that activates calcineurin23, 24, promoted TFEB nuclear translocation and this effect was blunted when an endogenous calcineurin inhibitor, CAIN, was electroporated into muscle cells25 (Fig. 1g). These loss-of-function and gain-of-function data demonstrate that calcineurin plays a crucial role in the regulation of TFEB subcellular localization, both in cell culture and in skeletal muscle, during energy-demanding conditions, such as starvation and physical exercise.
Figure 1.
Calcineurin regulates TFEB nuclear translocation. (a,b) siRNA-mediated inhibition of PPP3CB (siPPP3CB) suppresses starvation-induced nuclear translocation in HeLaTFEB-GFP cells. Scale bar 10µm. (b) The left plot represents the percentage of nuclear TFEB in starved cells (3h) silenced with each single PPP3CB oligonucleotide or the pool of the 3 oligonucleotides compared to starved control cells transfected with scramble (SCRMBL) siRNA oligonucleotides. qPCR analysis of PPP3CB mRNA levels confirms the efficacy of siRNA-mediated silencing (right plot). Bar graphs show mean ±s.d. for n=3 independent experiments. Scale bar 10µm. (c) Pharmacological inhibition of calcineurin reduces starvation-mediated TFEB nuclear localization. Images from a HC assay using normally fed or 1h starved HeLaTFEB-GFP cells treated with FK506 (5µM) and/or CsA (10µM). Plot represents the percentage of nuclear TFEB translocation compared with DMSO-treated fed and starved cells. n=3 independent experiments. Scale bar 10µm. (d) HeLaTFEB-GFP cells were transfected with a constitutive active form of calcineurin (HA-tagged-ΔCaN). The plot represents the percentage of TFEB nuclear translocation. Data shows the mean±s.d. of n=3 independent experiments. Scale bar 10µm. (e) HeLa cells were transfected with an empty vector or HA-tagged-ΔCaN. Subcellular localization of endogenous TFEB was analyzed for each condition using specific anti-TFEB antibodies. The plot represents the percentage of TFEB nuclear translocation. Bar graphs show mean ±s.d. of n=3 independent experiments. Scale bar 10µm. (f) Calcineurin overexpression upregulates TFEB target genes. Left panel: qPCR showing the efficiency of TFEB silencing (upper) and ΔCaN overexpression (lower). Right panel: overexpression of ΔCaN in HeLaTFEBGFP cells (ΔCaN) and in HeLaTFEBGFP cells transfected with an siRNA against TFEB (siTFEB+ΔCaN)• showing that calcineurin enhances the expression of TFEB target genes in a TFEB-dependent manner. n=3 independent experiments, mean ±s.d, *p<0.05. (g) Exercise induces TFEB nuclear translocation via calcineurin. Muscles were transfected by electroporation with TFEB-GFP (green), ΔCaN, its regulatory subunit (CnB) and the calcineurin inhibitor myc-CAIN. An antibody against dystrophin (Dys, red) was used to visualize the muscle fibers. The yellow arrowheads indicate TFEB nuclear translocation in fibers stimulated by ΔCaN+CnB and exercise, respectively. The plot shows the percentages of TFEB-positive nuclei in muscles under the following conditions: sedentary (control), sedentary with calcineurin (ΔCaN+CnB), exercised (run), exercised with CAIN (run + CAIN). The graph represents mean ±s.d. of n=3 mice. Scale bar 50µm. Source data are provided in Supplementary table 4.
Calcineurin binds and dephosphorylates TFEB
These data prompted us to investigate the relationship between calcineurin and TFEB. PPP3CB-TFEB interaction was detected by co-immunoprecipitation experiments, performed in both HeLaTFEB-GFP and in HEK-293TFEB-GFP cells (Supplementary Fig. 2a-c) and by a histidine-pulldown assay using purified His-TFEB and purified catalytic and regulatory calcineurin subunits (CaN) (Supplementary Fig. 2d). In the his-pulldown assay the interaction between purified TFEB and CaN plus calmodulin (CaM), a protein known to interact with calcineurin22, appeared to be stronger that the one between TFEB and CaN alone, while no interaction was detected between TFEB and CaM alone (Supplementary Fig. 2d). Notably, the interaction between PPP3CB and TFEB was detected both in fed and starved cells, suggesting that it is independent from the phosphorylation status of TFEB, as previously shown for the binding of calcineurin to NFAT19 (Supplementary Fig. 2a,c). Finally, a physical interaction between PPP3CB and TFEB was also detected by Proximity Ligation Assay (PLA)27 performed on endogenous proteins in non-transfected living cells (Fig. 2a). Similar data were obtained by PLA on exogenous overexpressed proteins (Fig. 2b). The interaction between mTOR and TFEB9 was used as a positive control of the assay, while the absence of interaction between the Golgi protein Golgin97 and TFEB was used as a negative control (Supplementary Fig. 3a). The specificities of the antibodies used in the PLA were tested by both immunofluorescence and immunoblotting (Supplementary Fig. 3b-d). These data demonstrate that calcineurin physically interacts with TFEB and suggest that TFEB is a calcineurin substrate.
Figure 2.
Calcineurin binds and de-phosphorylates TFEB. (a) Proximity Ligation Assay (PLA). HeLa cells were pre-treated 10 minutes before fixation with 1µM ER-tracker (green) to visualize the cytoplasm and with Hoechst 33258 (blue) to stain nuclei. After fixation, cells were processed to detect TFEB proximity interactions. Positive interactions between TFEB and PPP3CB (red dots) are clearly visible (white squares contain higher magnification images). n=3 independent experiments were performed. Scale bar, 10 µm. (b) PLA performed on Hela cells transfected with PPP3CB-myc, TFEB-GFP or both. PPP3CB and TFEB interactions are shown by the red dots. n=3 independent experiments were performed. Scale bar, 10µm. (c) Calcineurin knock-down reduces the starvation-induced downshift of endogenous TFEB electrophoretic mobility. HeLa cells expressing siRNA against PPP3CB were treated as indicated. Specific antibodies against TFEB were used to detect the endogenous protein. n=3 independent experiments were performed. (d) Calcineurin overexpression reduces TFEB electrophoretic mobility. TFEB-FLAG vector alone or in combination with a constitutively active form of calcineurin (HA- ΔCaN) was co-transfected in fed HeLa cells. Calcineurin overexpression was confirmed by using anti-HA antibodies. n=3 independent experiments were performed. (e) Calcineurin de-phosphorylates S142 and S211 of immunoprecipitated TFEB. Lambda phosphatase (λ-PPT) was used as positive control. Specific antibodies against S142-TFEB and 14.3.3 motif, which binds phosphorylated TFEB serine residue S211, were used to detect TFEB phosphorylation. Bar graphs show mean ±s.d. of n=3 independent experiments. (f) Calcineurin regulates TFEB downstream of mTOR. Nuclear translocation of TFEB is reduced in stable HeLaTFEB-GFP cells after silencing of PPP3CB, its essential regulatory subunit PPP3R1 or both genes (PPP3CB/R1). Representative images from HC assay of HeLaTFEB-GFP cells reverse transfected with SCRMBL, PPP3CB, PPP3R1 or PPP3CB/R1 siRNAs, and then subjected to the indicated conditions. The left plot shows the mean ±s.d. of the percentage of nuclear TFEB translocation in knock-down cells compared with their corresponding control-treated cells transfected with scramble siRNA oligonucleotides. n=3 independent experiments were performed. Scale bar 10µm. siRNA-mediated silencing was confirmed by qPCR analysis of PPP3CB and PPP3R1 mRNA levels (right plot). Uncropped scans of western blots are provided in Supplementary Figure 8. Source data are provided in Supplementary table 4.
Thus, we tested whether calcineurin de-phosphorylates TFEB. siRNA-mediated inhibition of calcineurin abolished the downshift in TFEB molecular weight that is associated with TFEB de-phosphorylation during starvation7 (Fig. 2c). In addition, expression of a constitutively active mutant form of calcineurin in normally fed cells resulted in TFEB molecular weight downshift (Fig. 2d). Calcineurin-mediated de-phosphorylation of TFEB was confirmed by an in vitro calcineurin phosphatase assay, which revealed that calcineurin de-phosphorylates two critical serine residues, S211 and S142 known to be involved in TFEB nuclear translocation9–12 (Fig. 2e). These result identify TFEB as a novel calcineurin substrate. Thus, calcineurin de-phosphorylates TFEB and induces its nuclear translocation by a mechanism similar to the one described for NFAT (REF).
Starvation and inhibition of mTOR have been shown to promote TFEB nuclear translocation 9–11. However, we found that siRNA-mediated inhibition of calcineurin significantly reduced the nuclear translocation of both endogenous and overexpressed TFEB in HeLa, HeLaTFEB-GFP, and MCF-7 cells, respectively, either starved or treated with Torin1, a strong mTOR inhibitor28, demonstrating that the regulation of TFEB subcellular localization by calcineurin is independent from mTOR activity and suggesting that calcineurin acts downstream of mTOR in the regulation of TFEB (Fig. 2f and Supplementary Fig. 3e).
Calcium modulates TFEB localization
Since calcineurin is activated by intracellular calcium20, 29, we assessed the calcium-dependence of TFEB regulation by artificially modulating cytoplasmic Ca2+ using Ca2+ chelators and ionophores. The Ca2+ chelator BAPTA-AM significantly reduced TFEB nuclear translocation in both starved and Torin1-treated cells (Fig. 3a). Both thapsigargin, an inhibitor of ER Ca2+-ATPase, and ionomycin, a Ca2+ ionophore, significantly induced TFEB nuclear translocation in normally fed cells and kept TFEB in the nucleus after starvation/re-feeding (Fig. 3b-d). Thapsigargin treatment also reduced the phosphorylation levels of TFEB serine residues S142 and S211 in fed cells (Fig. 3e,f). Importantly, thapsigargin-induced nuclear translocation of TFEB was suppressed when PPP3CB and its essential regulatory subunit PPP3R1 were silenced (Fig. 3g) and was blunted in PPP3R1−/− cells30 (Supplementary Fig. 3f), thus proving that the effects of Ca2+ on TFEB are mediated by calcineurin. Previous studies reported that long-term (24 hours) thapsigargin treatment inhibited mTOR activity via Ca2+-dependent induction of AMPK31. However, we found that up to 3 hours treatment with thapsigargin did not affect mTOR activity, measured by mTOR-mediated phosphorylation of P70S6 kinase (Supplementary Fig. 3g), indicating that intracellular Ca2+ elevation promotes TFEB nuclear translocation through the activation of calcineurin and further suggesting that calcineurin-dependent TFEB de-phosphorylation acts downstream of mTOR in the TFEB regulatory pathway.
Figure 3.
Ca2+ elevation induces TFEB nuclear translocation via calcineurin. (a) Chelation of intracellular Ca2+ suppresses TFEB nuclear translocation in both starvation (1h) and Torin1 treatment (1µM). The graph represents the percentage of TFEB nuclear translocation in starved and torin-treated HeLaTFEBGFP cells in presence or absence of BAPTA-AM (10µM) (mean ±s.d. for n=3 independent experiments). (b) Immunoblot of nuclear and cytosol fractions from HeLa cells transfected with TFEB-FLAG plasmid, starved for 1 hour or treated with thapsigargin or ionomycin in fed conditions, were incubated with anti-FLAG antibodies to detect TFEB localization. n=3 independent experiments. (c) Intracellular Ca2+ elevation inhibits nutrient-dependent relocation of TFEB from the nucleus to the cytosol. HeLaTFEBGFP cells were starved for 1h to induce TFEB nuclear translocation. Subsequently, the culture medium was changed to complete normal medium, which normally induces TFEB relocation to the cytosol, or complete medium plus calcium ionophores (1µM ionomycin or 300 nM thapsigargin) for 20 minutes. The plot shows the percentage of nuclear TFEB in treated cells compared with corresponding control cells (mean ±s.d. for n=3 independent experiments). (d) Thapsigargin induces endogenous TFEB translocation. Endogenous TFEB subcellular localization under feeding, starvation, and thapsigargin-treatment conditions was analyzed using a HC assay. The plot shows percentage of TFEB nuclear translocation compared with fed control cells (mean ±s.d., n=4 independent experiments). (e,f) Thapsigargin reduces TFEB phosphorylation at key serine residues S142 and S211. (e) Immunoblot analysis of TFEB phosphorylation using an anti-phospho S142-TFEB antibody. Protein lysates from HeLa cells transfected with TFEB-GFP vector and subjected to the indicated treatments were blotted against anti-phospho-S142-TFEB antibody, anti-GFP antibody was used to detect total TFEB. The plot shows the mean ± s.d. for n=4 independent experiments. (f) Indirect assessment of phosphorylation of TFEB residue S211 by immunoprecipitation of TFEB-GFP an immunoblot using an antibody against a specific 14-3-3 motif that recognizes phosphorylated proteins. The plot shows the mean ± s.d. for n=2 independent experiments. (g) Silencing of PPP3CB and its regulatory subunit PPP3R1 suppresses thapsigargin-induced nuclear translocation of TFEB. The graph represents the percentage of TFEB nuclear translocation in silenced cells vs cells transfected with a scramble oligonucleotide at the treatments indicated (mean ±s.d. for n=3 independent experiments). Scale bars 10µm. Uncropped western blots are provided in Supplementary Figure 8, and Source data in Supplementary table 4.
Starvation induces lysosomal Ca2+ release via MCOLN1
Our data indicate that starvation-induced TFEB nuclear translocation requires the activity of calcineurin and that an increase of cytosolic calcium induces TFEB nuclear translocation. Thus, we studied the effects of starvation on cytosolic Ca2+. Starvation did not induce changes in the overall levels of cytosolic Ca2+, as recorded by the aequorin-based probe localized in the bulk cytoplasm (Fig. 4a). Furthermore, while BAPTA-AM treatment inhibited TFEB nuclear translocation, a slower Ca2+-chelator (i.e. EGTA-AM)32 did not, suggesting that starvation rapidly triggers local calcium signals (Fig. 4b). The ER Ca2+ content was not significantly affected after starvation, as detected by measuring cytosolic calcium transients evoked by the IP3-mobilizing hormone histamine in fed and starved cells (Fig. 4a,c).
Figure 4.
Starvation induces lysosomal Ca2+ release via MCOLN1. (a) Starvation does not induce bulk cytosolic Ca2+ elevation in HeLa cells transfected with the Ca2+-sensitive probe aequorin. Bulk cytosolic [Ca2+] was monitored during perfusion with complete L-15 medium, HBSS and HBSS supplemented with 100µM histamine as indicated. n=8 coverslips from two independent transfections (b) Stable HeLaTFEB-GFP cells were left untreated or pretreated for 30 minutes with the Ca2+ chelators BAPTA-AM or EGTA-AM (5µM each). After washing, cells were left untreated or starved for 1 hr. After treatment, cells were fixed and a HC imaging analysis was performed. The plot shows the percentage of TFEB nuclear translocation in BAPTA-treated cells compared with untreated and EGTA treated (mean ± s.d., n=3 independent experiments). Scale bar 10µm. (c) Average [Ca2+]cyt evoked by maximal histamine stimulation in WT HeLa cells. Agonist stimulation was carried out in complete L-15 medium or after a three minutes starvation with HBSS (2.45 ± 0.19 µM, HeLa WT in L-15 medium; 2.406 ± 0.23 µM, HeLa WT in HBSS; n=8 coverslips from two independent transfections). (d) Representative traces of the cytosolic GCaMP6s and the perilysosomal ML1-GCaMP3 calcium probes. HeLa cells were transfected with the indicated probe and ratiometric imaging (474 and 410 nm excitation) was performed. Cells were continuously perfused with the indicated solutions. The plot in the middle represents the average perilysosomal calcium peak values induced by perfusion of the indicated buffer, as recorded by the GCaMP3-ML1 probe (R: 1.149 ± 0.051, L-15; 1.508 ± 0.060, HBSS; 7.500 ± 0.456, histamine; n= 18 cells from two independent transfections). The plot on the right represents the average cytosolic calcium peak values induced by perfusion of the indicated buffer, as recorded by the GCaMP6s probe (R: 0.404 ± 0.026, L-15; 0.389 ± 0.022, HBSS; 7.473 ± 0.428, histamine; n = 12 cells from two independent transfections). (e) Ca2+ release (measured with deltaF/F0 fluorescence intensity) was detected right after the L15 medium (containing 2 mM [Ca2+], aminoacids and 10% FBS) was switched to Tyrode’s solution (2mM [Ca2+]) in HEK293 cells stably expressing GCaMP3-ML136. The agonist of MCOLN1 ML-SA1 (10µM) was applied to induce MCOLN1-mediated Ca2+ release. n=3 independent experiments. (f) Similarly the lysosomotropic drug GPN blunted starvation-mediated calcium release detected by GCaMP3-ML1. n=3 independent experiments, *, p<0.05. Source data are provided in Supplementary table 4.
We postulated that a calcium release from the lysosome may be responsible for local calcineurin activation. Starvation clearly elicited a Ca2+ signal in HeLa cells transiently transfected with a MCOLN1 Ca2+ sensor linked to the lysosomal calcium channel mucolipin 1 (MCOLN1), also known as Transient Receptor Potential calcium channel MucoLipin subfamily member 1 (TRPML1), which is localized on the lysosomal surface (ML1-GCaMP3) (Figure 4d). Similar results were obtained in HEK293 cells stably overexpressing ML1-GCaMP3 (Figure 4e). Consistent with a lysosomal contribution to this calcium signaling mechanism, the increase in Ca2+ levels induced by starvation detected by ML1-GCaMP3 were not affected by the removal of extracellular Ca2+ (ruling out the contribution of plasma membrane calcium channels), but was blunted by treatment with the lysosomotropic agent GPN (Fig. 4f). Notably, neither the silencing of the lysosomal two-pore channel TPCN233 nor the overexpression of a critical regulator of mitochondrial Ca2+ uptake, MICU134 altered TFEB subcellular localization (Supplementary Fig. 4a,b). These data strongly suggest that starvation specifically induces lysosomal Ca2+ release via MCOLN1 and that this generates a lysosomal calcium microdomain that is responsible for calcineurin activation.
We then analyzed the involvement of MCOLN1 in the regulation of TFEB nuclear translocation35–38. ShRNA-mediated inhibition of MCOLN1 significantly reduced cytoplasm-to-nucleus shuttling of both endogenous and transiently transfected TFEB-GFP after starvation, further suggesting that this process requires Ca2+ release from the lysosome (Fig. 5a). Similar results were obtained by nucleus/cytoplasm fractionation (Fig. 5b). Consistently, depletion of MCOLN1 reduced the downshift of endogenous TFEB molecular weight during starvation, suggesting a defective de-phosphorylation (Supplementary Fig. 5a). Mutations in the MCOLN1 gene cause Mucolipidosis type 4 (MLIV), a lysosomal storage disease39, 40. We found that starvation-induced nuclear translocation of TFEB is inhibited in fibroblasts from MLIV patients (Fig. 5c). Furthermore, overexpression of wild type MCOLN1 and of gain of function MCOLN1 mutants in HeLaTFEBGFP cells promoted TFEB nuclear translocation (Fig. 5d) in a calcineurin-dependent fashion (Supplementary Fig. 5b) and similar results were obtained by using a recently described MCOLN1-specific agonist, SF5136 (Fig. 5e, Supplementary movies 1-3 and Supplementary Fig. 5c). Importantly, inhibition of MCOLN1 in skeletal muscles in vivo by transfection of shMCOLN1 suppressed the exercise-mediated nuclear translocation of TFEB (Fig. 5f), demonstrating the physiological relevance of lysosomal calcium signaling on TFEB regulation in vivo. Together these data suggest that while calcineurin-TFEB interaction may occur anywhere in the cytoplasm, focal Ca2+ release from the lysosome via MCOLN1 modulates TFEB activity via local calcineurin activation and suggest that MCOLN1 plays a crucial role in the activation of calcineurin near the lysosomal surface, through the generation of a lysosomal Ca2+ microdomain.
Figure 5.
MCOLN1-mediated calcium release induces TFEB nuclear translocation. (a-b) Silencing of MCOLN1 reduces starvation-mediated TFEB nuclear translocation. (a), HeLa cells or a HeLa cell line stably-transfected with a MCOLN1 shRNA were left untreated or transiently transfected with a TFEB-GFP plasmid. The left plot shows the percentage of TFEB-GFP nuclear translocation in starved (3hr) HeLa-shMCOLN1 cells compared with starved HeLa cells (n=3 independent experiments). Similarly, the middle plot shows the results on endogenous TFEB nuclear translocation (n=4 independent experiments). The right plot shows the efficiency of MCOLN1 gene silencing (n=3 independent experiments). The graphs show the mean ±s.d. (b) Silencing of MCOLN1 reduces starvation-mediated TFEB nuclear translocation. HeLa and HeLa-shMCOLN1cells were transfected with TFEB-3XFLAG construct. Following starvation, 50µg of nuclear and 100µg of cytosolic extracts were prepared and probed using anti-Flag antibody (n=3 independent experiments). (c) Endogenous TFEB nuclear translocation analysis of human- WT and mucolipidosis IV fibroblasts in fed conditions and after a 3 hr starvation of serum and aminoacids. The graph shows the percentage of TFEB nuclear translocation using (mean ±s.d of n=4 independent experiments). Scale bar 10µm. (d) Overexpression of MCOLN1 induces TFEB nuclear translocation. Stable HeLaTFEB-GFP were transfected with plasmids carrying wild type or constitutively active mutant forms of FLAG-tagged MCOLN1. TFEB subcellular localization was assessed in FLAG-positive (red-stained cells) and -negative cell populations. The graph shows the percentage of nuclear TFEB in the different transfected cell groups, compared to fed and starved (3hrs) conditions (mean ±s.d. for n=5 independent experiments). Scale bar 10µm. (e) Frames of time-lapse experiments in HeLaTFEB-GFP cells treated with the MCOLN1 agonist SF-51 (200µM) after the transfection of PPP3CB+PPP3R1 (siCaN), MCOLN1 or scramble siRNAs (Supplementary videos 1-3). Yellow arrowheads indicate TFEB nuclear localization. The plot shows the kinetics of TFEB nuclear localization during the time of recording (n=3 independent experiments). (f) Exercise induces TFEB nuclear translocation via MCOLN1. Muscles were electroporated with TFEB-GFP (green) and shRNAs against MCOLN1 (red). Antibodies against dystrophin (Dys, red) were used to reveal the muscle fibers. The arrowheads indicate TFEB nuclear localization in muscle fibers. The plot shows the percentages of TFEB-positive nuclei in muscles under the following conditions: sedentary (control), exercised (run), exercised with shMCOLN1 (run + shMCOLN1) (mean ±s.d of n=3 mice. Scale bar 50µm. Uncropped western blots and Source data are provided in Supplementary Figure 8 and Supplementary table 4.
Calcineurin and MCOLN1 regulate autophagy via TFEB
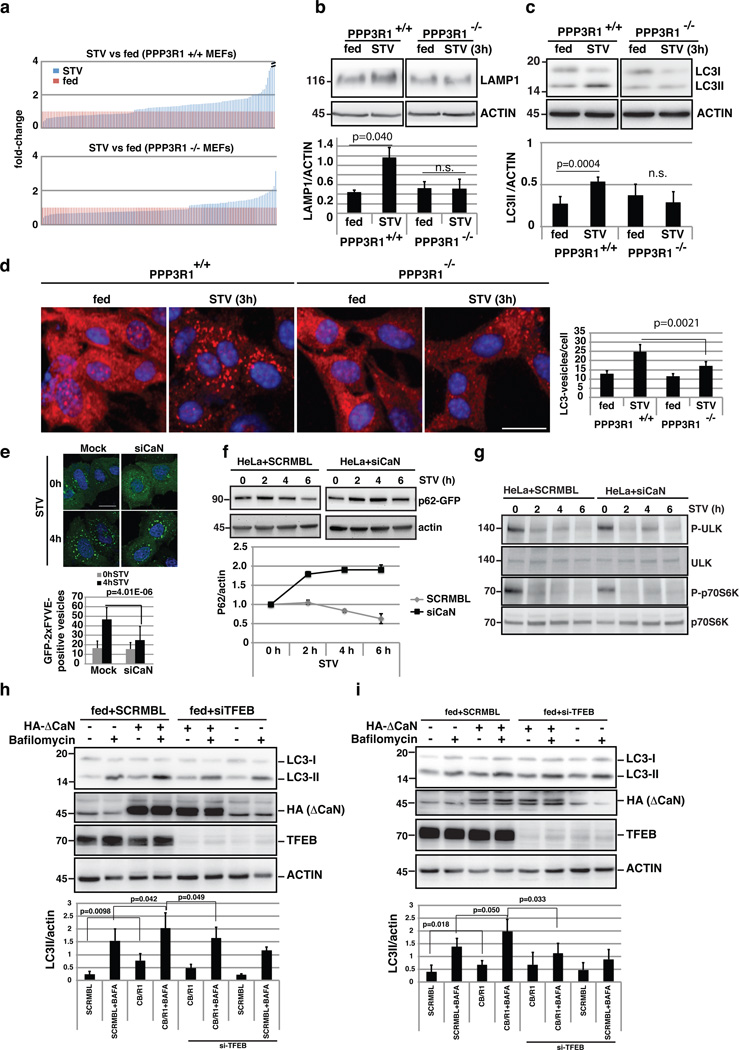
As TFEB transcriptionally regulates autophagy during starvation7, 41, we tested whether the transcriptional response of lysosomal/autophagic genes to starvation required calcineurin. Whole-genome gene expression profiling experiments revealed that starvation perturbed the transcriptomes of both wild type and PPP3R1−/− mouse embryonic fibroblasts (MEFs) in a statistically significant manner. We specifically analyzed the expression behavior of 322 lysosomal-autophagic genes (Supplementary Table 1) in the two cell lines. Remarkably, both the number of genes responsive to starvation and the amplitude of the response were significantly reduced in PPP3R1−/− compared to wild type cells (Fig. 6a, Supplementary Fig. 6a, Supplementary Table 2 and 3). Transcriptome analysis results were confirmed by real time PCR experiments on selected TFEB target genes (Supplementary Fig. 6b). Similar results were obtained in HeLa cells in which both PPP3R1 and PPP3CB were silenced (Supplementary Fig. 6c). Next we tested whether calcineurin-mediated TFEB activity had an effect on lysosomal/autophagic function. Lack of calcineurin in PPP3R1−/− cells suppressed the starvation-induced elevation of the lysosomal marker LAMP1 (Fig. 6b) and autophagosomal protein LC3, and blunted the increase in LC3-II levels and in the number of LC3-positive vesicles (Fig. 6c,d). Similar results were obtained by pharmacological inhibition of calcineurin (Supplementary Fig. 7a).
Figure 6.
Calcineurin regulates the lysosomal/autophagic pathway. (a) The transcriptional response of lysosomal/autophagic genes is reduced in PPP3R1−/− KO MEFS (bottom) compared to wild type (top) cells (ranked by fold-change). A value of 1 was assigned to expression levels in fed conditions (n=3 independent experiments). (b-c) Immunoblots against LAMP1 (b), and LC3 (c) from wild type and PPP3R1−/− MEFs in fed and starved (3 hours) conditions. The plot shows the quantification of Lamp1 and LC3-II proteins levels normalized by actin loading control (mean ±s.d for n=2 and n=5 independent experiments, respectively. (d) HC analysis of LC3-positive vesicles in wild type and PPP3R1−/− MEFs in fed and starved conditions. The bar-graph shows the mean ±s.d of LC3-positive vesicles in the different treatment conditions for n= 6 independent experiments. Scale bar 10µm. (e) Analysis of the overexpression of the autophagy-related PI(3)P reporter GFP-2xFYVE55 during starvation in (Mock) HeLa cells, and cells silenced for PPP3CB/PPP3R1. GFP-2xFYVE-positive vesicles were counted using ImageJ software. Approximately 50 cells per condition were analyzed by confocal imaging. Data shows the mean ±s.d for n=2 independent experiments. Scale bar 10µm. (f) Depletion of calcineurin reduces the autophagic flux. HeLa cells were transfected with SCRMBL siRNA or siRNA against both PPP3CB and PPP3R1 (siCaN). After 48 hours, cells were transfected with a plasmid carrying the autophagy substrate p62 fused to GFP for 24h, and treated as indicated. The graph shows the levels of p62-GFP normalized by actin at the different time-points. (g) Analysis of mTOR activity during starvation in normal cells and in cells silenced against calcineurin (siCaN). Two mTOR subtrates, phospho-ULK and phospho-p70S6K, as well as the total protein, were detected by immunoblotting using specific antibodies (n=2 independent experiments). (h,i) Overexpression of ΔCaN increased LC3II protein levels in a TFEB-dependent manner. (h) RPE-1 cells and (i) HeLa were transfected with TFEB or scramble siRNAs and after 48h they were transfected with ΔCaN for 24h. Cells were left in fed conditions and treated or not with bafilomycin. 50µg of protein extracts were then immunoblotted and tested for the amount of LC3-II using specific antibodies (n=4 independent experiments). Uncropped western blots and Source data are provided in Supplementary Figure 8 and Supplementary table 4.
In addition, the silencing of calcineurin, significantly reduced the number of phosphatidylinositol 3-phosphate (PI(3)P)-positive vesicles, indicating an impairment in autophagosome formation42, during starvation (Fig. 6e). Furthermore, silencing of both PPP3R1 and PPP3CB in HeLa cells resulted in a reduction of the autophagic flux, measured by the degradation of the autophagic substrate p62 (Fig. 6f). This autophagy impairment was not due to the inhibition of mTOR signaling, as we observed normal phosphorylation kinetics of two different mTOR substrates (ULK, and P70S6K)(Fig. 6g). Conversely, overexpression of ΔCaN increased LC3II protein levels in a TFEB-dependent manner (Fig. 6h,i). Together these data identify calcineurin as a novel regulator of the lysosomal-autophagic pathway.
Consistent with a role of MCOLN1 and lysosomal Ca2+ in the activation of calcineurin, the silencing of MCOLN1, significantly reduced the number of PI(3)P-positive vesicles (Fig.7a). Similar results were observed using immunofluorescence against endogenous WIPI protein, a marker of early steps in autophagosome formation, in cells silenced for MCOLN1 (Supplementary Fig. 7b). In addition, we found that MCOLN1 overexpression resulted in a significant increase in the levels of the autophagosome marker LC3, which were further increased by bafilomycin, indicating an induction of autophagy, rather than an impairment of autophagosome degradation (Fig. 7b,c). Thus, lysosomal calcium signaling via MCOLN1 controls autophagy through calcineurin-mediated activation of TFEB.
Figure 7.
Calcineurin regulates the lysosomal/autophagic pathway. (a) Representative images and analysis of the overexpression of the autophagy-related PI(3)P reporter GFP-2xFYVE55 during starvation in WT (Mock) HeLa cells, in cells silenced for MCOLN1. GFP-2xFYVE-positive vesicles were counted using ImageJ software. Approximately 50 cells were pooled per condition and were analyzed by confocal imaging. Data shows the mean ±s.d of n=2 independent experiments. Scale bar 10µm. (b) MCOLN1 overexpression leads to a significant increase in LC3 levels. HeLa cells were transfected with myc-tagged MCOLN1 or gain of function mutants R427P and V432P respectively. Immunofluorescence was performed using anti-LC3 antibodies. The number of LC3 spots per cell was quantified using HC imaging analysis. Mean values ± s.d are plotted for n=4 independent experiments. Scale bar 10µm. (c) 50 µg of protein extracts from HeLa cells treated with bafilomycin (BAFA) and overexpressing an empty vector of a myc-MCOLN1 plasmid for 24 h were immunoblotted against LC3 (n=3 independent experiments). (d,e) Overexpression of TFEB increases the effects of MCOLN1. Lysosomal patch-clamp was performed in both control (d) and TFEB overexpressing cells (e). Note that the effect of different concentrations of the MCOLN1 agonist ML-SA1 is strongly enhanced likely due to an increase in the number of MCOLN1 channels. n=3 independent experiments. Uncropped scans of western blots are provided in Supplementary Figure 8. Source data are provided in Supplementary table 4.
Interestingly, MCOLN1 is known to be a direct transcriptional target of TFEB6, 38, 41. This suggests the presence of a positive feedback loop by which TFEB regulates the expression of MCOLN1, which in turn promotes TFEB activity via Ca2+-mediated activation of calcineurin. Consistently, overexpression of TFEB dramatically increased the effects of MCOLN1 agonists, likely due to an increase in the number of MCOLN1 channels, as observed with lysosomal patch clamp (Fig. 7d,e).
DISCUSSION
The regulation of autophagy is thought to be a biphasic process, with a fast induction that relies on post-translational and protein-protein interaction events, and a sustained response that is mediated by transcriptional mechanisms17. Our results identify a calcium signaling pathway that originates from the lysosome and regulates autophagy by activating a TFEB-mediated transcriptional program. It has been well-established that autophagy is negatively regulated by the mTOR kinase43. The new pathway regulating autophagy identified in our study is mediated by the Ca2+-dependent phosphatase calcineurin and seems to operate independently from mTOR. Fig. 8 shows a model representing how starvation and exercise regulate TFEB via both the mTOR and calcineurin pathways. In normal feeding/sedentary conditions mTORC1 phosphorylates TFEB allowing its interaction with 14-3-3 proteins, thus preventing its nuclear translocation. Conversely, during starvation and physical exercise mTORC1 activity is inhibited and, at the same time, calcineurin activity is locally induced by lysosomal calcium release via MCOLN1. This leads to both a decreased rate of TFEB phosphorylation, via mTORC1 inhibition, and an induction of TFEB de-phosphorylation, via calcineurin induction. De-phosphorylated TFEB is free to travel to the nucleus and to induce the expression of lysosomal and autophagic genes (Fig. 8).
Figure 8.
Model of Ca2+ mediated regulation of TFEB. Under normal feeding conditions TFEB is phosphorylated on the lysosomal surface and is sequestered in the cytoplasm by the 14-3-3 proteins. During starvation and physical exercise Ca2+ is released from the lysosome via MCOLN1, thus establishing a Ca2+ microdomain. This leads to calcineurin activation and TFEB de-phosphorylation. De-phosphorylated TFEB is no longer able to bind 14-3-3 proteins and can freely translocate to the nucleus where it transcriptionally activates the lysosomal/autophagic pathway.
Previous studies reported that exercise induces autophagy in muscle 44–46. Our results provide a mechanistic link between calcineurin activation and autophagy induction during exercise. To date, the transcription factors of the NFAT family, master regulators of T cell activation47, have been considered the main transcriptional mediators of calcineurin function. The discovery that TFEB, a master controller of autophagy, is a calcineurin substrate expands the current view on the mechanism of action of calcineurin. Interestingly, we found that starvation induces calcineurin-mediated de-phosphorylation and nuclear translocation of both TFEB and NFAT, suggesting a coordinated regulation of the two different transcriptional networks. Previous studies implicated calcineurin in a variety of both physiological and pathological processes, such as neuronal synaptic transmission, adaptative immune response and muscle remodeling after physical exercise48, 49. Our data suggest that some of these processes may be linked to the lysosomal/autophagic pathway via TFEB. In addition, other calcineurin substrates may also play a role in autophagy.
Calcium plays a crucial role in the regulation of the activity of transcription factors via the induction of calcium-dependent kinases and phosphatases50. Until now conflicting results have been reported on the mechanism by which calcium regulates autophagy. Here we dissected a Ca2+-dependent pathway that originates from the lysosome and regulates autophagy at the transcriptional level. Recent studies have revealed the presence of calcium microdomains, mediated by different calcium channels, which activate calcineurin in the vicinity of different cellular compartments (i.e. sarcolemma, mitochondria, sarcoplasmatic reticulum and perinuclear sarcoplasmatic reticulum) in the heart51. Our study demonstrates that local calcineurin activation can also occur near the lysosome via the lysosomal calcium channel MCOLN1. In addition, our data also revealed a positive feedback loop by which the TFEB is activated by the calcium channel MCOLN1, which is known to be a TFEB transcriptional target41.
In conclusion, the identification of a mechanism by which lysosomal Ca2+ regulates autophagy via the activation of the phosphatase calcineurin and the transcription factor TFEB further support the role of the lysosome as a signaling hub. It is likely that future studies will lead to the identification of other signaling pathways originating from the lysosomal surface.
METHODS
Drugs and cellular treatments
The following drugs were used: Torin 1 (1µM, otherwise indicated) from Tocris Bioscience; FK506 (5 µM, otherwise indicated), cyclosporine A (10 µM, otherwise indicated), Chloroquine (100 µM) thapsigargin (300 nM), ionomycin (1µM), and EGTA from Sigma; BAPTA-AM (5-25 µM) from Invitrogen; EGTA-AM (5 µM) from Santa Cruz Biotechnology; Bafilomycin A1 (300 nM); SF51 (200 µM) from VITAS-M Laboratory, LTD. Starvation medium was HBSS, 10 mM Hepes.
Cell culture, plasmids and siRNA transfection
HeLa and MCF-7, HEK 293, RPE cells were purchased from ATCC and cultured in DMEM or RPMI 1640 media supplemented with 10% fetal bovine serum, 200 µM L-glutamine, 100 µM sodium pyruvate, 5% CO2 at 37 degrees. Cells were silenced with TFEB siRNA oligonucleotides (Thermo, Dhamacon D-009798-03), PPP3CB siRNA oligonucleotides (Ambion, 4390824), PPP3R1 siRNA oligonucleotides (Invitrogen, HSS108410-3, HSS183046-3, HSS108411-3), or non-targeting siRNAs (Thermo, D-001810-10-05), by direct or reverse transfection, using lipofectamine RNAiMAX reagent (Invitrogen) according to the protocol from manufacturer. siRNA-transfected cells were collected after 72 hours, if not otherwise stated.
Immortalized MEFs from conditional PPP3R1flox/flox (PPP3R1+/+) mice, kind gift from J. Molkentin (Cincinnati Children's Hospital Medical Center, USA) were cultured in DMEM, 10% CS, 200 µM L-glutamine, 100µM sodium pyruvate. PPP3R1flox/flox (PPP3R1+/+) MEFs were used as control, while lentiviral-mediated Cre transduction was used to generate PPP3R1−/− MEFs30.
Stably HeLashMCOLN1 cell line was described in Medina et al, 201131. Stably HeLaTFEB-GFP cell line was described in Settembre et al, 20129. ML-IV human fibroblasts were a kind gift of G.Borsani.
Human full-length TFEB-GFP, and TFEB-FLAG were previously described7. Human full length PPP3CB and MCOLN1 tagged with myc-FLAG were from OriGene. The plasmid caryinng a constitutively active form of human calcineurin catalytic subunit (ΔCan) was donated by B. Rothermel (The University of Texas Southwestern Medical Center, USA). NFATGFP was a gift from T. Baldari52; Micu1 plasmid was described in34. Cells were transfected using Lipofectamine LTX – Plus reagent (Invitrogen) or Trans-IT reagent (Mirus-Bio) according to the protocol from manufacturers.
Antibodies and Western blotting
For western blot the following antibodies were used: anti-FLAG (M2, cat. F1804, 1:2000), β-actin (AC-15, cat. A5441, 1:4000), β-tubulin (TUB 2.1, cat.T4026, 1:2000) and -ULK (cat. A7481, 1:1000) from Sigma-Aldrich; anti-LC3 (cat. NB100-2220, 1:1000) from Novus Biologicals; anti-PARP1 (cat. ALX-10-302, 1:4000) from ENZO Life Sciences; anti-TFEB (cat. 4240, 1:1000), -p-Ser 14-3-3 binding motif (cat. 9601, 1:1000), -p-P70S6K (cat. 108D2, 1:1000), -p-ULK (cat. 6888, 1:1000) from Cell Signaling Technologies; anti-HA (16B12, cat. MMS-101P, 1:7500) from Covance; anti-P70S6K (cat. KAP-CC035, 1:1000) from Stressgen; anti-PPP3CB (cat. TA308438, 1:1000) from OriGene; anti-pan 14-3-3 (B11, cat. sc-133232, 1:1000), -GAPDH (6C5, cat. sc-32233, 1:10000) and -c-Myc (9E10, cat. sc-40, 1:1000) from Santa Cruz Biotechnology; anti-GFP (1:10000) was provided by M. A. De Matteis (TIGEM); anti-p-Ser142-TFEB (1:1000) was produced as described previously9.
Total cell lysates were prepared by solubilisation in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl and 0.01 M sodium phosphate, at pH 7.2) containing protease (Roche) and phosphatase (Sigma) inhibitors. The soluble fraction was recovered in the supernatant after centrifugation at 12,000g for 15 min, and protein concentration was measured by the Bradford method. Nuclear/cytosolic fractions were isolated as previously described9.
After SDS–PAGE and immunoblotting, the proteins recognized by the specific antibodies were visualized by chemiluminescence methods (ECL Western Blotting Substrate, Pierce) using peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (Calbiochem). Membranes were developed using a Chemidoc UVP imaging system (Ultra-Violet Products Ltd) and densitometric quantification was performed in unsaturated images using ImageJ (NIH).
Proximity Ligation Assay
PLA assay was performed using the Duolink in situ reagents (Olink Biosciences) according to the manufacturer’s specifications.
Golgin 97 polyclonal antibody used for immunofluorescence was raised in rabbit by immunization with recombinant glutathione S-transferase (GST)-fusion full-length protein. The antibody was affinity purified over a column conjugated with a His fusion of the protein. Other antibodies used in this study were from the following sources: mAb TFEB (17, cat. sc-101532, 1:50) from Santa Cruz Biotechnology and anti- mTOR (cat. 7010, 1:200) from Cell Signaling Technology. Alexa-conjugated secondary antibodies and ER tracker were from Life Technologies.
Immunofluorescence experiments were performed as previously described53. Briefly, HeLa cells were fixed in 4% paraformaldehyde (PFA) for 10 minutes and permeabilized in 0.1% (w/v) saponin, 0.5% (w/v) BSA and 50 mM NH4Cl in PBS (blocking buffer). Cells were incubated with the indicated primary antibodies for 1 hour and subsequently incubated with secondary antibodies. For confocal imaging, the samples were examined under a Zeiss LSM 700 confocal microscope. Optical sections were obtained under a ×63 immersion objective at a definition of 1024 × 1024 pixels (average of eight or sixteen scans), adjusting the pinhole diameter to 1 Airy unit for each emission channel to have all the intensity values between 1 and 254 (linear range). For image analyses, Mock treated and TFEB-KD HeLa cells were fixed and stained as described above and cells acquired with the same laser parameters using the same magnification.
Co-immunoprecipitation assays
HeLa cells expressing the indicated tagged proteins were rinsed twice with ice-cold PBS and lysed in ice-cold lysis buffer (400 mM NaCl, 25 mM Tris-HCl, pH 7.4, 1 mM EDTA and 1% Triton X-100) containing protease and phosphatase inhibitors. The soluble fractions from cell lysates were isolated by centrifugation at 12000g for 15 min in a microfuge. For co-immunoprecipitations, each lysate was incubated with corresponding antibody in binding buffer (200 mM NaCl, 25 mM Tris-HCl, pH 7.4, 1mM EDTA) with constant rotation over-night at 4°C. Then, 40 µl of a 50% slurry of Protein-G beads (Sigma-Aldrich) were added to lysates and incubated with rotation for additional 2h at 4°C. After incubation, the resins were washed 5 times in binding buffer and the samples were eluted in Laemmli buffer and separated by SDS-PAGE.
Histidine pull-down assay
Purified His-TFEB (300 nM), CaN (ProAlt) and CaM (Enzo Life Sciences) and GST (100 nM each) were incubated in binding buffer (20 mM Tris, pH 8; 100 mM NaCl; 2mM CaCl2; 6 mM MgCl2; 0,2% Triton X-100; 30 mM imidazole; protease and phosphatase inhibitor) over-night at 4°C. His-TFEB was pulled-down by adding 20 µl of Ni-NTA resin (Qiagen) for 2 hrs at 4°C. After incubation, the resins were washed 5 times in binding buffer and the samples were eluted in Laemmli buffer and separated by SDS-PAGE.
In vitro calcineurin phosphatase assay
Phosphorylated FLAG-TFEB was obtained by immunoprecipitation from fed HeLa cells. Protein-G beads containing FLAG-TFEB were incubated for 40 min at 30°C in 24 µl of corresponding phosphatase reaction buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, 2 mM DTT, protease inhibitors with 1 mM CaCl2 or 10 mM EGTA) containing purified Calcineurin (100U; ENZO Life Sciences) and Calmodulin (4 mM; ENZO Life Sciences). Samples were eluted by adding 8 µl of 4X Laemmli buffer, resolved by SDS–PAGE and analysed by immunoblotting. Antibodies anti-p-Ser 14-3-3 binding motif10, anti-P-Ser142-TFEB9 and TFEB (Cell Signaling Technologies) were used to detect phosphorylated and total TFEB forms, respectively.
RNA extraction and quantitative PCR
Total RNA was extracted from cells using RNeasy Plus Mini Kit (Qiagen). Reverse transcription was performed using QuantiTect Rev Transcription Kit (Qiagen). Real-time quantitative Reverse Transcription PCR (qPCR) was performed using the LightCycler® System 2.0 (Roche Applied Science). HPRT was used for qRT-PCR as reference gene. The parameters of real-time qRT-PCR amplification were according to Roche recommendations.
The following primers were used in this study. Mouse primers: PGC1a; forward (fw) gaatcaagccactacagacaccg, reverse (rev) catccctcttgagcctttcgtg, MCOLN1; fw: gcgcctatgacaccatcaa, rev: tatcctggactgctcgat; ATP6V0D1; fw: gcatctcagagcaggaccttga, rev: ggataggacacatggcatcagc, ATP6V1H; fw: gttgctgctcacgatgttggag, rev: tgtagcgaacctgctggtcttc, CTSF; fw: acgcctatgcagccataaag, rev: cttttgccatctgtgctgag, CTSB; fw: ttagcgctctcacttccactacc, rev: tgcttgctaccttcctctggtta, TPP1; fw: aagccaggcctacatagtcaga, rev: ccaagtgcttcctgcagtttaga, DPP7; fw: cgccagcaatactgtctggatac, rev: aaatgatgttgctggctgcttta, WIPI; fw: acaggacgcctgaacttctc, rev: cggcagtttctggatcgt, TcFEB; fw: gcgagagctaacagatgctga, rev: ccggtcattgatgttgaacc, HPRT; fw: caagcttgctggtgaaaagg rev: gtcaagggcatatccaacaac, PPP3R1; fw: tgcctgagttacagcagaacc rev: tcgcctttgacactgaactg, ATG14; fw: gcttcgaaggtcacacatcc, rev: cttgaggtcatggcactgtc, PPP3CA; fw; atcccaagttgtcgacgacc, rev: acactttcttccagcctgcc. Human primers; PGC1a; fw: catgcaaatcacaatcacagg, rev: ttgtggcttttgctgttgac, MCOLN1; fw: gagtgggtgcgacaagtttc, rev: tgttctcttcccggaatgtc, ATP6V0E1; fw: cattgtgatgagcgtgttctgg, rev: aactccccggttaggaccctta, ATP6V1H; fw: ggaagtgtcagatgatcccca, rev: ccgtttgcctcgtggataat, CTSF; fw: acagaggaggagttccgcacta, rev: gcttgcttcatcttgttgcca, TFEB; fw: caaggccaatgacctggac, rev: agctccctggacttttgcag, HPRT; fw: tggcgtcgtgattagtgatg, rev: aacaccctttccaaatcctca, CTSD; fw: cttcgacaacctgatgcagc, rev: tacttggagtctgtgccacc, PPP3CA; fw: gctgccctgatgaaccaac, rev: gcaggtggttctttgaatcgg, DPP7; fw: gattcggaggaacctgagtg, rev: cggaagcaggatcttctgg, CTSB; fw: agtggagaatggcacacccta , rev: aagagccattgtcacccca, TPP1; fw: gatcccagctctcctcaatac, rev: gccatttttgcaccgtgtg, NEU1; fw: tgaagtgtttgcccctggac, rev: aggcaccatgatcatcgctg.
High content nuclear translocation assay
HeLaTFEBGFP cells were seeded in 96 or 384-well plates, incubated for 12 h, and treated as indicated in the text, washed, fixed, and stained with DAPI. For the acquisition of the images, at least 5 images fields were acquired per well of the 96/384-well plate by using confocal automated microscopy (Opera high content system; Perkin-Elmer). A dedicated script was developed to perform the analysis of TFEB localization on the different images (Harmony and Acapella software; Perkin-Elmer). The script calculates the ratio value resulting from the average intensity of nuclear TFEB–GFP fluorescence divided by the average of the cytosolic intensity of TFEB–GFP fluorescence. The results were normalized using negative (RPMI medium, or otherwise indicated) and positive (starvation, or otherwise indicated) control samples in the same plate54. At least two independent experiments, and up to 3000 individual cells per treatment from at least two independent wells are routinely analyzed. p-values were calculated on the basis of mean values from independent wells. The data are represented by the percentage of nuclear translocation vs the indicated control using Excel (Microsoft) or Prism software (GraphPad software).
The TFEB-GFP nuclear translocation analyses of different population of transfected cells (detected by immunofluorescence of tagged-plasmids) were performed by a modified script that calculates the ratio value resulting from the average intensity of nuclear TFEB–GFP fluorescence divided by the average of the cytosolic intensity of TFEB–GFP fluorescence on both positive-transfected and negative transfected cells.
High content LC3-autophagosome assay
Cells were seeded in 96-well plates, incubated for 12 h, and treated as indicated in the text, washed, fixed in ice-cold methanol, and immunofluorescence against LC3 (anti-LC3, Novus Biologicals, 1:400) was performed. After washing, the 96-well plate was analysed using confocal automated microscopy (Opera high content system; Perkin-Elmer). A dedicated script was developed to perform the analysis of the number of LC3-puncta on at least 5 different images per well (Harmony software; Perkin-Elmer).
Time-lapse
HeLaTFEBGFP were reverse transfected and plated in Mattek glass bottomed dishes. Time-lapse movies were acquired for 45’. One frame was acquired roughly every 20s with lasers set at 30% power or below. All imaging was performed with a x60 Plan Apo oil immersion lens using a Nikon Eclipse Ti Spinning Disk microscope, and images were annotated and 3D reconstructions made using the NIS Elements 4.20 software.
EGFP-2×FYVE Assay
HeLa cells were reverse transfected with PPP3CB/R1 or scramble control siRNA using RNAiMax. 48h later the cells transfected with the GFP-2xFYVE construct55 using Lipofectamine LTX and Plus (Invitrogen), and 24h later serum starved and fixed with 4% PFA in PBS. Imaging was performed with a 63x PlanApochromat NA 1.4 DIC oil immersion objective on an LSM700 confocal microscope (Zeiss). Image analysis (spot count per cell, minimum of 15 cells counted per treatment group) was performed using ImageJ.
Endolysosomal electrophysiology
Endolysosomal electrophysiology was performed in isolated endolysosomes using a modified patch-clamp method35–37. Cos-1 cells were transfected using Lipofectamine 2000 (Invitrogen) with TFEB-S211A fused to mCherry. To increase the size of isolated endolysosomes, cells were treated with 1 µM vacuolin-1, a lipid-soluble polycyclic triazine that can selectively increase the size of endosomes and lysosomes. Whole-endolysosome recordings were performed on isolated enlarged endolysosomes. In brief, a patch pipette (electrode) was pressed against a cell and quickly pulled away to slice the cell membrane35–37. Enlarged endolysosomes were released into a dish. TFEB-S211A-positive vacuoles were identified by monitoring mCherry fluorescence. Bath (internal/cytoplasmic) solution contained 140 mM K+-Gluconate, 4 mM NaCl, 1 mM EGTA, 2 mM Na2-ATP, 2 mM MgCl2, 0.39 mM CaCl2, 0.1 mM GTP and 10 mM HEPES (pH adjusted with KOH to 7.2; free [Ca 2+]i approximately 100 nM). The pipette (luminal) solution was pH 4.6 standard extracellular solution (modified Tyrode’s) with 145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 20 mM HEPES and 10 mM glucose (pH adjusted with NaOH). All bath solutions were applied by means of a fast perfusion system to achieve a complete solution exchange within a few seconds. Data were collected using an Axopatch 2A patch-clamp amplifier, Digidata 1440, and pClamp 10.0 software (Axon Instruments). Whole-endolysosome currents were digitized at 10 kHz and filtered at 2 kHz. All experiments were conducted at room temperature (21 – 23°C), and all recordings were analysed using pCLAMP10 (Axon Instruments) and Origin 8.0 (OriginLab). ML-SA1 was purchased from Princeton BioMolecular Research Inc.
GCaMP3 Ca2+ imaging in GCaMP3-ML1-HEK293 cells
GCaMP3 Ca2+ imaging was performed in HEK cells that stably express GCaMP3-ML1, which are lysosome-targeted genetically encoded Ca2+ sensors36. The fluorescence intensity at 488 nm (F488) was recorded with the spinning disk confocal imaging system, which consisted of an Olympus IX81 inverted microscope, a 60X or 100X objective (Olympus), a CSU-X1 scanner (Yokogawa), an iXon EM-CCD camera (Andor), and MetaMorph Advanced Imaging acquisition software v.7.7.8.0 (Molecular Devices). Live imaging of ML1-GCaMP3 was performed on a heated stage.
Ratiometric GCaMP3 Ca2+ imaging in HeLa cells
HeLa cells were grown on 24 mm coverslip and transfected with plasmid encoding plasmid for cytosolic-localized GCaMP6s or the perilysosomal-localized ML1-GCaMP3 calcium probes. After 24 or 48 hours, coverslips were placed in 1 mL of complete L-15 medium and imaging was performed on a Zeiss Axiovert 200 microscope equipped with a 40x/1.3 N.A. NeoFluar objective. Excitation was performed with a Deltaram V high speed monochromator (Photon Technology International) equipped with a 75W Xenon Arc lamp. Images were captured with a high sensitivity Evolve 512 Delta EMCCD (Photometrics). The system is controlled by Metamorph 7.5 and was assembled by Crisel Instruments. In order to be truly quantitative, we took advantage of the isosbestic point in GCaMP excitation spectrum: we experimentally determined in living cells that exciting GCaMP at 410 nm leads to fluorescence emission which is not Ca2+-dependent. As a consequence, the ratio between 474 and 410 nm excitation wavelengths is proportional to [Ca2+] while independent on probe expression. HeLa cells were thus alternatively illuminated at 474 and 410 nm and fluorescence was collected through a 515/30 nm bandpass filter (Semrock). For GCaMP6s experiments, exposure time was set to 50 ms at 474 nm and to 150 ms at 410 nm, in order to account for the low quantum yield at the latter wavelength. For the ML1-GCaMP3 experiments, exposure time was set to 300 ms at 474 nm and to 400 ms at 410 nm. Each field was acquired for 20 minutes (1 frame/s) under continuous perfusion with the indicated solutions. Analysis was performed with the Fiji distribution of ImageJ. Both images were background corrected frame by frame by subtracting mean pixel values of a cell-free ROI. Data are presented as the mean ± S.E.
Aequorin measurements
For the measurements of [Ca2+]cyt, HeLa cells grown on 13 mm round glass coverslips at 50% confluence were transfected with the cytosolic (cytAeq) probe. The coverslip with the cells was incubated at 37°C with 5 µM coelenterazine for 1-2 hours in L-15 medium (Life Technologies) supplemented with 10% FBS, and then transferred to the perfusion chamber. Aequorin measurements were carried out in either complete L-15 or HBSS (supplemented with CaCl2 and MgCl2, Life Technologies), as indicated. Agonists and other drugs were added to the same medium, as specified in the text. The experiments were terminated by lysing the cells with 100µM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+] and ionic strength, as previously described56. Data are presented as mean ± s.d. of the histamine-induced peak in [Ca2+].
Animals and in vivo transfection experiments
Animals were handled by specialized personnel under the control of inspectors of the Veterinary Service of the Local Sanitary Service (ASL 16 - Padova), the local officers of the Ministry of Health. All procedures are specified in the projects approved by the Italian Ministero Salute, Ufficio VI (authorization numbers C65). All experiments were performed on adult 2 month-old male CD1 mice (three mice per group, 28–32 g). In vivo transfection experiments were performed by intramuscular injection of expression plasmids in TA muscle followed by electroporation as previously described57. After 8 days from transfection mice were exercised on a treadmill. Mice performed concentric exercise on a treadmill (LE 8710 Panlab Technology 2B, Biological Instruments), with 10 degrees incline, till they were exhausted and then were sacrificed.
Histology and Fluorescence Microscopy
Immunofluorescence staining was performed on cryosections as previously described24 and then monitored with a fluorescence microscope. For nuclear localization studies, cyosections were stained with mouse-anti-Dystrophin-1 IgG2a antibodies (1:200; Novocastra) and DAPI to identify subsarcolemmal position of myonuclei.
Generation of the lysosomal-autophagic gene list
The complete gene list includes 322 members (Supplementary Table_S1): 98 lysosomal genes described previously41, 58; 9 novel genes co-localizing with the lysosomal marker LAMP258; the master regulator of lysosomal biogenesis and autophagy, TFEB; all the known lysosomal and not lysosomal protein coding genes with a role in the different LSDs selected by using the Online Mendelian Inheritance in Man (OMIM)59, 60; a subset of lysosomal and autophagy genes predicted by different bioinformatics tools. In details, three different bioinformatics tools were used: AmiGO61, Netview62 and uniPROT63.
Microarray data processing
The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO)64 and are accessible through GEO Series accession numbers GSE56671 (PPP3R1 WT Starved vs Normal medium) and GSE56672 (PPP3R1−/− Starved vs Normal medium). Statistical comparisons were made using analysis of variance (ANOVA). As threshold for statistically significance we used FDR< 0.05 followed by an additional threshold (p-value < 0.05).
Data analysis
Data are presented as the mean ± s.d. t-test statistical comparisons were made using analysis of variance. A p-value < 0.05 was considered statistically significant. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments.
Supplementary Material
Acknowledgments
We thank Jacopo Meldolesi, Tullio Pozzan, David Rubinsztein, and Roman Polishchuk for helpful suggestions and critical review of the manuscript. We thank Graciana Diez-Roux and Alexis Burton for their support in manuscript preparation. We are also grateful to Rossella De Cegli and Diego Carrella for their support in the statistical analysis of the results. The TIGEM Bioinformatic and High Content Screening Facilities are gratefully acknowledged for their technological contributions to the project. We also thank Jeffery D. Molkentin for the CanB KO MEFs, Beverly A. Rothermel for the HA-ΔCnA, and Pedro Aza-Blanc for suggestions on the reverse transfection protocol. We acknowledge the support of the Italian Telethon Foundation grant numbers TGM11CB6 (A.B.); the Beyond Batten Disease Foundation (A.B.); European Research Council Advanced Investigator grant no. 250154 (CLEAR) (A.B.); US National Institutes of Health (R01-NS078072) (A.B.); Telethon-Italy (TCP04009) (M.S.), European Research Council Consolidator grant no. 282310-(MyoPHAGY) (M.S.).
Footnotes
AUTHORS CONTRIBUTIONS
D.L.M., S.D.P., I.P., S.M., A.S.R., C.P., R.V., A.F., and C.S. performed all experiments in cultured cells. A.A. and M.S. performed in vivo experiments in the murine muscle. W.W., Q.G., D.D.S., H.X., and R.R. performed electrophysiology experiments. D.L.M., M.A.D. and A.B. designed the overall study. D.L.M. and A.B. supervised the work. All authors discussed the results and made substantial contributions to the manuscript.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 2.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nature Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 3.de Duve C. The lysosome turns fifty. Nature Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 4.Luzio JP, Parkinson MD, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem. Soc. Trans. 2009;37:1019–1021. doi: 10.1042/BST0371019. [DOI] [PubMed] [Google Scholar]
- 5.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 7.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martina JA, Puertollano R. RRAG GTPases link nutrient availability to gene expression, autophagy and lysosomal biogenesis. Autophagy. 2013;9:928–930. doi: 10.4161/auto.24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferron M, et al. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 18.Morgan AJ, Platt FM, Lloyd-Evans E, Galione A. Molecular mechanisms of endolysosomal Ca2+ signaling in health and disease. Biochem. J. 2011;439:349–374. doi: 10.1042/BJ20110949. [DOI] [PubMed] [Google Scholar]
- 19.Rizzuto R, Pozzan T. Microdomains of intracelular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- 20.Stewart AA IT, Manalan A, Klee CB, Cohen P. Discovery of a Ca2+- and calmodulin-dependent protein phosphatase: probable identity with calcineurin (CaM-BP80) FEBS Lett. 1982;11:80–84. doi: 10.1016/0014-5793(82)80319-0. [DOI] [PubMed] [Google Scholar]
- 21.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annual review of immunology. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 22.Hogan PG, Li H. Calcineurin. Current biology : CB. 2005;15:R442–R443. doi: 10.1016/j.cub.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. The EMBO journal. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell metabolism. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Lai MM, Burnett PE, Wolosker H, Blackshaw S, Snyder SH. Cain, a novel physiologic protein inhibitor of calcineurin. The Journal of biological chemistry. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 26.Loh C, et al. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. The Journal of biological chemistry. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 27.Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. The Journal of biological chemistry. 2012;287:9742–9752. doi: 10.1074/jbc.M111.304485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klee CBCTH, Krinks MH. Calcineurin: A calcium- and calmodulin-binding protein of the nervous system. Proc. Natl. Acad. Sci. U. S. A. 1979;76:6270–6273. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Wilkins BJ, Lee YJ, Ichijo H, Molkentin JD. Direct interaction and reciprocal regulation between ASK1 and calcineurin-NFAT control cardiomyocyte death and growth. Molecular and cellular biology. 2006;26:3785–3797. doi: 10.1128/MCB.26.10.3785-3797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoyer-Hansen M, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Molecular cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Morgan AJ, et al. Bidirectional Ca(2)(+) signaling occurs between the endoplasmic reticulum and acidic organelles. The Journal of cell biology. 2013;200:789–805. doi: 10.1083/jcb.201204078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calcraft PJ, et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459:596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patron M, et al. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Molecular cell. 2014;53:726–737. doi: 10.1016/j.molcel.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong XP, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nature communications. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nature communications. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samie M, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Developmental cell. 2013;26:511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina DL, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassi MT, et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. American journal of human genetics. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bargal R, et al. Identification of the gene causing mucolipidosis type IV. Nature genetics. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 41.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 42.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Current opinion in cell biology. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grumati P, et al. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamart C, Naslain D, Gilson H, Francaux M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. American journal of physiology. Endocrinology and metabolism. 2013;305:E964–E974. doi: 10.1152/ajpendo.00270.2013. [DOI] [PubMed] [Google Scholar]
- 47.Macian F. NFAT proteins: key regulators of T-cell development and function. Nature reviews. Immunology. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 48.Rusnak F, Mertz P. Calcineurin: form and function. Physiological reviews. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 49.Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 50.Mellstrom B, Naranjo JR. Ca2+-dependent transcriptional repression and derepression: DREAM, a direct effector. Semin. Cell Dev. Biol. 2001;12:59–63. doi: 10.1006/scdb.2000.0218. [DOI] [PubMed] [Google Scholar]
- 51.Heineke J, Ritter O. Cardiomyocyte calcineurin signaling in subcellular domains: from the sarcolema to the nucleus and beyond. J. Mol. Cell Cardiol. 2012;52:62–73. doi: 10.1016/j.yjmcc.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Plyte S, et al. Identification and characterization of a novel nuclear factor of activated T-cells-1 isoform expressed in mouse brain. The Journal of biological chemistry. 2001;276:14350–14358. doi: 10.1074/jbc.M007854200. [DOI] [PubMed] [Google Scholar]
- 53.Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9:2221–2235. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- 54.Sittampalam GS, Gal-Edd N, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Lemmon V, Li Z, McGee J, McManus O, Minor L, Napper A, Riss T, Trask OJ, Weidner J, editors. Assay Guidance Manual [Internet] Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004. [PubMed] [Google Scholar]
- 55.Pattni K, Jepson M, Stenmark H, Banting G. A PtdIns(3)P-specific probe cycles on and off host cell membranes during Salmonella invasion of mammalian cells. Current biology : CB. 2001;11:1636–1642. doi: 10.1016/s0960-9822(01)00486-9. [DOI] [PubMed] [Google Scholar]
- 56.Granatiero V, Patron M, Tosatto A, Merli G, Rizzuto R. The use of aequorin and its variants for Ca2+ measurements. Cold Spring Harbor protocols. 2014;2014:9–16. doi: 10.1101/pdb.top066118. [DOI] [PubMed] [Google Scholar]
- 57.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 59.Amberger J, et al. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37(Database issue):D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carbon S, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belcastro V, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic acids research. 2011;39:8677–8688. doi: 10.1093/nar/gkr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database : the journal of biological databases and curation, bar009. 2011 doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.