Abstract
We performed high-throughput mass spectrometry at high spatial resolution from individual regions (anterior cingulate and primary motor, somatosensory, and visual cortices) and layers of the neocortex (layers III, IV, and V) and cerebellum (granule cell layer), as well as the caudate nucleus in humans and chimpanzees. A total of 39 mass spectrometry peaks were matched with probable protein identifications in both species, allowing for direct comparison in expression. We explored how the pattern of protein expression varies across regions and cortical layers to provide insights into the differences in molecular phenotype of these neural structures between species. The expression of proteins differed principally in a region- and layer-specific pattern, with more subtle differences between species. Specifically, human and chimpanzee brains were similar in their distribution of proteins related to the regulation of transcription and enzyme activity but differed in their expression of proteins supporting aerobic metabolism. While most work assessing molecular expression differences in the brains of primates has been performed on gene transcripts, this dataset extends current understanding of differential molecular expression that may underlie human cognitive specializations.
Keywords: Brain energetics, chimpanzee, human evolution, mass spectrometry, proteomics, RRIDs: nif-0000-00377, nif-0000-02879, nif-0000-30108, nlx 156669
Graphical Abstract
Using region-specific mass spectrometry, the authors show that proteins supporting aerobic metabolism are more highly expressed in the human anterior cingulate cortex and caudate nucleus compared to the same regions in the chimpanzee.
Introduction
Despite being separated by only 6–8 million years of independent evolution (Chimpanzee Sequencing and Analysis Consortium, 2005; Langergraber et al., 2012; Prüfer et al., 2012), humans differ from chimpanzees, one of their closest living relatives, by an array of cognitive specializations, including the use of language, abstracting thinking, and the ability to understand the mental states of others (Sherwood et al., 2008). The human brain is distinguished from that of chimpanzees and other great apes by more than three-fold increase in neocortical size (Holloway, 1996) among other neuroanatomical features (Raghanti et al., 2008; Rilling et al., 2008; Schenker et al., 2010; Semendeferi et al., 2011; Bianchi et al., 2012; Spocter et al., 2012; Bauernfeind et al., 2013). New genomic sequencing technologies have revealed additional evolutionarily distinctive aspects of the human brain at the molecular level (Berezikov et al., 2006; Babbitt et al., 2010; Xu et al., 2010; Konopka et al., 2012). However, relatively little is known about how the abundance and distribution of specific proteins compare between human and chimpanzee brains.
Because the mammalian brain contains a vast diversity of cell types (Masland, 2004), studies investigating its molecular composition that employ homogenate samples dissected from relatively large anatomical regions are unable to detect biological signals that may be less abundant or spatially limited in expression (Geschwind, 2000; Kamme et al., 2003; Sugino et al., 2005). The human neocortex, for example, contains approximately 90 billion neurons (Herculano-Houzel, 2012) that vary in their size, density, connectivity patterns, and neurotransmitters according to laminar and regional distribution (DeFelipe, 1993; Zilles and Amunts, 2010; DeFelipe et al., 2013). To overcome this challenge, several studies have examined gene expression levels across cortical layers using high-throughput methods in mice (Lein et al., 2007; Belgard et al., 2011) and rhesus monkeys (Bernard et al., 2012). Currently, global regional gene expression levels is available for human brain but not at the level of individual layers (Kang et al., 2011; Hawrylycz et al., 2012). While these studies affirm the value of enhanced spatial resolution in molecular research, the expression levels of gene transcripts have proven to be relatively poor predictors of protein abundance with an average correlation of about 30% between the two molecules (Ramakrishnan et al., 2009; Schwanhäusser et al., 2011; Khan et al., 2013; Wu et al., 2013; Wilhelm et al., 2014). However, in addition to gene expression level, protein abundance is largely influenced by the gene-specific translation rate (Schwanhäusser et al., 2011; Low et al., 2013), which appears to be somewhat similar across tissue types (Wilhelm et al., 2014). Although very little is known about the concordance of gene and protein expression levels within discrete populations of cells, the variability of gene expression in individual neurons has been shown to be surprisingly diverse (McConnell et al., 2013), and the effect on protein expression is unknown.
Here, we present a comparative analysis of human and chimpanzee brains using a high spatial resolution and high-throughput proteomic technique. Mass spectrometry (MS) has been a useful tool for biologists to sample the relative quantities of many proteins simultaneously with a high degree of precision in measurement of the molecular weight (MW) of individual molecules (Walther and Mann, 2010). Furthermore, matrix-assisted laser desorption/ionization (MALDI) MS is able to achieve levels of spatial resolution not previously obtained by other proteomic methods (Stoeckli et al., 2001; Chaurand et al., 2006; Cornett et al., 2007). Using MALDI MS, we profiled brain tissue of adult humans and chimpanzees with a spatial resolution of 200 μm, such that individual layers of neocortex and cerebellum (CB) were sampled, representing the diversity of underlying cytoarchitecture and neuronal function. We analyzed protein expression profiles from neocortical regions, including anterior cingulate cortex (ACC), primary motor cortex (M1), somatosensory cortex (S1), and primary visual cortex (V1), as well as the caudate nucleus (CN) and CB. Our objective was to describe protein expression differences in humans and chimpanzees with a regional specificity not previously obtainable in order to characterize human-specific patterns.
Materials and Methods
Sample
Frozen brain samples from adult humans (n = 8; aged 27 to 50 years), who were free from neurological disorders, were obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD). Frozen brain samples from adult common chimpanzees, Pan troglodytes (n = 5; aged 18 to 45 years), were obtained from the Alamogordo Primate Facility (Holloman Air Force Base, Alamogordo, NM). The chimpanzees had been cared for according to Federal and Institutional Animal Care and Use guidelines and died of natural causes. All tissue was collected and stored at −80°C with postmortem intervals of less than 8 hours to diminish degradation of proteins. A detailed summary of the primate sample, including ages of individuals and regions sampled, is provided in Table 1. Additionally, 8 fresh frozen brains from Swiss Webster mice (8–10 wk old, mixed gender, meninges intact) were acquired from Pel-Freez (Rogers, AR). The CBs of these samples were used as a standard to monitor variation across runs.
Table 1.
Demographic Data for the Individuals in the Sample.
| Species | Individual | Age | Sex | Regions |
|---|---|---|---|---|
| Homo sapiens | 1 | 35.2 | M | All regions |
| Homo sapiens | 2 | 50.4 | F | All regions |
| Homo sapiens | 3 | 26.8 | F | CN |
| Homo sapiens | 4 | 34.3 | F | All regions |
| Homo sapiens | 5 | 45.5 | M | CN |
| Homo sapiens | 6 | 49.4 | M | ACC, M1, S1, V1, CB |
| Homo sapiens | 7 | 48.4 | F | All regions |
| Homo sapiens | 8 | 46.4 | F | All regions |
| Pan troglodytes | 1 | 28.9 | M | ACC, V1, CN, CB |
| Pan troglodytes | 2 | 25.4 | F | ACC, M1, S1, V1, CN |
| Pan troglodytes | 3 | 30.8 | F | M1, S1, CB |
| Pan troglodytes | 4 | 22.5 | F | All regions |
| Pan troglodytes | 5 | 34.5 | M | All regions |
Regions of interest at least 1 cm3 in volume were dissected from the left hemisphere of the brain without thawing. Cortical region assignments were confirmed on 10 μm-thick sections stained for Nissl substance with cresyl violet. The ACC was dissected near the genu of the corpus callosum, corresponding to Brodmann’s area 24. M1 was dissected from the precentral gyrus (Brodmann’s area 4) and was confirmed by the presence of Betz cells in layer V under microscopic examination. S1 was dissected from the postcentral gyrus (Brodmann’s area 3, 1, and 2) and was confirmed by the presence of a densely granular cortical layer IV. V1 (Brodmann’s area 17) was dissected from cortex surrounding the calcarine sulcus and was confirmed by the presence of its typical sublamination of layer IV. The CN was dissected from the head of the caudate nucleus. The CB was sampled from the lateral portion of the posterior cerebellar lobe.
Application of matrix and MS data acquisition
Tissue preparation and MS acquisition occurred at the Vanderbilt Mass Spectrometry Research Center Proteomics Laboratory (RRID:nlx 156669). Each frozen tissue block was sectioned into consecutive 10 μm-thick sections using a cryostat (Leica Biosystems, Buffalo Grove, IL). Every fifth section was mounted on a MS target plate immediately after sectioning and thawed. A section adjacent to the one prepared for MS was histologically stained using 0.05% cresyl violet (Sigma-Aldrich, St. Louis, MO). Human and chimpanzee tissue was plated on the same target plate so that possible drift in the signal of the instrument would affect both species equally.
A 25 mg/ml sinapinic acid (Sigma-Aldrich) matrix was made in 50:50 acetonitrile:H2O 0.1% trifluoroacetic acid solution. After coregistering a digital image of the adjacent histological section to a digital image of the section prepared for MS, discrete locations were selected within each region of interest prepared for MS. Using spatial coordinates to guide placement, the sinapinic acid matrix was deposited on the plated tissue within each identified region of interest with an acoustic robotic spotter (Labcyte, Sunnyvale, CA), which deposits the matrix on the tissue in uniform spots 200 μm in diameter (Aerni et al., 2006). Droplets were ejected at 10 Hz in a pattern of 13 drops/spot, which was repeated 6 times for a total of 78 drops/spot. Droplet volume was ~120 pl/drop, making the total volume deposited per spot ~9.4 nl. Once the matrix was applied to tissue, the proteins contained within that spot are considered to be mobile; thus, MS signals obtained from each spot are an average of the proteins within that anatomical location. Mass spectra were acquired using an Autoflex Speed time-of-flight mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a SmartBeam laser (Nd:YAG, 355 nm) and run using a positive ion linear mode acquisition method optimized for mass to charge (m/z) scores between 2 and 40 kDa. Data was acquired in an automated fashion from each discrete matrix spot, with a total of 400 laser shots acquired via random walk over the entire spot for each mass spectrum.
Analysis of mass spectra
Following MS, the plated tissue was stained with 0.05% cresyl violet (Sigma-Aldrich). In samples from both species, the locations of sinapinic acid spots were confirmed visually under microscopy. A matrix spot needed to be fully positioned within the region of interest in order for the mass spectrum it yielded to be included in further analysis. For neocortex and CB, the locations of sinapinic acid spots were only included if the spot was wholly within a single layer of neocortex or the granule cell layer (gcl) of the CB. From the spots with confirmed locations, at least 10 spectra from each individual were averaged arithmetically in ClinProTools software (version 2.2, Bruker Daltonics), for MS data analysis.
For humans and chimpanzees separately, the individual spectra for each region were arithmetically averaged to create a species mean expression spectrum. Because peak width changes with greater m/z scores, top hat baseline subtraction was performed using 10% baseline width to minimize baseline distortions. Spectra were normalized based on their total ion current. Recalibration was performed with a maximal peak shift of 100 parts per million (ppm) and a 30% match to calibrant peaks. Spectra that could not be recalibrated with a signal to noise threshold of 5 were excluded from analysis. Further information regarding these preprocessing procedures can be found elsewhere (Norris et al., 2007).
From the mean species expression spectrum for each region (different layers analyzed separately), peaks within the 2,000–30,000 m/z score range were considered for analysis. Initially, peaks were selected through the automated algorithm of ClinProTools using a resolution of 600 (a parameter corresponding to the peak mass/peak width). Peaks were manually defined if they had not been selected by the algorithm yet appeared to be distinct from noise. Because the peaks were selected from mean species spectra that were created from mean individual spectra, the contribution of noise to the signal was likely minimal.
MS data was collected over a period of 20 months. Sections of mouse CB (10 μm in thickness) were mounted on each target plate and used as a standard to compare variation across runs. MS spectra from the gcl of mouse CB were compared at 9 different times throughout the course of data collection. Mean m/z scores and intensities from each run (1 to 6 spots per run) were used to analyze signal drift over the course of data collection. Principal components analyses (PCAs) were run on the m/z scores and intensities of the15 largest peaks represented in each spectrum.
Assignment of protein identifications
We produced a list of proteins found in the human brain from two sources. First, we searched the UniProt database (http://www.uniprot.org; RRID:nif-0000-00377) on June 11, 2013 for the qualifying terms “human and brain”. The search produced 15,361 unique proteins and isoforms, of which 9,473 had reviewed entries with sufficient functional information to be included in this study. Next, we performed a literature search for studies that had identified and quantified proteins in the human brain using tryptic digestions of proteins followed by multidimensional separations by liquid chromatography or gel electrophoresis and subsequent MS analyses (Dumont et al., 2006; Pan et al., 2007; Ishii et al., 2009; Martins-de-Souza et al., 2009; Burkard et al., 2011). We used the ExPASy Bioinformatics Resource Portal (http://web.expasy.org/compute_pi/; RRID:nif-0000-30108) to find mean isotopic MWs for the list of proteins generated from the UniProt database and literature review. The MWs accounted for cleavages (including initiator methionines, transit peptides, and signal peptides), reflecting the weight of the mature protein. We compared these MWs to the m/z scores of the peaks found from our human spectra. We considered identification of a protein to be sufficient if it passed the following criteria:
MW: The identification of a particular protein was considered possible if the mean isotopic MW of the mature protein was within 0.05% of the MS peak. This criterion is equivalent to a mass change equivalent of one C12 atom per 2 kDa.
Prevalence in the brain: High-throughput proteomic techniques are biased toward detection of the most abundant proteins within a sample. As such, the detection of a protein by one or more other studies (Dumont et al., 2006; Pan et al., 2007; Ishii et al., 2009; Martins-de-Souza et al., 2009; Burkard et al., 2011) was considered to be evidence of a relatively high concentration of that protein in the brain and therefore, more likely to be detected in this study. The presence of a particular protein was further confirmed if the prevalence of its parent gene was shown to be abundantly expressed in GeneCards (http://www.genecards.org; RRID:nif-0000-02879).
z-charge: Although proteins detected by MALDI MS typically carry an electric charge of +1, we could not assume this was true for each peak. To ensure that an ion with a +2 or +3 charge had not created the peak, we divided the MW of each protein in our list by 2 and 3 in order to obtain MW for possible charged ions. If an m/z score from our spectra matched the criterion outlined in step 1 with any of the MWs of possible charged ions, we concluded that we did not have enough confidence for identification.
The chimpanzee peak list was compared to the human list to identify peaks that might represent homologous proteins. Using human proteins as a guide to identifying chimpanzee proteins was possible because 29% of coding sequences within the human and chimpanzee genomes are identical in sequence and most proteins differ by only 1 or 2 amino acids (Chimpanzee Sequencing and Analysis Consortium, 2005). Furthermore, the proteins detected are relatively small (~3.5 kDa–30 kDa), suggesting that changes in amino acid sequence affecting protein weight may be less likely than in larger proteins. The parent gene of each protein match was searched in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/gene/) to confirm conservation of gene sequences between humans and chimpanzees. If the sequences were not conserved, the Uniprot database was searched to see if the protein sequences were equivalent in their MW. For the peaks in the spectra to reflect probable homologues, we determined that the human and chimpanzee peaks should differ by no more than 0.12 % of their m/z score. This criterion is equivalent to a change in mass equivalent to one C12 atom per 833.3 Da. Considering that amino acids range in weight from 57 to 186 Da (glycine to tryptophan, respectively), this threshold allowed for no more than one change in amino acid sequence between humans and chimpanzees in the largest proteins represented in our spectra. The resulting homologous peaks were detectable in some or all of the regions under investigation in this study, including discreet cortical layers. Because MS peaks can be occluded from detection for a number of different reasons, it is impossible to say that a protein is absent from a sample. For example, proteins whose abundance measurements are absent from the chimpanzee sample (e.g., the homologues of UCR1 and COX7B2) were undetectable in our MS spectra but should not be interpreted as absent from the sample altogether. Validations of probable protein identifications using quantitative proteomic methods at a similar spatial resolution are not currently possible.
Data analysis
We opted to use peak intensity (peak height) as a proxy for protein abundance instead of peak area. When peak area is used to quantify abundance, adjacent peaks may influence the calculation due to the broader range of m/z scores considered (Zhang et al., 2010). Because a peak that is present in the spectra for one species may be absent in the spectra for the other, the possibility of nearby peaks biasing the interspecific protein comparisons seemed likely. Across each region of interest, interindividual means, standard deviations (SD), and coefficient of variations were found for the protein abundances. Anderson-Darling tests were performed to test for normality of the distribution of abundances across members of a species. Because the majority of these distributions were not normally distributed, nonparametric statistical tests were performed in all analyses. Kruskal-Wallis tests were used for the analysis of interindividual variation of abundances across members of a species. All further data analysis was performed on the mean expression levels for each species. Spearman rank correlation coefficients were computed on the expression levels of proteins that we were able to identify in at least half of the regions of interest in humans and chimpanzees together. Data analysis was performed in R (version 3; R Core Team, 2012).
A series of multivariate statistical methods were performed to describe the variation in how proteins were distributed across species and regions of interest. A PCA was performed on log-transformed protein expression to explore the amount of variation across the regions of interest. The analysis was performed using the ‘prcomp’ command in R, and the variables were zero-centered and scaled to unit variance. To describe the variation underlying the differences across species, regions of the brain, and layers of neocortex, a series of three canonical variate analyses (CVAs) were performed. Because proteins that were not observed in all regions of interest can influence the group assignments, these analyses were performed only on the 19 proteins that were measured in all regions of interest. The analyses were performed using the ‘lda’ command in R with species, brain area, or cortical layer given as the explanatory variable. Subsequently, discriminant function analyses were performed to examine whether the expression levels of the detected proteins were sufficient to reconstruct species identity or assignment to the sample’s brain region or neocortical layer of origin. The ‘predict’ function produced posterior probabilities for how appropriate the model was for the data. Finally, an unsupervised hierarchical cluster analysis of the expression of all identified proteins was performed using the unweighted pair-group method with the arithmetic mean of human and chimpanzee data to explore how regions of interest are alike or dissimilar in their protein expression profiles. The analysis was performed using the ‘hclust’ function in R.
Results
For the standard mouse CB run at 9 times throughout data collection, a PCA performed on the mean m/z scores found 100% of the variation to be described by PC1. The eigenvector for PC1 was equally loaded (0.33) across all 9 runs, demonstrating there was no drift in the measurement of molecular weights over time. A PCA for the mean peak intensities found 92.7% of the variation to be described by PC1. The loadings were similar (0.29–0.34) for each of the 9 runs in the eigenvector describing PC1. Consequently, we have confidence that the strength of the MS signal did not change markedly over time and likely did not contribute significantly to variation in peak intensities across other regions of interest.
MS spectra were collected from ACC (layers III, V), M1 (layers III, V), S1 (layers III, IV, V), V1 (layers III, IVA, IVB, IVC, and V), CN, and CB (gcl). Figure 1 displays examples of each of these regions from either human or chimpanzee. We identified 39 peaks representing homologous proteins in humans and chimpanzees that were detectable in at least 50% of the regions of interest (Fig. 2). Supplementary information on the proteins, the parent genes from which they are coded, and the gene ontology (GO) categories associated with their parent genes (Gene Ontology Consortium, 2000) can be found online (Supplementary Dataset 1). Table 2 includes the MWs of the proteins and information regarding the matching of homologous MS peaks. For simplicity, we refer to the parent gene name in this study, except in the case of the c-flanking peptide of neuropeptide Y (NPY), as this peptide has its own abbreviation, CPON. Because these proteins were not identified directly from the tissue samples themselves, our identifications remain “probable”. A total of 18 proteins were detected in all regions of interest in both species.
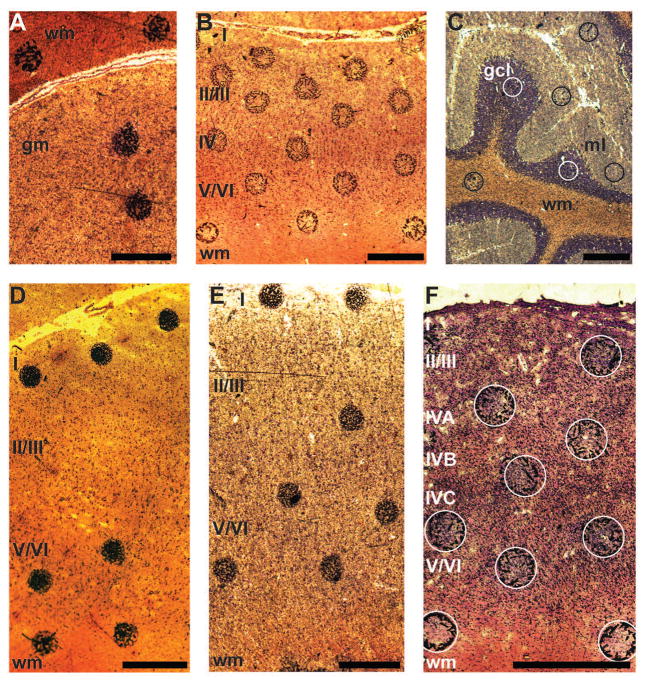
Figure 1.
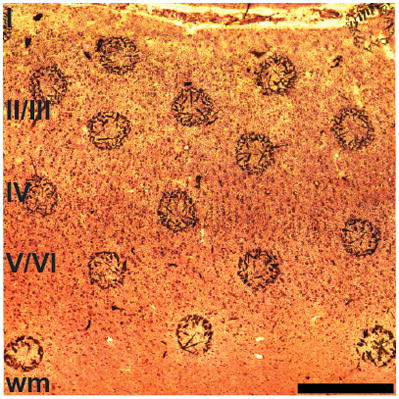
Nissl-stained sections of tissue with sinapinic acid matrix spots. A: caudate nucleus; Chimpanzee 2. B: somatosensory cortex; Chimpanzee 3. C: cerebellum; Human 7. D: anterior cingulate cortex; Human 1. E: primary motor cortex; Human 7. F: primary visual cortex; Chimpanzee 2. The sections are 10 μm-thick and mounted on a metal plate. Because Nissl stain is applied after the application of the matrix and mass spectrometry (MS) is performed, some matrix crystals move from their original locations. Circles are superimposed on the original positions of the matrix spots on the sections from V1 and CB. Neocortical layers and white matter (wm) are labeled in ACC, M1, S1, and V1. The gray matter (gm) of the CN is labeled as well as the surrounding wm. The granule cell layer (gcl), molecular layer (ml), and wm are identified in the CB. Each scale bar is 500 μm in length. The brightness and contrast of the panels were adjusted.
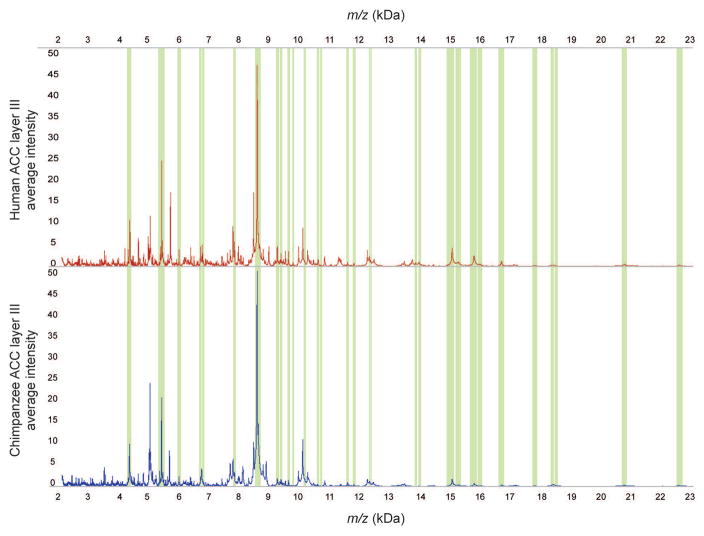
Figure 2.
Spectra from layer III of ACC. Mean spectra from layer III of ACC for human (top) and chimpanzee (bottom). The overlaid green bars highlight the integration areas of the peaks representing the 31 homologous proteins that are observed between the two species in this region.
Table 2.
Comparison of Molecular Weights for the Homologous Proteins of Humans (H.s.) and Chimpanzees (P.t.).
| Parent gene | UniProt ID | Protein name | Reference | Mean isotopic mass of the H.s. protein | Mean m/z score of MS peak | Percent difference between mass of protein and m/z score | Percent difference between H.s. and P.t. peaks | ||
|---|---|---|---|---|---|---|---|---|---|
| H.s. | P.t. | H.s. | P.t. | ||||||
| NPY | NPY_HUMAN_1 | C-flanking protein of neuropeptide Y (CPON) | NA | 4272.72 | 4271.82 | 4271.94 | 0.021 | 0.018 | 0.003 |
| TMSB10 | TYB10_HUMAN | Thymosin beta-10 | 5 | 4894.48 | 4894.11 | 4894.00 | 0.007 | 0.010 | 0.002 |
| COX7C | COX7C_HUMAN | Cytochrome c oxidase subunit 7C, mitochondrial | 5 | 5356.25 | 5356.77 | 5357.03 | 0.010 | 0.015 | 0.005 |
| NDUFC1 | NDUC1_HUMAN | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial | NA | 5937.89 | 5938.98 | 5941.49 | 0.018 | 0.061 | 0.042 |
| FAU | RS30_HUMAN | 40S ribosomal protein S30 | 1 | 6647.86 | 6649.40 | 6648.10 | 0.023 | 0.004 | 0.019 |
| COX7A2 | CX7A2_HUMAN | Cytochrome c oxidase subunit 7A2, mitochondrial | 2, 5 | 6721.81 | 6722.27 | 6721.09 | 0.007 | 0.011 | 0.018 |
| GNG10 | GBG10_HUMAN | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-10 | NA | 6907.93 | 6907.59 | 6908.18 | 0.005 | 0.004 | 0.009 |
| ATP5I | ATP5I_HUMAN | ATP synthase subunit e, mitochondrial | 1, 3, 4, 5 | 7802.02 | 7803.06 | 7803.75 | 0.013 | 0.022 | 0.009 |
| UBA52 | RL40_HUMAN | Ubiquitin-60S ribosomal protein L40 | 2, 3, 5 | 8564.84 | 8566.59 | 8567.51 | 0.020 | 0.031 | 0.011 |
| UQCRH | QCR6_HUMAN | Cytochrome b-c1 complex subunit 6, mitochondrial | 1 | 9246.09 | 9244.06 | 9244.97 | 0.022 | 0.012 | 0.010 |
| NDUFA4 | NDUA4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | 1, 3, 5 | 9369.86 | 9371.84 | 9372.08 | 0.021 | 0.024 | 0.003 |
| COX6A1 | CX6A1_HUMAN | Cytochrome c oxidase subunit 6A1, mitochondrial | NA | 9618.80 | 9620.20 | 9621.46 | 0.015 | 0.028 | 0.013 |
| UQCRQ | QCR8_HUMAN | Cytochrome b-c1 complex subunit 8 | 1, 5 | 9775.18 | 9778.30 | 9779.20 | 0.032 | 0.041 | 0.009 |
| SF3B5 | SF3B5_HUMAN | Splicing factor 3B subunit 5 | 1 | 10135.36 | 10136.39 | 10137.77 | 0.010 | 0.024 | 0.014 |
| SUMO3 | SUMO3_HUMAN | Small ubiquitin-related modifier 3 | 2 | 10524.83 | 10521.98 | 10523.81 | 0.027 | 0.010 | 0.017 |
| COX5B | COX5B_HUMAN | Cytochrome c oxidase subunit 5B, mitochondrial | 1, 2, 3, 5 | 10613.04 | 10613.60 | 10602.06 | 0.005 | 0.103 | 0.109 |
| CTSD | CATD_HUMAN_2 | Cathepsin D | 1, 3, 5 | 10679.98 | 10677.18 | 10678.77 | 0.026 | 0.011 | 0.015 |
| NDUFS6 | NDUS6_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | 3, 5 | 10698.97 | 10700.47 | 10702.45 | 0.014 | 0.033 | 0.019 |
| TXN | THIO_HUMAN | Thioredoxin | 1, 2, 4 | 11606.30 | 11606.29 | 11607.13 | 0.000 | 0.007 | 0.007 |
| GLRX | GLRX1_HUMAN | Glutaredoxin-1 | NA | 11644.55 | 11646.86 | 11649.30 | 0.020 | 0.041 | 0.021 |
| FKBP1A | FKB1A_HUMAN | Peptidyl-prolyl cis-trans isomerase FKBP1A | 1, 2 | 11819.51 | 11819.35 | 11820.20 | 0.001 | 0.006 | 0.007 |
| MIF | MIF_HUMAN | Macrophage migration inhibitory factor | 1, 5 | 12345.11 | 12345.59 | 12346.76 | 0.004 | 0.013 | 0.009 |
| NDUFS5 | NDUS5_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 | 5 | 12386.31 | 12382.10 | 12381.12 | 0.034 | 0.042 | 0.008 |
| COX5A | COX5A_HUMAN | Cytochrome c oxidase subunit 5A, mitochondrial | 1, 2, 3, 5 | 12501.17 | 12501.82 | 12499.77 | 0.005 | 0.011 | 0.016 |
| SNRPD3 | SMD3_HUMAN | Small nuclear ribonucleoprotein Sm D3 | 1 | 13916.25 | 13913.40 | 13918.68 | 0.020 | 0.017 | 0.038 |
| HIST1H2AB | H2A1B_HUMAN | Histone H2A type 1-B/E | 3, 4 | 14004.30 | 14005.54 | 14004.55 | 0.009 | 0.002 | 0.007 |
| HBA1 | HBA_HUMAN | Hemoglobin subunit alpha | 1, 2, 3, 4, 5 | 15126.36 | 15127.12 | 15128.67 | 0.005 | 0.015 | 0.010 |
| PIRT | PIRT_HUMAN | Phosphoinositide-interacting protein | NA | 15334.40 | 15336.00 | 15336.75 | 0.010 | 0.015 | 0.005 |
| CRABP1 | RABP1_HUMAN | Cellular retinoic acid-binding protein 1 | NA | 15434.37 | 15435.19 | 15407.68 | 0.005 | 0.173 | 0.178 |
| HBB | HBB_HUMAN_1 | Hemoglobin subunit beta | 2, 3, 4 5 | 15867.22 | 15868.28 | 15869.90 | 0.007 | 0.017 | 0.010 |
| MRPL27 | RM27_HUMAN | 39S ribosomal protein L27, mitochondrial | 1 | 16072.81 | 16074.96 | 16076.99 | 0.013 | 0.026 | 0.013 |
| EIF5A2 | IF5A2_HUMAN | Eukaryotic translation initiation factor 5A-2 | NA | 16793.20 | 16788.45 | 16790.20 | 0.028 | 0.018 | 0.010 |
| RPL26 | RL26_HUMAN | 60S ribosomal protein L26 | 1 | 17258.21 | 17254.21 | 17255.65 | 0.023 | 0.015 | 0.008 |
| PPIA | PPIA_HUMAN | Peptidyl-prolyl cis-trans isomerase A | 1, 2, 5 | 17881.30 | 17883.59 | 17842.11 | 0.013 | 0.219 | 0.232 |
| ACP1 | PPAC_HUMAN | Low molecular weight phosphotyrosine protein phosphatase | 1, 4 | 17911.29 | 17917.88 | 17923.80 | 0.037 | 0.070 | 0.033 |
| SKP1 | SKP1_HUMAN | S-phase kinase-associated protein 1 | 1, 2, 5 | 18526.81 | 18523.54 | 18528.19 | 0.018 | 0.007 | 0.025 |
| PMS2P3 | PM2P3_HUMAN | Putative postmeiotic segregation increased 2-like protein 3 | NA | 18715.80 | 18717.68 | 18726.45 | 0.010 | 0.057 | 0.047 |
| PEBP1 | PEBP1_HUMAN | Phosphatidylethanolamine-binding protein 1 | 1, 2, 3, 4, 5 | 20925.59 | 20918.62 | 20920.44 | 0.033 | 0.025 | 0.009 |
| MYL6B | MYL6B_HUMAN | Myosin light chain 6B | NA | 22763.99 | 22763.36 | 22739.00 | 0.003 | 0.110 | 0.107 |
| NPY | NPY_HUMAN_1 | Neuropeptide Y | NA | 4272.72 | 4271.82 | 4271.94 | 0.021 | 0.018 | 0.003 |
| TMSB10 | TYB10_HUMAN | Thymosin beta-10 | 5 | 4894.48 | 4894.11 | 4894.00 | 0.007 | 0.010 | 0.002 |
| COX7C | COX7C_HUMAN | Cytochrome c oxidase subunit 7C, mitochondrial | 5 | 5356.25 | 5356.77 | 5357.03 | 0.010 | 0.015 | 0.005 |
| NDUFC1 | NDUC1_HUMAN | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial | NA | 5937.89 | 5938.98 | 5941.49 | 0.018 | 0.061 | 0.042 |
The proteins identified support many different biological functions, including aerobic metabolism, cellular signaling, and protein synthesis, that are typically localized in the cytosol, mitochondrial membrane, or ribosome. The fact that many of the identified proteins were involved in metabolism is not surprising given that roughly 75% of a cell’s protein mass typically supports ‘housekeeping’ functions (Kim et al., 2014). In this study, the proteins with highest abundances tended to be those with m/z scores between 4–10 kDa. With this caveat in mind, the most abundantly measured protein in humans and chimpanzees was ubiquitin A-52 (UBA52), except for layer III of the chimpanzee ACC, where the most abundant proteins are the hemoglobin subunits, hemoglobin α1 (HBA1) and β (HBB). Because UBA52 was overwhelmingly the most abundant in all regions except layer III of chimpanzee ACC, this major difference likely had a significant impact on the grouping of this layer in the chimpanzee with the multivariate statistical analyses that follow. In humans, the vast majority of the identified proteins across all human regions of interest show significant differences in the expression levels across individuals (97% of p-values < 0.05 by Kruskal-Wallis test), and the same was true to a lesser extent in chimpanzees (67% of p-values < 0.05; Supplementary Dataset 1). Although most of the proteins in this study displayed similar amounts of variation between species, 20 proteins exhibited significant differences in the amount of interspecific variation between humans and chimpanzees in at least one region by the Brown-Forsythe test. This was true for each of the cytochrome c oxidase (COX) subunits, which is surprising given that proteins involved in metabolic functions are considered ‘housekeeping’ molecules and thought to contribute little to interindividual variation in both gene (Blekhman et al., 2008) and protein expression (Wu et al., 2013). Higher levels of interindividual variation in either species may ultimately drive differences in interspecific protein expression, an observation that has been made in gene expression studies (Khaitovich et al., 2006; Whitehead and Crawford, 2006; Gallego Romero et al., 2012).
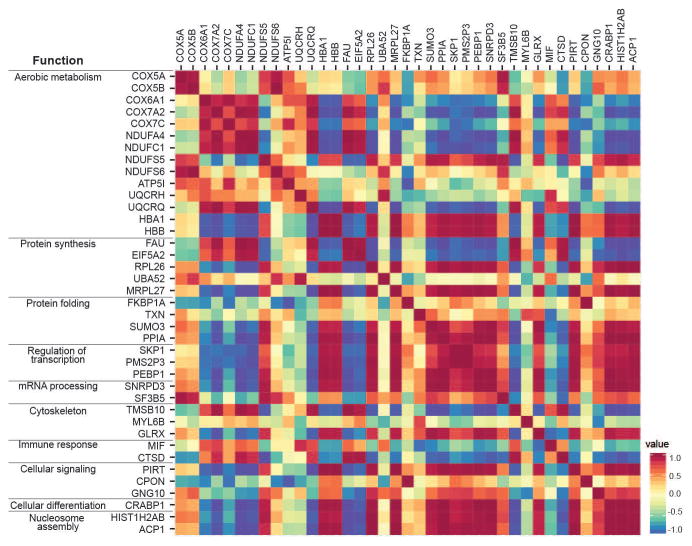
We expected that proteins with similar functions would have expression levels that would be highly correlated across samples. The correlation matrix of protein expression suggested that this was largely true (Fig. 3). HBA1 and HBB were perfectly correlated (r = 1.00). COX subunits, COX6A1, COX7A2, and COX7C, also displayed strong positive correlations in expression (r = 0.73–0.94). Interestingly, however, COX5A and COX5B did not show coordinated expression levels with the other COX proteins (r = −0.39–0.28), but they were positively correlated with each other (r = 0.95). Although comparative expression of COX5B has not been explored, upregulation in the expression of COX5A has been found in the prefrontal cortex of humans compared to chimpanzees (Uddin et al., 2008), which may change the relationship of COX5A to other COX proteins. Other groups of proteins whose constituents were highly positively correlated (r = 0.37–0.99) include those that support mRNA processing (small ubiquitin-related modifier 3 [SUMO3], peptidylprolyl isomerase A [PPIA]), regulation of transcription (S-phase kinase-associated protein 1 [SKP1], putative postmeiotic segregation increased 2-like protein 3 [PMS2P3], phosphatidylethanolamine-binding protein 1 [PEBP1]), and protein folding (small nuclear ribonucleoprotein D3 [SNRPD3], splicing factor 3B subunit 5 [SF3B5]). Performing correlations on humans and chimpanzees separately produced similar results.
Figure 3.
Spearman correlation matrix for the protein expression across all regions of humans and chimpanzees. The inset serves as a guide for the value of the correlation coefficients representing strongly positive correlations (red), no correlations (yellow), and strongly negative correlations (blue).
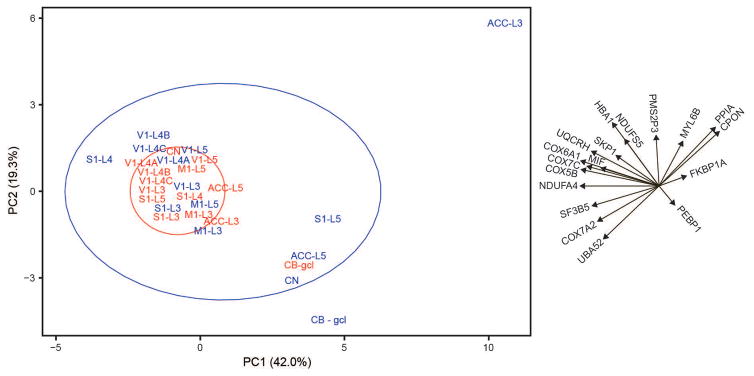
A PCA (Fig. 4) was performed to examine variation in protein expression across all regions of interest in humans and chimpanzees. Principal component (PC) 1 and PC2 accounted for 42.0 and 19.3% of the variation, respectively. A group of proteins comprising the electron transport chain, the final phase of aerobic metabolism, loaded heavily on PC1 (NDUFA4, UQCRH, COX5B, COX6A1, COX7A2, and COX7C). Several proteins that support regulatory functions drove differences along PC2. These proteins include PMS2P3, PPIA, PEBP1, UBA52, and CPON. PC1 and PC2 separate the CB of humans from the rest of the human regions of interest. The human neocortex and CN contributed a very small amount of variation to the analysis along either axis. While proteins supporting aerobic metabolism were associated with human neocortex and CN, expression of PEBP1 was more highly expressed in the human CB. The chimpanzee regions of interest displayed a much greater degree of variation than those of humans. Although many of the chimpanzee regions of interest clustered with the homologous human regions of interest, layer V of S1, layer V of ACC, CN, and the CB of the chimpanzee showed protein expression profiles that were more like that of human CB. Layer III of chimpanzee ACC was an outlier compared to all other regions of interest. We concluded that the unusual protein expression profile in this region is driven largely by biological variation, rather than measurement error (see the following paragraph). PC1 and PC2 separate this region from all others and appear to be primarily associated with higher expression levels of PPIA, CPON, and myosin light chain 6B (MYL6B).
Figure 4.
Principal component analysis of protein expression data. Human regions of interest are shown in red and those of the chimpanzee are in blue. The ellipses represent 68% probability for humans (red) and chimpanzees (blue). The text and arrows to the right of the graph display the loadings for each of the 18 proteins.
Because the protein expression in layer III of chimpanzee ACC proved to be an outlier in the PCA, we further explored the basis of this variation. The 3 chimpanzee ACC samples were analyzed on different days, but the intensity of the MS peaks from the 3 standard mouse CB that were analyzed contemporaneously displayed strong correlations across their spectra (each r > 0.71). Moreover, the same tissue specimen used to sample layer III was also used to sample layer V. Layer V of chimpanzee ACC clusters with the other cortical regions of both human and chimpanzee. Furthermore, most of the proteins that distinguish layer III of chimpanzee ACC from other regions (PPIA, MYL6B, PMS2P3, PEBP1, FK binding protein 1A [FKBP1A]) do not exhibit significant interindividual variation in protein expression (Kruskal-Wallis p = 0.19–0.95). However, the expression of CPON, a peptide with a conserved sequence in humans and chimpanzees, differed significantly across individuals in layer III of chimpanzee ACC (Kruskal-Wallis p < 0.001). Proteolytic processing of the NPY protein results in three peptide products, one of which is CPON. It is important to note that while the expression of CPON is measured in this study, the peptide is very strongly correlated with NPY in the mammalian nervous system (Allen et al., 1985; Gulbenkian et al., 1985), and therefore, comparisons in the expression of CPON to the expression of NPY in previous reports are appropriate. As a species, chimpanzee expression of CPON was significantly lower than that of humans (Mann-Whitney p = 0.015), an interesting finding as NPY has been implicated in learning and memory (Lewis et al., 2005). Previous research based on quantification of NPY-immunoreactive axon fibers found that in several cortical regions including ACC, NPY is not differentially expressed between humans and chimpanzees (Raghanti et al., 2013; Raghanti et al., 2014), although it is elevated relative to non-hominoid primates (Raghanti et al., 2014). Our data indicate that CPON exhibits differential species expression across layers, which may have been undetectable using other methods.
Because layer III of chimpanzee ACC influenced the results of the PCA by introducing a considerable degree of variation, we performed a second PCA (with the same parameters as the previous PCA) omitting this region. PC1 and PC2 accounted for 36.2 and 16.7% of the variation, respectively. As before, proteins supporting aerobic metabolism loaded heavily on PC1, while proteins involved in cellular structure (MYL6B), the regulation of transcription (PMS2P3), protein degradation (SKP1), in addition to aerobic metabolism (COX7A2) drove differences on PC2. Even without the inclusion of layer III of chimpanzee ACC, a greater degree of variation was exhibited among chimpanzee regions of interest along PC1 compared to those of humans, reflecting variability in the expression of proteins supporting aerobic metabolism in chimpanzees.
A CVA was performed to determine which proteins were most highly associated with differences between humans and chimpanzees (Table 3). The proteins that weighed most heavily in determining species assignment included PMS2P3 (coefficient of CV1 = 12.7), FKBP1A (−11.0), and COX5B (9.3). Higher expression levels of COX5B (Mann-Whitney test, p = 0.03) and PMS2P3 (p = 0.09) were associated with the human brain compared to the chimpanzee. Greater expression of COX5B in human brain regions may be expected due to the upregulation of proteins associated with aerobic metabolism in humans compared to chimpanzees, as previously reported (Uddin et al., 2004).
Table 3.
Results of the CVAs.
| Species assignment | Region assignment | Layer assignment (only cortical regions) | |||||
|---|---|---|---|---|---|---|---|
| CV1 | CV1 | CV2 | CV3 | CV4 | CV1 | CV2 | |
| Weight of linear determinant | 100.0% | 81.7% | 11.1% | 4.6% | 2.6% | 68.5% | 31.5% |
| Coefficients of linear discriminants: | |||||||
| COX5B | 9.3 | 14.1 | 4.4 | 3.4 | 4.1 | −4.8 | 7.0 |
| COX6A1 | −2.2 | −10.8 | −2.1 | 0.3 | −0.1 | 7.1 | −0.2 |
| COX7A2 | 2.9 | −0.5 | 1.8 | −2.8 | −1.9 | −1.7 | −0.8 |
| COX7C | −0.7 | 0.2 | −0.7 | 0.0 | 0.5 | −0.1 | 0.4 |
| NDUFA4 | 0.9 | 6.1 | 3.9 | 0.0 | −1.2 | −5.1 | −3.3 |
| NDUFS5 | −4.9 | 12.5 | −7.4 | −2.0 | 0.2 | −9.6 | −0.6 |
| UQCRH | 0.5 | 1.4 | 0.7 | −1.6 | −0.8 | −1.7 | 0.3 |
| HBA1 | 0.6 | 0.3 | 0.1 | 0.1 | −0.2 | −0.1 | −0.1 |
| UBA52 | −0.2 | −0.1 | −0.1 | 0.0 | 0.1 | 0.1 | 0.1 |
| FKBP1A | −11.0 | −14.1 | −12.2 | 4.2 | 4.7 | 14.3 | 8.0 |
| PPIA | −2.5 | −11.1 | 18.4 | −21.7 | −2.3 | −11.6 | −3.8 |
| SKP1 | −4.3 | 3.6 | −7.5 | 3.1 | 3.1 | 2.3 | 6.9 |
| PMS2P3 | 12.7 | −27.5 | 25.4 | −9.6 | −13.5 | 0.0 | −30.6 |
| PEBP1 | −0.3 | −29.7 | 8.9 | 1.1 | −5.1 | 15.8 | −12.6 |
| SF3B5 | 1.7 | 0.4 | 1.8 | −3.1 | −2.2 | −2.3 | −0.9 |
| MYL6B | −5.1 | −10.7 | −3.1 | 7.1 | 2.0 | 12.8 | 3.7 |
| MIF | 2.0 | −6.2 | 0.4 | 2.1 | −0.8 | 3.9 | −4.6 |
| NPY (CPON) | −0.1 | 3.4 | −3.3 | 2.4 | 1.1 | 0.4 | 1.9 |
The first analysis included all regions of interest and was used to predict the species based upon protein expression. The second analysis included all regions of interest and was used to predict the area of the brain from which the samples originated. The third analysis included only cortical regions and was used to predict the layer of cortex from which the sample originated. The results of each analysis predicted species, region, or neocortical layer assignment with 100% posterior probability.
Using a combined sample of humans and chimpanzees, a second CVA was performed to determine which proteins were most highly associated with different brain regions (Fig. 5a; Table 3). Four linear CVs described the brain region of origin (CV1-CV4 describe 81.7, 11.1, 4.6, and 2.6% of the vector, respectively; discriminant function analysis posterior probabilities p < 0.001). In this case, CV1 was mostly responsible for the separation of CN from the neocortical regions, while CV2 separated the CB from the other regions of interest. PEBP1 (coefficient of CV1 = −29.7), PMS2P3 (−27.5), FKBP1A (−14.1), COX5B (14.1), and NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 (NDUFS5) (12.5) are the proteins most related to CV1. Higher expression of COX5B and NDUFS5 supports aerobic metabolism and is associated with neocortical regions, while higher expression levels of PEBP1, PMS2P3, and FKBP1A were associated with the CN. PMS2P3 (coefficient of CV2 = 25.4), PPIA (18.4), and FKBP1A (−12.2) were the proteins that most strongly load on CV2. Higher expression of PMS2P3 and PPIA were associated with the CB, while higher expression levels of FKBP1A were associated with the CN and neocortical areas.
Figure 5.
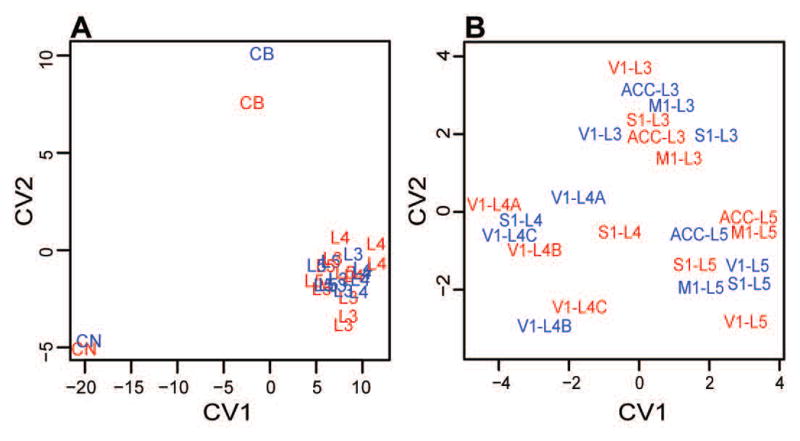
Canonical variate analyses (CVAs) of protein expression data. A: CVA of all regions of interest. Axis 1 separates CN from all other regions of interest, while axis 2 distinguished CB from the CN and cortical regions (shown here by neocortical layer). B: CVA of all regions with CN and CB removed. Axis 1 separates cortical layer IV (shown here as L4) from layer V (L5), while axis 2 separates cortical layer III (L3) from layer IV and layer V.
A final CVA was performed omitting CN and CB from the analysis in order to distinguish the cortical regions of interest from each other (Fig. 5b; Table 3). Two linear CVs separated each cortical region of interest into cortical layers III, IV, or V (CV1 and CV2 account for 68.5 and 31.5% of the vector, respectively; discriminant function analysis posterior probabilities p < 0.001). Cortical layers IV and V were separated by CV1, while layer III was differentiated from layers IV and V along CV2. PEBP1 (coefficient of CV1 = 15.8), FKBP1A (14.3), and MYL6B (12.8) are the proteins that had the most influence on CV1. Higher expression of PEBP1, FKBP1A, and MYL6B was associated with cortical layer V compared to layer IV. PMS2P3 (coefficient of CV2 = −30.6), PEBP1 (−12.6), FKBP1A (8.0), and COX5B (7.0) had the strongest influence on CV2. FKBP1A and COX5B, which function in protein folding and aerobic metabolism, respectively, were associated with higher expression levels in cortical layer III of all areas of the neocortex, while expression of PMS2P3 and PEBP1were higher in cortical layers IV and V. Like the CVA that included CN and CB, this analysis did not distinguish between human and chimpanzee regions of interest.
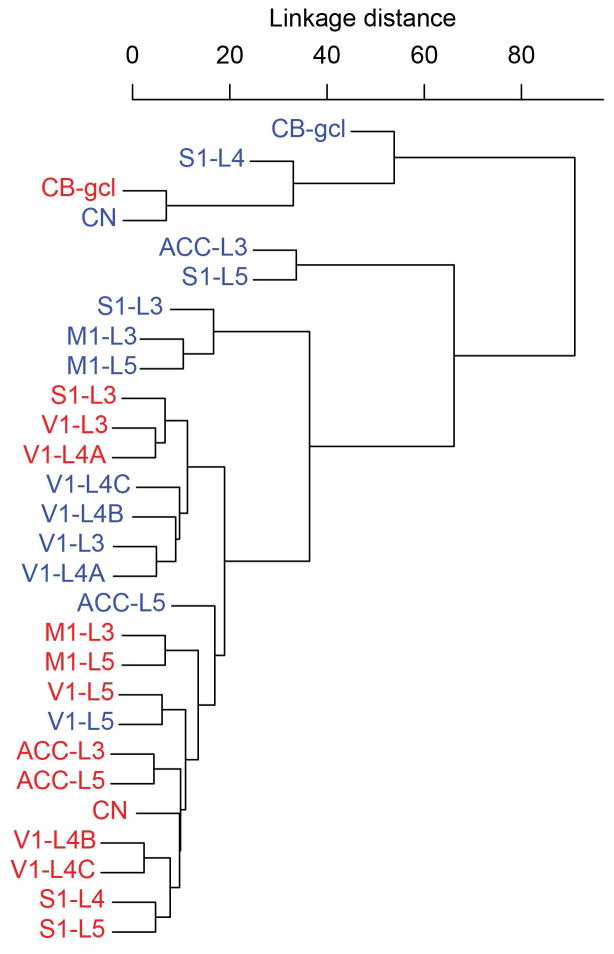
An unsupervised hierarchical cluster analysis revealed relationships in the expression of proteins across regions of interest in humans and chimpanzees (Fig. 6). Remarkably, the human and chimpanzee brain regions did not cluster separately according to species, but instead commonalities were observed in their regional patterns of protein expression. The gcl of CB in humans and chimpanzees were strongly differentiated from the other brain regions. Chimpanzee CN and layer IV of S1 were also included in this cluster. The other regions of the brain separated mostly by motor and sensory areas. Human and chimpanzee M1 were similar in their protein expression profiles. Sensory areas clustered mostly by supra- and infragranular layers designations. Human CN and ACC (layers III and V) clustered with the infragranular layers of sensory cortices, likely due to the high degree of connectivity of these regions to sensory cortices (Alexander et al., 1986; Lehéricy et al., 2004). While layer V of chimpanzee ACC also clustered with this group, layer III of chimpanzee ACC and chimpanzee CN did not.
Figure 6.
Cluster analysis of human and chimpanzee brain regions based on protein expression. Human (red) and chimpanzee brain (blue) regions are largely interrelated. Although layers III and V of human and chimpanzee M1 are very similar, other regions cluster largely according to supra- or infragranular layers of neocortex. The subdivisions of layer IV of V1 are divided among the supra- and infragranular clusters. Notably, the human CN and both layers III and V of ACC are found in the infragranular sensory cluster.
Discussion
This study provides a proteomic analysis comparable in spatial resolution to recent high-throughput studies of RNA transcripts (Lein et al., 2007; Belgard et al., 2011; Bernard et al., 2012; Hawrylycz et al., 2012) by providing relative protein quantifications from specific layers of the neocortex and CB. Until now, neither the transcriptomic nor the proteomic composition of the chimpanzee brain has been analyzed at a similar spatial resolution using a high-throughput technique to enable the study of variation in molecular expression between humans and their closest living relatives. While evolutionary divergence in protein expression levels have been hypothesized to be a major underlying cause of cognitive differences between humans and chimpanzees (King and Wilson, 1975), interspecific comparisons of protein abundances between these two species is imperative to assess the specific and unique differences in the human brain’s molecular phenotype.
To a large extent, proteins with similar biological functions were correlated in their expression levels across the total sample of human and chimpanzee brain regions. Within the groups of proteins supporting aerobic metabolism, regulation of transcription, mRNA processing, and nucleosome assembly, expression levels were especially highly correlated, which is suggestive of coordinated function of these molecules (Zhang and Horvath, 2005). However, proteins assigned to other broad biological functions, including cellular signaling and immune response, had expression profiles that were weakly correlated to the other proteins within their functional groups. Because coordinated expression levels among proteins is generally an effective predictor of molecular interactions (Fraser et al., 2004), this finding reflects the diversity of biological functions served by the proteins represented in this study.
In some ways, our results are consistent with previous research investigating the regional transcriptome of the brain. In humans and chimpanzees, we found greater variation in protein expression between the cerebral cortex and the CB and CN than among regions of the cerebral cortex itself. This result was anticipated due to the relatively homogenous transcriptomic expression in the human cerebral cortex compared to subcortical structures, and to a greater extent CB (Khaitovich et al., 2004; Hawrylycz et al., 2012). The pattern of protein expression was similar among supra- and infragranular layers of neocortex, reflecting consistency in the cytoarchitecture of cortical layers independent of region (DeFelipe et al., 2003). Comparable results have been reported for patterns of gene expression (Belgard et al., 2011; Bernard et al., 2012; Hawrylycz et al., 2012), indicating that layer-specific patterns of expression characterize both gene transcripts and proteins.
However, our results highlight important differences in region-specific expression between gene transcripts and proteins. First, transcriptional profiling in human brain has found similar expression levels between neighboring regions of neocortex (Bernard et al., 2012; Hawrylycz et al., 2012). Our investigation of proteins found no such pattern, suggesting that regulation of protein expression levels provides an additional level of regional specificity beyond that seen in gene transcripts. Second, while reports of gene expression have revealed a particularly unique biological signature in V1 in primates (Bernard et al., 2012; Hawrylycz et al., 2012), we did not find this to be true in protein expression. Both of these findings, however, may be the results of the limited number of proteins analyzed, and this effect may appear with a more comprehensive sampling.
Regions of the human neocortex and CN exhibited higher expression of proteins supporting aerobic metabolism, which differentiates them from the human and chimpanzee CB and from other regions of the chimpanzee brain. This finding is noteworthy due to the high neuronal density of the CB in both species compared to the neocortex (Herculano-Houzel, 2012); it is possible that brain regions with more tightly packed small neurons have lower mass-specific metabolic requirements based on reduced heat dissipation and reduced metabolic costs associated with neural transmission (Laughlin and Sejnowski, 2003). Additionally, human and chimpanzee protein expression profiles revealed by our analyses generally affirm the similarities in gene expression between cortical layers III and V that has been noted previously in human brain (Hawrylycz et al., 2012; Zeng et al., 2012) but is generally absent in the mouse (Zeng et al., 2012). Indeed, intracortical connections originating from neocortical layer III appear to have evolved during primate evolution and result in a more integrative cortical circuitry (Rockland and Pandya, 1979; Barbas, 1986; Hof et al., 1995). We find that supragranular layers of the neocortex appeared to have particularly high levels of proteins supporting aerobic metabolism compared to other layers of cortex. Such a result may be due to an increased density of glutamatergic corticocortical inputs in layer III compared to infragranular layers (DeFelipe et al., 2003), which may drive a higher metabolic demand locally.
One of our most noteworthy results is the unique protein expression profile in layer III of chimpanzee ACC relative to all other regions. The distinctiveness of this region was driven largely by relatively lower expression of proteins supporting metabolic function. Although proteins indicative of structural complexity, including those associated with synapses and receptors, as well as neurofilament proteins, are undetectable with the methods employed in this study due to their large size, other studies have found that the elaboration of dendritic arbors is correlated with increased metabolic demand (Jacobs et al., 2001; Liu et al., 2012). Therefore, the human ACC, with a higher level of proteins supporting aerobic metabolism than that of the chimpanzee, may be specialized for neuronal communication (Uddin et al., 2004), supporting the cognitive processing of arousal of the body state and working memory (Critchley et al., 2003). Likewise, the human CN also displayed higher levels of metabolic proteins compared to the chimpanzee CN, an important result considering the role of the CN in speech production (Jarvis, 2004; Crinion et al., 2006; Pfenning et al., 2014). Interestingly, our data revealed commonalities in the expression profiles of the human CN with ACC and S1, potentially reflecting the interconnectivity of these regions (Lehéricy et al., 2004) as regions with dense connections are thought to display similar patterns of molecular expression (Oldham et al., 2008). These results imply that human ACC and CN are specialized for the integration and interpretation of sensory information, such as that involved in empathy and language (Nimchinsky et al., 1999; Jarvis, 2004; Singer et al., 2004; Enard et al., 2009; Gu et al. 2013).
The study of protein expression provides value to comparative molecular biology that cannot be obtained by studies of gene expression alone. It has been shown that gene transcript expression levels only explain as little as 4 to as much as 40% of protein expression (de Sousa Abreu et al., 2009; Ramakrishnan et al., 2009; Schwanhäusser et al., 2011; Khan et al., 2013; Wu et al., 2013). While measurement noise likely accounts for some of the low correspondence, other biological factors account for the remaining variation (Vogel and Marcotte, 2012). For instance, protein stability, as measured by their biological half-life (the length of time between when a protein is produced and 50% of it is degraded), varies drastically based on function (Yen et al., 2008). In general, proteins integrated into the cell membrane or involved in signal transduction have short half-lives, while those supporting housekeeping functions or the cytoskeleton of the cell have long half-lives (Yen et al., 2008; Schwanhäusser et al., 2011). Therefore, the stability of mRNA and protein dictates their abundance and may cause divergence in the correlation of these molecules (Zhang et al., 2014). This fact suggests that a distinctive set of species-specific biological signals may be accessible by differential expression of proteins compared to transcripts. These considerations suggest a complementary relationship between proteomic and transcriptomic studies in determining molecular phenotype.
While the number of proteins detected in our study is small compared to studies of gene expression, our data support the idea that the regional phenotype of neurons in human and chimpanzee brains are the result of localized specificity in molecular expression (Cáceres et al., 2003; Johnson et al., 2009; Pontén et al., 2009; Hawrylycz et al., 2012). Currently, 13,000 proteins have been identified in the human brain (Lane et al., 2014), each engaging in remarkably complex protein-protein interactions (Choudhary and Mann, 2010). Quantification of a greater proportion of the brain’s proteins at a similar spatial resolution as produced here would enable systems biology approaches to explore the interacting networks of the constituent molecules, which has already produced significant insight into the biological function of the human brain (Oldham et al., 2006, 2008; Johnson et al., 2009; Winden et al., 2009). Because proteins provide a closer approximation of the functional phenotype than the transcriptome, comparative studies of localized proteins are critical to our understanding of species-specific molecular distinctiveness.
Acknowledgments
Acknowledgment of support:
This work was supported by the National Science Foundation (DGE-0801634, BCS-0827531), the James S. McDonnell Foundation (220020293), the Cosmos Club Foundation, The Journal of Experimental Biology Travelling Fellowship, and the Wenner-Gren Foundation for Anthropological Research.
The authors would like to thank the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Southwest National Primate Research Center at the Texas Biomedical Research Institute, and Alamogordo Air Force Base for providing tissue used in this study. Additionally, we thank Serena Bianchi for her insightful comments.
Footnotes
Conflict of Interest Statement
The authors declare no conflicting interests.
Author Contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: A.L.B., P.R.H., C.C.S. Acquisition of data: A.L.B., M.L.R. Analysis and interpretation of data: A.L.B., C.C.B., G.A.W., P.R.H., and C.C.S. Drafting of the manuscript: A.L.B. Critical revision of the manuscript for important intellectual content: A.L.B., C.C.B., G.A.W., P.R.H., and C.C.S. Statistical analysis: A.L.B. Obtaining funding: A.L.B. and C.C.S. Administrative, technical, and material support: A.L.B., M.L.R., R.M.C., J.J.E., P.R.H., C.C.S. Study supervision: A.L.B., M.L.R., R.M.C., C.C.S.
Literature Cited
- Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78:827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- Alexander GE, DeLong MR, Strick PL. Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu Rev Neurosci. 1986;9:357–381. doi: 10.1146/annurev.ne.09.030186.002041. [DOI] [PubMed] [Google Scholar]
- Allen JM, Polak JM, Bloom SR. Presence of the predicted c-flanking peptide of neuropeptide Y (CPON) in tissue extracts. Neuropeptides. 1985;6:95–100. doi: 10.1016/0143-4179(85)90100-3. [DOI] [PubMed] [Google Scholar]
- Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H. Pattern in the laminar origin of corticocortical connections. J Comp Neurol. 1986;252:415–422. doi: 10.1002/cne.902520310. [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, de Sousa AA, Avasthi T, Dobson SD, Raghanti MA, Lewandowski AH, Zilles K, Semendeferi K, Allman JM, Craig AD, Hof PR, Sherwood CC. A volumetric comparison of the insular cortex and its subregions in primates. J Hum Evol. 2013;64:263–279. doi: 10.1016/j.jhevol.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, García-Moreno F, Molnár Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RHA. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MME, Serikawa K, Lemon T, Morgan R, Copeland C, Smith K, Cullen V, Davis-Turak J, Lee C-K, Sunkin SM, Loboda AP, Levine DM, Stone DJ, Hawrylycz MJ, Roberts CJ, Jones AR, Geschwind DH, Lein ES. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi S, Stimpson CD, Bauernfeind AL, Schapiro SJ, Baze WB, McArthur MJ, Bronson E, Hopkins WD, Semendeferi K, Jacobs B, Hof PR, Sherwood CC. Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cereb Cortex. 2012;23:2429–2436. doi: 10.1093/cercor/bhs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLOS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard TR, Planyavsky M, Kaupe I, Breitwieser FP, Bürckstümmer T, Bennett KL, Superti-Furga G, Colinge J. Initial characterization of the human central proteome. BMC Syst Biol. 2011;5:17. doi: 10.1186/1752-0509-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MJ, Hof PR, Morrison JH. A subpopulation of primate corticocortical neurons is distinguished by somatodendritic distribution of neurofilament protein. Brain Res. 1991;539:133–136. doi: 10.1016/0006-8993(91)90695-r. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- Crinion J, Turner R, Grogan A, Hanakawa T, Noppeney U, Devlin JT, Aso T, Urayama S, Fukuyama H, Stockton K, Usui K, Green DW, Price CJ. Language control in the bilingual brain. Science. 2006;312:1537–1540. doi: 10.1126/science.1127761. [DOI] [PubMed] [Google Scholar]
- Critchley HD, Mathias CJ, Josephs O, O’Doherty J, Zanini S, Dewar B-K, Cipolotti L, Shallice T, Dolan RJ. Human cingulate cortex and autonomic control: converging neuroimaging and clinical evidence. Brain. 2003;126:2139–2152. doi: 10.1093/brain/awg216. [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb Cortex. 1993;3:273–289. doi: 10.1093/cercor/3.4.273. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, Arellano JI. Microstructure of the neocortex: comparative aspects. J Neurocytol. 2003;31:299–316. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, López-Cruz PL, Benavides-Piccione R, Bielza C, Larrañaga P, Anderson S, Burkhalter A, Cauli B, Fairén A, Feldmeyer D, Fishell G, Fitzpatrick D, Freund TF, González-Burgos G, Hestrin S, Hill S, Hof PR, Huang J, Jones EG, Kawaguchi Y, Kisvárday Z, Kubota Y, Lewis DA, Marín O, Markram H, McBain CJ, Meyer HS, Monyer H, Nelson SB, Rockland K, Rossier J, Rubenstein JLR, Rudy B, Scanziani M, Shepherd GM, Sherwood CC, Staiger JF, Tamás G, Thomson A, Wang Y, Yuste R, Ascoli G. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont D, Noben J-P, Verhaert P, Stinissen P, Robben J. Gel-free analysis of the human brain proteome: application of liquid chromatography and mass spectrometry on biopsy and autopsy samples. Proteomics. 2006;6:4967–4977. doi: 10.1002/pmic.200600080. [DOI] [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, Brückner MK, Schreiweis C, Winter C, Sohr R, Becker L, Wiebe V, Nickel B, Giger T, Müller U, Groszer M, Adler T, Aguilar A, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Favor J, Fuchs H, Gailus-Durner V, Hans W, Hölzlwimmer G, Javaheri A, Kalaydjiev S, Kallnik M, Kling E, Kunder S, Mossbrugger I, Naton B, Racz I, Rathkolb B, Rozman J, Schrewe A, Busch DH, Graw J, Ivandic B, Klingenspor M, Klopstock T, Ollert M, Quintanilla-Martinez L, Schulz H, Wolf E, Wurst W, Zimmer A, Fisher SE, Morgenstern R, Arendt T, de Angelis MH, Fischer J, Schwarz J, Pääbo S. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Wall DP, Eisen MB. Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci USA. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH. Mice, microarrays, and the genetic diversity of the brain. Proc Natl Acad Sci USA. 2000;97:10676–10678. doi: 10.1073/pnas.97.20.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Hof PR, Friston KJ, Fan J. Anterior insular cortex and emotional awareness. J Comp Neurol. 2013;521:3371–3388. doi: 10.1002/cne.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbenkian S, Wharton J, Hacker GW, Varndell IM, Bloom SR, Polak JM. Co-localization of neuropeptide tyrosine (NPY) and its C-terminal flanking peptide (C-PON) Peptides. 1985;6:1237–1243. doi: 10.1016/0196-9781(85)90456-5. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc Natl Acad Sci USA. 2012;109:10661–10668. doi: 10.1073/pnas.1201895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Nimchinsky EA, Morrison JH. Neurochemical phenotype of corticocortical connections in the macaque monkey: quantitative analysis of a subset of neurofilament protein-immunoreactive projection neurons in frontal, parietal, temporal, and cingulate cortices. J Comp Neurol. 1995;362:109–133. doi: 10.1002/cne.903620107. [DOI] [PubMed] [Google Scholar]
- Holloway RL. Toward a synthetic theory of human brain evolution. In: Changeux JP, Chavaillon J, editors. Origins of the human brain. Oxford: Clarendon Press; 1996. pp. 42–54. [Google Scholar]
- Ishii A, Dutta R, Wark GM, Hwang SI, Han DK, Trapp BD, Pfeiffer SE, Bansal R. Human myelin proteome and comparative analysis with mouse myelin. Proc Natl Acad Sci USA. 2009;106:14605–14610. doi: 10.1073/pnas.0905936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs B, Schall M, Prather M, Kapler E, Driscoll L, Baca S, Jacobs J, Ford K, Wainwright M, Treml M. Regional dendritic and spine variation in human cerebral cortex: a quantitative Golgi study. Cereb Cortex. 2001;11:558–571. doi: 10.1093/cercor/11.6.558. [DOI] [PubMed] [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovi D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran D-T, Zhu J, Luo L, Bittner A, Guo H-Q, Miller N, Wan J, Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do H-H, Weiss G, Enard W, Heissig F, Arendt T, Nieselt-Struwe K, Eichler EE, Pääbo S. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang T-C, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang G-Z, Luo R, Preuss TM, Geschwind DH. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane L, Bairoch A, Beavis RC, Deutsch EW, Gaudet P, Lundberg E, Omenn GS. Metrics for the Human Proteome Project 2013–2014 and strategies for finding missing proteins. J Proteome Res. 2014;13:15–20. doi: 10.1021/pr401144x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber KE, Prüfer K, Rowney C, Boesch C, Crockford C, Fawcett K, Inoue E, Inoue-Muruyama M, Mitani JC, Muller MN, Robbins MM, Schubert G, Stoinski TS, Viola B, Watts D, Wittig RM, Wrangham RW, Zuberbühler K, Pääbo S, Vigilant L. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci USA. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin SB, Sejnowski TJ. Communication in neuronal networks. Science. 2003;301:1870–1874. doi: 10.1126/science.1089662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehéricy S, Ducros M, Van De Moortele PF, Francois C, Thivard L, Poupon C, Swindale N, Ugurbil K, Kim DS. Diffusion tensor fiber tracking shows distinct corticostriatal circuits in humans. Ann Neurol. 2004;55:522–529. doi: 10.1002/ana.20030. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong H-W, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf K-R, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, Li N, Hu Y, Chen W, Qiu Z, Pääbo S, Khaitovich P. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low TY, van Heesch S, van den Toorn H, Giansanti P, Cristobal A, Toonen P, Schafer S, Hübner N, van Breukelen B, Mohammed S, Cuppen E, Heck AJR, Guryev V. Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 5:1469–1478. doi: 10.1016/j.celrep.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Maccarrone G, Reckow S, Falkai P, Schmitt A, Turck CW. Shotgun mass spectrometry analysis of the human thalamus proteome. J Sep Sci. 2009;32:1231–1236. doi: 10.1002/jssc.200900008. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal cell types. Curr Biol. 2004;14:R497–R500. doi: 10.1016/j.cub.2004.06.035. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Gilissen E, Allman JM, Perl DP, Erwin JM, Hof PR. A neuronal morphologic type unique to humans and great apes. Proc Natl Acad Sci USA. 1999;96:5268–5273. doi: 10.1073/pnas.96.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom. 2007;260:212–221. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Shi M, Jin J, Albin RL, Lieberman A, Gearing M, Lin B, Pan C, Yan X, Kashima DT, Zhang J. Proteomics identification of proteins in human cortex using multidimensional separations and MALDI tandem mass spectrometer. Mol Cell Proteomics. 2007;6:1818–1823. doi: 10.1074/mcp.M700158-MCP200. [DOI] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mouncastle J, Moseley MA, Thompson JW, Soderblom EJ, Iriki A, Kato M, Gilbert MTP, Zhang G, Bakken T, Bongaarts, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontén F, Gry M, Fagerberg L, Lundberg E, Asplund A, Berglund L, Oksvold P, Björling E, Hober S, Kampf C, Navani S, Nilsson P, Ottosson J, Persson A, Wernérus H, Wester K, Uhlén M. A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, Knight JR, Mullikin JC, Meader SJ, Ponting CP, Lunter G, Higashino S, Hobolth A, Dutheil J, Karakoç E, Alkan C, Sajjadian S, Catacchio CR, Ventura M, Marques-Bonet T, Eichler EE, André C, Atencia R, Mugisha L, Junhold J, Patterson N, Siebauer M, Good JM, Fischer A, Ptak SE, Lachmann M, Symer DE, Mailund T, Schierup MH, Andrés AM, Kelso J, Pääbo S. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Conley T, Sudduth J, Erwin JM, Stimpson CD, Hof PR, Sherwood CC. Neuropeptide Y-immunoreactive neurons in the cerebral cortex of humans and other haplorrhine primates. Am J Primatol. 2013;75:415–424. doi: 10.1002/ajp.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Edler MK, Meindl RS, Sudduth J, Bohush T, Erwin JM, Stimpson CD, Hof PR, Sherwood CC. Humans and great apes share increased neocortical neuropeptide Y innervation compared to other haplorhine primates. Front Hum Neurosci. 2014;8:101. doi: 10.3389/fnhum.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Stimpson CD, Marcinkiewicz JL, Erwin JM, Hof PR, Sherwood CC. Cortical dopaminergic innervation among humans, chimpanzees, and macaque monkeys: a comparative study. Neuroscience. 2008;155:203–220. doi: 10.1016/j.neuroscience.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP, Wang R. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics. 2009;25:1397–1403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TEJ. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Rockland KS, Pandya DN. Laminar origins and terminations of cortical connections of the occipital lobe in the rhesus monkey. Brain Res. 1979;179:3–20. doi: 10.1016/0006-8993(79)90485-2. [DOI] [PubMed] [Google Scholar]
- Schenker NM, Hopkins WD, Spocter MA, Garrison AR, Stimpson CD, Erwin JM, Hof PR, Sherwood CC. Broca’s area homologue in chimpanzees (Pan troglodytes): probabilistic mapping, asymmetry, and comparison to humans. Cereb Cortex. 2010;20:730–742. doi: 10.1093/cercor/bhp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex. 2011;21:1485–1497. doi: 10.1093/cercor/bhq191. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: tracing evolutionary changes in brain and cognition. J Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Seymour B, O’Doherty J, Kaube H, Dolan RJ, Frith CD. Empathy for pain involves the affective but not sensory components of pain. Science. 2004;303:1157–1162. doi: 10.1126/science.1093535. [DOI] [PubMed] [Google Scholar]
- Spocter MA, Hopkins WD, Barks SK, Bianchi S, Hehmeyer AE, Anderson SM, Stimpson CD, Fobbs AJ, Hof PR, Sherwood CC. Neuropil distribution in the cerebral cortex differs between humans and chimpanzees. J Comp Neurol. 2012;520:2917–2929. doi: 10.1002/cne.23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2005;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Uddin M, Opazo JC, Wildman DE, Sherwood CC, Hof PR, Goodman M, Grossman LI. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Mann M. Mass spectrometry-based proteomics in cell biology. J Cell Biol. 2010;190:491–500. doi: 10.1083/jcb.201004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Mol Ecol. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Wilhem M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzman L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina, Boese J-H, Bantsceff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Geschwind DH. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu AG, He L, Li Z, Xu Y, Li M, Fu X, Yan Z, Yuan Y, Menzel C, Li N, Somel M, Hu H, Chen W, Pääbo S, Khaitovich P. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput Biol. 2010;6:e1000843. doi: 10.1371/journal.pcbi.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, Smith KA, Ebbert AJ, Swanson B, Kuan L, Page DT, Overly CC, Lein ES, Hawrylycz MJ, Hof PR, Hyde TM, Kleinman JE, Jones AR. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJC, Carr SA, Tabb DL, Coffey RJ, Slebos RJC, Liebler DC National Cancer Institute Clinical Proteomics Tumor Analysis Consortium. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ueberheide BM, Waldemarson S, Myung S, Molloy K, Eriksson J, Chait BT, Neubert TA, Fenyö D. Protein quantification using mass spectrometry. Methods Mol Biol. 2010;673:211–222. doi: 10.1007/978-1-60761-842-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centenary of Brodmann’s map — conception and fate. Nat Rev Neurosci. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]