Abstract
Pneumonia is the leading cause of death among children less than 5 years old worldwide. A wide range of viral, bacterial and fungal agents can cause pneumonia: although viruses are the most common etiologic agent, the severity of clinical symptoms associated with bacterial pneumonia and increasing antibiotic resistance makes bacterial pneumonia a major public health concern. Bacterial pneumonia can follow upper respiratory viral infection and complicate lower respiratory viral infection. Secondary bacterial pneumonia is a major cause of influenza-related deaths. In this review, we evaluate the following hypotheses: (i) respiratory viruses influence the etiology of pneumonia by altering bacterial community structure in the upper respiratory tract (URT) and (ii) respiratory viruses promote or inhibit colonization of the lower respiratory tract (LRT) by certain bacterial species residing in the URT. We conducted a systematic review of the literature to examine temporal associations between respiratory viruses and bacteria and a targeted review to identify potential mechanisms of interactions. We conclude that viruses both alter the bacterial community in the URT and promote bacterial colonization of the LRT. However, it is uncertain whether changes in the URT bacterial community play a substantial role in pneumonia etiology. The exception is Streptococcus pneumoniae where a strong link between viral co-infection, increased carriage and pneumococcal pneumonia has been established.
Keywords: co-infection, ecology, pneumonia, influenza, Streptococcus pneumoniae
INTRODUCTION
Pneumonia is the leading cause of death in children <5 years worldwide, which is responsible for one million deaths each year [1]. The burden is greatest in developing countries, at an estimated 0.22 episodes per child-year, but remains a major public health concern even among developed countries where there are an estimated 0.015 episodes per child-year [2]. In the USA, pneumonia is the second only to newborn infant births as the most common reason for hospital admissions (36 cases per 10 000 persons [3]) and causes nearly 50 000 deaths each year [4].
A wide range of viral, bacterial and fungal agents can cause pneumonia when aspirated into the lungs. The Centers for Disease Control and Prevention Etiology of Pneumonia in the Community (EPIC) study identified viruses as the most commonly identified etiologic agent in children and adults hospitalized with pneumonia. An etiologic agent was detected in 81% of 2222 children <18 years of age: 66% had one or more viral pathogens, 8% one or more bacterial pathogens and 7% both bacterial and viral pathogens [5]. Among 2259 adults, an etiologic agent was detected in 38%: 23% had one or more viral pathogens, 11% one or more bacterial pathogens and 3% both bacterial and viral pathogens [6]. However, virtually all these serious pneumonia cases were treated with antibiotics, as secondary bacterial infection can complicate lower respiratory viral infection. Therefore, even in cases determined to be pneumonia solely of viral etiology, bacterial interactions of virus and bacteria may play some role.
Explanatory Box 1. Challenges in determining the etiology of pneumonia
Even in countries where pneumonia surveillance is routinely conducted such as the USA, no information on microbial etiology is recorded for approximately 65–85% of hospitalized pneumonia cases [11, 12]. Severely ill patients often are not included in surveillance, organisms on the causal pathway may have been cleared by the time that the patient presents clinically or prior to testing because of rapid treatment with antibiotics when pneumonia is suspected, and autopsies are infrequently done on the elderly. To optimally determine etiology, direct sampling via bronchoalveolar lavage is required, but usually detection of causal agents is conducted on blood, sputum and urine because of ease of collection, ethical issues and costs. Bacteremia is observed in only 7%–13% of adult pneumonia cases and 1–5% in child pneumonia cases, sputum can potentially be contaminated by bacteria in the URT and is difficult to obtain from children, and blood and urine antigen assays require further validation or are limited to adults and specific to only a few pathogens (e.g. Streptococcus pneumoniae and Legionella species) (reviewed by Murdoch et al. [13]). Although modern molecular biologic techniques make it feasible to conduct untargeted screens for all bacterial, viral and fungal species present, it is still difficult to distinguish between infection, colonization or contamination [14]. Continued efforts are needed to develop more accurate methods to determine the etiology of pneumonia, and thus maximize treatment and prevention efforts.
The large proportion of pneumonia cases without a detected pathogen underscores the limitations of current surveillance and detection methods and how they frame our understanding of pneumonia etiology (Box 1). EPIC study results suggest we may not be detecting the full panel of pathogens in cases we currently define as viral pneumonia nor considering the potential role of bacteria on the pathogenic potential of viruses. Bacterial causes of pneumonia are associated with more severe clinical symptoms and increasing antibiotic resistance complicates treatment [2, 7–10], making bacterial causes of pneumonia a major concern.
In this review, we examine two hypotheses that argue the etiology of bacterial pneumonia is a consequence of ecologic selection influenced by the interaction of respiratory viruses and bacteria within the host: (i) respiratory viruses influence the etiology of pneumonia by altering bacterial carriage structure in the upper respiratory tract (URT) and (ii) respiratory viruses promote or inhibit colonization of the lower respiratory tract (LRT) by certain bacterial species residing in the URT. We begin by describing the normal processes of bacterial selection in the upper and LRTs and then present evidence on how these processes can potentially be altered by respiratory viruses.
METHODS
We conducted a systematic literature search in PubMed for studies published between 1 January 1990 and 9 December 2015. We restricted studies to those conducted in the USA to minimize potential geographic variation of associations. The following search string was used: ‘(bacteria[All Fields] OR bacterial[All Fields]) AND (virus[All Fields] OR viral[All Fields]) AND (lower respiratory tract infection[All Fields] OR LRTI[All Fields] OR lower respiratory tract[All Fields] OR LRT[All Fields] OR lower respiratory infection[All Fields] OR LRI[All Fields] OR pneumonia[All Fields] OR bronchitis[All Fields]) AND (“1990/01/1”[PDAT]: “2015/12/09”[PDAT]) AND United States[All Fields] AND (time[All Fields] OR temporal[All Fields] OR season*[All Fields])’. Among 464 articles written in English, exclusions were made based on titles, abstracts and full articles. We excluded reviews, in vivo and in vitro experiments, and studies of immunocompromised populations. Nine articles were retrieved from the literature search and three additional studies were selected from the reference list of retrieved articles.
BACTERIAL SELECTION IN THE URT
Bacterial pneumonia is primarily caused by the commensal bacteria normally residing in the URT [11, 12]. The most common causes of bacterial pneumonia for children <5 years of age are Streptococcus pneumoniae, followed by Haemophilus influenzae and Staphylococcus aureus [11] although this varies over time and space. From a rudimentary ecological perspective, the human respiratory tract can be defined as an ecosystem with two distinct niches: the URT, characterized by regular asymptomatic carriage of commensal bacteria, and the LRT, which is inhabited at a low abundance by bacteria in healthy individuals [13]. During the first year after birth, the nasopharynx is rapidly colonized [14] and URT carriage is established via ongoing synergistic and antagonistic interactions among commensal bacteria [15]. Although pneumonia is an infection of the lungs, microbial selection in the URT may play an important role in etiology as bacterial strains in the URT can be readily aspirated into the LRT. For example, URT carriage is believed to be a necessary precursor of pneumonia due to S. pneumoniae [16, 17].
Numerous epidemiologic studies describe synergistic and antagonistic relationships among various commensal bacteria [18–33] and, although the exact biological mechanisms remain unclear, in vivo and in vitro experiments suggest potential mechanisms involve either direct interaction between bacterial species or indirect interactions via the host immune system (Table 1). A number of population studies suggest that S. pneumoniae carriage is positively associated with H. influenzae [18–27] and Moraxella catarrhalis [18, 23–28] carriage but negatively associated with S. aureus [19, 20, 24–27, 29–32]. Furthermore, S. aureus carriage is generally negatively associated with H. influenzae and M. catarrhalis [19, 24, 31] carriage, whereas H. influenzae and M. catarrhalis are believed to be positively associated [18, 22, 24, 33]. Nevertheless, our understanding is limited, as the dynamics of niche competition likely consist of complex relationships between multiple species [31, 34] and strains [15, 35], further influenced by host and environmental factors [22, 36]. As carriage is an important precursor of respiratory infections for certain bacterial species [12], unraveling the complex system of bacterial interactions that determine URT microbiota may be key factor for understanding the etiology of pneumonia.
Table 1.
Known interactions and potential mechanisms for observed associations between primary bacterial colonizers of the nasopharynx
| Organism 1 | Organism 2 | Interactiona | Potential mechanismsb |
|---|---|---|---|
| S. pneumoniae | S. aureus | Antagonism [19, 20, 24–27, 29–32] | Hydrogen peroxide production [107] |
| Catalase [108] | |||
| Pilus [109] | |||
| Immune-mediated competition [110, 111] | |||
| S. pneumoniae | H. influenzae | Synergism [18–27, 112] | Provision of nutrients [15] |
| Production of β-lactamase [113] | |||
| Formation of biofilms [113] | |||
| Phosphorychlorine expression [12] | |||
| S. pneumoniae | H. influenzae | Antagonism [31] | Hydrogen peroxide production [114] |
| Catalase [114] | |||
| Desialylation [115] | |||
| Immune-mediated competition [15, 116, 117] | |||
| S. pneumoniae | M. catarrhalis | Synergism [18, 23–28] | Passive antibiotic protection [118, 119] |
| S. pneumoniae | M. catarrhalis | Antagonism | Hydrogen peroxide production [114] |
| S. aureus | H. influenzae | Synergism [22] | Provision of nutrients [15] |
| S. aureus | H. influenzae | Antagonism [19, 24, 31] | |
| S. aureus | M. catarrhalis | Antagonism [24] | |
| H. influenzae | M. catarrhalis | Synergism [18, 22, 24, 33] | Outer membrane vesicles [120] |
Epidemiologic studies.
In vitro and in vivo experiments
BACTERIAL SELECTION IN THE LRT
Lung microbiome studies suggest that bacteria colonizing the LRT overlap with those found in the URT, but that the abundance of organisms is quite low [13], and their role in pneumonia etiology has yet to be explored. To colonize the LRT, an organism must overcome mucociliary clearance and phagocytosis by resident alveolar macrophages, neutrophils and monocyte-derived macrophages [37, 38], but many URT pathogens have developed strategies to overcome these barriers. H. influenzae, Mycoplasma pneumoniae and Bordetella pertussis resist mucociliary clearance by impairing ciliary function. Streptococcus pyogenes, Streptococcus agalactiae, H. influenzae, Neisseria meningitidis and S. pneumoniae possess capsules that resist phagocytosis [37]. Streptococcus pneumoniae, the leading cause of pneumonia [39], is characterized by over 90 serotypes differentiated by variations in the bacterial polysaccharide capsule [40, 41] and associated with different propensities of invasive potential [42]. In addition to protecting against phagocytosis, the capsule prevents clearance by mucous secretion and restricts autolysis [43]. Other species, including S. aureus, release anti-opsonizing proteins and possess surface protein A to evade phagocytosis. Furthermore, S. aureus secretes leukotoxins that lyse leukocytes and express superantigens that hinder immune response (reviewed by Naber et al. [44]).
The crucial role these various mechanisms play in determining respiratory disease is demonstrated by contrasting M. catarrhalis with S. pneumoniae. Similar to S. pneumoniae, M. catarrhalis is a primary carriage species estimated to colonize between 31% and 50% of children <2 years in the USA [45] and frequently causes URT infections, such as acute otitis media. However, unlike S. pneumoniae, M. catarrhalis rarely causes pneumonia [46], suggesting that differences in mechanisms of pathogenicity may be the explanation.
RISK FACTORS OF BACTERIAL PNEUMONIA
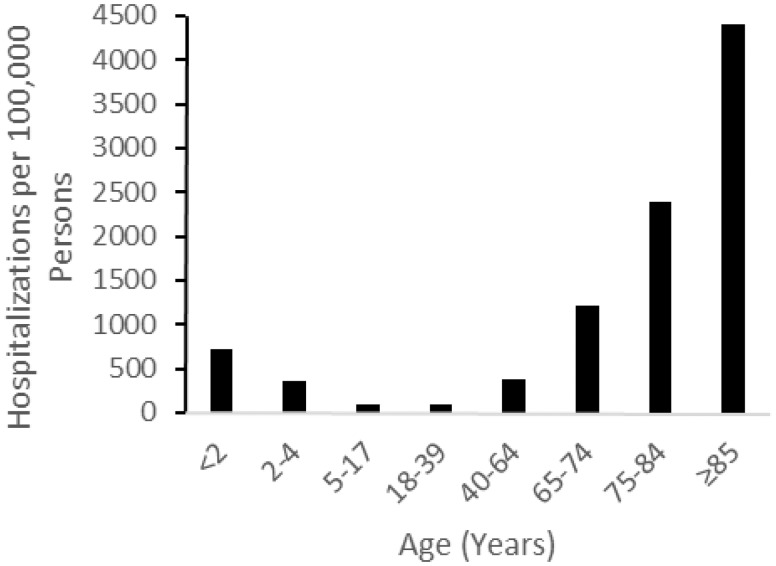
Various other factors—including underlying medical conditions and smoking—can increase the risk of pneumonia by compromising pulmonary clearance mechanisms and the host immune response [47], potentially influencing the selection of pathogens in both the URT and LRT . Age plays a major role in pneumonia risk. In developed countries, such as the USA, the risk of pneumonia is highest in individuals who are 65 years or over (Fig. 1) [48]. The elevated risk in the elderly is likely due to impaired host defenses and an increase in comorbidities—heart failure, liver disease and underlying lung disease—which increase risk of aspiration pneumonia that can occur from dysphagia and gastroesophageal reflux disease (reviewed by Akgün et al. [49]). In developing countries, the burden of pneumonia is greatest in young children [2] due to their inability to physically remove and immunologically deal with bacterial pathogens (reviewed by Siegrist [50]). Very young children also have the greatest prevalence in the nasopharynx of common bacterial pneumonia pathogens: S. pneumoniae, H. influenzae and M. catarrhalis [23, 51]. Increased carriage may be an important risk factor for pneumonia if the URT bacterial community structure is a determinant of pneumonia etiology. Unfortunately, the majority of carriage studies have been conducted among children <5 years of age, which limits our ability to establish the role of nasopharyngeal carriage in other age groups.
Figure 1.
Rate of hospitalization for pneumonia; the USA, 2007–2009. Adapted from Griffin et al. [48]
Regardless of age, viral infection is an important risk factor for bacterial pneumonia. Viruses can lead to rapid, drastic increases in morbidity and mortality in all age groups as seen in historic influenza epidemics and pandemics [52], making it a major public health concern.
TEMPORAL ASSOCIATIONS BETWEEN VIRUSES AND BACTERIA
The 1918 Spanish flu pandemic resulted in ∼50 million deaths worldwide: most of the deaths were caused by secondary bacterial pneumonia [53, 54]. During the 2009 H1N1 pandemic, bacterial co-infection was detected in 18–34% of influenza cases (reviewed by Chertow and Memoli [8]) with vulnerability peaking ∼1 week after influenza infection [55]. The association of viral infection and bacterial pneumonia is not limited to influenza although that interaction has been most studied: adenovirus, human metapneumovirus, respiratory syncytial virus (RSV), and other viruses have been temporally associated with an increased risk of pneumococcal pneumonia and invasive pneumococcal disease (IPD), defined as the isolation of S. pneumoniae from a normally sterile site, in the USA (Table 2) [56–67]. The majority of US studies suggest strong associations between S. pneumoniae infections (both pneumonia and IPD) and influenza virus and RSV, with potential effect modification by age. Temporal associations with other viruses are less supported and limited to IPD. We did not find any studies in the USA that examined temporal associations between viruses and bacterial species other than S. pneumoniae. Six studies conducted in other developed countries examined temporal associations between respiratory viruses and IPD [68–73]. Three out of five studies that examined influenza virus found associations with IPD in UK, The Netherlands and Sweden [70, 71, 73]. Among four studies that examined RSV in other countries, two indicated associations with IPD in all age groups [70, 71], one found an association only among children [68] and the last observed an association only in individuals 2 years or older [69].
Table 2.
Temporal associations between respiratory viruses and S. pneumoniae, the USA
| Study | Virus | Outcome | Age group | Temporal association |
|---|---|---|---|---|
| Kim et al. [56] | ADV | IPD | All | Yes |
| IV | Yes | |||
| PCV | No | |||
| PIV | No | |||
| RSV | Yes | |||
| All | Yes | |||
| except | ||||
| IV | ||||
| Talbot et al. [57] | IV | IPD | All | Yes |
| RSV | Yes | |||
| Ampofo et al. [58] | ADV | IPD | <18 years | No |
| hMPV | Yes | |||
| IV | Yes | |||
| PIV | No | |||
| RSV | Yes | |||
| Murdoch and Jennings [59] | IV | IPD | All | Yes |
| PIV1 | No | |||
| PIV2 | No | |||
| PIV3 | Yes | |||
| RSV | Yes, only in < 5 years | |||
| Nelson et al. [60] | IV | IPD | All | Yes |
| Walter et al. [61] | IV | Pneumonia | All | Yes |
| Zhou et al. [62] | IV | Pneumonia | All | Varies by season |
| RSV | Varies by season | |||
| Weinberger et al. 2012 [63] | 2009 H1N1 season | Pneumonia | All | Yes |
| Shrestha et al. [64] | Influenza seasons | Pneumonia | All | Yes |
| Fleming-Dutra et al. [65] | 2009 H1N1 season | Pneumonia | All | Yes |
| Weinberger et al. 2014 [66] | RSV | Pneumonia | <7 years | Yes |
| Weinberger et al. 2015 [67] | RSV | Pneumonia | <1 years | Yes |
| 1 to < 2 years | Yes | |||
| IV | <1 years | No | ||
| 1 to < 2 years | Yes |
Abbreviations: ADV (adenovirus), hMPV (human metapneumovirus), IV (influenza virus) and PCV (picornavirus)
Temporal associations provide evidence of virus–bacterial interactions, but do not necessarily prove these interactions exist. Many viral infections are seasonal, as is pneumonia infection, so the temporal associations may merely reflect the influence of other seasonal phenomena, environmental or host, that are shared by both viral infection and pneumonia [74]. However, evidence for true virus–bacterial interactions is supported by population studies that estimate a high prevalence of viral co-infection during pneumonia [5, 6] and animal models that suggest increased susceptibility to pneumonia and increased disease severity during viral co-infection [75]. In the USA, ∼47% of children and 19% of adults with bacterial pneumonia are co-infected with one or more viruses [5, 6]. Further, vaccination for S. pneumoniae reduced pneumonia associated with RSV, influenza A and parainfluenza (PIV) types 1-3 [76]. Influenza vaccine probe studies may provide additional insight to the burden of influenza co-infection on bacterial pneumonia.
RESPIRATORY VIRUS ALTERS ASYMPTOMATIC CARRIAGE OF KNOWN BACTERIAL PATHOGENS
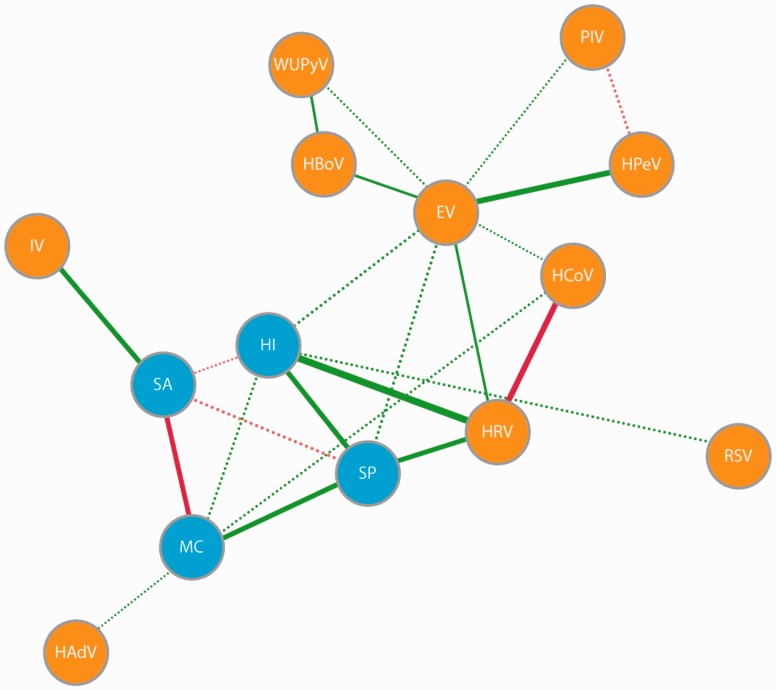
Consistent with our first hypothesis, viral infection frequently has been associated with carriage of common pneumonia pathogens. In a cross-sectional analysis of aboriginal and non-aboriginal children in Western Australia, Jacoby et al. [18] observed positive associations between rhinovirus and S. pneumoniae, H. influenzae and M. catarrhalis and a positive association between adenovirus and M. catarrhalis in the nasopharynx. In a US study, children with a viral URT co-infection not associated with otitis media had a higher prevalence of nontypeable H. influenzae and M. catarrhalis relative to healthy children. Furthermore, children with viral co-infection associated with acute otitis media had an increased prevalence of S. pneumoniae, nontypeable H. influenzae and M. catarrhalis but a decreased prevalence of α-hemolytic Streptococci [77]. van den Bergh et al. [26] assessed the prevalence of 20 respiratory viruses and the main commensal bacteria in the nasopharynx of 433 healthy Dutch children aged 6–24 months. In their study, rhinovirus was positively associated with S. pneumoniae and H. influenzae, RSV was positively associated with H. influenzae, coronaviruses and adenovirus were positively associated with M. catarrhalis, and influenza virus was positively associated S. aureus (Fig. 2). However, as the associations found in the above-mentioned studies are based on cross-sectional analyses, we cannot determine whether viruses influenced carriage structure, bacterial carriage influenced host susceptibility to viruses or if bidirectional interactions occurred. Prospective studies are required to resolve this temporal ambiguity.
Figure 2.
Network of interactions between virus and bacteria in the upper respiratory tract. Figure 1A in van den Bergh et al. [26] used under the Creative Commons Attribution License. Green lines indicate synergistic associations and red lines indicate antagonistic associations. Solid lines indicate associations with P < 0.01 and dashed lines indicate associations with P between 0.01 and 0.05 for associations between species. enterovirus (EV), H. influenzae (HI), human adenovirus human (HAdV), bocavirus (HBoV), human coronavirus (HCov), human parechovirus (HPeV), human rhinovirus (HRV), influenza virus (IV), M. catarrhalis (MC), S. aureus (SA), S. pneumoniae (SP) and WU polyomavirus (WUPyV)
Although the impact of the host microbiota on viral infections is an important consideration (reviewed by Wilks et al. [78]), the majority of in vivo experiments pertaining to virus–bacterial interactions in the URT focus on the role of viruses on the host microbiota. The results of these studies suggest that viruses can alter carriage structure by promoting the colonization of certain commensals. In both animal models and human adults, infection with influenza A virus showed increased colonization by S. pneumoniae and H. influenzae in the URT [79–83]. Similarly, infecting rats and chinchillas with RSV led to increased colonization by nontypeable H. influenzae [84, 85]. Collectively, epidemiologic studies and laboratory experiments suggest that the introduction of a virus to the URT niche can substantially alter the bacterial community present [26].
THE MISSING LINK BETWEEN BACTERIAL CARRIAGE STRUCTURE AND PNEUMONIA
Although there is substantial evidence that viral infection influences the URT bacterial community, whether these changes are reflected in the LRT and ultimately in pneumonia etiology is unclear, which weakens our first hypothesis (i.e. respiratory viruses can influence the etiology of pneumonia by altering bacterial carriage structure in the URT). Studies that examine the joint effects of viral co-infection, bacterial carriage and bacterial pneumonia would provide one strategy for filling this gap. However, we found only two such studies. In a South African hospital-based surveillance study of severe acute respiratory illness, 969 nasopharyngeal-oropharyngeal specimens were tested for S. pneumoniae and a panel of respiratory viruses. A high pneumococcal colonization density in the nasopharynx and oropharynx was associated with both respiratory virus co-infection and pneumococcal pneumonia [86]. A second hospital-based case–control study compared nasopharyngeal carriage among 274 radiologically confirmed cases of pneumonia, 276 cases of other LRT infections and 350 controls in Vietnam. Their findings for S. pneumoniae were similar to that of the South African study. However, the investigators also studied H. influenzae and M. catarrhalis and found no clear association between viral co-infection, nasopharyngeal bacterial load and pneumonia for these species [87]. As noted above, M. catarrhalis rarely causes pneumonia, but H. influenzae is second only to S. pneumoniae. Although there appears to be a persuasive argument for a link between viral co-infection, carriage and pneumonia for S. pneumoniae, whether or why the interaction is not true for other URT bacteria needs further exploration. In particular, studies that can directly test whether viral infection led to bacterial colonization or overgrowth by a potential pathogen, which led to bacterial pneumonia by that pathogen, are in order. In conclusion, there is no definitive answer to our first hypothesis. Epidemiologic studies and experiments indicate viruses alter the bacterial community in the URT, but they do not yet adequately address whether these changes in the URT bacterial community play a significant role in pneumonia etiology.
MECHANISMS OF INTERACTION SUGGEST THAT VIRUS CAN ALTER BACTERIAL SELECTION IN THE LRT
There are several studies that support our second hypothesis, that respiratory viruses can promote bacterial colonization of the LRT by certain commensals in the URT. Viruses interact with bacteria and the host at various stages along the pathologic pathway to promote bacterial pneumonia (Table 3). For example, virus can increase shedding of URT bacteria into the LRT: in vitro biofilm and murine studies suggest influenza A virus infection can lead to the dispersion of S. pneumoniae biofilms, releasing virulent pneumococci for subsequent secondary infections in the LRT [88, 89]. When in a biofilm, S. pneumoniae is less virulent; capsule polysaccharide and pneumolysin production are reduced and synthesis of the bacterial adhesin phosphorylcholine increased [90, 91].
Table 3.
Mechanisms of synergistic virus-bacteria Interaction
| Mechanism | Virus | Bacteria |
|---|---|---|
| Biofilm dispersion | IAV | S. pneumoniae [88, 89] |
| Increased expression of cell surface receptors | ADV | S. pneumoniae [121] |
| IAV | S. pneumoniae [122] | |
| PIV | H. influenzae [100, 123] | |
| RSV | S. pneumoniae [100, 123] | |
| H. influenzae [100, 123] | ||
| S. pneumoniae [100, 123] | ||
| Direct binding of virus and bacteria | RSV | S. pneumoniae [124, 125] |
| Damaged and inhibited repair of respiratory epithelium cells | IAV | S. aureus [26] |
| S. pneumoniae [75] | ||
| Decreased mucociliary velocity | IAV | S. pneumoniae [127] |
| Viral neuraminidase | IAV | S. pneumoniae [95, 96] |
| Impairment of leukocytes (i.e. neutrophils) response | IAV | S. aureus [128] |
| RSV | S. pneumoniae [104, 105, 129, 130] | |
| S. pneumoniae [131] | ||
| Impairment of alveolar macrophage response | IAV | S. aureus [101, 132–134] |
| Impairment of monocytes | IAV | S. aureus [128] |
| RSV | M. catarrhalis [135] | |
| NTHi [135] | ||
| S. pneumoniae [135] | ||
| Reduced natural killer cell recruitment | IAV | S. aureus [136] |
| Exacerbation of inflammatory mediators and tissue damage | hMPV | S. pneumoniae [137] |
| IAV | S. pneumoniae [138–141] |
Abbreviations: ADV (adenovirus), IAV (influenza A virus), hMPV (human metapneumovirus), NTHi (nontypeable H. influenzae), PIV (parainfluenza virus), and RSV (respiratory syncytial virus).
Viral infections also can promote bacterial adhesion to host cells [92–94]. Influenza and PIV promote bacterial adhesion with respiratory epithelium cells by cleaving sialic acid and exposing receptors on host cell oligosaccharide chains [95, 96]. In vitro and in vivo experiments suggest free sialic acid released by viral neuraminidase can behave as signaling molecules promoting pneumococcal biofilm formation, nasopharyngeal colonization and bacterial spread to the lungs [97]. Free sialic acid is believed to play a role in invasion by nontypeable H. influenza as it is an important component of the biofilm matrix and incorporated into the bacterial capsular polysaccharide to evade host defense mechanisms [98]. Although literature is scarce, the relationship may be bilateral as bacterial neuraminidase can promote virus survival during treatment with neuraminidase inhibitors [99]. In addition, viruses can promote bacterial adhesion by upregulating cell surface receptors for pathogenic bacteria. For example, RSV and PIV-3 infection can lead to upregulation of receptors intracellular adhesion molecule 1 (ICAM-1), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), and platelet-activating factor receptor (PAF-r) to promote binding of nontypeable H. influenzae and S. pneumoniae to epithelial cells [100].
Respiratory viral infection can damage and impede the repair of respiratory epithelial cells leading to reduced mucociliary clearance. Consequently, bacteria can more easily enter the lungs to cause pneumonia [75]. Many of the virus–bacteria interaction mechanisms involve viral compromise of the innate immune system. These include impairment and depletion of resident alveolar macrophages [101–103] and neutrophils, which are necessary for bacterial clearance, mediated by induction of type I interferons [104] and desensitization to Toll-like receptor ligands [105]. Detailed descriptions of potential biological pathways involved in these mechanisms are discussed in earlier reviews by Robinson et al. [55] and McCullers [9]. Finally, excessive inflammation in the lungs due to virus-initiated exacerbation of inflammatory mediators, cytokines and chemokines, can cause tissue damage [6], which increases susceptibility to secondary bacterial infections.
Despite the considerable literature on potential mechanisms of viral–bacterial interactions that may lead to pneumonia, most studies are limited to experiments conducted in animal models using select viral and bacterial strains, which may not reflect what is occurring in human populations. Furthermore, the interactions between virus and bacteria are undoubtedly far more complex than identified in animal models, and likely consists of a complex web of interactions between different viruses and bacteria with viruses similar to that described in the URT [12, 26]. Even after considering these limitations, the overwhelming evidence for the existence of multiple biological mechanisms under various conditions supports our second hypothesis that respiratory viruses can alter bacterial selection in the LRT and is an important factor in pneumonia etiology.
CONCLUSIONS
In this review, we discussed how the respiratory tract is an ecosystem with two niches, the URT and the LRT; each with ecological and microbial pressures that determine bacterial selection. We hypothesized that viruses influence bacterial selection in the URT leading to colonization of the LRT and sometimes pneumonia. There appears to be a complex network of interactions among viruses and bacteria in the URT that responds to viral introduction by altering what bacteria are present or modifying their relative abundance. For a least one species, S. pneumoniae, viruses can increase nasopharyngeal carriage density and increase risk of pneumococcal pneumonia. Whether this is true for other URT bacteria that cause pneumonia is uncertain. We also proposed that bacterial selection in the LRT could be altered by viral infection. The LRT is normally inhabited by low density of microbes, a state maintained by local host defenses and bacterial mechanisms of evasion. In vitro and in vivo studies suggest viruses can promote entry and colonization of the LRT for select bacterial species via a range of biological mechanisms including URT biofilm dispersion, increased bacterial adhesion to host epithelial cell by upregulation of cell receptors, reduced pulmonary clearance, impairment of multiple components of the innate immune response and changes in inflammatory response. Although there are limitations in interpreting the results of experiments, evidence of numerous mechanisms observed under various conditions strongly suggest that viruses also play an important role in the selection of bacteria in the LRT and pneumonia etiology.
The greatest difficulty in addressing our hypotheses was our inability to determine the relative contributions of URT bacterial community structure and local host defenses on bacterial selection into the LRT. In the simplest case, how much is the risk of pneumonia following viral infection attributable to the presence of a known bacterial pneumonia pathogen (such as S. pneumoniae) in the URT? To determine this, studies must examine time-dependent carriage of bacteria, species-specific pneumonia outcomes and the effects of viral co-infection among other known risk factors—which, to the best of our knowledge, do not currently exist. Nonetheless, the literature strongly supports the presence of an interaction between viral infection and secondary bacterial pneumonia; the failure to fully understand the mechanisms should act as a spur for future studies while continuing current efforts to reduce the worldwide burden of pneumonia.
CONFLICTS OF INTEREST
Dr. Gordon has received consultancy fees from Abt Associates. All other authors report no potential conflicts of interest.
REFERENCES
- 1.Liu L, Oza S, Hogan D. et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. The Lancet 2015;385:430–40. [DOI] [PubMed] [Google Scholar]
- 2.Rudan I, O’Brien KL, Nair H. et al. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health 2013;3:010401 DOI: 10.7189/jogh.03.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfuntner A, Wier LM, Stocks C, Most Frequent Conditions in U.S. Hospitals, 2011. HCUP Statistical Brief #162. Rockville, MD: Agency for Healthcare Research and Quality; 2013. [PubMed] [Google Scholar]
- 4.Murphy SL, Kochanek KD, Jiaquan X. et al. Deaths: Final Data for 2012. Natl Vital Stat Rep 2015;63. [PubMed] [Google Scholar]
- 5.Jain S, Williams DJ, Arnold SR. et al. Community-acquired pneumonia requiring hospitalization among U.S. Children. N Engl J Med 2015;372:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain S, Self WH, Wunderink RG. et al. Community-acquired pneumonia requiring hospitalization among U.S. Adults. N Engl J Med 2015;373:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beadling C, Slifka MK. How do viral infections predispose patients to bacterial infections? Curr Opin Infect Dis 2004;17:185–91. [DOI] [PubMed] [Google Scholar]
- 8.Chertow DS, Memoli MJ. Bacterial coinfection in influenza: a grand rounds review. JAMA 2013;309:275–82. [DOI] [PubMed] [Google Scholar]
- 9.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 2006;19:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization, 2014. [Google Scholar]
- 11.Rudan I, Boschi-Pinto C, Biloglav Z. et al. Epidemiology and etiology of childhood pneumonia. Epidemiol. Etiol Neumonía En Niñez 2008;86:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosch AATM, Biesbroek G, Trzcinski K. et al. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 2013;9:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res J Lab Clin Med 2012;160:258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faden H, Duffy L, Wasielewski R. et al. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis 1997;175:1440–5. [DOI] [PubMed] [Google Scholar]
- 15.Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol 2010;10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogaert D, De Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 2004;4:144–54. [DOI] [PubMed] [Google Scholar]
- 17.Simell B, Auranen K, Käyhty H. et al. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 2012;11:841–55. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby P, Watson K, Bowman J. et al. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine 2007;25:2458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiri T, Nunes MC, Adrian PV. et al. Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother-child dyads. BMC Infect Dis 2013;13:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chien Y-W, Vidal JE, Grijalva CG. et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 2013;32:72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdullahi O, Karani A, Tigoi CC. et al. The prevalence and risk factors for pneumococcal colonization of the nasopharynx among children in Kilifi District, Kenya. PLoS ONE 2012;7:e30787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jourdain S, Smeesters PR, Denis O. et al. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect 2011;17:907–14. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie GA, Leach AJ, Carapetis JR. et al. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis 2010;10:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae S, Yu J-Y, Lee K. et al. Nasal colonization by four potential respiratory bacteria in healthy children attending kindergarten or elementary school in Seoul, Korea. J Med Microbiol 2012;61:678–85. [DOI] [PubMed] [Google Scholar]
- 25.Tsai M-H, Huang S-H, Chen C-L. et al. Pathogenic bacterial nasopharyngeal colonization and its impact on respiratory diseases in the first year of life: The PATCH Birth Cohort Study. Pediatr Infect Dis J 2015;34:652–8. [DOI] [PubMed] [Google Scholar]
- 26.Van den Bergh MR, Biesbroek G, Rossen JWA. et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLoS ONE 2012;7:e47711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwambana BA, Barer MR, Bottomley C. et al. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect Dis 2011;11:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedel V, Zilora S, Bogaard D. et al. Five-year prospective study of paediatric acute otitis media in Rochester, NY: modelling analysis of the risk of pneumococcal colonization in the nasopharynx and infection. Epidemiol Infect 2014;142:2186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogaert D, van Belkum A, Sluijter M. et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. The Lancet 2004;363:1871–2. [DOI] [PubMed] [Google Scholar]
- 30.Regev-Yochay G, Dagan R, Raz M. et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. Jama 2004;292:716–20. [DOI] [PubMed] [Google Scholar]
- 31.Pettigrew MM, Gent JF, Revai K. et al. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis 2008;14:1584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Žemlicková H, Urbášková P, Adámková V. et al. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus isolated from the nasopharynx of healthy children attending day-care centres in the Czech Republic. Epidemiol Infect 2006;134:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhaegh SJC, Snippe ML, Levy F. et al. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology 2011;157:169–78. [DOI] [PubMed] [Google Scholar]
- 34.Pettigrew MM, Laufer AS, Gent JF. et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 2012;78:6262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auranen K, Mehtälä J, Tanskanen A. et al. Between-strain competition in acquisition and clearance of pneumococcal carriage–epidemiologic evidence from a longitudinal study of day-care children. Am J Epidemiol 2010;171:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adegbola RA, DeAntonio R, Hill PC. et al. Carriage of Streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS ONE 2014;9:e103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAdam AJ, Milner DA, Sharpe AH. Infectious diseases In: Kumar V, Abbas AK (eds). Robbins and Cotran Pathologic Basis of Disease. 9th ed Philadelphia, PA: Elsevier/Saunders, 2015, 341–402. [Google Scholar]
- 38.Smith AM, McCullers JA, Adler FR. Mathematical model of a three-stage innate immune response to a pneumococcal lung infection. J Theor Biol 2011;276:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien KL, Wolfson LJ, Watt JP. et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 2009;374:893–902. [DOI] [PubMed] [Google Scholar]
- 40.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol 1995;33:2759–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park IH, Pritchard DG, Cartee R. et al. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol 2007;45:1225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babl FE, Pelton SI, Theodore S. et al. Constancy of distribution of serogroups of invasive pneumococcal isolates among children: experience during 4 decades. Clin Infect Dis 2001;32:1155–61. [DOI] [PubMed] [Google Scholar]
- 43.Van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 2009;374:1543–56. [DOI] [PubMed] [Google Scholar]
- 44.Naber CK. Staphylococcus aureus bacteremia: epidemiology, pathophysiology, and management strategies. Clin Infect Dis 2009;48:S231–7. [DOI] [PubMed] [Google Scholar]
- 45.Xu Q, Almudervar A, Casey JR. et al. Nasopharyngeal bacterial interactions in children. Emerg Infect Dis 2012;18:1738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy TF, Parameswaran GI. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis 2009;49:124–31. [DOI] [PubMed] [Google Scholar]
- 47.Musher DM. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin Infect Dis 1992;14:801–7. [DOI] [PubMed] [Google Scholar]
- 48.Griffin MR, Zhu Y, Moore MR. et al. U.S. hospitalizations for pneumonia after a decade of pneumococcal vaccination. N Engl J Med 2013;369:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akgün KM, Crothers K, Pisani M. Epidemiology and management of common pulmonary diseases in older persons. J Gerontol a Biol Sci Med Sci 2012;67:276–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegrist C-A. Neonatal and early life vaccinology. Vaccine 2001;19:3331–46. [DOI] [PubMed] [Google Scholar]
- 51.García-Rodríguez JÁ, Martínez MJF. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother 2002;50:59–74. [DOI] [PubMed] [Google Scholar]
- 52.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis 2006;6:303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taubenberger JK, Reid AH, Fanning TG. The 1918 influenza virus: a killer comes into view. Virology 2000;274:241–5. [DOI] [PubMed] [Google Scholar]
- 54.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008;198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson KM, Kolls JK, Alcorn JF. The immunology of influenza virus-associated bacterial pneumonia. Curr Opin Immunol 2015;34:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim PE, Musher DM, Glezen WP. et al. Association of invasive pneumococcal disease with season, atmospheric conditions, air pollution, and the isolation of respiratory viruses. Clin Infect Dis 1996;22:100–6. [DOI] [PubMed] [Google Scholar]
- 57.Talbot TR, Poehling KA, Hartert TV. et al. Seasonality of invasive pneumococcal disease: temporal relation to documented influenza and respiratory syncytial viral circulation. Am J Med 2005;118:285–91. [DOI] [PubMed] [Google Scholar]
- 58.Ampofo K, Bender J, Sheng X. et al. Seasonal invasive pneumococcal disease in children: role of preceding respiratory viral infection. Pediatrics 2008;122:229–37. [DOI] [PubMed] [Google Scholar]
- 59.Murdoch DR, Jennings LC. Association of respiratory virus activity and environmental factors with the incidence of invasive pneumococcal disease. J Infect 2009;58:37–46. [DOI] [PubMed] [Google Scholar]
- 60.Nelson GE, Gershman KA, Swerdlow DL. et al. Invasive pneumococcal disease and pandemic (H1N1) 2009, Denver, Colorado, USA. Emerg Infect Dis 2012;18:208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter ND, Taylor TH, Shay DK. et al. Influenza circulation and the burden of invasive pneumococcal pneumonia during a non-pandemic period in the United States. Clin Infect Dis 2010;50:175–83. [DOI] [PubMed] [Google Scholar]
- 62.Zhou H, Haber M, Ray S. et al. Invasive pneumococcal pneumonia and respiratory virus co-infections. Emerg Infect Dis 2012;18:294–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberger DM, Simonsen L, Jordan R. et al. Impact of the 2009 influenza pandemic on pneumococcal pneumonia hospitalizations in the United States. J Infect Dis 2012;205:458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shrestha S, Foxman B, Weinberger DM. et al. Identifying the interaction between influenza and pneumococcal pneumonia using incidence data. Sci Transl Med 2013;5:191ra84–191ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fleming-Dutra KE, Taylor T, Link-Gelles R. et al. Effect of the 2009 influenza A(H1N1) pandemic on invasive pneumococcal pneumonia. J Infect Dis 2013;207:1135–43. [DOI] [PubMed] [Google Scholar]
- 66.Weinberger DM, Grant LR, Steiner CA. et al. Seasonal drivers of pneumococcal disease incidence: impact of bacterial carriage and viral activity. Clin Infect Dis off Publ Infect Dis Soc Am 2014;58:188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinberger DM, Klugman KP, Steiner CA. et al. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med 2015;12:e1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson M, Gilmour R, Menzies R. et al. The association of respiratory viruses, temperature, and other climatic parameters with the incidence of invasive pneumococcal disease in Sydney, Australia. Clin Infect Dis 2006;42:211–5. [DOI] [PubMed] [Google Scholar]
- 69.Stensballe LG, Hjuler T, Andersen A. et al. Hospitalization for respiratory syncytial virus infection and invasive pneumococcal disease in Danish children aged <2 years: a population-based cohort study. Clin Infect Dis off Publ Infect Dis Soc Am 2008;46:1165–71. [DOI] [PubMed] [Google Scholar]
- 70.Nicoli EJ, Trotter CL, Turner KME. et al. Influenza and RSV make a modest contribution to invasive pneumococcal disease incidence in the UK. J Infect 2013;66:512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansen AGSC, Sanders EaM, VAN DER Ende A. et al. Invasive pneumococcal and meningococcal disease: association with influenza virus and respiratory syncytial virus activity? Epidemiol Infect 2008;136:1448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toschke AM, Arenz S, von Kries R. et al. No temporal association between influenza outbreaks and invasive pneumococcal infections. Arch Dis Child 2008;93:218–20. [DOI] [PubMed] [Google Scholar]
- 73.Grabowska K, Högberg L, Penttinen P. et al. Occurrence of invasive pneumococcal disease and number of excess cases due to influenza. BMC Infect Dis 2006;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feikin DR, Scott JAG, Gessner BD. Use of vaccines as probes to define disease burden. The Lancet 2014;383:1762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kash JC, Walters K-A, Davis AS. et al. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio 2011;2:e00172–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madhi SA, Klugman KP. The vaccine trialist group. a role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med 2004;10:811–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedel V, Chang A, Wills J. et al. Impact of respiratory viral infections on α-hemolytic streptococci and otopathogens in the nasopharynx of young children. Pediatr Infect Dis J 2013;32:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilks J, Beilinson H, Golovkina TV. Dual role of commensal bacteria in viral infections. Immunol Rev 2013;255:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCullers JA, McAuley JL, Browall S. et al. Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J Infect Dis 2010;202:1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diavatopoulos DA, Short KR, Price JT. et al. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. Faseb J 2010;24:1789–98. [DOI] [PubMed] [Google Scholar]
- 81.Wadowsky RM, Mietzner SM, Skoner DP. et al. Effect of experimental influenza A virus infection on isolation of Streptococcus pneumoniae and other aerobic bacteria from the oropharynges of allergic and nonallergic adult subjects. Infect Immun 1995;63:1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wren JT, Blevins LK, Pang B. et al. Influenza A virus alters pneumococcal nasal colonization and middle ear infection independently of phase variation. Infect Immun 2014;82:4802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirano T, Kurono Y, Ichimiya I. et al. Effects of influenza A virus on lectin-binding patterns in murine nasopharyngeal mucosa and on bacterial colonization. Otolaryngol Head Neck Surg 1999;121:616–21. [DOI] [PubMed] [Google Scholar]
- 84.McGillivary G, Mason KM, Jurcisek JA. et al. Respiratory syncytial virus-induced dysregulation of expression of a mucosal beta-defensin augments colonization of the upper airway by non-typeable Haemophilus influenzae. Cell Microbiol 2009;11:1399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patel J, Faden H, Sharma S. et al. Effect of respiratory syncytial virus on adherence, colonization and immunity of non-typable Haemophilus influenzae: implications for otitis media. Int J Pediatr Otorhinolaryngol 1992;23:15–23. [DOI] [PubMed] [Google Scholar]
- 86.Wolter N, Tempia S, Cohen C. et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis 2014;210:1649–57. [DOI] [PubMed] [Google Scholar]
- 87.Vu HTT, Yoshida LM, Suzuki M. et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 2011;30:11–8. [DOI] [PubMed] [Google Scholar]
- 88.Marks LR, Davidson BA, Knight PR. et al. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 2013;4:e00438–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pettigrew MM, Marks LR, Kong Y. et al. Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza A virus infection. Infect Immun 2014;82:4607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chao Y, Marks LR, Pettigrew MM. et al. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect Microbiol 2014;4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanchez CJ, Kumar N, Lizcano A. et al. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS ONE 2011;6:e28738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Selinger DS, Reed WP, McLaren LC. Model for studying bacterial adherence to epithelial cells infected with viruses. Infect Immun 1981;32:941–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davison VE, Sanford BA. Adherence of Staphylococcus aureus to influenza A virus-infected Madin-Darby canine kidney cell cultures. Infect Immun 1981;32:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.George RC, Broadbent DA, Drasar BS. The effect of influenza virus on the adherence of Haemophilus influenzae to human cells in tissue culture. Br J Exp Pathol 1983;64:655–9. [PMC free article] [PubMed] [Google Scholar]
- 95.McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis 2003;187:1000–9. [DOI] [PubMed] [Google Scholar]
- 96.Peltola VT, Murti KG, McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 2005;192:249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trappetti C, Kadioglu A, Carter M. et al. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis 2009;199:1497–505. [DOI] [PubMed] [Google Scholar]
- 98.Johnston JW, Apicella MA. Sialic acid metabolism and regulation by Haemophilus influenzae: potential novel antimicrobial therapies. Curr Infect Dis Rep 2008;10:83–4. [DOI] [PubMed] [Google Scholar]
- 99.Nishikawa T, Shimizu K, Tanaka T. et al. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS ONE 2012;7:e45371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Avadhanula V, Rodriguez CA, DeVincenzo JP. et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol 2006;80:1629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nickerson CL, Jakab GJ. Pulmonary antibacterial defenses during mild and severe influenza virus infection. Infect Immun 1990;58:2809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kleinerman ES, Daniels CA, Polisson RP. et al. Effect of virus infection on the inflammatory response. Depression of macrophage accumulation in influenza-infected mice. Am J Pathol 1976;85:373–82. [PMC free article] [PubMed] [Google Scholar]
- 103.Astry CL, Jakab GJ. Influenza virus-induced immune complexes suppress alveolar macrophage phagocytosis. J Virol 1984;50:287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shahangian A, Chow EK, Tian X. et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest 2009;119:1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Didierlaurent A, Goulding J, Patel S. et al. Sustained desensitization to bacterial Toll-like receptor ligands after resolutionof respiratory influenza infection. J Exp Med 2008;205:323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun J, Madan R, Karp CL. et al. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med 2009;15:277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Regev-Yochay G, Trzcinski K, Thompson CM. et al. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 2006;188:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park B, Nizet V, Liu GY. Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J Bacteriol 2008;190:2275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Regev-Yochay G, Lipsitch M, Basset A. et al. The pneumococcal pilus predicts the absence of Staphylococcus aureus co-colonization in pneumococcal carriers. Clin Infect Dis 2009;48:760–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lijek RS, Weiser JN. Co-infection subverts mucosal immunity in the upper respiratory tract. Curr Opin Immunol 2012;24:417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lijek RS, Luque SL, Liu Q. et al. Protection from the acquisition of Staphylococcus aureus nasal carriage by cross-reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci 2012;109:13823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Odutola A, Antonio M, Owolabi O. et al. Comparison of the prevalence of common bacterial pathogens in the oropharynx and nasopharynx of gambian infants. PLoS ONE 2013;8:e75558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weimer KED, Juneau RA, Murrah KA. et al. Divergent mechanisms for passive pneumococcal resistance to β-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis 2011;203:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pericone CD, Overweg K, Hermans PW. et al. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 2000;68:3990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shakhnovich EA, King SJ, Weiser JN. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect Immun 2002;70:7161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lysenko ES, Ratner AJ, Nelson AL. et al. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 2005;1:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weiser JN, Shchepetov M, Chong ST. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect Immun 1997;65:943–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Budhani RK, Struthers JK. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob Agents Chemother 1998;42:2521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perez AC, Pang B, King LB. et al. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis 2014;70:280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tong TT, Mörgelin M, Forsgren A. et al. Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 2007;195:1661–70. [DOI] [PubMed] [Google Scholar]
- 121.Håkansson A, Kidd A, Wadell G. et al. Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect Immun 1994;62:2707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 2002;186:341–50. [DOI] [PubMed] [Google Scholar]
- 123.Avadhanula V, Wang Y, Portner A. et al. Nontypeable Haemophilus influenzae and Streptococcus pneumoniae bind respiratory syncytial virus glycoprotein. J Med Microbiol 2007;56:1133–7. [DOI] [PubMed] [Google Scholar]
- 124.Smith CM, Sandrini S, Datta S. et al. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am J Respir Crit Care Med 2014;190:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hament J-M, Aerts PC, Fleer A. et al. Direct binding of respiratory syncytial virus to pneumococci: a phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model. Pediatr Res 2005;58:1198–203. [DOI] [PubMed] [Google Scholar]
- 126.Lee M-H, Arrecubieta C, Martin FJ. et al. A postinfluenza model of Staphylococcus aureus pneumonia. J Infect Dis 2010;201:508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pittet LA, Hall-Stoodley L, Rutkowski MR. et al. Influenza virus infection decreases tracheal mucociliary velocity and clearance of Streptococcus pneumoniae. Am J Respir Cell Mol Biol 2010;42:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Abramson JS, Mills EL, Giebink GS. et al. Depression of monocyte and polymorphonuclear leukocyte oxidative metabolism and bactericidal capacity by influenza A virus. Infect Immun 1982;35:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McNamee LA, Harmsen AG. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect Immun 2006;74:6707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cobb NK, Byron MJ, Abrams DB. et al. Novel Nicotine Delivery Systems and Public Health: The Rise of the “E-Cigarette.” Am J Public Health 2010;100:2340–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stark JM, Stark MA, Colasurdo GN. et al. Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J Med Virol 2006;78:829–38. [DOI] [PubMed] [Google Scholar]
- 132.Jakab G. Immune impairment of alveolar macrophage phagocytosis during influenza virus pneumonia. Am Rev Respir Dis 1982;126:778–82. [DOI] [PubMed] [Google Scholar]
- 133.Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol 2013;191:1250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-γ during recovery from influenza infection. Nat Med 2008;14:558–64. [DOI] [PubMed] [Google Scholar]
- 135.Raza MW, Blackwell CC, Elton RA. et al. Bactericidal activity of a monocytic cell line (THP-1) against common respiratory tract bacterial pathogens is depressed after infection with respiratory syncytial virus. J Med Microbiol 2000;49:227–33. [DOI] [PubMed] [Google Scholar]
- 136.Small C-L, Shaler CR, McCormick S. et al. Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J Immunol 2010;184:2048–56. [DOI] [PubMed] [Google Scholar]
- 137.Kukavica-Ibrulj I, Hamelin M-È, Prince GA. et al. Infection with human metapneumovirus predisposes mice to severe pneumococcal pneumonia. J Virol 2009;83:1341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McAuley JL, Hornung F, Boyd KL. et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2007;2:240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith MW, Schmidt JE, Rehg JE. et al. Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia after influenza. Comp Med 2007;57:82. [PMC free article] [PubMed] [Google Scholar]
- 140.Van der Sluijs KF, van Elden LJR, Nijhuis M. et al. IL-10 Is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J Immunol 2004;172:7603–9. [DOI] [PubMed] [Google Scholar]
- 141.Li W, Moltedo B, Moran TM. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of T cells. J Virol 2012;86:12304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.American Lung Association. Trends in Pneumonia and Influenza Morbidity and Mortality. Chicago: American Lung Association; 2010. [Google Scholar]
- 143.Smith SB, Ruhnke GW, Weiss CH. et al. Trends in pathogens among patients hospitalized for pneumonia from 1993 to 2011. JAMA Intern Med 2014;174:1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murdoch DR, O’Brien KL, Driscoll AJ. et al. laboratory methods for determining pneumonia etiology in children. Clin Infect Dis 2012;54:S146–52. [DOI] [PubMed] [Google Scholar]
- 145.Bhat N, O’Brien KL, Karron RA. et al. Use and evaluation of molecular diagnostics for pneumonia etiology studies. Clin Infect Dis off Publ Infect Dis Soc Am 2012;54:S153–8. Suppl [DOI] [PMC free article] [PubMed] [Google Scholar]