Abstract
The immediate early gene c-fos has long been known as a molecular marker of neural activity. The neuron’s activity is transformed into intracellular calcium influx through NMDA receptors and L-type voltage sensitive calcium channels. For the transcription of c-fos, neural activity should be strong enough to activate mitogen-activated protein kinase (MAPK) signaling pathway which shows low calcium sensitivity. Upon translation, the auto-inhibition by Fos protein regulates basal Fos expression. The pattern of external stimuli and the valence of the stimulus to the animal change Fos signal, thus the signal reflects learning and memory aspects. Understanding the features of multiple components regulating Fos signaling is necessary for the optimal generation and interpretation of Fos signal.
Keywords: c-fos, Fos, neuron
INTRODUCTION
Extracellular stimuli evoke very rapid transcription of an array of immediate early genes such as c-fos, Egr1/Zif268, Arc/Arg3.1 and Homer1a (Herdegen & Leah, 1998). Among the immediate early genes, c-fos was discovered very early and has been studied in detail subsequently (Durchdewald et al., 2009). Egr1 is similar to c-fos as transcription factor, but basal expression level is high compared to c-fos and the time course of expression is different (Zangenehpour & Chaudhuri, 2002; Veyrac et al., 2014). Unlike c-fos and Egr1, Arc is not a transcription factor, but the mRNA and protein are localized in dendrites at sites of synaptic activity mediating synaptic plasticity ("effector" immediateearly gene) (Bramham et al., 2008). Homer1a is similar to Arc in localization and function, but slower in mRNA expression than Arc (Vazdarjanova et al., 2002; Hu et al., 2010).
In addition to the detection of mRNA and protein, recently other molecular markers or functional tools could be attached to the Fos gene to amplify the Fos signal detection or to provide other manipulations including optogenetic and chemogenetic stimulations (Kawashima et al., 2014). However, all of these tools still depend on the experimental condition of c-fos expression. Thus, the nature of Fos signal should be kept in mind. Fos has long been studied in detail and the literature is vast. The purpose of this brief review is to provide very basic properties of Fos signal as an activity marker. Further information can be found from excellent reviews on Fos and other immediate early genes (Lyons & West, 2011; Nestler, 2015).
Calcium influx
Many external stimuli can induce c-fos by increasing neuronal activity (Morgan & Curran, 1991; Herdegen & Leah, 1998). However, intracellular calcium is the common molecule that mediates synaptic activity to c-fos gene expression (Ghosh et al., 1994). Because of the diverse effects of calcium, basal neuronal intracellular calcium is kept very low by extrusion to the extracellular space and by uptake into calcium stores (Cohen & Greenberg, 2008). During neuronal activity, intracellular calcium concentration increases due to calcium influx mainly from two routes, a type of glutamate receptor (NMDA receptor, NMDAR) and L-type voltage-sensitive calcium channel (VSCC) (Chaudhuri et al., 2000).
NMDARs can be activated by coincident presynaptic glutamate release and postsynaptic depolarization. As a glutamate receptor-channel complex, the NMDAR is highly permeable to calcium (Lau & Zukin, 2007). The calcium influx through NMDARs can modulate c-fos gene expression (Cohen & Greenberg, 2008). In support of this notion, activation of NMDARs induces c-fos mRNA and Fos protein (Liste et al., 1995; Berretta et al., 1997; Parthasarathy & Graybiel, 1997). In contrast, NMDAR antagonists block Fos expression (Berretta et al., 1992; Ghosh et al., 1994; Berretta et al., 1997; Vanhoutte et al., 1999). In addition to NMDAR, glutamate can also activate fast-acting AMPA receptors and metabotropic glutamate receptors (mGluRs). AMPA receptors can also allow calcium influx especially during early development, but in mature neurons the calcium influx through AMPA receptors is limited in function (Derkach et al., 2007). Still, AMPARs indirectly can contribute to the calcium influx by membrane depolarization to activate NMDARs and VSCCs. Group I mGluRs can increase calcium influx, but the effect on Fos expression is not strong (Wang et al., 2007).
Voltage dependent calcium channels open with membrane depolarization allowing for calcium influx. The L-type VSCC is involved with immediate early gene expression (Rajadhyaksha et al., 1999). Basal expression of c-fos is rapidly suppressed by exposure to L-type VSCC antagonists and increased by a VSCC agonist (Murphy et al., 1991). As an alternative source of intracellular calcium, calcium can be released from calcium stores after neuronal activity, but more studies are needed to understand its contribution to Fos expression (Lyons & West, 2011).
Signaling pathway
After the local calcium increase around the pore of the channels, the specific functional consequences of calcium influx is determined by the signaling molecules physically associated with the NMDAR and VSCC (Cohen & Greenberg, 2008). The increased calcium can activate multiple kinase pathways including MAPK, CaMKIV, CaMKII and PKA. However, Fos expression is mainly mediated by MAPK pathway which is distinct from other pathways (Chaudhuri et al., 2000). Compared with CaMKIV pathway, MAPK pathway activation is relatively slow, requires strong external stimulation and higher calcium increase (Wu et al., 2001; Deisseroth & Tsien, 2002). This property can tune MAPK pathway to large and continued neural activity (Murphy et al., 2002; Deisseroth et al., 2003). As a result, strong external stimuli preferentially activate the MAPK pathway to result in Fos expression while weak stimuli are less likely to be translated into Fos signal.
The MAPK activates two transcription factors binding to the c-fos promoter in the nucleus (Lyons & West, 2011). First, CREB (cyclin AMP-response element-binding protein) is phosphorylated via ribosomal S6 kinase. Phosphorylated CREB binds to the calcium response element (or cyclic AMP response element) in the c-fos promotor. Second, phosphorylated Elk-1 in association with serum response factor (SRF) bind to the serum response element in the cfos promotor (Cruz et al., 2014). Fos protein translated in the cytoplasm moves back into nucleus. Inside the nucleus, Fos and Jun make up the AP-1 complex that acts as a transcription factor for "late" genes.
Use of Fos as an activity marker
At the basal level, low constitutive levels of c-fos is maintained. This is due to instability of c-fos mRNA and auto-repression of c-fos transcription by the Fos protein itself (Lucibello et al., 1989; Morgan & Curran, 1991). The other possibility is the presence of active repression of the promoter because transcription factors and the RNA polymerase II complex are bound already before neural activity (Carrion et al., 1999; Lyons & West, 2011).
These molecular conditions suggest the optimal conditions for eliciting the best Fos signal as a neural activity marker in vivo. Before an experiment, the animal's behavioral activity should be suppressed. For example, sensory deprivation can be given in the form of social isolation in a singlyhoused animals before stimulus presentation. The isolation duration can be 1 to 7 days. To maximize the signal, a novel stimulus is recommended. In contrast, the condition to avoid is a prolonged stimulation. It will produce negligible result or less than optimal signal.
In the same line of thinking, repetitive stimulation is expected to reduce the signal. But this issue becomes complex because learning can both sensitize and desensitize the neural circuit depending on the nature of the stimulus to the organism. For example, in negative stimulations such as social defeat from other animals, multiple attacks over days can make the individual animals either depressed or resilient (Wang et al., 2014). It is difficult to predict what the Fos signal will be like and how to interpret the signal at the level of single cells.
An interesting example was reported in the comparison of acute cocaine injection and a test injection after prior repeated cocaine injection in Fos-GFP transgenic mice (Koya et al., 2012). In this mouse, Fos-GFP fusion protein under c-fos promoter is expressed after strong neural activation (Barth et al., 2004). After repeated cocaine injection, test cocaine injection increased Fos signal as GFP positive (GFP+) cell count compared to acute cocaine injection after repeated saline. The increase of Fos signal may reflect increased neural circuit activity. If this is true, increased synaptic strength and increased synaptic transmissions are predicted. In GFP negative cells (GFP–), the result is consistent with this prediction. Synaptic strength was higher and excitatory postsynaptic current (EPSC) frequency was increased in the case of repeated cocaine injections. However, in GFP+ positive cells, synaptic strength was lower than GFP– cells and EPSC frequency was lower than GFP– cells. Because the number of GFP– cells are much more than GFP+ cells, in randomly selected cells the overall results reflected those of GFP– cells.
Though GFP+ cells are small in number, they are not less important than GFP– cells. GFP+ cells are responsible for the cocaine mediated behavioral effect. This is an example where Fos as neural activity marker represented overall activity correctly, but the prediction did not extend to a functionally important subset of neurons (ensemble). It is intriguing to speculate whether similar results will be found in negative behaviors such as social defeat.
Multiple social defeat (e.g. 10 episodes) can lead to depression-like behavior in defeated animals (Krishnan & Nestler, 2011). In general, Fos was decreased after repeated social defeat in multiple brain regions (Martinez et al., 2002). When the cue associated with social defeat was presented 1 day or 7 days later, the cue increased Fos signal in medial prefrontal cortex (Bourne et al., 2013). In this case, the Fos signal is used as an indicator of longterm memory. In another study c-fos mRNA in medial prefrontal cortex did not change, but Arc and Zif268 mRNA were decreased (Covington et al., 2010).
The physiological properties of the Fos positive cells in this negative valence is an interesting question. In learned helpless behavior training in Fos-GFP transgenic mice, Fos+ cells of helpless mice show increased evoked gluta matergic transmission compared with Fos– cells. In resilient mice, Fos+ cells show reduced glutamatergic transmission compared to Fos– cells (Wang et al., 2014). In this study, it was not tested if the Fos+ signal changed over repeated training sessions.
The change of one immediate early gene signal over the course of a repeated motor task was tested in Arc-GFP mice (Cao et al., 2015). Though not Fos, this study can give a clue on Fos signal change, too. In repeated rotarod learning over 3 days, overall GFP+ cell number and signal intensity decreased. At individual cell level, those cells with strong GFP expression on day 3 were those exhibiting strong expression previously on day 1. It is unknown why those cells are chosen as a persistent ones. Cellular recordings from that specific class of cells were not done in this study, although testing the unique properties of those persistent cells is technically possible (Cossell et al., 2015). It is a question whether c-fos will behave similarly to Arc, or if the pattern will be quite different.
Fos as a learning marker
Fos signal has been used as neural activity marker for a long time. However, the directional change of Fos signal as Fos+ cell number or intensity to repeated stimuli can be a cellular marker for learning or memory (Fig. 1). Repeated stimuli of positive value such as cocaine increase Fos signal over repetition. In contrast, repeated stimuli of negative value (e.g. social defeat) decrease Fos signal over time. It is a question what changes at the cell's property or synaptic connections. One way is to capture those Fos+ cells and compare the property Fos+ vs Fos– cells. If the captured cells can be identified and tested after variable time intervals (e.g. days, weeks or months), cellular basis of long term memory can be tested.
Fig. 1. Fos signal as a learning marker.
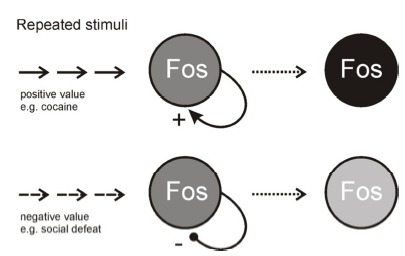
Repeated stimuli can increase or decrease Fos signal according to the positive or negative value of the stimuli to the organism. Capturing and studying of individual Fos positive cells before and after the stimuli provides an opportunity to study learning or memory for specific valences. Increased or decreased Fos signal of cell number or signal intensity was indicated as darkness or lightness, respectively.
Future questions
The utility of c-fos as a neural activity marker is still in progress with many, new useful tools to be employed. But there are questions to be addressed to use them optimally. First, in repeated stimuli presentation over multiple days, what does the Fos signal represent? How can we separate the Fos change itself from experimental main effect? Second, beyond the role of activity marker, what is the inherent role of Fos in behaving animals (Zhang et al., 2002; Durchdewald et al., 2009)? What is the role of "late" genes that Fos as a transcription factor serves to induce in the nervous system? Third, after neuronal activity, multiple immediate early genes are expressed including c-fos (Lyons & West, 2011). Why are they expressed so quickly? What is the overlapping or unique role of the "late" genes from multiple immediate early genes as transcription factors?
REFERENCES
- Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. Journal of Neuroscience. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S, Robertson HA, Graybiel AM. Dopamine and glutamate agonists stimulate neuron-specific expression of Fos-like protein in the striatum. Journal of Neurophysiology. 1992;68:767–777. doi: 10.1152/jn.1992.68.3.767. [DOI] [PubMed] [Google Scholar]
- Berretta S, Parthasarathy HB, Graybiel AM. Local release of GABAergic inhibition in the motor cortex induces immediate-early gene expression in indirect pathway neurons of the striatum. Journal of Neuroscience. 1997;17:4752–4763. doi: 10.1523/JNEUROSCI.17-12-04752.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne AR, Mohan G, Stone MF, Pham MQ, Schultz CR, Meyerhoff JL, Lumley LA. Olfactory cues increase avoidance behavior and induce Fos expression in the amygdala, hippocampus and prefrontal cortex of socially defeated mice. Behavioural Brain Research. 2013;256:188–196. doi: 10.1016/j.bbr.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. Journal of Neuroscience. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao VY, Ye Y, Mastwal S, Ren M, Coon M, Liu Q, Costa RM, Wang KH. Motor learning consolidates Arc-expressing neuronal ensembles in secondary motor cortex. Neuron. 2015; 6:1385–1392. doi: 10.1016/j.neuron.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A, Zangenehpour S, Rahbar-Dehgan F, Ye F. Molecular maps of neural activity and quiescence. Acta Neurobiologiae Experimentalis. 2000;60:403–410. doi: 10.55782/ane-2000-1359. [DOI] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossell L, Iacaruso MF, Muir DR, Houlton R, Sader EN, Ko H, Hofer SB, Mrsic-Flogel TD. Functional organization of excitatory synaptic strength in primary visual cortex. Nature. 2015;518:399–403. doi: 10.1038/nature14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, Lobo MK, 3rd, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, Neve RL, Deisseroth K, Nestler EJ. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. Journal of Neuroscience. 2010;30:16082–16090. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Javier Rubio F, Hope BT. Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. Brain research. 2014. http://www.dx.doi.org/10.1016/j.brainres.2014.11.005 . [DOI] [PMC free article] [PubMed]
- Deisseroth K, Tsien RW. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron. 2002;34:179–182. doi: 10.1016/s0896-6273(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Mermelstein PG, Xia H, Tsien RW. Signaling from synapse to nucleus: the logic behind the mechanisms. Current Opinion in Neurobiology. 2003;13:354–365. doi: 10.1016/s0959-4388(03)00076-x. [DOI] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nature Reviews Neuroscience. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- Durchdewald M, Angel P, Hess J. The transcription factor Fos: a Janus-type regulator in health and disease. Histology and Histopathology. 2009;24:1451–1461. doi: 10.14670/HH-24.1451. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Ginty DD, Bading H, Greenberg ME. Calcium regulation of gene expression in neuronal cells. Journal of Neurobiology. 1994;25:294–303. doi: 10.1002/neu.480250309. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Research Reviews. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Hu JH, Park JM, Park S, Xiao B, Dehoff MH, Kim S, Hayashi T, Schwarz MK, Huganir RL, Seeburg PH, Linden DJ, Worley PF. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 2010;68:1128–1142. doi: 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Okuno H, Bito H. A new era for functional labeling of neurons: activity-dependent promoters have come of age. Frontiers in Neural Circuits. 2014;8 doi: 10.3389/fncir.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Cruz FC, Ator R, Golden SA, Hoffman AF, Lupica CR, Hope BT. Silent synapses in selectively activated nucleus accumbens neurons following cocaine sensitization. Nature Neuroscience. 2012;15:1556–1562. doi: 10.1038/nn.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. Animal models of depression: molecular perspectives. Current Topics in Behavioral Neurosciences. 2011;7:121–147. doi: 10.1007/7854_2010_108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nature Reviews Neuroscience. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Liste I, Rozas G, Guerra MJ, Labandeira-Garcia JL. Cortical stimulation induces Fos expression in striatal neurons via NMDA glutamate and dopamine receptors. Brain Research. 1995;700:1–12. doi: 10.1016/0006-8993(95)00958-s. [DOI] [PubMed] [Google Scholar]
- Lucibello FC, Lowag C, Neuberg M, Muller R. Transrepression of the mouse c-fos promoter: a novel mechanism of Fos-mediated trans-regulation. Cell. 1989;59:999–1007. doi: 10.1016/0092-8674(89)90756-3. [DOI] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Progress in Neurobiology. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Calvo-Torrent A, Herbert J. Mapping brain response to social stress in rodents with c-fos expression: a review. Stress. 2002;5:3–13. doi: 10.1080/102538902900012369. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annual Review of Neuroscience. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nature Cell Biology. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- Murphy TH, Worley PF, Baraban JM. L-type voltagesensitive calcium channels mediate synaptic activation of immediate early genes. Neuron. 1991;7:625–635. doi: 10.1016/0896-6273(91)90375-a. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. FosB: a transcriptional regulator of stress and antidepressant responses. European Journal of Pharmacology. 2015;753:66–72. doi: 10.1016/j.ejphar.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy HB, Graybiel AM. Cortically driven immediate-early gene expression reflects modular influence of sensorimotor cortex on identified striatal neurons in the squirrel monkey. Journal of Neuroscience. 1997;17:2477–2491. doi: 10.1523/JNEUROSCI.17-07-02477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajadhyaksha A, Barczak A, Macias W, Leveque JC, Lewis SE, Konradi C. L-Type Ca(2+) channels are essential for glutamate-mediated CREB phosphorylation and c-fos gene expression in striatal neurons. Journal of Neuroscience. 1999;19:6348–6359. doi: 10.1523/JNEUROSCI.19-15-06348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoutte P, Barnier JV, Guibert B, Pages C, Besson MJ, Hipskind RA, Caboche J. Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinasedependent pathway in brain slices. Molecular and Cellular Biology. 1999;19:136–146. doi: 10.1128/mcb.19.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. Journal of Neuroscience. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrac A, Besnard A, Caboche J, Davis S, Laroche S. The transcription factor Zif268/Egr1, brain plasticity, and memory. Progress in Molecular Biology and Translational Science. 2014;122:89–129. doi: 10.1016/B978-0-12-420170-5.00004-0. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Fibuch EE, Mao L. Regulation of mitogenactivated protein kinases by glutamate receptors. Journal of Neurochemistry. 2007;100:1–11. doi: 10.1111/j.1471-4159.2006.04208.x. [DOI] [PubMed] [Google Scholar]
- Wang M, Perova Z, Arenkiel BR, Li B. Synaptic modifications in the medial prefrontal cortex in susceptibility and resilience to stress. Journal of Neuroscience. 2014;34:7485–7492. doi: 10.1523/JNEUROSCI.5294-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nature Neuroscience. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
- Zangenehpour S, Chaudhuri A. Differential induction and decay curves of c-fos and zif268 revealed through dual activity maps. Brain Research Molecular Brain Research. 2002;109:221–225. doi: 10.1016/s0169-328x(02)00556-9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang D, McQuade JS, Behbehani M, Tsien JZ, Xu M. c-fos regulates neuronal excitability and survival. Nature Genetics. 2002;30:416–420. doi: 10.1038/ng859. [DOI] [PubMed] [Google Scholar]


