Abstract
Stabilization of the hypoxia-inducible factor-1 (HIF-1) increases lifespan and healthspan in nematodes through an unknown mechanism. We report that neuronal stabilization of HIF-1 mediates these effects in C. elegans through a cell non-autonomous signal to the intestine resulting in activation of the xenobiotic detoxification enzyme flavin-containing monooxygenase-2 (FMO-2). This pro-longevity signal requires the serotonin biosynthetic enzyme TPH-1 in neurons and the serotonin receptor SER-7 in the intestine. Intestinal FMO-2 is also activated by dietary restriction (DR) and necessary for DR-mediated lifespan extension, suggesting that this enzyme represents a point of convergence for two distinct longevity pathways. FMOs are conserved in eukaryotes and induced by multiple lifespan-extending interventions in mice, suggesting that these enzymes may play a critical role in promoting health and longevity across phyla.
In nematodes, as in mammals, hypoxia-inducible factor (HIF) proteins have a central role in responding to changes in environmental oxygen (1). HIF proteins are transcription factors regulated by oxygen-dependent proteasomal degradation and are stabilized under low oxygen conditions to modulate expression of hundreds of target genes to produce the hypoxic response (2). In mammals, constitutive stabilization of HIF through loss of the E3 ubiquitin ligase von Hippel-Lindau (VHL) protein leads to a disease characterized by angiomas and renal carcinomas (3), while in Caenorhabditis elegans, loss of the VHL homolog gene, vhl-1, improves proteostasis and increases lifespan (4, 5). This difference likely reflects the fact that somatic cells of adult C. elegans are post-mitotic, with little or no potential for tumor development, and raises the possibility that specific targets of HIF-1 that promote healthy aging in C. elegans may function similarly in mammals.
To understand how hypoxic signaling slows aging in worms, we identified genes downstream of HIF-1 that promote longevity and healthspan. We took advantage of the large reduction in age-associated autofluorescence observed in vhl-1 knockout animals (4) to screen for known HIF-1 target genes required for this phenotype (fig. S1). Our screen identified 24 RNAi clones that substantially increased autofluorescence in vhl-1 animals, eight of which also reduced the long lifespan of vhl-1 mutant animals (table S1 and fig. S2). Six of these RNAi clones had no effect on the lifespan of the wild-type reference strain (N2 Bristol), indicating that they may function specifically to enhance longevity when HIF-1 is stabilized.
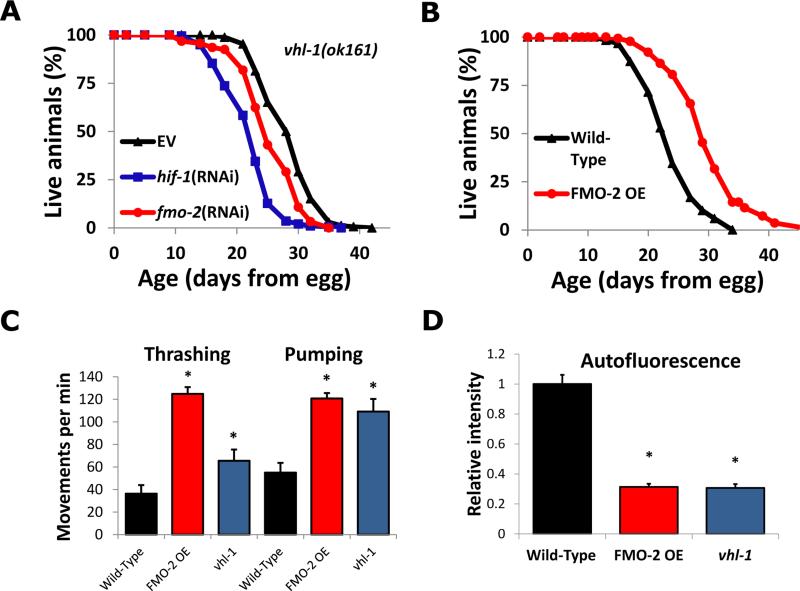
Having established a set of HIF-1-target genes necessary for the full longevity effect of activation of HIF-1, we tested whether any of these genes were sufficient to enhance longevity and healthspan. We used the Mos1 transposase-mediated single copy insertion system (6) to overexpress a single copy of each of the six genes from the ubiquitous eft-3 promoter (fig. S3). Depletion of the xenobiotic detoxification enzyme Flavin-containing monooxygenase-2 (fmo-2) by RNAi showed it to be required for full lifespan extension in vhl-1 knockout animals (Fig. 1A). FMO-2 was also sufficient to extend lifespan on its own (Fig. 1B, fig. S3). Ubiquitous FMO-2 overexpression (FMO-2 OE) also improved multiple measures of healthspan, including enhanced maintenance of motility (measured by the ability to swim, or thrash, in liquid), pharyngeal pumping, and decreased age-associated autofluorescence (Fig. 1C and D, fig. S4). FMO-2 OE animals did not show the decreased brood size or delay in development observed in animals lacking vhl-1, thus these negative consequences of HIF-1 activation likely result from other HIF-1 targets and are separable from lifespan and healthspan extension (fig. S4).
Fig. 1. A screen for age-associated autofluorescence identifies FMO-2 as a modulator of longevity and healthspan in the hypoxic response pathway.
(A) Lifespans of vhl-1(ok161) animals on empty vector (EV), fmo-2 RNAi, or hif-1 RNAi. (B) Lifespans of Wild-Type worms and worms overexpressing FMO-2 ubiquitously (eft-3 promoter, FMO-2 OE). (C, D) Thrashing, pumping and autofluorescence measurements of Wild-Type, FMO-2 overexpression worms, and vhl-1(ok161) mutant worms during adulthood (day 10, 13 and 5, respectively). * indicates statistical difference (p<0.05) from Wild-Type by individual t-test for each strain. Error bars represent SEM, N ≥ 3 for all experiments.
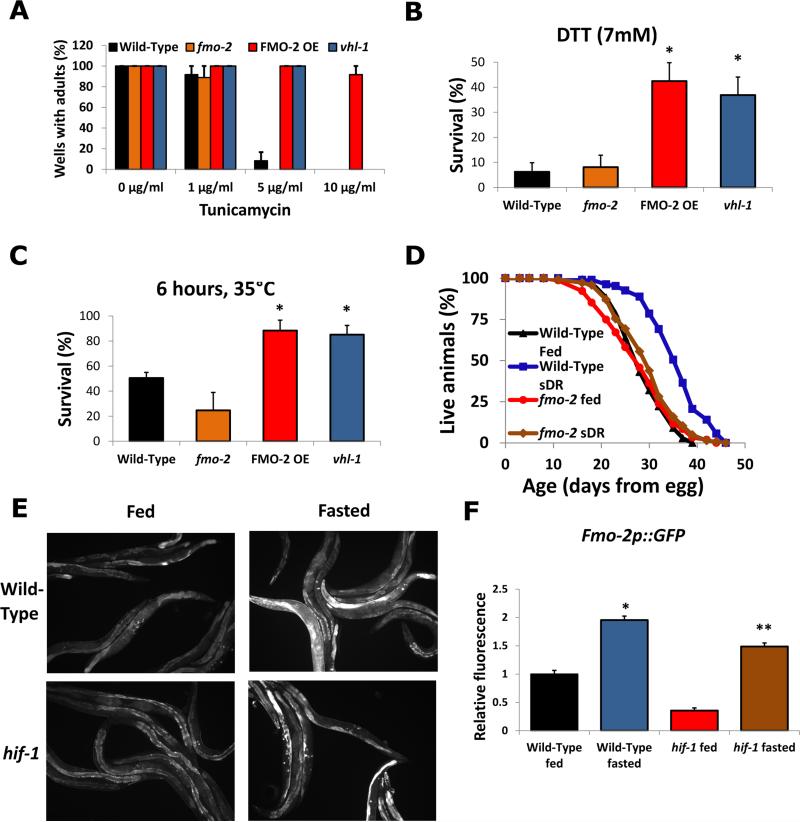
Maintaining proteostasis is critical for healthy aging (7), and both dietary restriction (DR) and stabilization of HIF-1 enhance proteostasis in C. elegans (4, 8). To determine whether FMO-2 enhances proteostasis, we examined the effect of FMO-2 OE on resistance to proteotoxic stress. The most notable effect of FMO-2 OE was resistance to proteotoxic stress within the endoplasmic reticulum (ER), as evidenced by reduced growth inhibition in response to treatment of animals with tunicamycin (up to 10μg/ml) and reduced mortality of animals treated with dithiothreitol (DTT, 7mM) (Fig. 2A, B). FMO-2 OE animals were also resistant to general proteotoxic stress induced by high temperature (Fig. 2C), reductive proteotoxic stress from 2-carboxyethyl phosphine hydrochloride (TCEP) treatment, and transgenic expression of an aggregation-prone polyglutamine peptide fused to yellow fluorescent protein (Q35::YFP) (9) (fig. S5).
Fig. 2. FMO-2 modulates proteostasis and longevity downstream of HIF-1 and DR.
(A-C) Control, fmo-2(ok2147), vhl-1(ok161), and FMO-2 overexpression (FMO-2 OE) resistance to tunicamycin (growth from egg), dithiothreitol (survival at L4), and heat (survival at L4). (D) Wild-Type and fmo-2(ok2147) lifespans on dietary restriction (sDR). (E) fmo-2p::GFP reporter worms on fed and fasted conditions (F) Quantitative measurements of fmo-2 fluorescence shown in (E). * indicates statistical difference (p<0.05) from Wild-Type (** from hif-1) by individual t-test for each strain. Error bars represent SEM, N ≥ 3 for all experiments.
We examined the interaction between fmo-2 and other important longevity pathways. Lifespan extension from stabilization of HIF-1 is genetically distinct from that regulated by both the insulin-like signaling pathway and dietary restriction (4, 5, 10). Life extension in FMO-2 OE animals appears not to require the rest of the hypoxic response pathway, insulin-like signaling, or the phase II detoxification pathway because it was not lost in hif-1, daf-16, or skn-1 mutants, respectively (fig. S6). Thus, FMO-2 does not act through these transcription factors to promote longevity. Similarly, fmo-2 appears not to be necessary for lifespan extension produced by known aging-related pathways because loss of fmo-2 alone had only a modest effect on lifespan and did not prevent lifespan extension in response to reduced insulin-like signaling caused by daf-2 RNAi or inhibition of mitochondrial respiration caused by isp-1 RNAi (fig. S7). However, fmo-2 was required for lifespan extension induced by dietary restriction, using the technique of periodic feeding and fasting, or sDR (11) (Fig. 2D). To further explore the possibility that FMO-2 acts in both the hypoxic response and DR, we confirmed that fmo-2 is transcriptionally induced by food deprivation by monitoring a reporter for fmo-2 transcription (fmo-2p::GFP) (Fig. 2E and F). Unlike that caused by hypoxia (12), induction of fmo-2 in response to fasting was not dependent upon HIF-1 (Fig. 2E-F). This is consistent with our observation that lifespan extension from DR does not require hif-1 (4) and raises the possibility that DR and the hypoxic response converge on FMO-2 to promote longevity through distinct signal transduction pathways.
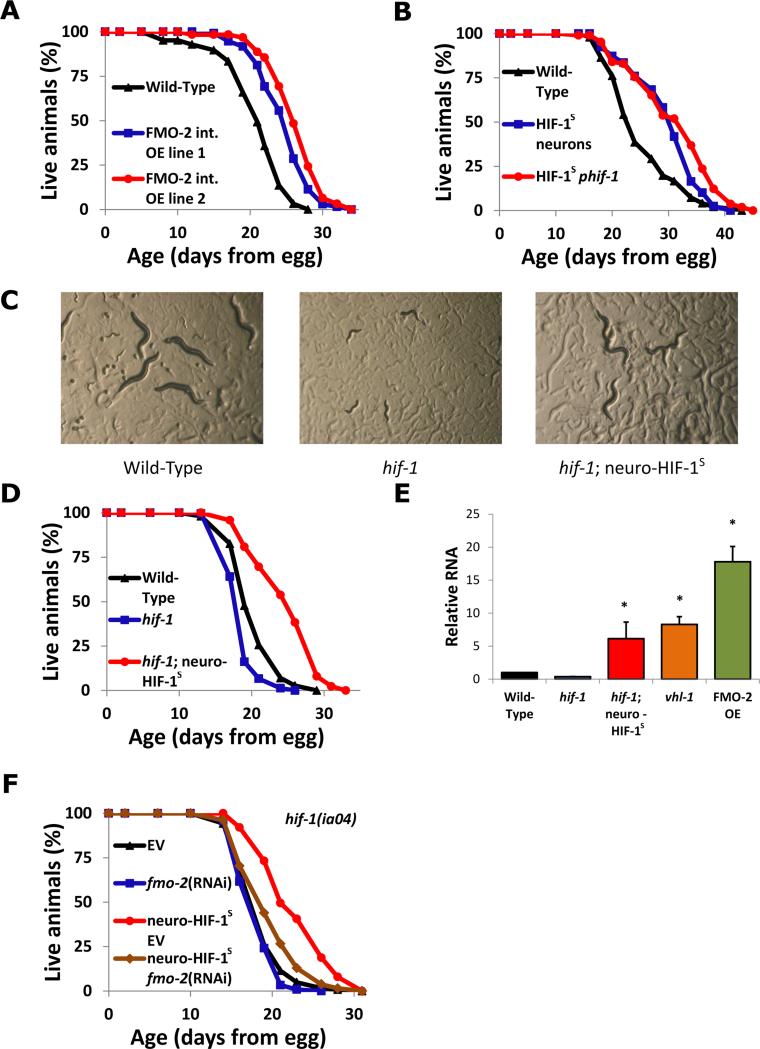
The simplest way HIF-1 might increase fmo-2 expression is to bind the fmo-2 promoter directly and promote transcription. Previous reports, and our results with transcriptional reporters, both indicate that FMO-2 is expressed predominantly in the intestine (13). In agreement with this, overexpression of FMO-2 under an intestinal promoter was sufficient to promote longevity (Fig 3A). To test whether HIF-1 also acts in the intestine to promote longevity, we used transgenic nematodes with a non-degradable HIF-1 variant (14), referred to hereafter as HIF-1S. Intestinal HIF-1S had no effect on longevity (fig. S8), whereas neuronal HIF-1S was sufficient to increase lifespan (Fig. 3B). Expressing HIF-1S in neurons was also sufficient to rescue additional defects in hif-1 knockout animals including failure to develop in hypoxia (0.5% oxygen) (Fig. 3C, fig. S9), loss of vulval integrity during aging (fig. S10), and lifespan extension from hypoxia during adulthood (fig. S11) (15, 16). Neuronal HIF-1S in animals lacking HIF-1 in other tissues was also sufficient to extend lifespan (Fig. 3D), indicating that stabilization of HIF-1 in neurons alone is sufficient to extend lifespan in C. elegans, even without HIF-1 in other cell types.
Fig. 3. Neuronal HIF-1 activates intestinal fmo-2 to increase longevity.
(A) Lifespans of worms overexpressing FMO-2 under an intestinal (vha-6p) promoter. (B) Lifespans of control worms and worms expressing non-degradable HIF-1 (HIF-1S) under neuronal (unc-14p) and ubiquitous (hif-1p) promoters. (C) Growth in hypoxia (0.5% oxygen) of wild-type, hif-1(ia04), and hif-1(ia04)::neuro-HIF-1S worms after 6 days from egg. (D) Lifespans of control, hif-1(ia04), and hif-1(ia04) worms with stabilized neuronal HIF-1. (E) QPCR measurement of fmo-2 transcript in multiple strains. (F) Lifespans of hif-1(ia04) and hif-1(ia04) worms with stabilized neuro-HIF-1S in control (EV) and fmo-2 RNAi. * indicates statistical difference (p<0.05) from Wild-Type (** from hif-1) by individual t-test for each strain. Error bars represent SEM, N ≥ 3 for all experiments.
Neuronal overexpression of fmo-2 had no detectable effect on longevity (fig. S8). Thus HIF-1 and FMO-2 appear to promote longevity and healthspan by acting in distinct tissues: HIF-1 in neurons and FMO-2 in intestine. Consistent with this model, transcription of fmo-2 was significantly induced in the intestine by stabilization of HIF-1 in neurons in a background where hif-1 is knocked out in all other tissues (hif-1(ia04); neuro- HIF-1S) as measured by both quantitative RT-PCR and by fluorescence in a reporter strain (Fig. 3E, fig. S12). In agreement with these results, depletion of fmo-2 with RNAi prevented lifespan extension in this strain (Fig. 3F) despite the inefficiency of RNAi in neurons (17), indicating that neuronal HIF-1 signaling to intestinal FMO-2 is probably necessary for the longevity benefit.
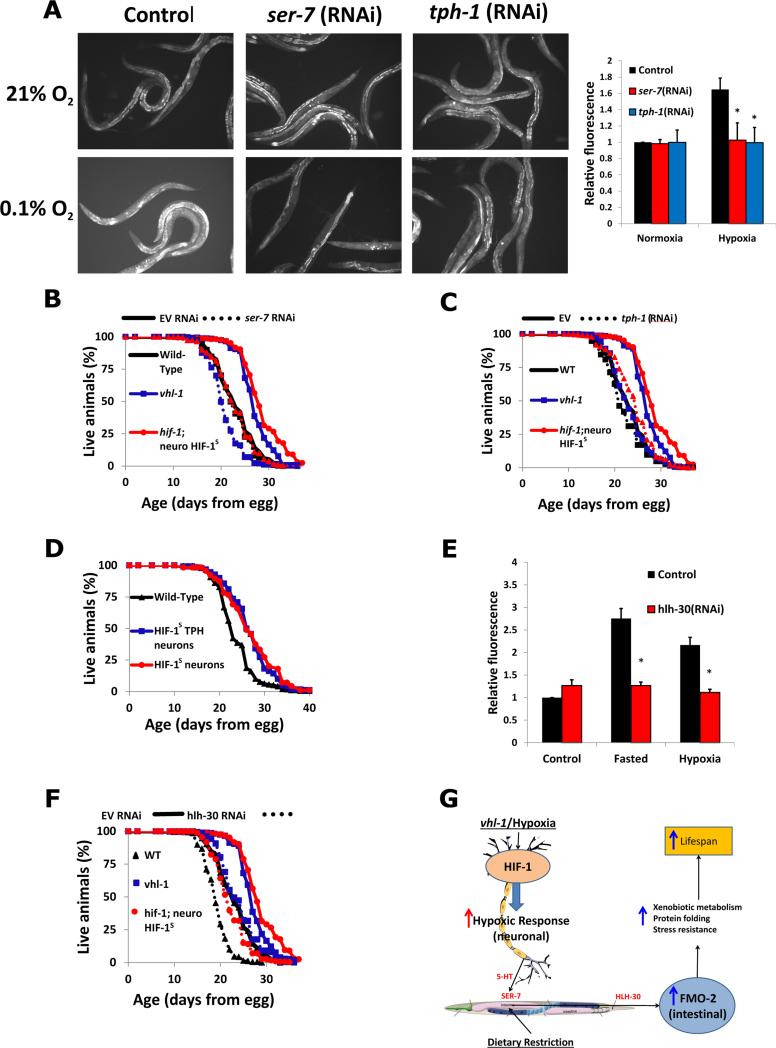
Having established a connection between neuronal HIF-1 signaling and intestinal FMO-2 activation, we explored potential signal transduction pathways by depleting signaling components and transcription factors chosen on the basis of previous reports and in silico promoter analysis (18, 19). Although most of the factors examined had no effect (fig. S13), the serotonergic signaling pathway was both necessary and sufficient for the cell non-autonomous effect of HIF-1 signaling in neurons on expression of FMO-2 in intestine and subsequent longevity benefit of FMO-2. The 5-hydroxytryptamine7 receptor ser-7 and the rate limiting enzyme in serotonin production, tph-1, were both required for the activation of FMO-2 in hypoxia and the longevity benefit from neuronal HIF-1S or vhl-1 mutation (Fig. 4A-C, fig. S14). In agreement with this, HIF-1S expressed under the serotonergic tph-1 promoter was sufficient to improve longevity to an extent comparable to that of pan-neuronal expression (Fig. 4D). A transcription factor with predicted binding to the fmo-2 promoter, HLH-30, was necessary for either hypoxia or starvation to fully induce transcription of an FMO-2 reporter in the intestine (Fig. 4E). HLH-30 is necessary for lifespan extension by DR (20), and our results indicate it is also required to achieve maximal lifespan extension from HIF-1 stabilization (Fig 4F), although expression of HIF-1S or deletion of vhl-1 still partially increase lifespan in animals depleted of hlh-30.
Fig. 4. The signaling pathway from neuronal HIF-1 to intestinal FMO-2 involves serotonin and HLH-30.
(A) Fluorescence images and quantification of fmo-2p::GFP reporter worms in normoxia (~21% O2) and hypoxia (0.1% O2) on control, tph-1, and ser-7 RNAi. (B, C) Lifespans of wild-type, vhl-1(ok161) mutant, and hif-1(ia04) with stabilized neuro-HIF-1S worms on control (EV, solid lines), ser-7 RNAi (B, dashed lines) and tph-1 RNAi (C, dashed lines). (D) Lifespans of worms expressing HIF-1S under the panneuronal (unc-54p) and serotonergic (tph-1p) promoters. (E) Expression of fmo-2p::GFP reporter in fed, fasted and hypoxic conditions under control and hlh-30 RNAi. (F) Lifespans of wild-type, vhl-1(ok161) mutant, and hif-1(ia04) with stabilized neuro-HIF-1S worms on control (EV, solid lines) and hlh-30 RNAi (dashed lines) (G) Model of hypoxic response and dietary restriction converging on intestinal FMO-2. * indicates statistical difference (p<0.05) from Wild-Type by individual t-test for each strain. Error bars represent SEM, N ≥ 3 for all experiments.
Our results support a model in which the flavin-containing monooxygenase FMO-2 functions in the intestine to increase lifespan, improve healthspan, and enhance proteostasis in animals undergoing the hypoxic response or DR. Further, intestinal fmo-2 is regulated cell non-autonomously through serotonergic signaling originating in neurons, and subsequent activation of the transcription factor HLH-30 in the intestine (Fig. 4G). FMO-2 is thus an enzyme both necessary and sufficient for a majority of the beneficial effects of either of these longevity pathways. The FMO-2 substrates important for healthy aging in C. elegans remain unknown. It will also be of interest to directly assess whether FMOs may function in mammalian aging. There are five mammalian FMO proteins (FMO1-5) (21), similar to the five C. elegans FMOs (13), and all of these proteins came from a single ancestral FMO (22). In mammals, there is relatively limited information on the specific functions of each FMO beyond tissue-specific expression patterns and the role of FMO3 in a single human disease, fish-odor syndrome (23). Mammalian FMOs also have a major role in regulating cholesterol and fat metabolism (24, 25). Abundance of FMO proteins is increased in the tissues, particularly liver, of several long-lived mouse models including Snell dwarf mice, Ames dwarf mice, growth-hormone receptor knockout mice, Little mice, dietary restricted mice, and rapamycin-fed mice (26). Indeed, FMO3 mRNA is the most consistently induced mRNA under dietary restriction in mouse liver (27). Taken with the data presented here, these observations raise the possibility that activation of FMOs may be a conserved mechanism for enhancing protein homeostasis, improving healthspan, and extending lifespan, and that appropriate activation of FMOs might promote healthy aging in mammals and people.
Supplementary Material
Acknowledgments
We thank C. Dolphin for the vector to make the fmo-2p::GFP reporter strains and A. Mendenhall for advice and help with injecting worms. Strains were provided by the Caenorhabditis Genetics Center. This work was supported by NIH grant R01AG038518 and an award from the Samsung Advanced Institute of Technology, Samsung Electronics Co. to MK and NIH grant K99AG045200 to SFL. SFL and FJR were supported by NIH Training Grant T32AG000057. SFL was also supported by an AFAR Post-doctoral fellowship. Additional support was provided by the UW Healthy Aging and Longevity Research Institute, the UW Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH grant P30AG013280), and an award to MK from the M. J. Murdock Charitable Trust. DLM is an Ellison Medical Foundation New Scholar in Aging and receives support from NIA R00AGA0033050. Additional data available in Supplementary Materials (Methods, references 28-33, figs. S1-S14, Tables S1-S3).
References and Notes
- 1.Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 3.Ivan M, Kaelin WG. The von Hippel-Lindau tumor suppressor protein. Curr Opin Genet Dev. 2001;11:27–34. doi: 10.1016/s0959-437x(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 4.Mehta R, et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller RU, et al. The von Hippel Lindau tumor suppressor limits longevity. J Am Soc Nephrol. 2009;20:2513–2517. doi: 10.1681/ASN.2009050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labbadia J, Morimoto RI. Proteostasis and longevity: when does aging really begin? F1000prime reports. 2014;6:7. doi: 10.12703/P6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinkraus KA, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7:394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem. 2005;280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- 13.Petalcorin MI, Joshua GW, Agapow PM, Dolphin CT. The fmo genes of Caenorhabditis elegans and C. briggsae: characterisation, gene expression and comparative genomic analysis. Gene. 2005;346:83–96. doi: 10.1016/j.gene.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Pocock R, Hobert O. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nature neuroscience. 2008;11:894–900. doi: 10.1038/nn.2152. [DOI] [PubMed] [Google Scholar]
- 15.Leiser SF, Fletcher M, Begun A, Kaeberlein M. Life-span extension from hypoxia in Caenorhabditis elegans requires both HIF-1 and DAF-16 and is antagonized by SKN-1. J Gerontol A Biol Sci Med Sci. 2013;68:1135–1144. doi: 10.1093/gerona/glt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller DL, Roth MB. C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr Biol. 2009;19:1233–1237. doi: 10.1016/j.cub.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci. 2010;13:610–614. doi: 10.1038/nn.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapierre LR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez D, Janmohamed A, Chandan P, Phillips IR, Shephard EA. Organization and evolution of the flavin-containing monooxygenase genes of human and mouse: identification of novel gene and pseudogene clusters. Pharmacogenetics. 2004;14:117–130. doi: 10.1097/00008571-200402000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Dolphin CT, Janmohamed A, Smith RL, Shephard EA, Phillips IR. Missense mutation in flavin-containing mono-oxygenase 3 gene, FMO3, underlies fish-odour syndrome. Nat Genet. 1997;17:491–494. doi: 10.1038/ng1297-491. [DOI] [PubMed] [Google Scholar]
- 24.Veeravalli S, et al. The phenotype of a flavin-containing monooyxgenase knockout mouse implicates the drug-metabolizing enzyme FMO1 as a novel regulator of energy balance. Biochem Pharmacol. 2014;90:88–95. doi: 10.1016/j.bcp.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Shih DM, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res. 2015;56:22–37. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinbaugh MJ, Sun LY, Bartke A, Miller RA. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am J Physiol Endocrinol Metab. 2012;303:E488–495. doi: 10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.