Significance
Toll-like receptors (TLRs) activate the innate immune system in response to invading pathogens. The single-stranded RNA (ssRNA) sensor TLR8 requires proteolytic cleavage at the loop region (Z-loop) for its activation. Biochemical and biophysical studies have revealed that the uncleaved Z-loop prevents formation of the TLR8 dimer, which is essential for its activation. Crystallographic analysis demonstrated that the uncleaved Z-loop located on the ascending lateral face prevents the approach of the dimerization partner by steric hindrance. Like TLR8, TLR7 and TLR9 also contain the Z-loop, and thus these proteins also might have this autoinhibition mechanism.
Keywords: Toll-like receptor, innate immunity, proteolytic processing, X-ray crystallography, Z-loop
Abstract
Toll-like receptor 8 (TLR8) senses single-stranded RNA (ssRNA) and initiates innate immune responses. TLR8 requires proteolytic cleavage at the loop region (Z-loop) between leucine-rich repeat (LRR) 14 and LRR15 for its activation. However, the molecular basis of Z-loop processing remains unknown. To elucidate the mechanism of Z-loop processing, we performed biochemical and structural studies of how the Z-loop affects the function of TLR8. TLR8 with the uncleaved Z-loop is unable to form a dimer, which is essential for activation, irrespective of the presence of agonistic ligands. Crystallographic analysis revealed that the uncleaved Z-loop located on the ascending lateral face prevents the approach of the dimerization partner by steric hindrance. This autoinhibition mechanism of dimerization by the Z-loop might be occurring in the proteins of the same subfamily, TLR7 and TLR9.
Toll-like receptors (TLRs) constitute a family of innate immune receptors that recognize pathogen-associated molecular patterns (1). The TLR molecule is a type I transmembrane protein characterized by an extracellular leucine-rich repeat (LRR) domain, a transmembrane helix, and an intracellular Toll/interleukin-1 receptor (TIR) homology domain (2). The typical TLR molecule is considered to be monomeric in the absence of ligands, transforming into an activated dimer form on ligand binding, which allows for dimerization of the intracellular TIR domain and subsequent signaling (2).
The TLR subfamily comprising TLR7, TLR8, and TLR9 recognizes single-stranded (ss) nucleic acids from viruses and bacteria (3). Specifically, TLR7 and TLR8 recognize uridine- and guanosine-rich single-stranded RNA (ssRNA) (4–11), whereas TLR9 recognizes ssDNA containing the unmethylated cytosine-phosphate-guanine (CpG) dideoxynucleotide motif (12). Furthermore, TLR7 and TLR8 are also activated by synthetic chemical compounds (13, 14), such as imiquimod (TLR7-specific), resiquimod (R848; both TLR7 and TLR8), and CL075 (both TLR7 and TLR8).
Certain regulation mechanisms of the functions of the TLR7–9 subfamily members are shared because of a high degree of sequence similarities (3). They reside on the endosomal membrane, and their transportation from endoplasmic reticulum (ER) to endolysosomes is mediated by the ER membrane protein Unc93B1 (15). Moreover, TLR7–9 possess a long inserted loop region (Z-loop), consisting of ∼30 amino acid residues, between LRR14 and LRR15, and the processing by proteolytic cleavage at the Z-loop is believed to be indispensable for their function (16–21). Specifically, the processing at the Z-loop of human TLR8 mediated by furin-like proprotein convertase and cathepsins produces functional TLR8 capable of ligand binding and signaling in endolysosomes. In addition, the cleaved form of TLR8 has been found to be predominant in immune cells (16). Recent structural studies demonstrate that the N- and C-terminal halves of TLR8 after Z-loop cleavage associate with each other, and that both fragments are cooperatively involved in ligand binding (22). Moreover, a recent study revealed that the latter half of the cleaved Z-loop interacts with LRRs to stabilize the TLR8 structure and contributes to ssRNA recognition by TLR8 (23).
Although accumulating evidence illustrates the functional importance of Z-loop processing at the cellular level, mechanistic insights into this processing in the regulation of TLR8 function at the molecular level are lacking. Here, to unveil the mechanistic role of Z-loop processing of TLR8, we present the results of a combined structural and biochemical investigation of TLR8 with the uncleaved Z-loop.
Results
Construction of Mutant TLR8 with the Uncleaved Z-Loop.
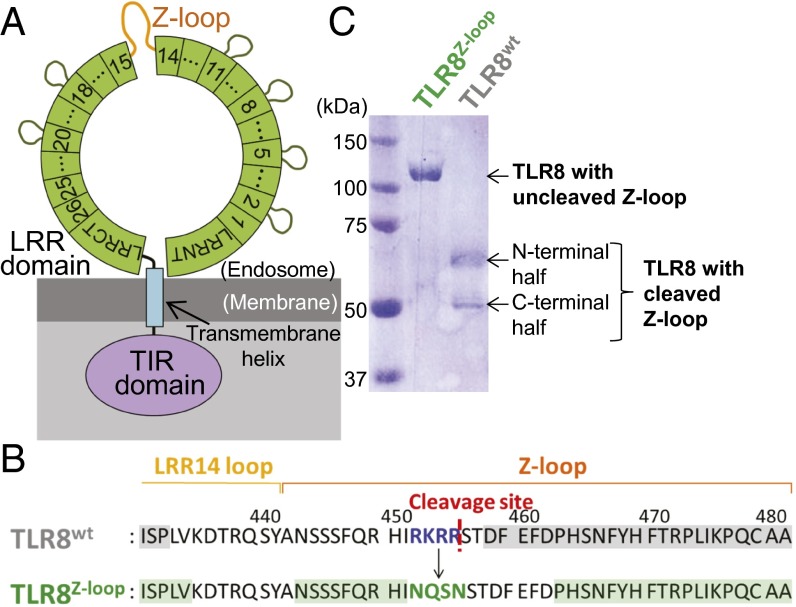
The ectodomain of human TLR8 consists of 26 LRRs with the Z-loop (residues 442–481) between LRR14 and LRR15 (Fig. 1 A and B). Wild-type TLR8 (TLR8wt) is already cleaved at the Z-loop by an unknown protease when expressed in S2 cells, resulting in two bands, at 60 kDa and 50 kDa, on SDS/PAGE, corresponding to the N-terminal and C-terminal fragments, respectively (22) (Fig. 1C, right lane). N-terminal peptide sequencing of the C-terminal fragment indicated that the cleavage occurred after Arg455 (Fig. 1B). The tetrabasic amino acid sequence before the cleavage site, Arg452-Lys453-Arg454-Arg455, suggested that a furin-like protease might be involved in the cleavage (24). To obtain TLR8 with an uncleaved Z-loop, we designed a mutant TLR8 in which RKRR (residues 452–455) was replaced by NQSN (Fig. 1B) to avoid proteolytic cleavage. After expression and purification of this mutant TLR8, no cleaved TLR8 was detected, demonstrating that we successfully obtained TLR8 with an uncleaved Z-loop (Fig. 1C, left lane). Hereinafter, we refer to the mutant TLR8 with the uncleaved Z-loop as TLR8Z-loop.
Fig. 1.
Z-loop cleavage of human TLR8. (A) Schematic representation of the domain organization of human TLR8. The ring (green), rectangular box (blue), and oval (purple) represent the extracellular LRR domain, transmembrane helix, and intracellular TIR domain, respectively. LRRs are indicated by numbered boxes. The Z-loops are shown in orange. (B) Sequence of TLR8wt (Top) and TLR8Z-loop (Bottom). The cleavage site in TLR8wt is shown as a red dashed line. Mutation sites are shown in blue (TLR8wt) and green (TLR8Z-loop). The residues visible in the electron density maps are highlighted in gray (TLR8wt) and green (TLR8Z-loop). (C) SDS/PAGE (10% polyacrylamide) analysis of the purified TLR8Z-loop (Right) and TLR8wt (Left) under reducing conditions.
TLR8Z-loop Is Monomeric and Inactive.
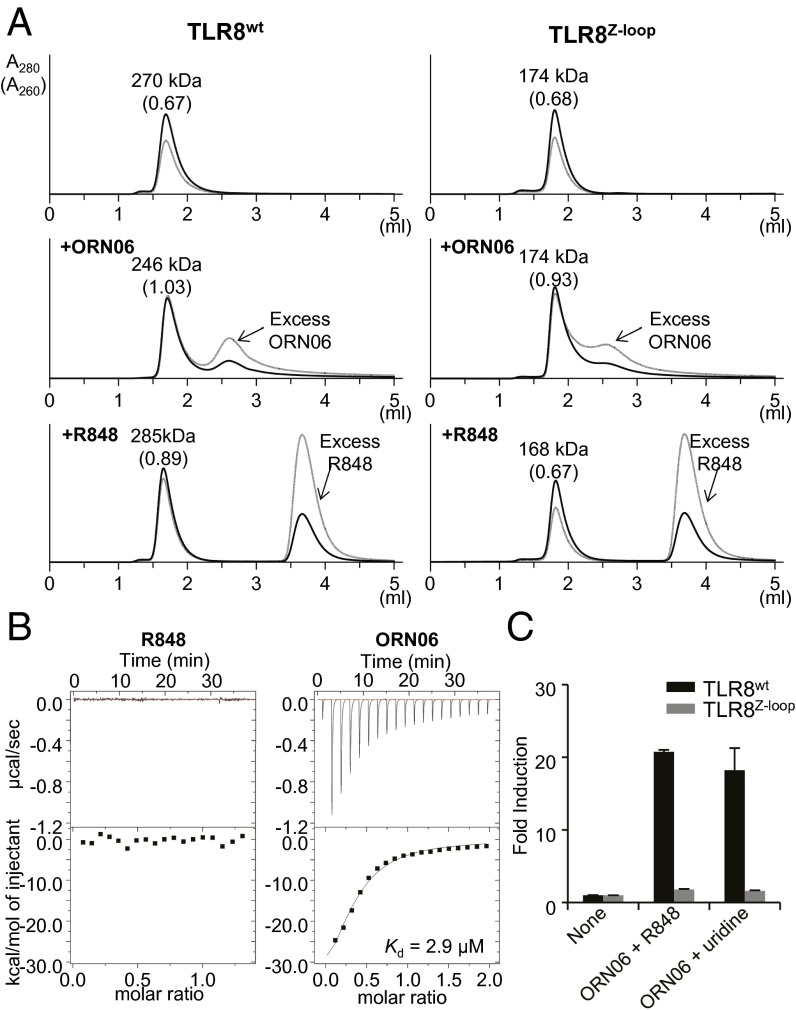
To determine the oligomerization state of TLR8Z-loop, we performed gel-filtration chromatography analyses of TLR8wt and TLR8Z-loop in the absence and presence of agonistic ligands (chemical ligand R848 and ssRNA ORN06). TLR8wt was eluted as a dimer irrespective of the presence of ligands (Fig. 2A), as was previously shown by crystallography using the ectodomain and cellular experiments using full-length protein (22, 25). In contrast, TLR8Z-loop was eluted at a greater retention volume, corresponding to a monomer, in both the absence and the presence of agonistic ligands (Fig. 2A). Of note, the ratio of absorbance values at 260 nm and 280 nm increased in the presence of ORN06, indicating that TLR8Z-loop could bind ssRNA as TLR8wt could. This result coincides well with the fact that an oligonucleotide binds to the second site irrespective of dimerization, as discussed below.
Fig. 2.
Oligomerization state and ligand binding of TLR8Z-loop. (A) Gel filtration chromatography of TLR8wt and TLR8Z-loop. TLR8wt (Left, Top), TLR8wt/ORN06 (Left, Middle), TLR8wt/R848 (Left, Bottom), TLR8Z-loop (Right, Top), TLR8Z-loop/ORN06 (Right, Middle), and TLR8Z-loop/R848 (Right, Bottom) were analyzed by Superdex 200 gel filtration chromatography. The black and gray lines show absorbance at 280 nm (A280) and 260 nm (A260), respectively. Molecular weights estimated from elution volume are shown above the peak. The ratios of A260 to A280 are given in parentheses. (B) ITC analysis of TLR8Z-loop titrated with R848 (Left) and ORN06 (Right). (C) NF-κB activation by TLR8WT and TLR8Z-loop. The response to agonistic ligand under synergistic conditions was assessed using NF-κB–dependent luciferase assay. To induce synergy, cells were treated for 6 h by indicated TLR8 ligands. Data represent the mean fold induction of NF-κB activity, calculated as the RLU of stimulated cells divided by the RLU of nonstimulated cells. Data from three independent experiments are shown.
We evaluated whether Z-loop cleavage affects ligand binding through isothermal titration calorimetry (ITC) analyses (Fig. 2B). TLR8Z-loop exhibited an affinity for ORN06 with a dissociation constant (Kd) value of 2.9 μM, comparable to that of TLR8wt (4.8 μM) (23); however, we did not detect any heat release or absorption when R848 was titrated into TLR8Z-loop (Fig. 2B), indicating a very weak binding affinity of R848 to TLR8Z-loop compared with that of TLR8wt for R848 (Kd = 0.20 μM) (23).
To confirm the activity of TLR8Z-loop, we next performed luciferase assays using nuclear factor κB (NF-κB) reporter plasmid. Because TLR8 demonstrates enhanced activation by uridine or chemical ligands in a synergistic manner in the presence of oligonucleotides (23), we examined the ability of TLR8 to activate NF-κB under synergistic conditions. TLR8Z-loop could not activate NF-κB even under synergistic conditions (Fig. 2C), demonstrating that TLR8Z-loop was completely inactive.
Crystal Structure of TLR8Z-loop.
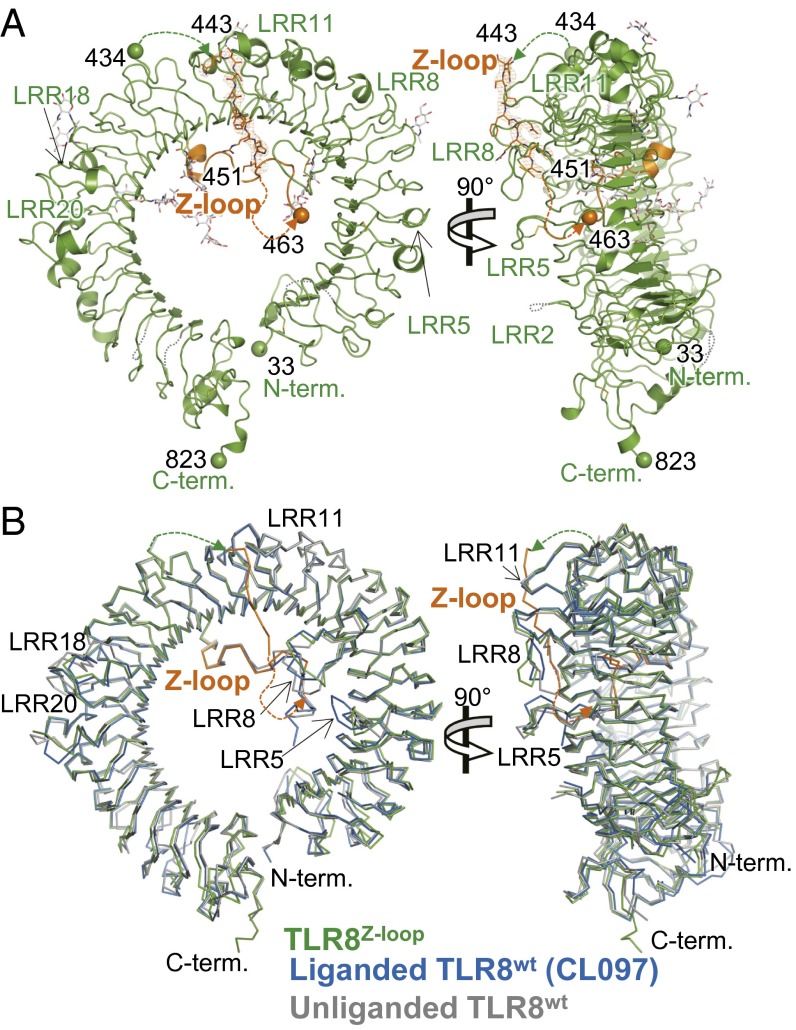
To gain structural insight into TLR8Z-loop, we determined its crystal structure at 2.6 Å resolution (Table 1). In agreement with the gel filtration analysis, TLR8Z-loop was monomeric in the crystal. The structure of TLR8Z-loop was ring-shaped, and N and C termini interacted directly in a manner similar to that seen in TLR8wt (22) (Fig. 3A). The overall structure of TLR8Z-loop was essentially the same as that of TLR8wt; superpositions of the TLR8Z-loop with unliganded [Protein Data Bank (PDB) ID code 3W3G] and liganded (TLR8wt/CL097; PDB ID code 3W3J) forms of TLR8wt yielded rmsd values of 1.0 Å and 0.9 Å, respectively (Fig. 3B). However, LRR8 and LRR18 of TLR8Z-loop exhibited large structural differences compared with the unliganded form of TLR8wt, possibly due to the absence of the dimerization partner or the presence of the uncleaved Z-loop.
Table 1.
Data collection and refinement statistics
| Crystal | TLR8Z-loop |
| Data collection | |
| X-ray source | PF-AR NE3A |
| Wavelength, Å | 1.0000 |
| Space group | R32 |
| Unit cell parameters, Å | |
| a | 171.5 |
| b | 171.5 |
| c | 301.3 |
| Resolution, Å | 2.6 |
| Completeness, % | 100.0 (100.0) |
| Redundancy | 19.5 (19.5) |
| Rmerge, I* | 0.127 (1.146) |
| Average, I/σ(I) | 17.3 (3.1) |
| Refinement | |
| Resolution range, Å | 133.2–2.6 |
| No. of reflections used | 49,947 |
| Model | 1×TLR8 |
| R, %† | 20.6 |
| Rfree, %‡ | 23.9 |
| rmsd | |
| Bond length, Å | 0.012 |
| Bond angles, ° | 1.77 |
Values in parentheses are for the shell with the highest resolution.
Rmerge(I) = Σ|I − <I>|/ΣI, where I is the diffraction intensity.
R = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure amplitudes, respectively.
Rfree is an R value for a 5% subset of all reflections, but was not used in the refinement.
Fig. 3.
Crystal structure of TLR8Z-loop. (A) Front (Left) and side (Right) views of TLR8Z-loop structure. The LRR structure and Z-loop are shown in green and orange, respectively. N-glycans and disulfide bonds are shown as gray and yellow sticks, respectively. The N and C termini of each fragment are shown as spheres. The dashed lines show unmodeled regions in the structure. (B) Superposition of TLR8Z-loop with TLR8wt. TLR8Z-loop (green), one protomer from unliganded TLR8wt dimer (gray; PDB ID code 3W3G), and one protomer from liganded TLR8wt dimer (blue, TLR8/CL097; PDB ID code 3W3J) are superposed and shown in Cɑ trace models. The Z-loop in TLR8Z-loop is shown in orange.
In the structure of TLR8wt, electron density for the first half of the Z-loop was missing, implying that this region was disordered (22). In sharp contrast, in the structure of TLR8Z-loop, the electron density corresponding to a part of the Z-loop was observed on the ascending lateral face near the loop region of LRR11 and was assigned as residues Asn443 to Ile451 (NSSSFQRHI) (Figs. 1B and 3A). This part of the Z-loop was sandwiched by LRR8 and LRR11–LRR13 of one monomer in an asymmetric unit, and by LRR18–LRR19 of its crystallographic symmetry molecule. Thus, the Z-loop was confined to this position. The high average B-factor of 113 Å2 of this region of the Z-loop suggests that it was not tightly stuck to the protein surface. Because this part of the Z-loop was not visible in the TLR8wt structure (Fig. 1B), in an earlier study we could not define which side of the lateral face of TLR8 the Z-loop passed (22); however, the present structural study allows us to confidently establish that the uncleaved Z-loop connecting residues 434–463 passed the ascending lateral face, as shown in Fig. 3A. The latter half of the Z-loop (from residue 463) was observed in the same position as TLR8wt (Fig. 3B). The remaining regions of the Z-loop (residues 435–442 and 452–462) were not visible in the electron density map in TLR8Z-loop.
The Uncleaved Z-Loop Prevents Dimerization of TLR8.
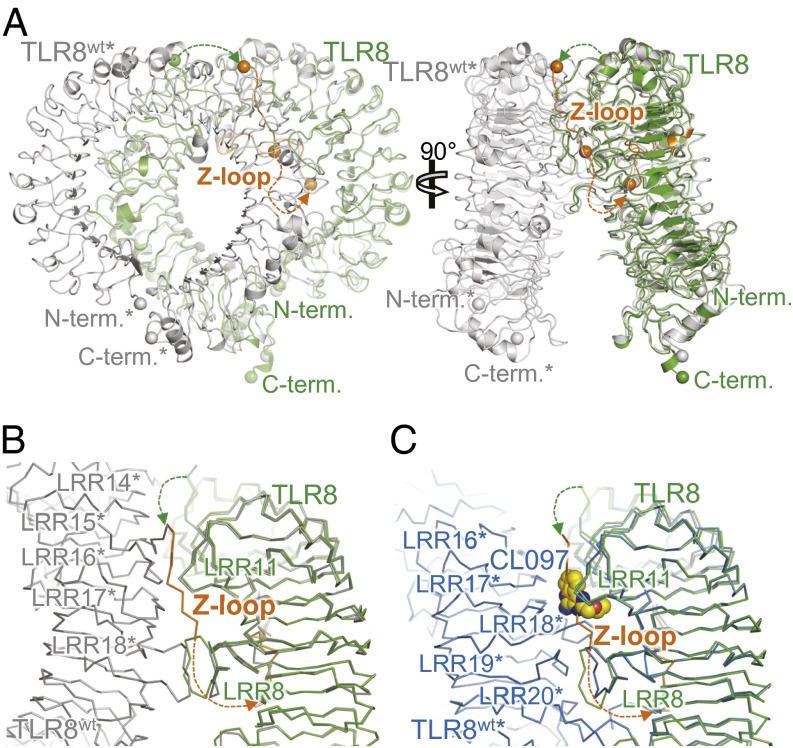
The fact that the uncleaved Z-loop passed the ascending lateral face of TLR8, which was engaged in the dimerization of TLR8, clearly allows us to explain why uncleaved TLR8 is unable to dimerize. Heavy steric clashes with the dimerization partner of TLR8wt* (the asterisk denotes the dimerization partner) occur when TLR8Z-loop is superimposed with TLR8wt in both the unliganded and liganded forms (Fig. 4A). When TLR8Z-loop was superposed with the unliganded form of TLR8wt, a part of the Z-loop (residues 443–451) of TLR8Z-loop clashed with LRR14* and LRR15* of TLR8wt*. In addition, LRR8 clashed with LRR18*, whereas LRR8 interacted with LRR18* (and LRR18* with LRR8) in the unliganded form of TLR8wt (Fig. 4B). These steric clashes prohibited TLR8 from dimerization, resulting in the inactive form.
Fig. 4.
The Z-loop inhibits TLR8 dimerization. (A) Superposition of TLR8Z-loop (green) and unliganded TLR8wt dimer (gray) structures. (B) Close-up view of LRR14–LRR18 in TLR8Z-loop (green) superimposed on unliganded TLR8wt dimer (gray) in Cɑ trace models. (C) Close-up view of LRR8–LRR11 TLR8Z-loop (green) superimposed on liganded TLR8wt dimer (blue, TLR8/CL097) in Cɑ trace models. The CL097 molecule is shown in a space-filling representation.
Similarly, when TLR8Z-loop was superposed with the liganded form of TLR8wt (TLR8wt/CL097 complex), a part of the Z-loop (residues 443–451) of TLR8Z-loop clashed with LRR18* of TLR8wt* (Fig. 4C). Moreover, this part of the Z-loop overlapped completely with the chemical ligands.
Reconstruction of Functional TLR8 by N- and C-Terminal Fragments.
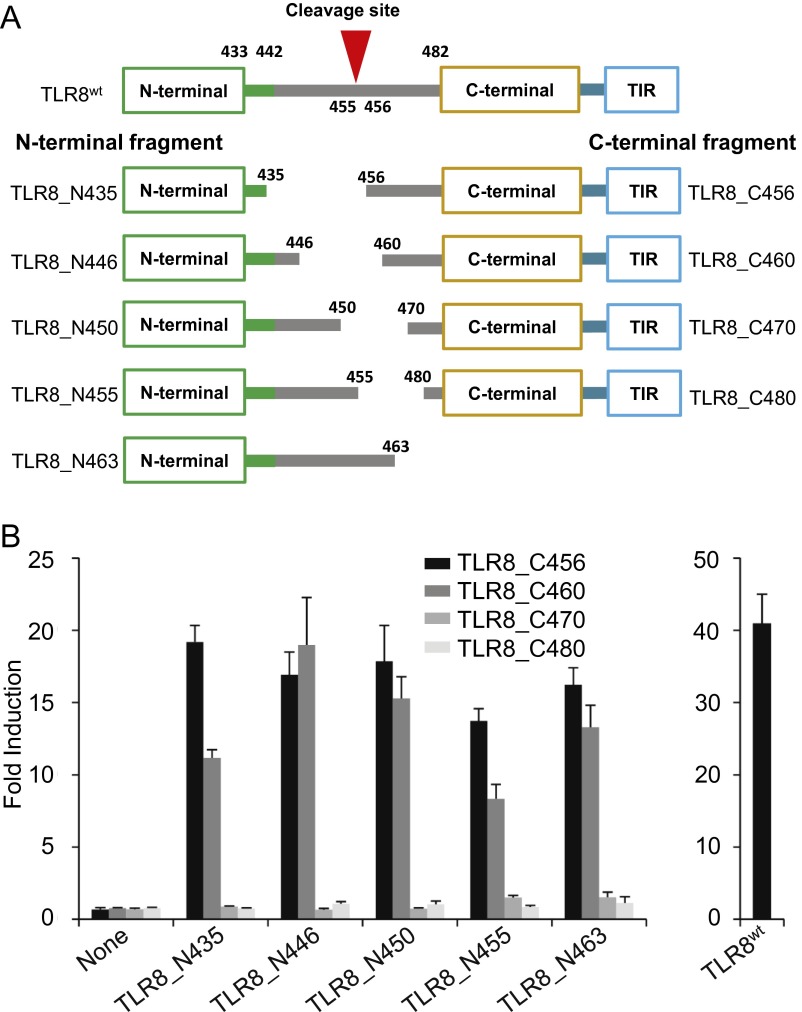
Our structural study demonstrated that an uncleaved TLR8 is unable to dimerize and signal. To strengthen our observations, we generated deletion mutants of TLR8 that independently encode the N-terminal fragment (TLR8_N435, TLR8_N446, TLR8_N450, TLR8_N455, TLR8_N463) or the C-terminal fragment (TLR8_C456, TLR8_C460, TLR8_C470, TLR8_C480) (Fig. S1A). These fragments were cotransfected into HEK293T cells, and the reconstruction of functional TLR8 was assessed by the response to the TLR8 ligand using an NF-κB–dependent luciferase reporter assay. All N-terminal fragments combined with TLR8_C456 and TLR8_C460 showed responsiveness to the ligand, whereas those combined with TLR8_C470 and TLR8_C480 failed to respond to the ligand (Fig. S1B).
Fig. S1.
Reconstruction of functional TLR8 by N- and C-terminal fragments. (A) Schematic representation of various N- and C-terminal deletion mutants of human TLR8 used in the present study. The numbers denote the amino acid positions in each construct. The LRR14 loop (residues 433–441) and Z-loop (residues 442–481) are depicted in green and gray lines, respectively. (B) NF-κB activation in HEK293T cells expressing indicated human TLR8 fragments and TLR8WT. The response was assessed using an NF-κB–dependent luciferase assay stimulated by 5 mM uridine and 25 μg/mL ssRNA40 complexed with DOTAP. Data represent the mean fold induction of NF-κB activity, calculated as the RLU of stimulated cells divided by the RLU of nonstimulated cells. Data from six independent samples are shown.
Structural studies have demonstrated that the latter half of the Z-loop (after Asp458) contributes in part to stabilization and ligand recognition by making contact with the concave face of the LRR structure (22, 23). Because TLR8_C470 and TLR8_C480 lack this region, activities were not observed for these mutants. These results are consistent with structural observation that N- and C-terminal portions of the cleaved TLR8 remain associated after proteolytic processing (22). Also consistently, Ishii et al. (16) demonstrated a noncovalent association of the N-terminal half of TLR8 with the C-terminal half detected by immunoprecipitation assay in RAW macrophages stably expressing processed human TLR8.
Discussion
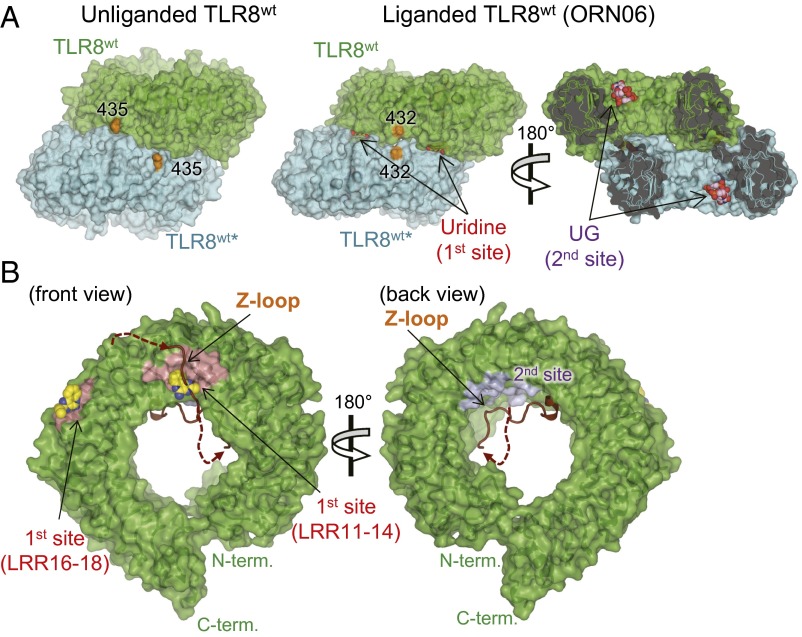
The functional importance of Z-loop processing in TLR7–9 subfamily members at the cellular level has been demonstrated by several groups (16–21) and has been widely accepted. However, mechanistic insights into Z-loop processing at the molecular level have not been provided thus far, because of the lack of biochemical and structural studies using purified protein. We successfully obtained purified TLR8 with an uncleaved Z-loop and characterized it biochemically and structurally. We found that TLR8Z-loop was monomeric irrespective of the presence of agonistic ligands (Fig. 2A), unlike TLR8wt, which forms a dimer in both the unliganded and liganded forms (22). On structural analysis, the uncleaved Z-loop was located in front of the ascending lateral face of TLR8, which was involved in the dimerization, as we had anticipated in an earlier study (26) (Fig. 3A). Uncleaved TLR8 is unable to form a dimer, because there is no space to accommodate the Z-loop between the two protomers of the dimer in both the unliganded and liganded forms (Fig. 5A).
Fig. 5.
Surface representations of TLR8Z-loop and TLR8wt. (A) Surface representations of the unliganded TLR8wt (Left) and liganded TLR8wt (Right, TLR8/ORN06; PDB ID code 4R07). TLR8wt protomers are shown in green (TLR8wt) and cyan (TLR8wt*). Uridine (first site) and UG (second site) molecules are shown in a space-filling representation. The C atoms of the ligands are colored yellow (uridine) or purple (UG), and the O, N, and P atoms of the ligands are colored red, blue, and orange, respectively. The end of the LRR14 loop is in orange. (B) Surface representations of front (Left) and back (Right) views of TLR8Z-loop. The LRR14 loop and the Z-loop are shown in brown in a cartoon model. The first and second sites are pink and purple, respectively. The chemical ligand (CL097) in TLR8WT/CL097 shown as a sphere is overlaid on TLR8Z-loop structure.
A recent crystallographic study demonstrated that TLR8 has two ligand-binding sites (first and second sites) (23) (Fig. 5B). The first site, composed of LRR11–14 and LRR16*–18* from two protomers of TLR8, accommodates small molecules such as chemical ligands and uridine mononucleoside. The second site, located on the concave surface of the ring-like structure of one TLR8 protomer, accommodates ssRNA longer than 2-mer. Dimerization of TLR8 is necessary for the binding of small molecules to the first site, but not for ssRNA to the second site. This is in agreement with our observations that TLR8Z-loop could bind to ssRNA but not to the chemical ligand, as shown by gel filtration chromatography and ITC analyses (Fig. 2). The monomeric TLR8Z-loop no longer interacts with chemical ligands. On the other hand, the second site is outside the dimerization interface and is not affected by Z-loop cleavage (Fig. 5A); therefore, TLR8Z-loop still interacts with ssRNA at the second site with an affinity similar to that for TLR8wt.
The autoinhibition mechanism of dimerization by the uncleaved Z-loop should apply to TLR7 and TLR9. In agreement with the results of TLR8, TLR9 with an uncleaved Z-loop is unable to dimerize, although it binds agonistic DNA, whereas TLR9 cleaved at the Z-loop dimerizes in response to agonistic DNA (27). With respect to the regulation of dimerization, the precise position of Z-loop cleavage might be only marginally important, considering the fact that the sequences of Z-loops among TLR7–9 are not conserved at all and multiple proteases are involved in Z-loop cleavage (16–21). For instance, TLR9 artificially cleaved at the Z-loop by V8 protease can form the activated form of the receptor (27).
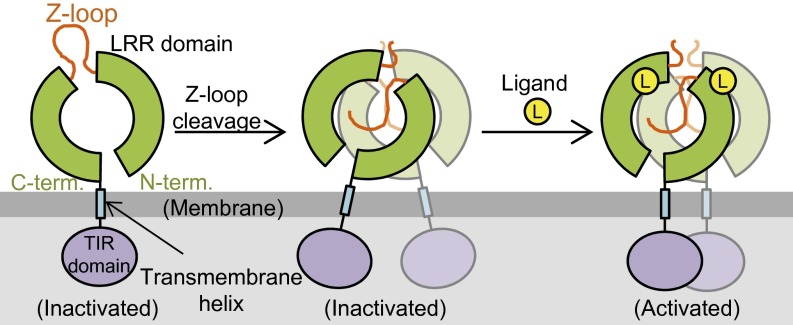
In conclusion, TLR8 with the intact Z-loop cannot form the preformed and subsequently activated dimer owing to the steric hindrance of the Z-loop located on the ascending lateral face. Thus, TLR8 with the intact Z-loop is inactive. On Z-loop processing by proteolytic cleavage, the autoinhibition of TLR8 is relieved to allow the dimerization of TLR8 (Fig. 6). This study represents an important step forward in understanding the regulatory mechanism for activation of the TLR7–9 subfamily.
Fig. 6.
Proposed activation mechanism of TLR8 by Z-loop cleavage. TLR8 with an uncleaved Z-loop is a monomer (Left, inactive). On Z-loop cleavage, it forms an unliganded dimer (Middle, inactive). By binding to agonistic ligands, the preformed dimer transforms into the activated dimer (Right, active).
Materials and Methods
Purification and Crystallization.
The extracellular domain of human TLR8 (TLR8wt, residues 27–827) was prepared as described previously (22). The extracellular domain of TLR8 with mutations RKRR to NQSN at residues 452–455 (TLR8Z-loop) was prepared similarly with minor modifications in the purification steps. TLR8Z-loop protein was purified by IgG Sepharose affinity chromatography, Protein A tag cleavage by thrombin, saccharide trimming by Endo Hf, and Superdex 200 gel filtration chromatography followed by HiTrap SP cation-exchange chromatography. The purified TLR8Z-loop was concentrated to 8 mg/mL in 10 mM MES, pH 5.5, and 50 mM NaCl. Crystals of TLR8Z-loop were obtained using the sitting-drop vapor-diffusion method at 293 K by mixing the protein solution with an equal volume of reservoir solution (2.1 M ammonium sulfate, 0.1 M sodium acetate, pH 4.8, and 20–30% glycerol).
Data Collection and Structure Determination.
Diffraction datasets were collected on beamlines PF-AR NE3A (Ibaraki, Japan) under cryogenic conditions at 100 K. Crystals of TLR8Z-loop were soaked in a cryoprotectant solution containing 1.5 M ammonium sulfate, 75 mM sodium acetate pH 4.8, 8 mM MES pH 5.5, 75 mM NaCl, and 35% glycerol before flash-cooling.
The dataset was processed using XDS (28). The structure of TLR8Z-loop was determined by a molecular replacement method using the Molrep program (29) using the TLR8wt/CL097 structure (PDB ID code 3W3J). The structural model was refined with stepwise cycles of manual model building using Coot (30), and restrained refinement using REFMAC (31) until the R factors converged. The quality of the final structure was evaluated with PROCHECK (32). The statistics of the data collection and refinement are summarized in Table 1. The figures representing structures were prepared with PyMOL (33). The coordinates and structure-factor data have been deposited in the PDB (PDB ID code 5HDH).
Gel Filtration Chromatography.
Gel filtration chromatography experiments were done in a buffer composed of 10 mM MES, pH 5.5, and 150 mM NaCl using Superdex 200 Increase 5/150 GL column (GE Healthcare). The injection samples (total volume, 50 μL) contained 0.1 mM TLR8wt or TLR8Z-loop with/without 0.1 mM ORN06 or 0.5 mM R848. The molecular weights were calculated based on the elution volumes of molecular weight standards, including apoferritin (440 kDa), β-amylase (200 kDa), BSA (67 kDa), and cytochrome c (12 kDa).
ITC.
ITC experiments were conducted in a buffer composed of 10 mM MES, pH 5.5, and 100 mM NaCl at 298 K in a MicroCal iTC200 ITC system (GE Healthcare). The titration sequence included a single 0.4-μL injection followed by 18 injections of 2 μL each, with an interval of 120 s between injections. The titration conditions were as follows: 100 μM R848 into 15 μM TLR8Z-loop and 300 μM ORN06 into 30 μM TLR8Z-loop. OriginLab software (GE Healthcare) was used to analyze the raw ITC data.
Luciferase Reporter Gene Assay.
For this assay, 1 × 106 HEK293T cells seeded in collagen-coated six-well plates were transiently transfected with wild type or mutant human TLR8 cDNAs in pMX-puro-IRES-rat CD2 (2 μg) and pELAM1-luc reporter plasmid (5 ng), using Polyethylenimine Max (Polysciences). At 20 h after transfection, transfected cells were reseeded in collagen-coated flat 96-well plates (Corning) at a density of 1 × 105 cells per well. After preincubation for 6 h in 96-well plates, cells were stimulated for 6 h with 1 μg/mL R848 and 25 μg/mL ORN06, or 1 mM uridine and 25 μg/mL ORN06 complexed with DOTAP, and subjected to a luciferase assay using the Promega Luciferase Assay System. Relative light unit (RLU) chemiluminescence values was measured with a luminometer (MiniLumat LB9506; Berthold).
SI Materials and Methods
pMX-based retroviral expression vectors (kindly provided by Dr. T. Kitamura, Tokyo, Japan) were used to express N- and C-terminal fragments of human TLR8 in HEK293T cells. C-terminal human TLR8 fragments corresponding to amino acids 456–827 (TLR8_C456), 460–827 (TLR8_C460), 470–827 (TLR8_C470), or 480–827 (TLR8_C480) with amino acids 1–26, the signal peptide, and subsequent five amino acids (EENFS) on the N terminus were tagged with Flag-6×His and cloned into pMXpuro vector. Human TLR8 fragments corresponding to amino acids 1–435 (TLR8_N435), 1–446 (TLR8_N446), 1–450 (TLR8_N450), 1–455 (TLR8_N455), or 1–463 (TLR8_N463) were cloned into pMX_ IRES_rat CD2 vector.
We coexpressed all combinations of N- and C-terminal fragments in HEK293T cells and detected NF-κB activation by the luciferase reporter assay. In brief, 2 × 105 HEK293T cells seeded in collagen-coated 24-well plates were transiently transfected with N- and C-terminal human TLR8 cDNAs (200 ng each), together with mouse Unc93B1 in pMX vector (200 ng) and pELAM1-luc reporter plasmid (2 ng), using Polyethylenimine Max (Polysciences). At 24 h after transfection, transfected cells, reseeded in collagen-coated flat 96-well plates (Corning) at a density of 1 × 105 cells per well, were stimulated with 5 mM uridine and 25 μg/mL ssRNA40 complexed with DOTAP for 12 h, and then subjected to a luciferase assay using the Promega Luciferase Assay System. The RLU of luciferase activity was measured with the Promega GloMax Explorer System.
Acknowledgments
We thank the beamline staff members at the Photon Factory for their assistance with data collection. This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to H.T., U.O., K.M., and T. Shimizu) and by funding from the Japan Science and Technology Agency’s Core Research for Evolutional Science and Technology (CREST) Program (T. Shibata and T. Shimizu), the Takeda Science Foundation (U.O. and T. Shimizu), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (U.O.), and the Daiichi Sankyo Foundation of Life Science (U.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.M.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5HDH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516000113/-/DCSupplemental.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol Rev. 2012;250(1):216–229. doi: 10.1111/j.1600-065X.2012.01167.x. [DOI] [PubMed] [Google Scholar]
- 3.Wagner H. The immunobiology of the TLR9 subfamily. Trends Immunol. 2004;25(7):381–386. doi: 10.1016/j.it.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Wang JP, et al. Toll-like receptor–mediated activation of neutrophils by influenza A virus. Blood. 2008;112(5):2028–2034. doi: 10.1182/blood-2008-01-132860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsbach A, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180(6):3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 6.Diebold SS, et al. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36(12):3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 7.Triantafilou K, et al. TLR8 and TLR7 are involved in the host’s immune response to human parechovirus 1. Eur J Immunol. 2005;35(8):2416–2423. doi: 10.1002/eji.200526149. [DOI] [PubMed] [Google Scholar]
- 8.Barrat FJ, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202(8):1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heil F, et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 11.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 12.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 13.Jurk M, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3(6):499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 14.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3(2):196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 15.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 16.Ishii N, Funami K, Tatematsu M, Seya T, Matsumoto M. Endosomal localization of TLR8 confers distinctive proteolytic processing on human myeloid cells. J Immunol. 2014;193(10):5118–5128. doi: 10.4049/jimmunol.1401375. [DOI] [PubMed] [Google Scholar]
- 17.Hipp MM, et al. Processing of human Toll-like receptor 7 by furin-like proprotein convertases is required for its accumulation and activity in endosomes. Immunity. 2013;39(4):711–721. doi: 10.1016/j.immuni.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewald SE, et al. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208(4):643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sepulveda FE, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31(5):737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Park B, et al. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9(12):1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewald SE, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456(7222):658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science. 2013;339(6126):1426–1429. doi: 10.1126/science.1229159. [DOI] [PubMed] [Google Scholar]
- 23.Tanji H, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat Struct Mol Biol. 2015;22(2):109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- 24.Hosaka M, et al. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266(19):12127–12130. [PubMed] [Google Scholar]
- 25.Zhu J, et al. Characterization of bovine Toll-like receptor 8: Ligand specificity, signaling essential sites and dimerization. Mol Immunol. 2009;46(5):978–990. doi: 10.1016/j.molimm.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Ohto U, Tanji H, Shimizu T. Structure and function of Toll-like receptor 8. Microbes Infect. 2014;16(4):273–282. doi: 10.1016/j.micinf.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Ohto U, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520(7549):702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 33.Delano WL. 2002. The PyMOL molecular graphics system. Available at www.pymol.org. Accessed February 19, 2016.