Abstract
Identifying the functional motions of membrane proteins is difficult because they range from large-scale collective dynamics to local small atomic fluctuations at different timescales that are difficult to measure experimentally due to the hydrophobic nature of these proteins. Elastic Network Models, and in particular their most widely used implementation, the Anisotropic Network Model (ANM), have proven to be useful computational methods in many recent applications to predict membrane protein dynamics. These models are based on the premise that biomolecules possess intrinsic mechanical characteristics uniquely defined by their particular architectures. In the ANM, interactions between residues in close proximity are represented by harmonic potentials with a uniform spring constant. The slow mode shapes generated by the ANM provide valuable information on the global dynamics of biomolecules that are relevant to their function. In its recent extension in the form of ANM-guided molecular dynamics (MD), this coarse-grained approach is augmented with atomic detail. The results from ANM and its extensions can be used to guide experiments and thus speedup the process of quantifying motions in membrane proteins. Testing the predictions can be accomplished through (a) direct observation of motions through studies of structure and biophysical probes, (b) perturbation of the motions by, e.g., cross-linking or site-directed mutagenesis, and (c) by studying the effects of such perturbations on protein function, typically through ligand binding and activity assays. To illustrate the applicability of the combined computational ANM—experimental testing framework to membrane proteins, we describe—alongside the general protocols—here the application of ANM to rhodopsin, a prototypical member of the pharmacologically relevant G-protein coupled receptor family.
Keywords: Anisotropic network model (ANM), Normal mode analysis (NMA), Structural dynamics, Molecular dynamics (MD) simulations, Structure prediction, Conformational changes, Ensembles of structures, G-protein coupled receptors, Multiscale models and methods
1. Introduction
1.1. The Functional Roles of Motions in Membrane Proteins
While the structure of a protein provides some insights into its function, it only yields static information. Detailed information on the dynamics of a protein is necessary for a complete understanding of its function. However, it is not a trivial problem to resolve the functionally relevant motions of biomolecular systems. The timescales involved in different events range from femtoseconds to seconds or even longer, and usually more than a single experimental technique or computational model and method is required to span such a wide range. The spatiotemporal resolutions of the computational models or experimental techniques need to be optimally selected to explore the particular time and length scales of the systems and process of interest.
Computational approaches can visualize molecular events at timescales and resolutions that may not be readily examined by experimental techniques, thus complementing the information acquired from experiments and providing deeper insights into biomolecular mechanisms of function. Not surprisingly, computational biology tools and biophysical theories have been advantageously employed in the last two decades for investigating the dynamics of biomolecular systems. Among them, molecular dynamics (MD) simulations, and normal mode analysis (NMA) found broad utility in biomolecular applications (1–4), and especially in investigating the dynamics of membrane proteins in recent years (5–8). Membrane proteins are particularly important as they serve as targets for the majority of drugs, and their molecular motions remain largely unknown due to scarcity of structural data. It is now widely recognized that the motions of membrane proteins underlie key functional events, such as receptor–ligand binding and signaling, ion channeling and gating, folding and translocation, and the allosteric control of them. An assessment of the type and size of these motions is critically important for designing modulators, agonists, or antagonists of membrane proteins’ functions.
1.2. Overview of Structural Data for Membrane Proteins
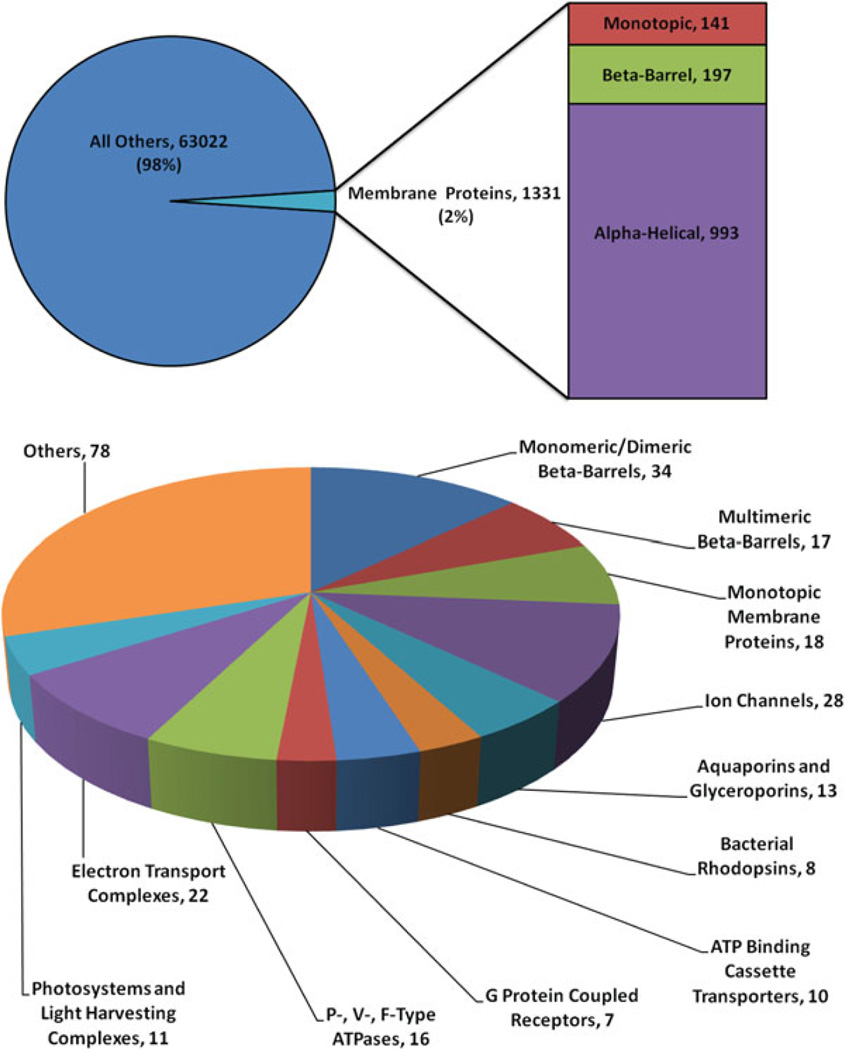
Obtaining structural data for membrane proteins experimentally is challenging for a number of reasons. First, structural techniques such as X-ray crystallography and NMR spectroscopy require large quantities of proteins purified to homogeneity. For membrane proteins, both the production and purification steps are very difficult. Membrane proteins usually have very low expression levels and are thermally unstable when removed from their native membrane environment and tend to aggregate. Secondly, the need for mimicking membranes requires addition of lipids or detergents that interfere with structural techniques, for example by creating large background signals in NMR and increasing the effective molecular weight of the membrane protein–detergent mixed micellar complex. Thus, determining membrane protein structures has been a challenge, a fact which is reflected by the limited number of available membrane protein structures resolved and deposited to date in the Protein Data Bank (PDB) (9). As of the end of November 2010 there have been only 1,331 membrane protein structures deposited in the PDB, which corresponds to only 2% of all structures in the PDB (Fig. 1a). Moreover, only 262 out of these 1,331 are unique membrane protein structures, as defined by a sequence cutoff of 95%. Thus, only a small number of unique membrane proteins are available as examples for different functional and structural categories that membrane proteins engage in (Fig. 1b).
Fig. 1.
Fraction and composition of membrane proteins in the PDB. (a) The pie chart shows that of all structurally reserved proteins, 2% correspond to membrane proteins. The chart on the right provides the breakdown of membrane proteins into different structural categories. (b) Structural and functional distribution of unique membrane protein structures. Data was retrieved on 26 November 2010 from the curated membrane protein database available at http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
1.3. Need for Complementary Predictive Computational Approaches to Study the Dynamics of Membrane Proteins
The general challenges faced in obtaining structures of membrane proteins experimentally described above, also impact the ability to determine membrane protein motions. Even for those proteins for which a structure exists, it is most often only in one functional state. Thus, there is a great need and opportunity to utilize the protein structures that are available to predict functional motions by computational techniques. These predicted dynamics can then be tested experimentally by targeting specific regions in the protein or types of motions through biophysical approaches. A large battery of techniques is available in which attachment points such as cysteine replacements are introduced at specific sites that allow application of specific techniques, where the choice of technique depends on the timescale and scale of motions to be tested. This approach allows obtaining site-specific information and is ideally suited to target specific predictions made by computational models. In this chapter, we will review this combined experimental–computational approach.
1.4. State of the Art in Computational Modeling of Protein Dynamics: Application to Membrane Proteins
1.4.1. Molecular Dynamics as the Gold Standard for Simulating Motions at Atomic Resolution, but Limited to Short Times and Small Systems
The most widely used atomic-level approach in modeling protein dynamics is molecular dynamics (MD) simulations. The interactions of atoms are described therein by complex potential energy functions and parameters, which define the instantaneous forces exerted on each atom. Knowledge of these forces permits us to calculate the dynamic behavior of the system (composed of the protein with its substrates/ligands, and the environment—mainly water, lipid molecules and ions) using the classical Newton’s equations of motion (1). MD simulations thus provide atomic-level detail with high temporal resolution, which is usually viewed as the gold standard.
On the other hand, it is also widely known that MD simulations have limitations. First, the trajectories are accurate to the extent that force fields themselves are. Second, the size of the time steps of these simulations, usually 1–2 fs, implies that millions of steps are required to reach the nanosecond timescale; and nanoseconds can provide a good description of local events at best.
The rapid growth in computer power and new developments in parallelizable MD codes designed for high-performance simulations now enable us to simulate large biomolecular systems at hundreds of microseconds or even milliseconds scales (5, 10, 11). These long simulations confirm that the “global” motions of proteins (e.g., interconversions between substates) are several orders of magnitude slower than local motions. For example, the global backbone rearrangements of BPTI, a small protein of 58 residues, occur on a timescale of 10 µs, whereas side-chain motions are faster than 10 ns (11). Clearly, MD simulations on the millisecond timescale can investigate such global motions, including folding and unfolding events for small proteins only. Application to larger proteins, and to membrane proteins in particular, incurs serious convergence problems (6, 12).
1.4.2. Normal Mode Analysis with Elastic Network Models: A Useful Approach for Identifying Global Motions, Albeit at Low Resolution
A method for exploring global motions accessible under equilibrium conditions is NMA, a technique introduced several decades ago by computational chemists for describing small molecules’ vibrational dynamics (13). NMA is a harmonic approximation and assumes that the conformational energy surface can be characterized by a single energy minimum. Classical, full-atomic NMA requires reaching a minimum on the potential energy surface. Hence, a stringent energy minimization of the potential energy function is required before performing NMA, or before evaluating the Hessian matrix (the 3n × 3n matrix of the second derivatives of the potential energy function with respect to the 3n degrees of freedom for a structure of n atoms, for atomic NMA). The low-frequency modes obtained by NMA often correlate well with experimentally observed changes between different functional states (e.g., open and closed form of a ligand-free and -bound enzyme). However, energy minimization and numerical evaluation of the Hessian matrix using a full-fledged force field makes full-atomic NMA a computationally expensive method for large biomolecules (1, 14).
The investigation of cooperative motions in large systems requires the adoption of coarse-grained models, and possibly analytical methods, which brings us to the focus of our review, the use of elastic network models (ENMs). NMA has seen a revival in recent years, with the realization that it can be advantageously used in conjunction with simplified models such as ENMs for studying the global motions of proteins (15, 16). A breakthrough in the field was the demonstration by Tirion that the global modes obtained by the detailed NMA could be almost identically reproduced by using a single parameter harmonic potential for the force field (17). This study led to the introduction of the Gaussian Network Model (GNM) (18, 19) as a simplified network representation of the protein structure, amenable to the investigation of its dynamics.
In the GNM, the collective dynamics of the protein (of N residues) is fully defined by an N × N Kirchhoff matrix that describes the spatial connectivity (or topology) of the structure at the level of α-carbons. The physical basis and the analytical method for examining the statistical mechanics of such a mass-spring network bear close similarities to the elasticity theory of random polymer networks (20).
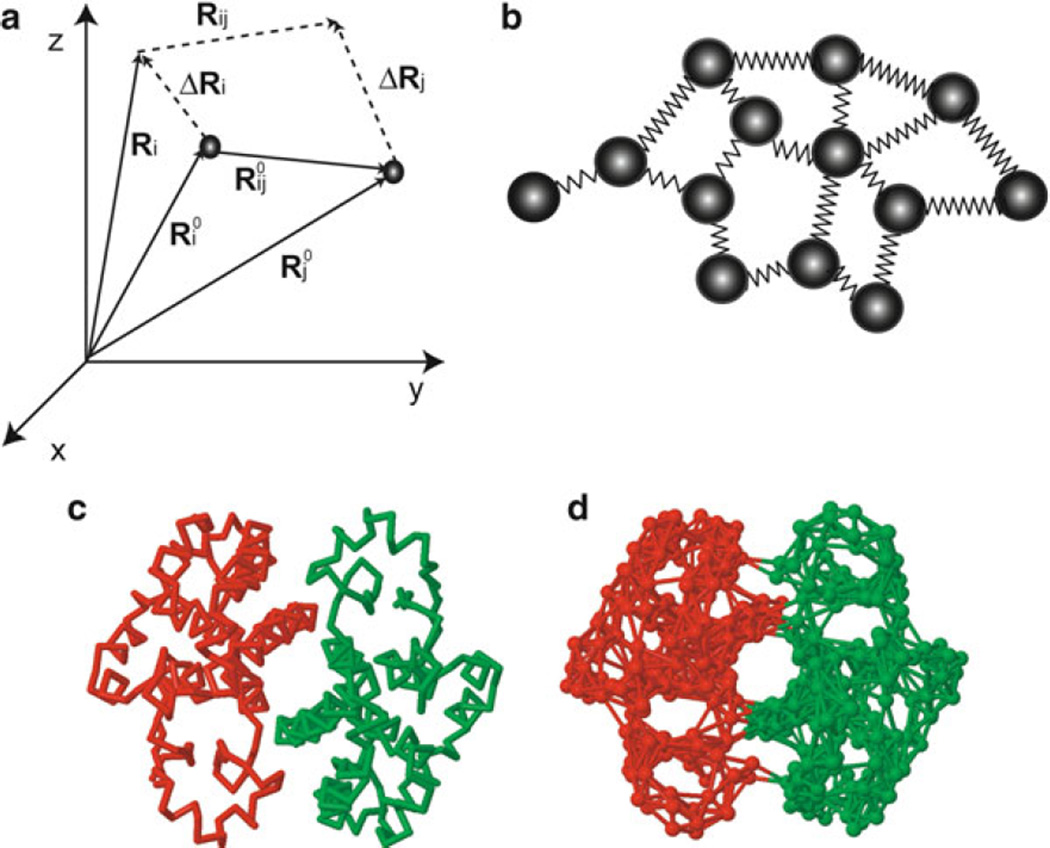
The same level of coarse-graining, i.e., residue-level description with the position of network nodes identified by that of the α-carbons, has been adopted in the Anisotropic Network Model (ANM) (21–25). The main advantage of performing NMA using the ANM, shortly referred to here as ANM analysis, is the possibility of computing an analytical, unique solution for the collective modes of motion accessible to the examined system. The basic ingredient in the model is the topology of inter-residue contacts in the native structure (as the GNM), which turns out to be a major determinant of equilibrium dynamics. Any deformation from the native state coordinates is resisted by linear springs that associate closely neighboring residues, bonded and nonbonded (Fig. 2). Despite its simplicity, the ANM results proved to be in remarkable agreement with experiments. The global modes obtained with the ANM analysis have been shown in numerous applications to be highly robust (26) and consistent with experimental data in terms of the shapes and mechanisms of motions. This includes, for example, the directions of domain movements and their correlations, the location of hinge-bending sites, the most likely collective rearrangements, or allosteric switches triggered by ligand/substrate binding. The ANM server (27) is being broadly used by the molecular structural and computational biology communities for rapid assessment and visualization of the intrinsic dynamics of known protein structures.
Fig. 2.
Representation of a given structure by the ANM. Panel (a) displays the equilibrium position of node i, its instantaneous fluctuation ΔRi, and instantaneous position Ri. The fluctuation from the mean position is given by . For a pair of interacting residues i and j, we denote the equilibrium inter-node distance vector extending from residue i to residue j as and the instantaneous change in distance vector is . Panel (b) displays the node and spring representation where each node represents an amino acid, and springs connect interacting nodes (bonded or nonbonded pairs of amino acids with an interaction cutoff distance of Rc). Panels (c) and (d) display the backbone (stick), and elastic network (ANM) representations (based on a cutoff distance of 10 Å) for an example protein, glutathione transferase (PDB code 2A2R) (109). Note that the ANM also includes springs at the interface between the two monomers.
However, ENMs, including the GNM and ANM, also have their own limitations. The information provided lacks atomic precision and may in some instances yield unphysical bond lengths or bond angles at the atomic scale (28). Moreover, no absolute timescale and size of motions can be deduced, only the shape of motions. Also, neither the specificity of amino acids nor the effects of a protein’s environment (e.g., lipid bilayer in membrane proteins, or water molecules in general) is taken into consideration. In parallel with all NMAs, the approach is in principle limited to movements within a single energy well, and becomes most accurate in the immediate vicinity of the energy well. This means that transitions that involve passage over energy barriers or jumps between minima cannot be predicted. On the other hand, the coarse-graining of the structure and thereby smoothing out of the energy landscape in the ANM presumably help merge substates separated by low energy barriers, thus allowing for sampling of substates separated by low energy barriers.
The basic assumptions and theory underlying the ANM, and its use in predicting structure-based motions as well as its limitations, will be described in detail in Subheading 3.1. From the brief introduction above, it is clear that both extremes, MD simulations and ANM analysis, have limitations, but they also provide complementary information. In particular, they allow for exploring different timescales and events at different resolutions, the former being powerful in examining events in the picoseconds to hundreds of nanoseconds regime, and the latter successfully describing the global machinery of supramolecular systems. Clearly, methods that encompass both regimes are highly desirable, and we will present in Subheading 3.1.3. the development and use of such a method, ANM-steered MD recently applied to rhodopsin (29). First, we provide a brief description of approaches undertaken for extending the range of MD simulations.
1.4.3. Recent Progress in Atomic Simulations: Steered MD, Targeted MD and ANM-Guided MD
Various methods have been developed to accelerate MD simulations and increase their sampling efficiency. Steered MD (SMD) is one of them. The idea is to observe large conformational changes by applying external forces to facilitate such movements (30). A similar approach, Targeted MD (TMD), applies time-dependent, purely geometrical constraints to induce conformational changes for attaining a known target structure at physiological temperatures. Additionally, guiding MD simulations by collective coordinates has proven to be useful for efficient sampling and expansion of the accessible conformational space. Berendsen and coworkers developed a method in which essential dynamics analysis (EDA) or principle component analysis (PCA) of an ensemble of snapshots obtained from MD trajectories are used to generate collective modes (31). These collective modes are used as constraints in MD simulations to sample the conformational space efficiently (31–33). Collective coordinates have been combined with ensemble sampling by Abheser and Nilges. Their method uses an additional biasing potential defined along the collective modes used as restraints in a set of independent MD trajectories (34). Another technique, Amplified Collective Motion couples the low frequency motions obtained from the ANM to a higher temperature by using a weak coupling method (35).
More recently, we introduced the ANM-guided MD methodology (29), a hybrid methodology that will be explained in detail in Subheading 3.1.3. The method, originally introduced for exploring the dynamics of rhodopsin (29) is, in principle, applicable to all membrane proteins. ANM-guided (or ANM-restrained) MD is useful for modeling small molecule binding to proteins and flexible docking of proteins in particular. It has also been recently adopted for studying the dynamics of biological systems and protein-protein interactions, in conjunction with MM-PBSA (Molecular Mechanic/Poisson-Boltzmann Surface Area) calculations (36). It also proved useful in establishing the significance of the conformational change induced by thrombosbondin-1 binding to the calreticulin structure, essential to signaling focal adhesion disassembly (36). In the present review, we will provide a detailed description of the methodology, and illustrate its utility in generating conformers that may be advantageously exploited for flexible docking of inhibitors.
1.4.4. Structure-Based Coarse-Grained Computations for Membrane Proteins
Structure-based coarse-grained models have been used for several decades in computational biology with the understanding that these may help explore different time and length scales of biological interest, albeit at low resolution. Pioneering studies in this field are the work of Michael Levitt (37, 38) or the virtual bond model based on α-carbons widely used by Flory and coworkers in delineating the statistical mechanics of biopolymers. Coarse-grained models for membrane proteins, on the other hand, are relatively more recent, presumably due to lack of structural data. As recently reviewed by Sansom and collaborators (39), coarse-grained simulations of membrane proteins are particularly useful for exploring large-scale changes in conformation including the diffusion and reorientation of peptides or insertion of membrane proteins across the lipid bilayer, and self-assembly of membrane protein–detergent micelles. A typical self-assembly of a protein-bilayer requires simulating at least 0.1 µs, and such simulations become prohibitively expensive, if not statistically inaccurate (due to convergence problems), with full atomic models and force fields. In particular, the MARTINI force field introduced by Marrink and coworkers for lipids (40) has found wide applications in recent years and have been successfully extended by the Sansom and Schulten labs to protein–lipid systems (41–43).
In parallel to coarse-grained MD simulations, ANM-based methods have also been exploited for examining the functional motions of membrane proteins, as we have recently reviewed (7, 8). Notable applications include the pore opening of potassium channels (44), the dynamics of the mechanosensitive channel (MscL) (45), the allosteric global torsional motions of nicotinic acetyl choline receptor (46), cooperative domain movements of ABC transporter BtuC (47), and identification of core amino acids and intrinsic dynamics of rhodopsin (44, 48), as will be described in detail below.
2. Materials
Hardware and software to run ANM and ANM-restrained MD
ANM-restrained MD simulations of rhodopsin were performed at four machines each with eight 2.6 GHz Intel Xeon processors and 32 GB memory. The simulations described above complete in approximately 10 h. The molecular dynamics simulations and energy minimization sections of the protocol were run using Nanoscale Molecular Dynamics Software (10). ANM calculations are performed in Fortran 90. The protocol is fully automated using a tcl code that enables the communication of the different sections. This code is available by contacting the authors.
The ANM code is available in both Fortran 90 and C programming languages. ANM calculations of rhodopsin take approximately 1 min on one of the above processors. The Web site implementation of ANM is also available and described in Note 1.
3. Methods
3.1. Methods I: Theory and Modeling
3.1.1. Overview
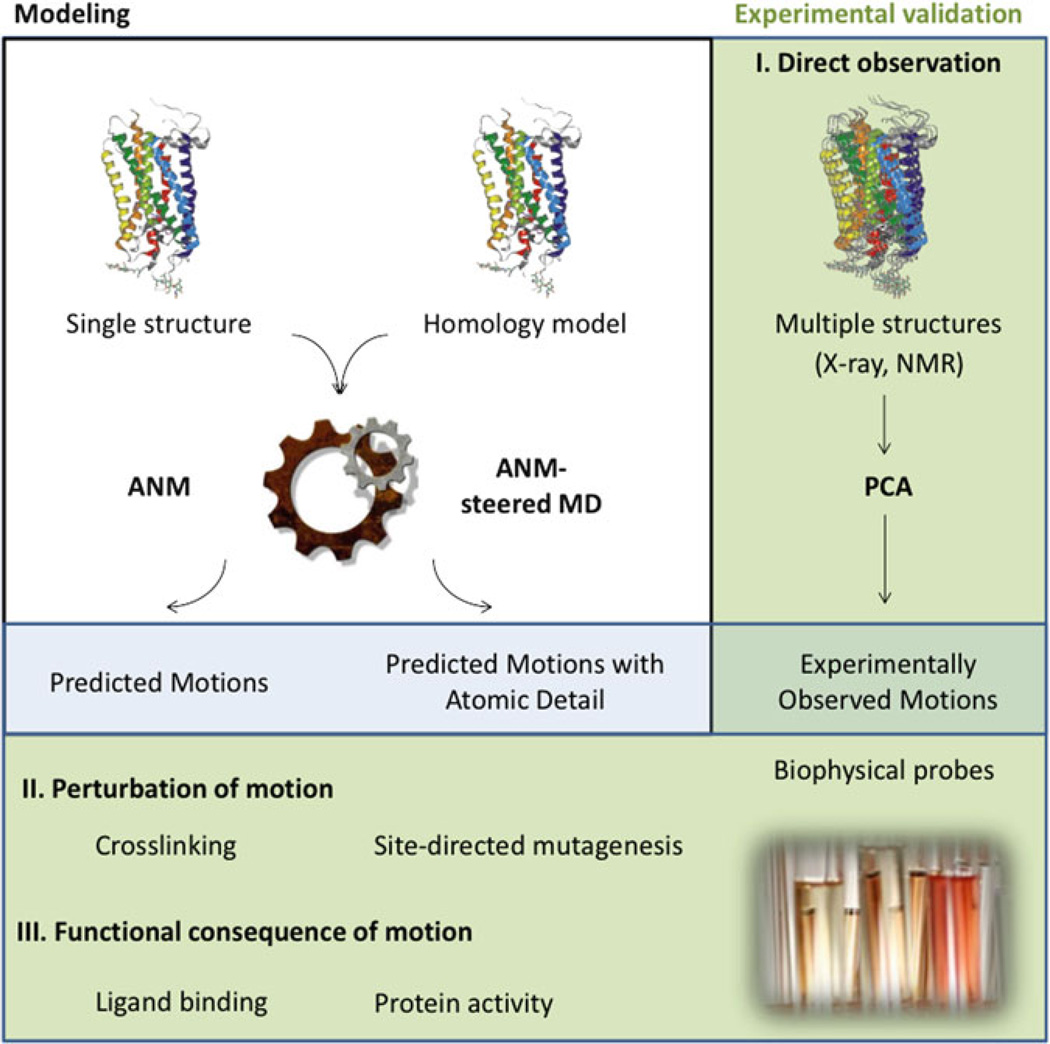
The diagram in Fig. 3 provides an overview of the overall approach, where modeling of single structures are outlined in the white box and experimental studies in the green boxes. When multiple structures are available, modeling is also used to help in the analysis (green box, right), while other experiments may be driven by the modeling (green box, bottom). The inputs and outputs of these approaches are summarized below and described in detail in the subsequent sections.
Fig. 3.
Schematic overview of the combined experimental–computational approach. Using either single structures or homology (or other) models as an input, ANM or ANM-steered MD simulations are used to predict motions without or with atomic detail, respectively (white box, left top). If available, the input into computations can also be multiple structures that can be used to validate predictions as well as help better analyze them (“direct observation,” green box right). The predictions can be used to design experiments to validate them, through perturbation of motion or study of the functional consequences of motions (green box, bottom).
Two types of inputs may be used in ANM calculations: a known structure accessible in the PDB, or a structural model generated by the user, e.g., using structure determination techniques or computational modeling such as homology modeling (also see below). For example, the ANM server requires as input either the PDB identifier of the protein of interest (which is directly downloaded from the PDB), or a four-letter filename for the structural model in PDB format (found by browsing and retrieval from the local desktop) (27). In either case, to generate/predict the equilibrium motions of a protein using the ANM, one structure file, or the three-dimensional coordinates of the residues forms the input, even at the coarse-grained scale (see below). As a typical example, we will illustrate below the application of ANM to rhodopsin, which permitted us to explain 93% of experimentally observed effects for 119 rhodopsin mutants (49). In the GNM, the input may even be simply the list of “interacting” residue pairs without information on their detailed coordinates. The main ingredient of the model is the N × N Kirchhoff matrix that describes the presence or absence of contacts between the N residues of the proteins.
On the other hand, if multiple structures are available for a given protein (its mutant, different bound forms, or family members), and if the structures are sufficiently variable/heterogeneous, one can take advantage of the available data to perform a PCA of the ensemble of structures and extract the dominant types of structural variations. This type of information, of purely experimental origin, helps test/validate the theoretically predicted ANM modes. It can also be used to help understand which of the modes predicted by the ANM are exploited by the protein for which types of functional events (50).
If there are no structural data available for the protein of interest, homology (or other) models could be built, which might be used as templates for further refinement and analysis in conjunction with the ANM studies. Currently, there are structural data available for a quarter of all known single-domain families in the protein universe (of all organisms sequenced to date), and >70% of all known sequences can be partially modeled using existing structural data as templates/models (51). Evidently, membrane proteins are the most challenging proteins to predict due to scarcity of structural data ((52) ; see also Fig. 1). Homology models constructed using ANM modes have been successfully employed to demonstrate the preferential binding of allosteric ligands to active and inactive conformations of metabotropic glutamate receptors (53).
3.1.2. Modeling I: ANM Theory, Assumptions, and Implementation
ANM Protocol
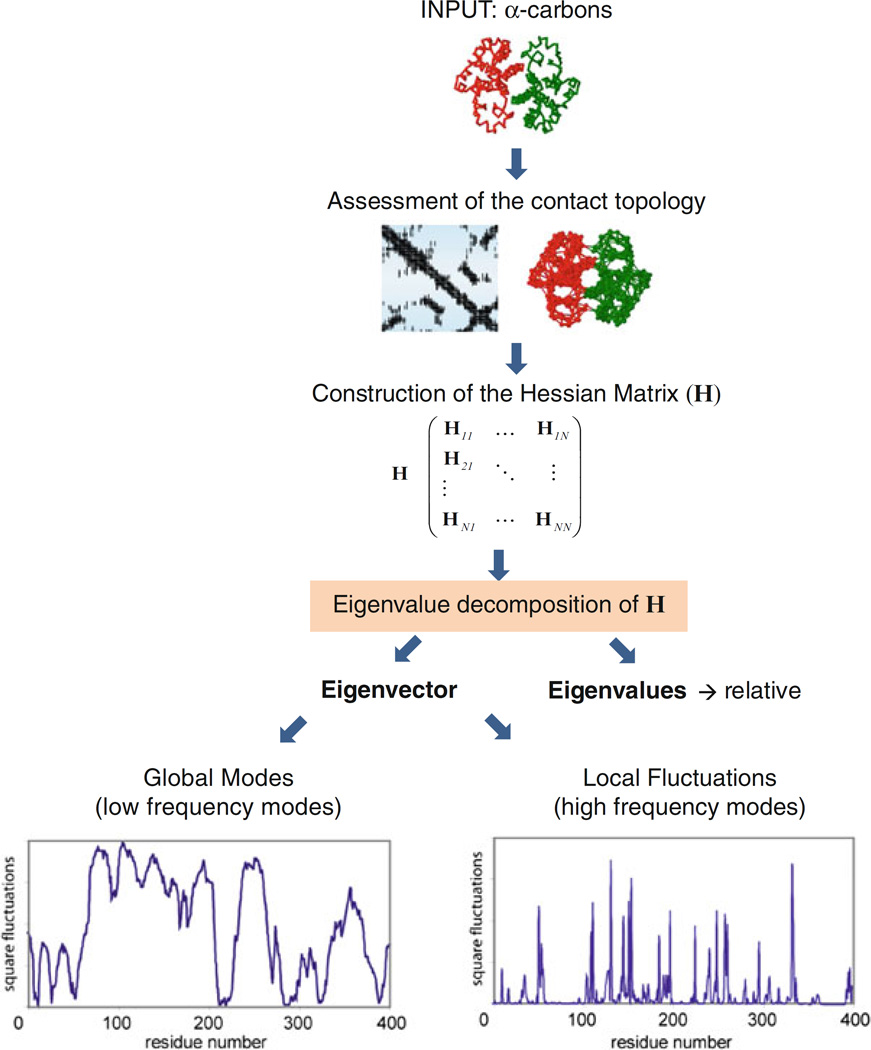
We present below a detailed description of the steps required for performing the ANM analysis of a given structure. A schematic description is provided in Fig. 4.
Fig. 4.
Schematic representation of ANM calculation steps and released outputs.
Input
ANM analysis requires knowledge of α-carbon coordinates. These define the positions of the “nodes” in the elastic network. The coordinates are usually retrieved directly from the PDB file corresponding to the examined structure (e.g., the ANM server directly accepts as input the PDB identifier); or alternatively one can feed the coordinates of a model (e.g., homology model or snapshot from MD simulations) in PDB format. This brings into consideration the first assumption implicit in the ANM analysis: The coordinates in the PDB file are accepted to be the equilibrium coordinates without the need to perform further energy minimization. This is in contrast to classical NMA which requires the energy minimization of the PDB structure or any conformer from simulations prior to performing NMA. The most widely used ANM adaptations use α-carbon coordinates as the node positions, corresponding to a single-site-per-residue description. Other variants use β-carbon coordinates or the lower resolution models with one-site-per-m residues, where m may be as sparse as m = 40 (54). Such lower resolution models have been successfully used for representing supramolecular systems such as viral capsids and the ribosome. Such larger systems have been reviewed previously (26, 55).
Assessment of the topology of contacts
The topology of contacts here refers to the list of amino acid pairs, or nodes, that are “connected” by an elastic spring in the network. By definition, all pairs bonded or nonbonded within a certain cutoff distance Rc are assumed to be connected. Previous analyses demonstrated that Rc = 12–15 Å provides a good description of the native contact topology. Thus, the first task is to determine all node pairs within this distance range. This is readily accomplished by taking the difference
| (1) |
between all pairs of nodes i and j (or all pairs of α-carbons) (Fig. 2a). The pairs of amino acids whose α-carbons are separated by a distance smaller than Rc are thus assumed to interact via a harmonic potential that stabilizes their equilibrium distance At the end of this step we know all pairs of nodes that are connected.
Construction of the Hessian matrix
The Hessian matrix, H (Fig. 4), is the only ingredient that the model needs. H provides a complete description of the topology of contacts in the original structure. How do we construct H? In classical NMA, H is by definition a 3 N × 3 N matrix for a system of N interaction sites, composed of the second derivatives of the total potential energy with respect to the 3 N degrees of freedom (usually the x-, y-, z- components of the position vectors Ri for all atoms).
Eigenvalue decomposition of H
Eigenvalue decomposition of H yields 3 N − 6 nonzero normal modes, the frequencies and shapes of which are defined by the nonzero eigenvalues and corresponding eigenvectors of H. The eigenvalue decomposition may be written as
| (2) |
Here Λ is the diagonal matrix of the eigenvalues. Λ is composed of six zero eigenvalues (due to translational and rotational invariance of H) and 3 N − 6 nonzero eigenvalues{λ1, λ2, …, λ3 N−6} organized in ascending order (i.e., λ1 ≤ λ2 ≤ …. λ3 N−6). U is a 3 N × 3 N matrix composed of 3 N-dimensional eigenvectors written as columns u1, u2, …, u3 N−6 (preceded by the six eigenvectors corresponding to the zero eigenvalues).
What is the physical meaning of eigenvectors and eigenvalues? Each eigenvector represents a direction of motion in the 3 N-dimensional conformational space. It may also be viewed as a supervector, the super-elements of which are each three-dimensional vectors corresponding to the displacements of individual nodes, listed in sequential order along the protein sequence. The kth eigenvector, for example, is composed of N blocks (each being a three-dimensional vector) representing the relative displacement of the N α-carbons along the kth mode. The kth eigenvalue, on the other hand, provides a measure of the frequency, and size, of the motion as it scales with the square frequency of the kth mode and the size of the fluctuations are inversely proportional to the frequency. Note that the eigenvectors provide information on the relative movements of residues, not their absolute size, since each eigenvector is normalized (sum of elements squared is equal to zero). The size of the motion scales with the reciprocal square root of eigenvalues, as will be further clarified below.
Eigenvalue decomposition is the most time-consuming step of ANM. A suitable subroutine, e.g., BLZPACK (http://crd.lbl.gov/~osni/marques.html#BLZPACK) which is an implementation of the block Lanczos algorithm to solve large, sparse matrices, is used for this purpose. For molecules with 300 residues represented by a 900 × 900 Hessian matrix, the solution is obtained within 2 s. For proteins with thousands of residues it is obtained within a few minutes.
Mode shapes
The kth mode shape, also called kth mode profile, refers to the relative square displacements of residues, [(ΔRi)2]k = [(ΔXi)2]k + [(Δϒi)2]k + [(ΔZi)2]k along mode k as a function of residue index 1 ≤ i ≤ N. This profile is readily evaluated from the trace (sum of the diagonal elements) of each diagonal super element of the matrix [uk ukT]. The mode shape is normalized, i.e., the terms plotted as a function of residue index i sum up to unity. Therefore, the kth mode shape represents the distribution of residue motions driven by mode k.
Global modes, hinge sites, and recognition sites
Global motions usually involve large portions of the molecule, i.e., they are highly cooperative, and have been shown in numerous applications to be relevant to biological function (16). The slowest mode shape indeed provides valuable information on the global dynamics of the molecule. Minima refer to sites that serve as hinge sites, or anchors: these sites play a key role in mediating the collective dynamics of the entire molecule. Not surprisingly, residues located at such sites are usually conserved. Maxima in the global mode shapes, on the other hand, usually refer to substrate recognition sites. Their intrinsic mobility facilitates optimal recognition and binding of substrates. Both, hinge regions as well as substrate recognition sites are in general critical for biological function. For example, the highly conserved shallow pockets that serve as receptor-binding sites in influenza virus hemagglutinin A, or the antigen binding hypervariable loops of immunoglobulins, form maxima in the slowest mode shape, whereas linkers or interfacial regions between domains subject to anticorrelated motions form minima (15, 28, 55, 56) It is important to note that the global modes are insensitive to the details of the models and energy parameters used in normal mode analyses; they are essentially defined by the distribution of inter-residue contacts, which is rigorously accounted for in the ANM.
High frequency modes
In contrast to global modes, the high frequency modes are highly localized, and as such they are sensitive to the detailed interactions at the atomic level. They usually contain white noise contributions that need to be filtered out in order to extract physically meaningful information. Not surprisingly, these modes have been referred to as “uninteresting modes” (31) when extracted from MD trajectories. They usually drive isolated fluctuations, as opposed to the correlated ones that underlie the intramolecular communication.
The ANM results differ from those extracted from MD simulations or from full atomic NMA, in that they are devoid of random noise effects; and they are uniquely determined for the given topology of native contacts. The high frequency modes identified by the GNM, in particular, proved to be “interesting”: they indicate the most strongly constrained sites in the presence of the intricate coupling between all residues. The peaks emerging in these mode shapes are usually implicated in folding nuclei, or key tertiary contacts stabilizing the overall fold. As a consequence, they are evolutionarily conserved among different members of a given family (57, 58).
Application of ANM to Rhodopsin
To illustrate the application of ANM to membrane proteins, we describe here its application to rhodopsin, the vertebrate dim-light photoreceptor. Rhodopsin is the prototypic member of the largest known superfamily of cell surface membrane receptors, the G-protein coupled receptors (GPCRs) (59). These receptors perform diverse functions including responses to light, odorant molecules, neurotransmitters, hormones and a variety of other signals. All GPCRs contain a bundle of seven transmembrane (TM) helices (H1–H7) (60). In rhodopsin, this TM bundle contains the chromophore, 11-cis-retinal, covalently bound to the ε-amino group of Lys296 in H7. The capture of a photon by rhodopsin isomerizes 11-cis-retinal into all-trans-retinal. The structural perturbation in the TM helical bundle results in formation of a series of distinct photointermediates that ultimately lead to tertiary structure changes in the cytoplasmic (CP) domain that are the hallmark of the active state of rhodopsin, Meta II (61, 62). Meta II binds the heterotrimeric G-protein, transducin, which is activated via exchange of GDP to GTP (63).
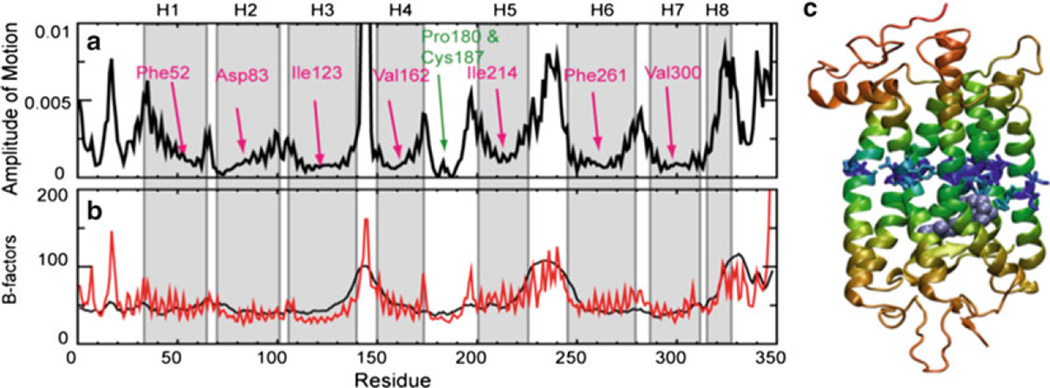
The aim of the protocol described in this chapter is to integrate experimental and computational data to better understand the structural changes that lead to the Meta II state. The global mode profile generated by the ANM for rhodopsin is presented in Fig. 5a. Panel b compares the theoretically predicted (red, ANM) and experimentally observed (X-ray crystallographic; black) B-factors Bi = (8π2 / 3) < (ΔRi)2 >. Note that the relative displacements of the different structural elements become more distinctive upon extraction of the global mode (panel a). Figure 5c maps the latter information onto a color-coded ribbon diagram. Minima in the mode shape curve are colored blue, and maxima, orange. Seven minima are identified and their centers are labeled in Fig. 5. The ribbon diagram reveals that they all occupy a position in the middle of the TM helices, about halfway between the extracellular (EC) and CP sides. As such, the loci of minima serve as a hinge plane about which the two halves of the molecule undergo anticorrelated motions.
Fig. 5.
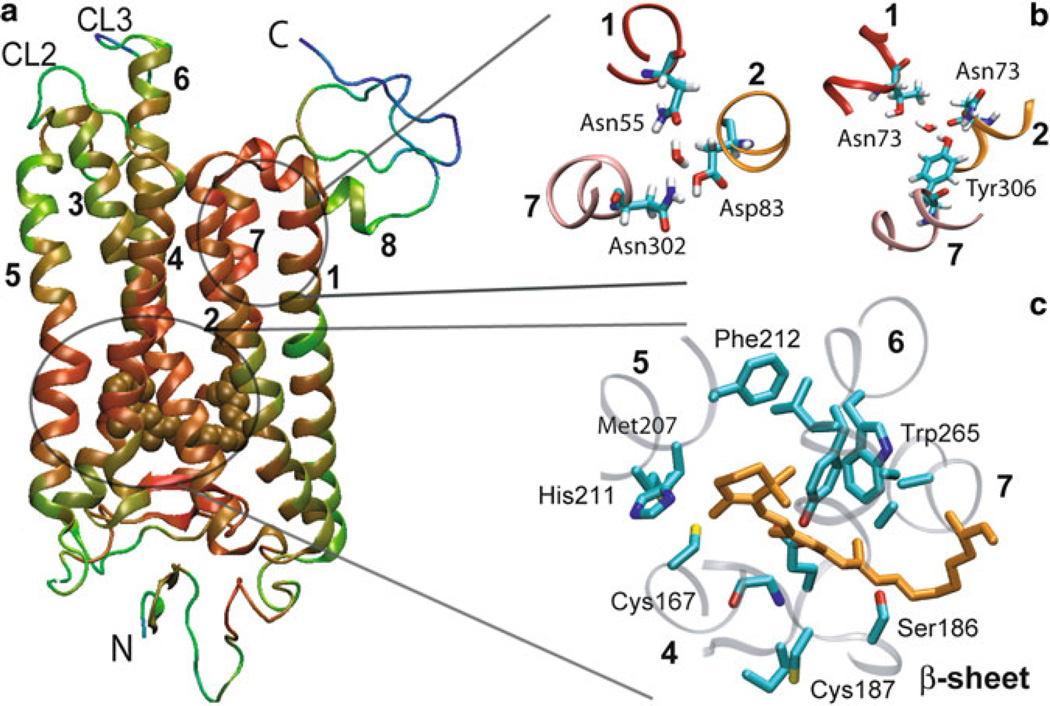
Rhodopsin motions predicted by the ANM. (a) The distribution of square displacements of residues along ANM mode 1. The non-TM regions exhibit higher mobilities in general, especially CL2 (between H3 and H4) and CL3 (between H5 and H6). Residues acting as hinge centers (minima) are labeled. Most lie at the TM helices centers, except for two additional minima: Pro180 and Cys187 near the EC entrance to the chromophore binding pocket. (b) Experimental (black) and predicted B-factors from the ANM (dashed blue). (c) Ribbon diagram of rhodopsin color-coded according to the relative motions in panel (a) in the order of increasing mobility blue (lowest mobility), cyan, green, yellow, orange (highest mobility). Side chains are shown for the seven hinge residues labeled in panel (a) and 11-cis-retinal is shown in light blue spacefilling representation. The hinge site divides the protein into two anti-correlated regions, one on the CP side and the other containing the chromophore binding pocket on the EC side. The image was generated using VMD (110). More details are described in (49).
Largest amplitude motions are predicted for the loop regions between the TM helices and the CP loops tend to undergo larger size motions than the EC loops (Fig. 5a). The EC loop 2 between H4 and H5 is much less mobile than the shorter CP loops 2 and 3 between H3 and H4, and H5 and H6, respectively. This suggests there is a possibly larger conformational motion upon retinal isomerization on the CP side than the EC side.
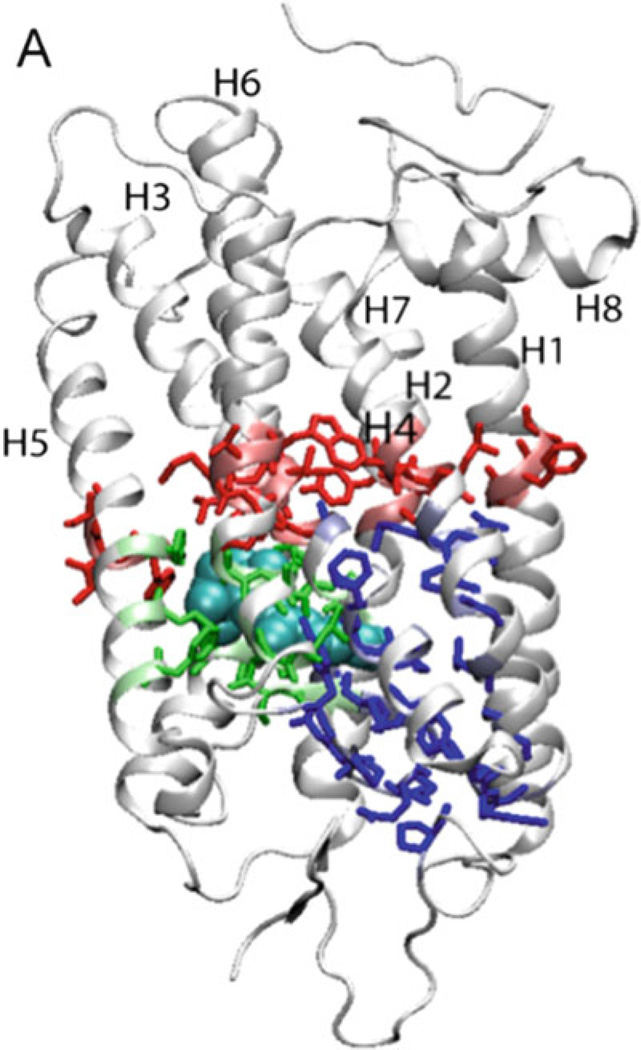
The ANM analysis of rhodopsin led to the identification of three groups of residues of interest overall summing to 61 residues implicated in functional dynamics (49) : (1) those participating in the global hinge region (colored red in Fig. 6), (2) those directly affected by retinal isomerization (blue), and (3) those emerging as peaks in ANM fast modes, i.e., distinguished in the high frequency modes (green). The complete list of residues in the three categories is given in Table 1 of our previous work (49).
Fig. 6.
Residues identified by ANM to play a key role in the activation of rhodopsin. Three sets of residues are highlighted: global hinge sites in red, amino acids affected by retinal isomerization in blue, and peaks in high frequency modes in green (Adapted from Isin et al., 2006) (49).
3.1.3. Modeling II: ANM-Guided MD
ANM-Guided MD Protocol
The intrinsic dynamics of proteins has been shown in many applications to be dominated by the protein architecture itself (15, 64, 65) and the same property appears to hold for many membrane proteins as well (8). On the other hand, the interactions with the environment such as the lipid bilayer, water molecules, ions and other ligands may also play a role in defining the detailed mechanisms of structural changes in membrane proteins. We developed a method referred to as ANM-restrained-MD, or ANM-guided MD, for explicitly taking into account the specificities of residues and their interactions with the environment. We applied the methodology to rhodopsin (29). The idea is to use iteratively the deformations derived from ANM analysis as restraints in MD trajectories. This permits us to guide the MD trajectories so as to be able to sample the cooperative motions that are otherwise beyond reach.
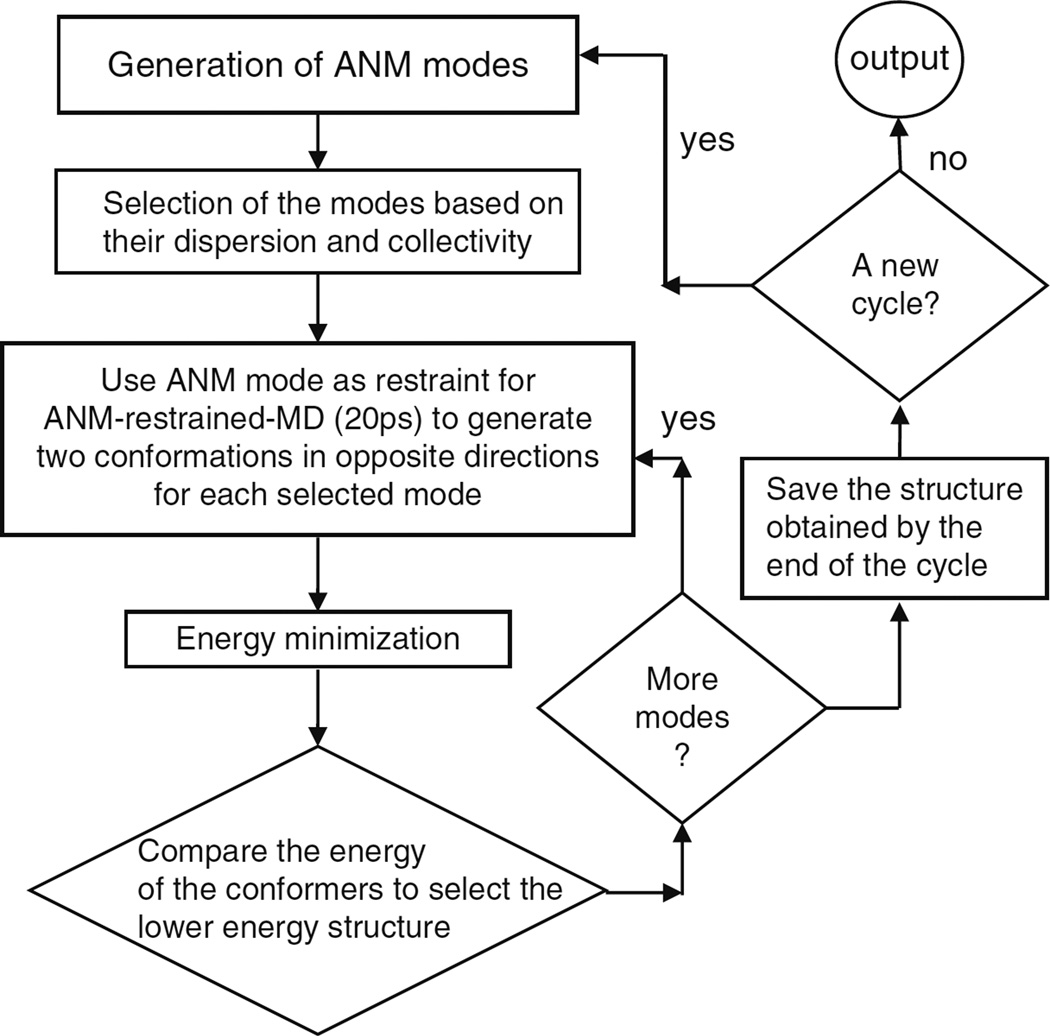
In ANM-guided-MD, an ensemble of ANM modes is used in an iterative scheme, as depicted in Fig. 7. Essentially, the algorithm consists of two loops: The first (inner) loop generates a succession of conformations using ANM modes as harmonic restraints in MD runs, succeeded in each case by a short energy minimization algorithm to allow the molecule to settle in a local energy minimum (The details of mode selection criteria are given in Notes 2). To this aim, we select from a pool of low frequency ANM modes. Then, for each mode we define two target conformations and we run short MD simulations (20 ps) in the presence of harmonic restraints that favor these target structures. Since the restraints may lead to unrealistic strains in the structure, we perform a short energy minimization succeeding each run and choose from the two alternative structures the one that is energetically favored. After a sufficient number of iterations by screening all modes selected by collectivity and eigenvalue dispersion criteria (detailed in the following sections) a new cycle (outer loop) is initiated with the updated ANM modes corresponding to the structure reached by the end of the first cycle, and this procedure is terminated after a certain number of cycles when the targeted conformation with experimentally identified residue interactions or the final acceptable root-mean-square deviation (RMSD) from the target is reached. The underlying assumption in this protocol is that ANM-derived restraints drive the excursion of the molecule toward a direction that would otherwise be naturally selected at much longer times.
Fig. 7.
Schematic description of the ANM-guided MD methodology.
We developed a fully automated Tcl code for the implementation of ANM-restrained-MD protocol in the NAMD software. The following sections provide the details on the individual steps of the protocol.
Selection of distinctive and cooperative modes
The lowest frequency modes of ANM are chosen to drive the MD simulation at each cycle as they represent the functional motions. Two criteria were considered: mode frequency dispersion (or eigenvalue distribution) and degree of collectivity to further determine the most relevant lowest frequency modes. The details of these criteria are given in the Notes section see Note 2.
Preparing the target conformations
Since each mode corresponds to a fluctuation between two oppositely directed motions, both directions being equally probable, two sets of deformations are considered for each mode, referred to as “plus” or “minus” displacements along the particular mode axis. The corresponding “target” conformation at a given cycle is required to have an RMSD in Cα-atom positions lower than 1.5 Å compared to the starting conformer, if the reconfiguration is performed along mode 1. The allowed upper RMSD becomes 1.5(λi/λ1)1/2 Å for mode i, consistent with the relative size (λi/λ1)1/2 of mode i with respect to mode 1.
Application of harmonic restraints along the ANM modes in MD
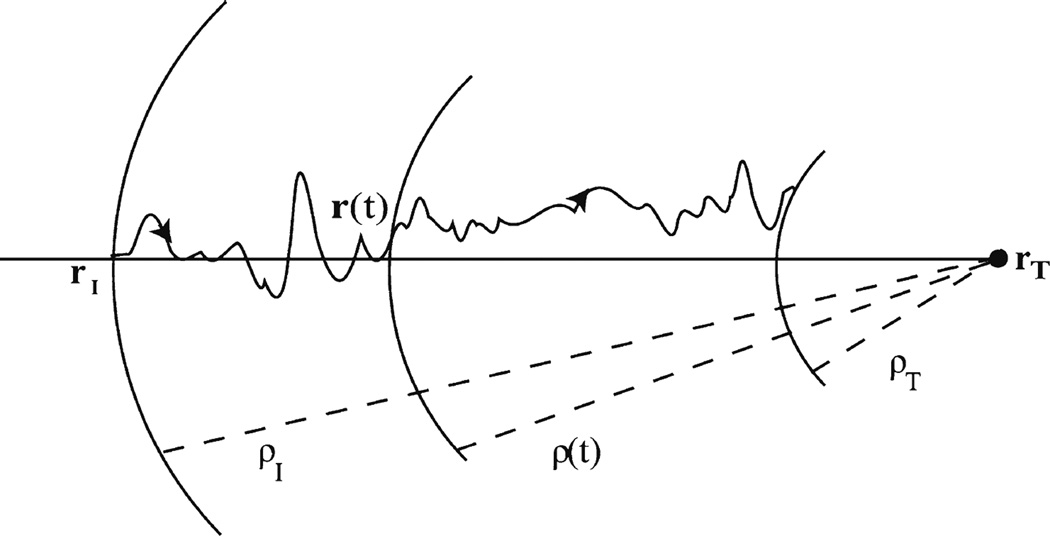
Each Cα-atom is harmonically restrained to approach the target conformations for 20 ps. Figure 8 schematically describes the application of harmonic restraints in MD simulations. The instantaneous distance ρ(t) of each configuration from the target configuration can be written as:
| (3) |
where r(t) designates the 3 N-dimensional instantaneous conformation vector of the biomolecule, rT is its target conformation; ri(t) is the 3 × 1 instantaneous position vector of the atom i at time t; and rTi is its target conformation.
Fig. 8.
Applying harmonic restraints along the ANM modes in MD. Two dimensional schematic representation of a pathway that the molecule follows while applying harmonic restraints along the ANM modes in MD. The distance r(t) from the target structure is gradually decreased during the course of simulation. Dashed lines show the distances of the molecule from the target structure at the initial conformation (rI), at time t(r (t)), and at the final (rT) conformation.
The implementation of the restraints is achieved by the following steps:
Set the distance ρI between the initial and target conformations as ρI = |rI − rT|
Solve the equation of motion containing the additional force for the restrains, by assigning initial coordinates using the initial conformation and an appropriate set of initial velocities.
At each succeeding timestep, decrease the distance by where ts is the simulation time and ρT is the distance of the conformation from the target structure at the end of the simulation, which should be as small as possible.
Energy minimization
After reaching the two target structures for a given mode, both are subjected to 1,000 steps of energy minimization using the steepest descent algorithm to relieve possible unrealistic distortions and to select the lower energy conformer among the two.
Application of ANM-Guided MD to Rhodopsin
ANM-guided MD was applied to rhodopsin to obtain atomic detail on the simulation (29). The results helped confirm and further refine the precise location of the previously identified hinge site (highlighted in red Fig. 6a). We observed that the hinge residues at H1, H2, and H7 are relatively closer to the CP region where two water molecules are found to further stabilize the hinge site, details that are not possible to obtain with ANM alone. Inclusion of explicit water in our model thus contributed to refine the precise location of the global hinge region, as well as identify a hydrogen bond network consolidated by water molecules. Several new interactions were also observed to contribute to the mechanism of signal propagation from the retinal-binding pocket to the G-protein binding sites in the CP domain (29).
Figure 9a displays the rhodopsin ribbon diagram color-coded by the RMSDs observed by the end of the ANM-guided-MD simulations, from red (least mobile) to blue (most mobile). Except for Pro23, all residues that exhibit minimal displacements (colored red in Fig. 9a) are clustered in two regions, the chromophore binding pocket and the CP end of H1, H2, and H7, which we shortly refer to as chromophore-binding and CP sites. These two sites are enclosed by ellipses and displayed from different perspectives in the respective panels b and c. Below we present more details on these two sites.
Fig. 9.
Rhodopsin dynamics and key interactions near all-trans-retinal obtained by ANM-guided MD. (a) Ribbon diagram of the structure color-coded by the mobilities observed in simulations (red, most constrained; green/blue, most mobile). Non-TM regions exhibit higher mobilities in general, especially CL2 (between H3 and H4) and CL3 (between H5 and H6). The all-trans retinal is colored brown. Residues that exhibit the lowest RMSDs are clustered in two regions: around the chromophore, and in the CP portion of helices 1, 2, and 7. The CP ends of H3, H4, H5, and H6, including the loops CP2 and CP3, exhibit high RMSDs. (b) CP end of H1, H2 and H7, viewed from the CP region. Two water molecules are found be connected to highly conserved residues throughout the simulations. The first forms hydrogen bonds with D55 (H1), N302 (H7) and D83 (H2) (left), and the second forms hydrogen bonds with T62 (H1), N73 (H2) and Y306 (H7) (right). (c) Hinge residues in the vicinity of the chromophore viewed from the CP regions. These are residues distinguished by their high stability (low mobility). They include Cys167 on H4, Ser186 and Cys187 on the β-sheet, Met207, His211, and Phe212 on H5, and Trp265 on H6. All-trans-retinal is shown in orange (Adapted from Isin et al., 2008).
CP ends of H1, H2 and H7
There exists an extensive interhelical hydrogen-bond network between H1, H2 and H7. This network includes highly conserved residues in the GPCR family including the interhelical N–D pair (Asn55 on H1, Asp83 on H2) and the NPXXY motif between Asn302 and Tyr306 on H7. We have found that two residues belonging to the NPXXY motif on H7, Asn302, and Tyr306, are connected to H1 and H2 through water molecules located in the cavity between helices H1, H2 and H7. Notably, in the resulting conformation, Asn55 (H1), Asp83 (H2), and Asn302 (H7) are hydrogen bonded to a central water molecule (Fig. 9b); a second water molecule interacts closely with Thr62 (H1), Asn73 (H2), and Tyr306 (H7) (Fig. 9b). Overall ~ 20 water molecules interacting with H1, H2, H3, H4, and H7 residues were observed in our simulations to span the helical bundle from the EC to the CP region and some exchanged neighbors during the simulations.
Chromophore Binding Pocket, and Rearrangements to Accommodate All-Trans-Retinal
The chromophore binding pocket is highly packed. Therefore, small conformational changes in the retinal are sufficient to significantly affect interactions within the binding site. We have observed that the isomerization of the retinal to all-trans state and the accompanying flip of the β-ionone ring cause steric clashes with the surrounding residues (49). These clashes are relieved upon rearrangements in the positions and orientations of the surrounding helices, which in turn induce a redistribution of contacts in the chromophore binding pocket. We note in particular that the number of atom–atom contacts between Trp265 and all-trans retinal (based on interatomic distance cutoff of 4.5 Å) is significantly lower than those made in the cis form (Fig. 9c). Furthermore, Phe261, Tyr268 and Ala269 on H6 make contacts with the β-ionone-ring to stabilize 11-cis-retinal in the dark state (not shown), while these contacts are lost during ANM-guided MD runs. Instead, new amino acids, Cys167 on H4 and two residues Phe203, Met207 and His211 on H5, and Thr118 on H3 line the chromophore binding pocket.
3.2. Methods II: Experimental Validation
3.2.1. Experimental Validation I. PCA of Multiple Structures
PCA Protocol
The PCA of structural ensembles permits us to visualize the major structural differences by identifying the dominant directions of conformational variations. PCA may be performed for an ensemble of PDB structures resolved for the same protein in the presence of different substrates, for NMR models obtained for the same protein (66, 67), or MD snapshots from essential dynamics analysis (EDA). The first principal component axis displays the greatest variance in the dataset, and followed by the second principal component axis, and so on. Briefly, PCA is achieved by designating for each conformer s in the ensemble of M conformers, the conformation vector
| (4) |
or the 3 N-dimensional fluctuation vector
| (5) |
that describes the departure in the position vectors of the N sites from their equilibrium position The equilibrium positions are identified by the average over all conformers (i.e., optimally superimposed PDB structures or snapshots from MD trajectories). The covariance matrix is expressed in terms of these fluctuations as
| (6) |
Comparison of ANM results with those disclosed by the PCA of experimental data thus reduces to the comparison of the covariances from the two approaches, or the comparison of the two sets of eigenvalues/eigenvectors that describe the dominant motions (ANM) or structural variations (PCA).
Application of PCA to Rhodopsin
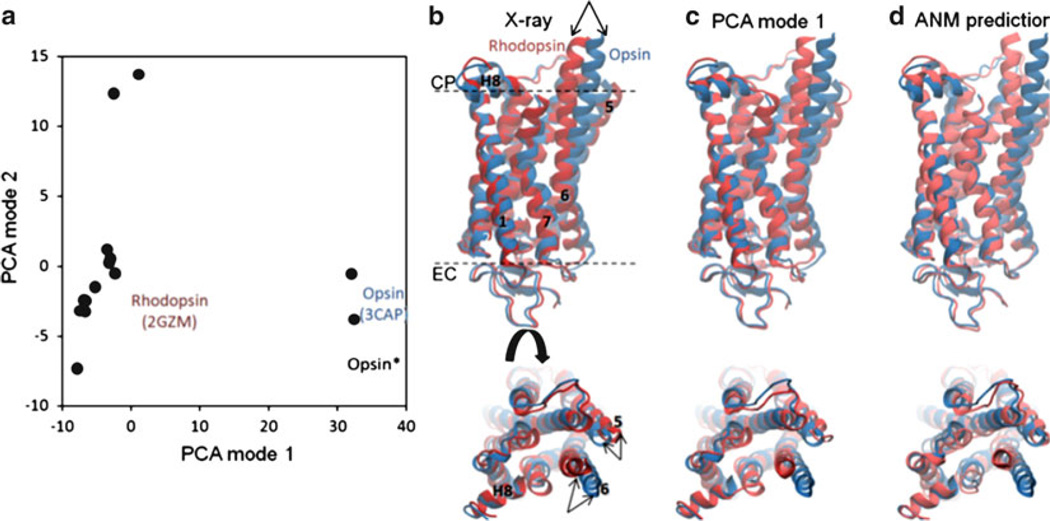
Recent application of this type of comparison to GPCR family proteins yielded the results presented in Fig. 10 (7, 8). In this study, we used all available rhodopsin structures in the ground and photoactivated states and two opsin structures (7, 8). In panel a, these 16 known structures that project onto the conformational subspace spanned by the two dominant PCA axes (corresponding to PCA modes 1 and 2) are displayed. These two modes account for 62% and 12% of the structural variability in the dataset (see Eq. 3.14). Comparison of panels c and d demonstrates that the dominant structural changes observed in experiments (PCA mode 1) for rhodopsin agree well with the conformational changes predicted by the ANM, in accord with the results obtained (50) for other proteins.
Fig. 10.
PCA and ANM calculations for rhodopsin. (a) Distribution of 16 X-ray structures in the conformation subspace spanned by the PCA modes 1 and 2. PCA1 differentiates the inactive and (putative) activated structures; PCA2 further differentiates between the structures in the cluster of inactive rhodopsins (b) Superimposition of experimentally determined rhodopsin and opsin structures, indicated by the labels on panel (a). (c) Rhodopsin structure generated by deforming the opsin structure along PCA1. (d) Rhodopsin conformation predicted by deforming the opsin structure along the 20 lowest frequency ANM modes. Calculations have been performed for Cα atoms; the remaining backbone atoms were reconstructed with BioPolymer module of Sybyl 8.3 (Tripos). ANM calculations were performed using a relatively short cutoff distance (Rc = 8 Å) so as to release interhelical constraints.
3.2.2. Experimental Validation II. Biophysical Probes
When direct structure determination is not feasible, introduction of biophysical probes can be a suitable alternative to probe the local environment at a particular site, especially when combined with ANM predictions. The predictions allow the design of specific experiments, where individual sites can be chosen with more insight. In the absence of any predictions, a large numbers of mutants need to be screened to identify the regions of conformational change. With the predictions, biophysical probes can be introduced site-specifically to probe for changes in the environment at sites where such changes have been predicted by ANM.
Biophysical Probes Protocols
A simple and very specific way to introduce biophysical probes is through cysteine mutagenesis followed by reaction with derivatization reagents that carry the functional group to be investigated with the goal of probing its environment. The free sulfhydryl group of the cysteine is amenable for chemical derivatization with different reagents which can then be characterized by different spectroscopic methods. For example, cysteine accessibility can be studied by 4,4′-dithiodipyridine (4-PDS) labeling. 4-PDS reacts with free sulfhydryl groups in cysteines to produce stoichiometric amounts of 4-thiopyridone that absorbs at 323 nm (68). The reaction rates and extents of a cysteine in a protein with 4-PDS can be used as a biophysical probe for the local environment of that cysteine. A step-by-step protocol for 4-PDS labeling of purified rhodopsin can be found in (69).
Another popular approach is application of EPR spectroscopy to a spin-labeled protein. This is a highly informative approach to study conformational changes in a protein. The native cysteines or the introduced cysteines in the proteins are typically derivatized with a nitroxide label. The most commonly used nitroxide label is methanethiosulfonate and the corresponding side chain derivatized is designated as R1 (70). EPR can be used to measure (1) dynamics of the side chain by the spectral line shape, (2) distance between the paramagnetic labels via dipolar interactions and (3) solvent accessibility of side chains via accessibility parameters. For studying relative domain movements, two R1 labels are suitably introduced in the proteins and the distance between the labels is measured under different conditions. A step-by-step protocol for EPR studies of rhodopsin can be found in (71).
Cysteine accessibility and EPR spectroscopy are only examples of the methods that can be used, others include fluorescence spectroscopy, NMR spectroscopy, and others.
Application of Biophysical Probes to Rhodopsin Identified by ANM and ANM-Guided MD Simulations
To understand the conformational changes accompanying the activation of rhodopsin and other GPCRs, extensive cysteine-scanning mutagenesis experiments were conducted in combination with site-directed spin labeling followed by EPR analysis of mobility, accessibility, spin-spin interactions, sulfhydryl reactivity, and disulfide cross-linking rates (62, 72–79). Upon isomerization, the mobility of spin-labeled side chains at the buried surfaces of H1, H2, H3, H6, and H7 were found to increase. Furthermore, experiments suggest that the CP ends of helices, especially H3, H5, and H6, need to be highly flexible to bind and activate the G-protein The increased accessibility of Val250 and Thr251 upon activation (72) and the changes in the spin-spin distances with respect to Val139Cys (77) were attributed to the movement of H6 away from the helical bundle such that its interaction with H3 and H7 was weakened. These observations were later confirmed by the recent crystal structures in the active forms of rhodopsin and other GPCRs such as β2- and ;1-adrenergic receptors (80–82). We have previously shown that the first two global modes of ANM drive the relative rearrangements of helices H3, H4, H5, and H6 (49) ; the ANM-guided MD simulations closely reproduce these modes at the CP ends of these helices. On the other hand, the CP ends of H1, H2, and H7 are presently found to closely maintain their dark state structure (Fig. 9a, b), consistent with the relatively smaller conformational changes observed experimentally in this region (73, 74, 79, 83–86).
Interaction and stabilization of conserved motifs through water molecule network in the transmembrane region
It has been noted that water molecules within the TM region could play critical roles in regulating the activity of GPCRs and the spectral sensitivity in visual pigments (87). One of the two water molecules that mediates the hydrogen bond network between the NPXXY motif on H7 and N–D pair on H1 and H2 is already in contact with these residues in the X-ray structure (87). Both water molecules remain at the same position despite the implementation of ten cycles of ANM-restraints in MD simulations. Later, the role of water molecules facilitating the hydrogen bond network within the TM region of another GPCR, β2-adrenergic receptor, was also shown by the MD simulations (88–90) and the recent crystal structures of this protein (80, 81).
Activation of rhodopsin by redistribution of residue contacts with its ligand
Cross-linking experiments by using photoactivatable analogs of 11-cis-retinal (91, 92) showed that in the dark state, 11-cis retinal cross-links to Trp265, while the all-trans-retinal cross-links to Ala169 instead of Trp265. Further investigation by high-resolution solid-state NMR measurements also showed that Trp126 and Trp265 interact more weakly with retinal in the active state (93). Additionally, the NMR data also determined that both the side chain of Glu122 and the backbone carbonyl of His211 are disrupted by the orientation of the β-ionone ring of all-trans-retinal in Meta II (93). The authors in (93) further proposed that the contact of the ionone ring with H5 near His211 moves H5 to an active state orientation.
These findings are consistent with the redistribution of contacts between the chromophore and rhodopsin we have observed during our simulations. We have found that the β-ionone ring of 11-cis-retinal in the dark state is almost parallel to the aromatic ring of Trp265 on H6. Furthermore, Phe261, Tyr268, and Ala269 on H6 make contacts with the β-ionone-ring to stabilize 11-cis-retinal in the dark state (not shown). These contacts are either lost or drastically reduced during ANM-guided MD simulations, and new contacts with other amino acids are made, including Cys167 on H4 and Phe203, Met207 and His211 on H5, and Thr118 on H3 (Fig. 9c). We note that these new interactions also provide insights about the activation mechanism of other GPCRs. Site-directed mutagenesis studies of the β2-adrenergic receptor have previously showed that Ser203 and Ser207 on H5 are critically important for binding catechol hydroxyl moieties from the aromatic ring of the agonists and hence activation of the protein (94).
3.2.3. Experimental Validation III: Perturbation of Motions
Perturbation of Motions Protocols
A convenient method to identify if motions at specific domains or regions of proteins play a functional role is to restrict these motions and measure the effect of this restriction on ligand binding or on activation. One way to achieve this is by suitably placing a pair of cysteines on the protein and to monitor the formation of a disulfide bond between them. Once a disulfide bond has formed, the relative motion between the two cysteines and the regions connecting them is restricted. The step-by-step protocol for measuring disulfide bond formation in rhodopsin can be found elsewhere (84). Site-directed mutagenesis is a technique to selectively introduce or delete specific amino acid(s) in protein by mutating bases in the corresponding nucleotide sequence. Many protocols have been prepared, and commercial kits are available.
Application of Perturbation of Motions to Rhodopsin
Disulfide bonds have been strategically introduced in rhodopsin to study their effects on G protein activation, Meta II decay and phosphorylation by rhodopsin kinase (95). Different effects on function were observed, depending on where the disulfide bonds were formed, although all regions were shown by biophysical probes to undergo conformational changes upon light activation. In particular, a disulfide tethering helix VIII to the first CP loop does not affect any of these functional characteristics, in contrast to a disulfide bond cross-linking the ends of helices III and VI, while a disulfide bond tethering the C-terminus to the CP loops actually enhanced G protein activation, while fully abolishing phosphorylation by rhodopsin kinase. These results show that some motions are dispensable for function.
3.2.4. Experimental Validation IV: Functional Consequences of Motions
Functional Consequences of Motions Protocols
Introducing mutations in proteins or restricting their motions through disulfide bond formation (described in Subheading 3.2.3., Perturbation of motions) can be used to investigate the significance of a particular motion for the function of a protein. The functional assay best suited to study different proteins depends on what the function of the protein is. In the case of GPCRs, binding and activation of the G protein is a hallmark of its function. For receptors other than GPCRs, other signaling proteins affected by receptor activation may be suitable. For receptors with enzymatic activity such as kinase activity, auto-phosphorylation may be the best assay. For other membrane proteins like ion channels yet other functional assays are available. Because the functional assay differs for each protein under study, we do not provide specific protocols here.
In addition to G protein activation, another common functional assay in receptors relate to binding of ligand. In rhodopsin, the retinal binding assayed by absorbance spectroscopy is the method of choice. Radioligand assays are more generally used to study other GPCRs. Retinal binding in rhodopsin is often used to not only assess ligand binding itself, but also as an indirect readout to study folding and misfolding, which is relevant to the retinal degeneration disease, Retinitis pigmentosa. Finally, loss of retinal binding after rhodopsin activation is also used as a functional assay, referred to as “Meta II decay.” The half-life of the increase in tryptophan fluorescence upon retinal loss is used as a measure of stability of the activated state.
Application of Functional Consequences of Motions to Rhodopsin Found by ANM and ANM-Guided MD Simulations
G protein activation
In order for the G protein to optimally bind and be activated by rhodopsin, the CP loops and ends of helices have to become more accessible to the aqueous environment and thus the G protein. The ANM predictions can thus be validated by surface accessibility measurements. Indeed, the CP ends of H3, H4, H5, and H6, and the connecting loops CL2 and CL3 at the CP region, are highly mobile as evident by their high RMS deviation values (Figs. 5 and 9), in agreement with spin labeling and cross-linking experiments (77, 78, 91, 92, 96). The surface accessibility of the G-protein contact site near the ERY motif is found to increase during the simulations.
Mutations associated with autosomal dominant retinitis pigmentosa
Two members of residues identified to take part in the global hinge site, Cys187 and Cys110, form a disulfide bridge, critical for folding and stability of rhodopsin (97). Mutation of both residues was also found to be associated with autosomal dominant retinitis pigmentosa (ADRP), a hereditary progressive blinding disease (98, 99). This is most probably caused by the destabilization of the opsin structure near the chromophore binding site in Meta II and dark state of rhodopsin. During ANM-guided MD simulations, the positions of Cys187 and Cys110 do not change and these residues have the lowest RMS deviation from the original structure.
In addition to the two regions indicated in Fig. 9a, another deep minimum is observed which contains only one residue, Pro23, near the soluble N-terminus at the extracellular region. It has been shown that Pro23His mutation results in severe misfolding of the entire protein, made irreversible by the formation of a wrong disulfide bond (100). This mutation has been associated with the most frequently occurring form of ADRP (101). The pathogenicity of human mutant Pro23His causing retinal degeneration was also confirmed by transgenic mice strain experiments (102, 103).
Meta II decay rates
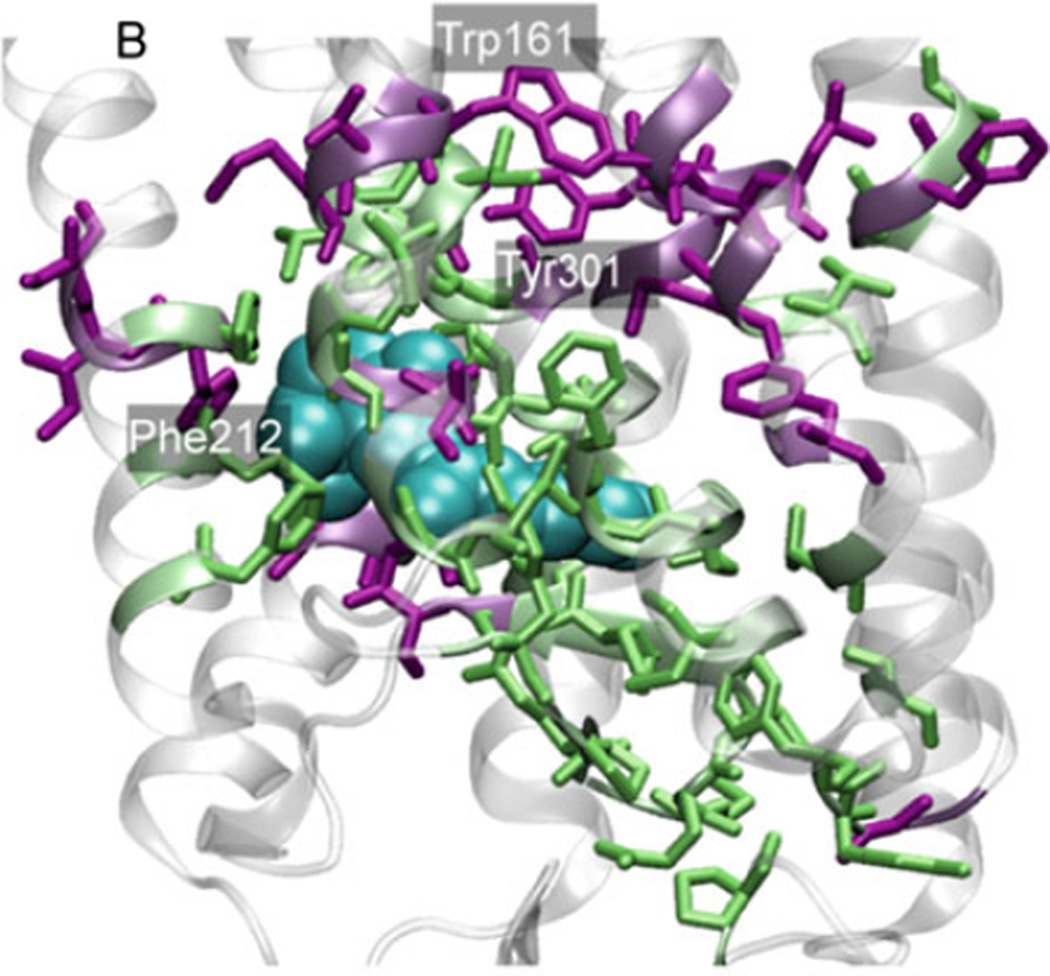
Meta II stability of rhodopsin mutants is characterized by quantifying Meta II decay rates (104). This method has been useful to estimate the role of a given amino acid in structure and function of the protein. We used our compiled comprehensive list for Meta II decay rates of rhodopsin mutants (49) to determine the effect of the hinge residues on Meta II stability. 97 of reported 228 amino acid replacements corresponded to unique positions in the rhodopsin sequence. In all cases, Meta II decay was determined using the fluorescence assay developed by Farrens and Khorana (104). ANM correctly predicts 93% of the experimentally observed effects in rhodopsin mutants for which the decay rates have been reported. The observed validation with experimental data, whenever available, strongly supports the use of GNM/ANM for guiding experiments. In particular we note among them a few aromatic residues, Tyr301, Trp161, and Phe212, which appear to play a central role in stabilizing the global hinge region (Fig. 11).
Fig. 11.
Comparison of ANM results with experimental data. Key residues predicted by the ANM (see Fig. 6, colored residues) and confirmed by Meta II and folding experiments to play a critical role are colored green, and the rest, purple. Both structures refer to the dark state conformation with 11-cis retinal shown in space-filled model colored cyan.
Acknowledgments
This work was in part supported by the National Science Foundation grant CCF-1144281, a CAREER grant CC044917 and by the National Institutes of Health Grant 5R01LM007994-06 (IB) and NIAID U01 AI070499 (ZNO).
Footnotes
Web site implementations of GNM and ANM
We have developed the following software and database servers for performing and retrieving the types of calculations described in the present chapter: ANM server (27) for ANM calculations; GNM server (105) and GNM database (106) for performing online GNM calculations and releasing precomputed data, respectively; PCA_NEST server for performing PCA of NMR structures (67), and ProDy software (107) for performing PCA of ensembles of structures and comparison with ANM predictions.
Choosing ANM modes for guiding MD simulations
References
- 1.Becker OM, MacKerell ADJ, Roux B, Wanatabe M. Computaional biochemistry and biophysics. New York: Marcel Dekker; 2001. [Google Scholar]
- 2.Cui Q, Bahar I. Theory and applications to biological and chemical systems. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2006. Normal mode analysis. [Google Scholar]
- 3.Leach AR. Molecular modelling: principles and applications. Upper Saddle River, NJ: Prentice Hall; 2001. [Google Scholar]
- 4.Schlick T. Molecular modeling and simulation: an interdisciplinary guide. New York: Springer; 2002. [Google Scholar]
- 5.Dror RO, Jensen MO, Borhani DW, Shaw DE. Exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. J Gen Physiol. 2010;135:555–562. doi: 10.1085/jgp.200910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu Rep Comput Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahar I. On the functional significance of soft modes predicted by coarse-grained models for membrane proteins. J Gen Physiol. 2010;135:563–573. doi: 10.1085/jgp.200910368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahar I, Lezon TR, Bakan A, Shrivastava IH. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem Rev. 2010;110:1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 12.Grossfield A, Feller SE, Pitman MC. Convergence of molecular dynamics simulations of membrane proteins. Proteins. 2007;67:31–40. doi: 10.1002/prot.21308. [DOI] [PubMed] [Google Scholar]
- 13.Brooks B, Karplus M. Normal modes for specific motions of macromolecules: application to the hinge-bending mode of lysozyme. Proc Natl Acad Sci U S A. 1985;82:4995–4999. doi: 10.1073/pnas.82.15.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berendsen HJ, Hayward S. Collective protein dynamics in relation to function. Curr Opin Struct Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 15.Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 18.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 19.Haliloglu T, Bahar I, Erman B. Gaussian dynamics of folded proteins. Phys Rev Lett. 1997;79:3090–3093. [Google Scholar]
- 20.Flory PJ. Statistical thermodynamics of random networks. Proc R Soc London A. 1976;351:351–380. [Google Scholar]
- 21.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doruker P, Atilgan AR, Bahar I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: application to alpha-amylase inhibitor. Proteins. 2000;40:512–524. [PubMed] [Google Scholar]
- 23.Isin B, Doruker P, Bahar I. Functional motions of influenza virus hemaglutinin: a structure-based analytical approach. Biophys J. 2002;82:569–581. doi: 10.1016/S0006-3495(02)75422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keskin O, Durrell SR, Bahar I, Jernigan RL, Covell DG. Relating molecular flexibility to function. A case study of tubulin. Biophys J. 2002;83:663–680. doi: 10.1016/S0006-3495(02)75199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Tama F, Brooks CL. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu Rev Biophys Biomol Struct. 2006;35:115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- 27.Eyal E, Yang LW, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22:2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 28.Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Isin B, Schulten K, Tajkhorshid E, Bahar I. Mechanism of signal propagation upon retinal isomerization: insights from molecular dynamics simulations of rhodopsin restrained by normal modes. Biophys J. 2008;95:789–803. doi: 10.1529/biophysj.107.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isralewitz B, Baudry J, Gullingsrud J, Kosztin D, Schulten K. Steered molecular dynamics investigations of protein function. J Mol Graph Model. 2001;19:13–25. doi: 10.1016/s1093-3263(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 31.Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 32.Amadei A, Linssen AB, de Groot BL, van Aalten DM, Berendsen HJ. An efficient method for sampling the essential subspace of proteins. J Biomol Struct Dyn. 1996;13:615–625. doi: 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- 33.de Groot BL, Amadei A, Scheek RM, van Nuland NA, Berendsen HJ. An extended sampling of the configurational space of HPr from E. coli. Proteins. 1996;26:314–322. doi: 10.1002/(SICI)1097-0134(199611)26:3<314::AID-PROT7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Abseher R, Nilges M. Efficient sampling in collective coordinate space. Proteins. 2000;39:82–88. doi: 10.1002/(sici)1097-0134(20000401)39:1<82::aid-prot9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Shi Y, Liu H. Molecular dynamics simulations of peptides and proteins with amplified collective motions. Biophys J. 2003;84:3583–3593. doi: 10.1016/S0006-3495(03)75090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Q, Murphy-Ullrich JE, Song YH. Structural insight into the role of thrombospondin-1 binding to calreticulin in calreticulin-induced focal adhesion disassembly. Biochemistry. 2010;49:3685–3694. doi: 10.1021/bi902067f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levitt M, Warshel A. Computer simulation of protein folding. Nature. 1975;253:694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 38.Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- 39.Sansom MS, Scott KA, Bond PJ. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem Soc Trans. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- 40.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 41.Bond PJ, Holyoake J, Ivetac A, Khalid S, Sansom MS. Coarse-grained molecular dynamics simulations of membrane proteins and peptides. J Struct Biol. 2007;157:593–605. doi: 10.1016/j.jsb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Psachoulia E, Fowler PW, Bond PJ, Sansom MS. Helix-helix interactions in membrane proteins: coarse-grained simulations of glycophorin a helix dimerization. Biochemistry. 2008;47:10503–10512. doi: 10.1021/bi800678t. [DOI] [PubMed] [Google Scholar]
- 43.Shih AY, Freddolino PL, Arkhipov A, Schulten K. Assembly of lipoprotein particles revealed by coarse-grained molecular dynamics simulations. J Struct Biol. 2007;157:579–592. doi: 10.1016/j.jsb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Shrivastava IH, Bahar I. Common mechanism of pore opening shared by five different potassium channels. Biophys J. 2006;90:3929–3940. doi: 10.1529/biophysj.105.080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valadie H, Lacapcre JJ, Sanejouand YH, Etchebest C. Dynamical properties of the MscL of Escherichia coli: a normal mode analysis. J Mol Biol. 2003;332:657–674. doi: 10.1016/s0022-2836(03)00851-9. [DOI] [PubMed] [Google Scholar]
- 46.Taly A, Delarue M, Grutter T, Nilges M, Le NN, Corringer PJ, Changeux JP. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys J. 2005;88:3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng J, Ma J, Fan K, Wang W. The conformational coupling and translocation mechanism of vitamin B12 ATP-binding cassette transporter BtuCD. Biophys J. 2008;94:612–621. doi: 10.1529/biophysj.107.110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rader AJ, Anderson G, Isin B, Khorana HG, Bahar I, Klein-Seetharaman J. Identification of core amino acids stabilizing rhodopsin. Proc Natl Acad Sci U S A. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isin B, Rader AJ, Dhiman HK, Klein-Seetharaman J, Bahar I. Predisposition of the dark state of rhodopsin to functional changes in structure. Proteins. 2006;65:970–983. doi: 10.1002/prot.21158. [DOI] [PubMed] [Google Scholar]
- 50.Bakan A, Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci U S A. 2009;106:14349–14354. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganapathiraju M, Jursa CJ, Karimi HA, Klein-Seetharaman J. TMpro web server and web service: transmembrane helix prediction through amino acid property analysis. Bioinformatics. 2007;23:2795–2796. doi: 10.1093/bioinformatics/btm398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanamala N, Tirupula KC, Klein-Seetharaman J. Preferential binding of allosteric modulators to active and inactive conformational states of metabotropic glutamate receptors. BMC Bioinformatics. 2008;9(Suppl 1):S16. doi: 10.1186/1471-2105-9-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doruker P, Jernigan RL, Bahar I. Dynamics of large proteins through hierarchical levels of coarse-grained structures. J Comput Chem. 2002;23:119–127. doi: 10.1002/jcc.1160. [DOI] [PubMed] [Google Scholar]
- 55.Chennubhotla C, Rader AJ, Yang LW, Bahar I. Elastic network models for understanding biomolecular machinery: from enzymes to supramolecular assemblies. Phys Biol. 2005;2:S173–S180. doi: 10.1088/1478-3975/2/4/S12. [DOI] [PubMed] [Google Scholar]
- 56.Ming D, Kong YF, Lambert MA, Huang Z, Ma JP. How to describe protein motion without amino acid sequence and atomic coordinates. Proc Natl Acad Sci U S A. 2002;99:8620–8625. doi: 10.1073/pnas.082148899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahar I, Atilgan AR, Demirel MC, Erman B. Vibrational dynamics of folded proteins: significance of slow and fast motions in relation to function and stability. Phys Rev Lett. 1998;80:2733–2736. [Google Scholar]
- 58.Demirel MC, Atilgan AR, Jernigan RL, Erman B, Bahar I. Identification of kinetically hot residues in proteins. Protein Sci. 1998;7:2522–2532. doi: 10.1002/pro.5560071205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: insights from recent structural studies. Annu Rev Biophys Biomol Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 60.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 61.Klein-Seetharaman J. Dynamics in rhodopsin. Chembiochem. 2002;3:981–986. doi: 10.1002/1439-7633(20021004)3:10<981::AID-CBIC981>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 62.Meng EC, Bourne HR. Receptor activation: what does the rhodopsin structure tell us? Trends Pharmacol Sci. 2001;22:587–593. doi: 10.1016/s0165-6147(00)01825-3. [DOI] [PubMed] [Google Scholar]
- 63.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 angstrom crystal structure of a heterotrimeric G protein46. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 64.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 65.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 66.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 67.Yang LW, Eyal E, Bahar I, Kitao A. Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics. Bioinformatics. 2009;25:606–614. doi: 10.1093/bioinformatics/btp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grassetti DR, Murray JF., Jr Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch Biochem Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 69.Dutta A, Tirupula KC, Alexiev U, Klein-Seetharaman J. Characterization of membrane protein non-native states 1. Extent of unfolding and aggregation of rhodopsin in the presence of chemical denaturants. Biochemistry. 2010;49:6317–6328. doi: 10.1021/bi100338e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 71.Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG. Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 72.Altenbach C, Yang K, Farrens DL, Farahbakhsh ZT, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: a site-directed spin-labeling study. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 73.Altenbach C, Klein-Seetharaman J, Hwa J, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 59–75 connecting helices I and II in rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7945–7949. doi: 10.1021/bi990014l. [DOI] [PubMed] [Google Scholar]
- 74.Altenbach C, Cai K, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 306–322 extending from helix VII to the palmitoylation sites in rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 75.Cai K, Langen R, Hubbell WL, Khorana HG. Structure and function in rhodopsin: topology of the C-terminal polypeptide chain in relation to the cytoplasmic loops. Proc Natl Acad Sci U S A. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farahbakhsh ZT, Ridge KD, Khorana HG, Hubbell WL. Mapping light-dependent structural changes in the cytoplasmic loop connecting helices C and D in rhodopsin: a site-directed spin labeling study. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 77.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 78.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 79.Klein-Seetharaman J, Hwa J, Cai K, Altenbach C, Hubbell WL, Khorana HG. Probing the dark state tertiary structure in the cytoplasmic domain of rhodopsin: proximities between amino acids deduced from spontaneous disulfide bond formation between Cys316 and engineered cysteines in cytoplasmic loop 1. Biochemistry. 2001;40:12472–12478. doi: 10.1021/bi010746p. [DOI] [PubMed] [Google Scholar]
- 80.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 81.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altenbach C, Cai K, Klein-Seetharaman J, Khorana HG, Hubbell WL. Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 65 in helix TM1 and residues in the sequence 306–319 at the cytoplasmic end of helix TM7 and in helix H8. Biochemistry. 2001;40:15483–15492. doi: 10.1021/bi011546g. [DOI] [PubMed] [Google Scholar]
- 84.Cai K, Klein-Seetharaman J, Farrens D, Zhang C, Altenbach C, Hubbell WL, Khorana HG. Single-cysteine substitution mutants at amino acid positions 306–321 in rhodopsin, the sequence between the cytoplasmic end of helix VII and the palmitoylation sites: sulfhydryl reactivity and transducin activation reveal a tertiary structure. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 85.Cai K, Klein-Seetharaman J, Altenbach C, Hubbell WL, Khorana HG. Probing the dark state tertiary structure in the cytoplasmic domain of rhodopsin: proximities between amino acids deduced from spontaneous disulfide bond formation between cysteine pairs engineered in cytoplasmic loops 1, 3, and 4. Biochemistry. 2001;40:12479–12485. doi: 10.1021/bi010747h. [DOI] [PubMed] [Google Scholar]
- 86.Klein-Seetharaman J, Hwa J, Cai K, Altenbach C, Hubbell WL, Khorana HG. Single-cysteine substitution mutants at amino acid positions 55–75, the sequence connecting the cytoplasmic ends of helices I and II in rhodopsin: reactivity of the sulfhydryl groups and their derivatives identifies a tertiary structure that changes upon light-activation. Biochemistry. 1999;38:7938–7944. doi: 10.1021/bi990013t. [DOI] [PubMed] [Google Scholar]
- 87.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X- ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han DS, Wang SX, Weinstein H. Active state-like conformational elements in the beta2-AR and a photoactivated intermediate of rhodopsin identified by dynamic properties of GPCRs. Biochemistry. 2008;47:7317–7321. doi: 10.1021/bi800442g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Romo TD, Grossfield A, Pitman MC. Concerted interconversion between ionic lock substrates of the beta(2) adrenergic receptor revealed by microsecond timescale molecular dynamics. Biophys J. 2010;98:76–84. doi: 10.1016/j.bpj.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Borhan B, Souto ML, Imai H, Shichida Y, Nakanishi K. Movement of retinal along the visual transduction path. Science. 2000;288:2209–2212. doi: 10.1126/science.288.5474.2209. [DOI] [PubMed] [Google Scholar]
- 92.Nakayama TA, Khorana HG. Orientation of retinal in bovine rhodopsin determined by cross-linking using a photoactivatable analog of 11-cis-retinal. J Biol Chem. 1990;265:15762–15769. [PubMed] [Google Scholar]
- 93.Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Coupling of retinal isomerization to the activation of rhodopsin. Proc Natl Acad Sci U S A. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RA. Identification of two serine residues involved in agonist activation of the beta-adrenergic receptor. J Biol Chem. 1989;264:13572–13578. [PubMed] [Google Scholar]
- 95.Cai K, Klein-Seetharaman J, Hwa J, Hubbell WL, Khorana HG. Structure and function in rhodopsin: effects of disulfide cross-links in the cytoplasmic face of rhodopsin on transducin activation and phosphorylation by rhodopsin kinase. Biochemistry. 1999;38:12893–12898. doi: 10.1021/bi9912443. [DOI] [PubMed] [Google Scholar]
- 96.Bourne HR. How receptors talk to trimeric G proteins. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 97.Hwa J, Reeves PJ, Klein-Seetharaman J, Davidson F, Khorana HG. Structure and function in rhodopsin: further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc Natl Acad Sci U S A. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Richards JE, Scott KM, Sieving PA. Disruption of conserved rhodopsin disulfide bond by Cys187Tyr mutation causes early and severe autosomal dominant retinitis pigmentosa. Ophthalmology. 1995;102:669–677. doi: 10.1016/s0161-6420(95)30972-4. [DOI] [PubMed] [Google Scholar]
- 99.Vaithinathan R, Berson EL, Dryja TP. Further screening of the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. Genomics. 1994;21:461–463. doi: 10.1006/geno.1994.1301. [DOI] [PubMed] [Google Scholar]
- 100.Hwa J, Klein-Seetharaman J, Khorana HG. Structure and function in rhodopsin: mass spectrometric Identification of the abnormal intradiscal disulfide bond in misfolded retinitis pigmentosa mutants. Proc Natl Acad Sci U S A. 2001;98:4872–4876. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berson EL. Retinitis pigmentosa. The Friedenwald lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- 102.Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 103.Wang M, Lam TT, Tso MO, Naash MI. Expression of a mutant opsin gene increases the susceptibility of the retina to light damage. Vis Neurosci. 1997;14:55–62. doi: 10.1017/s0952523800008750. [DOI] [PubMed] [Google Scholar]
- 104.Farrens DL, Khorana HG. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 105.Yang LW, Liu X, Jursa CJ, Holliman M, Rader AJ, Karimi HA, Bahar I. iGNM: a database of protein functional motions based on Gaussian Network Model. Bioinformatics. 2005;21:2978–2987. doi: 10.1093/bioinformatics/bti469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang LW, Rader AJ, Liu X, Jursa CJ, Chen SC, Karimi HA, Bahar I. oGNM: online computation of structural dynamics using the Gaussian Network Model. Nucleic Acids Res. 2006;34:W24–W31. doi: 10.1093/nar/gkl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bruschweiler R. Collective protein dynamics and nuclear-spin relaxation. J Chem Phys. 1995;102:3396–3403. [Google Scholar]
- 109.Tellez-Sanz R, Cesareo E, Nuccetelli M, Aguilera AM, Baron C, Parker LJ, Adams JJ, Morton CJ, Lo BM, Parker MW, Garcia-Fuentes L. Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1. Protein Sci. 2006;15:1093–1105. doi: 10.1110/ps.052055206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Biol. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]