Significance
Copper nitrite reductase (CuNiR) is involved in denitrification of the nitrogen cycle. Synchrotron X-rays rapidly reduce copper sites and decompose the substrate complex structure, which has made crystallographic studies of CuNiR difficult. Using femtosecond X-ray free electron lasers, we determined intact structures of CuNiR with and without nitrite. Based on the obtained structures, we proposed a redox-coupled proton switch model, which provides an explanation for proton-coupled electron transfer (PCET) in CuNiR. PCET is widely distributed through biogenic processes including respiratory and photosynthetic systems and is highly expected to be incorporated into bioinspired molecular devices. Our study also establishes the foundation for future studies on PCET in other systems.
Keywords: copper, bioinorganic chemistry, free electron laser, SAD phasing, damage-free structure
Abstract
Proton-coupled electron transfer (PCET), a ubiquitous phenomenon in biological systems, plays an essential role in copper nitrite reductase (CuNiR), the key metalloenzyme in microbial denitrification of the global nitrogen cycle. Analyses of the nitrite reduction mechanism in CuNiR with conventional synchrotron radiation crystallography (SRX) have been faced with difficulties, because X-ray photoreduction changes the native structures of metal centers and the enzyme–substrate complex. Using serial femtosecond crystallography (SFX), we determined the intact structures of CuNiR in the resting state and the nitrite complex (NC) state at 2.03- and 1.60-Å resolution, respectively. Furthermore, the SRX NC structure representing a transient state in the catalytic cycle was determined at 1.30-Å resolution. Comparison between SRX and SFX structures revealed that photoreduction changes the coordination manner of the substrate and that catalytically important His255 can switch hydrogen bond partners between the backbone carbonyl oxygen of nearby Glu279 and the side-chain hydroxyl group of Thr280. These findings, which SRX has failed to uncover, propose a redox-coupled proton switch for PCET. This concept can explain how proton transfer to the substrate is involved in intramolecular electron transfer and why substrate binding accelerates PCET. Our study demonstrates the potential of SFX as a powerful tool to study redox processes in metalloenzymes.
Since the invention of the Haber–Bosch process, the amount of fixed nitrogen in soils and waters has been increasing, and this trend has significant impact on the global environment (1, 2). Fixed nitrogen is oxidized to nitrite (NO2−) or nitrate (NO3−) by nitrification and then converted to gaseous dinitrogen (N2) by microbial denitrification, which closes the nitrogen cycle. Microorganisms involved in denitrification couple their respiratory systems to stepwise reduction of nitrogen oxides to N2 (NO3− → NO2− → NO → N2O → N2) (3, 4). The reduction of NO2− to toxic nitric oxide (NO2− + 2H+ + e− → NO + H2O) is referred to as the key step in denitrification and catalyzed by either cd1-heme nitrite reductase (cd1NiR) or copper nitrite reductase (CuNiR) (3, 4). Although the catalytic mechanism of cd1NiR is well understood (5, 6), that of CuNiR is controversial (7). CuNiR is a homotrimeric protein containing two distinct Cu sites per monomer (SI Appendix, Fig. S1). Type 1 Cu (T1Cu) with a Cys–Met–His2 ligand set is an electron acceptor incorporated near the molecular surface, whereas type 2 Cu (T2Cu) with a His3 ligand set is a catalytic center, which is ∼12 Å distant from the molecular surface and located between two adjacent monomers (7, 8). Spaced ∼12.5 Å apart, the two Cu sites are linked by a Cys–His bridge and a sensor loop. Whereas the Cys–His bridge is an electron pathway, the sensor loop is thought to control electron distribution between T1Cu and T2Cu (9).
Two conserved residues, Asp98 and His255 (Alcaligenes faecalis numbering), are located above the T2Cu site and bridged by a water molecule called bridging water (SI Appendix, Fig. S1). They are essential to the CuNiR activity because they assist proton transfer (PT) to the substrate (10–12). Although intramolecular electron transfer (ET) from T1Cu to T2Cu can occur in the resting state (RS) (13, 14), the differences in the redox potentials of T2Cu minus T1Cu are small and sometimes negative in the absence of NO2−, meaning that intramolecular ET before NO2− binding is not energetically favorable (15, 16). By contrast, intramolecular ET is dramatically accelerated in the presence of NO2− (15, 17). An explanation for this gating-like phenomenon is that substrate binding raises the redox potential of T2Cu and shifts the equilibrium of the ET reaction (16). However, pH dependence of intramolecular ET in the presence of NO2− cannot be explained by such a change of redox potentials (15). Instead, Kobayashi et al. (15) proposed that reduction-induced structural change of His255 is responsible for the gating-like mechanism. Because it has been recently proven that intramolecular ET in CuNiR is accompanied by PT and hence proton-coupled ET (PCET) (17, 18), one can readily speculate that intramolecular ET contributes PT to NO2− and that the structural change of His255 is involved in PCET. Crystal structures of CuNiR from Rhodobacter sphaeroides (RhsNiR) implies this possibility because His287 in RhsNiR, which corresponds to His255, seems to show pH- and redox-dependent conformational changes (19, 20). However, presumably because of X-ray radiation damages implied by rerefinement of RhsNiR structures (21), electron density around His287 was so unusual that interpretation of it is difficult (SI Appendix, Fig. S2).
Crystal structures determined by synchrotron radiation crystallography (SRX) have provided insights into the enzymatic mechanism of CuNiR (22–25), and these studies are summarized elsewhere (7). High-resolution nitrite complex (NC) structures revealed an O-coordination of NO2− showing a near face-on binding mode (22, 23), whereas Cu(II)-NO2− model complexes show a vertical binding mode (7, 26–29). The near face-on coordination manner is thought to facilitate its conversion to side-on NO, which was observed in the crystal structures of CuNiR exposed to NO (22, 23, 25). Skeptical eyes have, however, been cast on these CuNiR structures because SRX data might be affected by some problems connected to the high radiation dose delivered on the crystals. First, strong synchrotron X-rays cause not only radiation damages to amino acid residues but also photoreduction of metalloproteins (30, 31). Although a comparison between oxidized and reduced states is necessary to closely investigate redox reactions, completely oxidized structures are almost impossible to determine by SRX. Indeed, the Cu centers in CuNiR are rapidly reduced by exposure to synchrotron X-rays (21, 32). Second, following the photoreduction of T2Cu, NO2− is easily reduced and produces NO and water in SRX (21). Consequently, electron density at the catalytic site of an NC structure is derived from the mixture of both substrate and product, making interpretation of data complicated and unreliable. Third, cryogenic manipulations for reducing radiation damages in SRX have also been focused as a factor that changes the population of amino acid residues (33, 34) and enzyme–substrate complexes (35). Crystallographic (36), computational (37), and spectroscopic (38–40) studies actually show that binding modes of NO2− and NO in CuNiR crystal structures can differ from those in physiological environments.
We here ventured to use photoreduction in SRX to initiate a chemical reaction and to trap an enzymatically produced intermediary state (30, 31). Furthermore, to visualize intact CuNiR structures in the resting and NC states, we applied serial femtosecond crystallography (SFX) with X-ray free electron lasers (XFELs) (41), which enables damage-free structural determination of metalloproteins (42, 43) and evaluation of the native conformational population at room temperature (RT) (44). By comparing SRX and SFX data, we discuss PCET and nitrite reduction in CuNiR.
Results and Discussion
RS Structures Determined by SFX and SRX.
The SFX and cryogenic SRX structures of CuNiR from A. faecalis (AfNiR) (45, 46) in RS were refined to 2.03- and 1.20-Å resolution, respectively (SFX RS and SRX RS, SI Appendix, Tables S1 and S2). We also collected SRX data at 293 K, which is the temperature in the SFX experiment, and the structure was determined at 1.56-Å resolution (SRX RSRT, SI Appendix, Table S2). Although the T1Cu site is rapidly reduced by synchrotron X-rays (21, 32), there is no significant difference in the geometry between the SRX and SFX structures (SI Appendix, Table S3). Because the typical differences of the T1Cu geometries between the reduced and oxidized states are <0.1 Å (47), higher-resolution data are necessary for closer evaluation. The apical positions of the T2Cu site in the cryogenic and RT SRX structures were occupied by water (SI Appendix, Fig. S3 A and B), whereas that in the SFX structure was occupied by a chloride ion (SI Appendix, Fig. S3C) because of the difference of the purification method (SI Appendix). Except for the T2Cu–His135 bond, the distances between His residues and T2Cu did not show significant differences (SI Appendix, Table S3). The T2Cu–His135 bonds in the SRX RS structures (cryogenic: 2.00 ± 0.02 Å, RT: 2.03 ± 0.02 Å) were shorter than that in the SFX RS structure (2.12 ± 0.06 Å). Although this difference was subtle and the resolution of the SFX RS structure was too low for further judgment, it is noteworthy that His135 constitutes the Cys–His bridge for intramolecular ET.
SRX NC Structure.
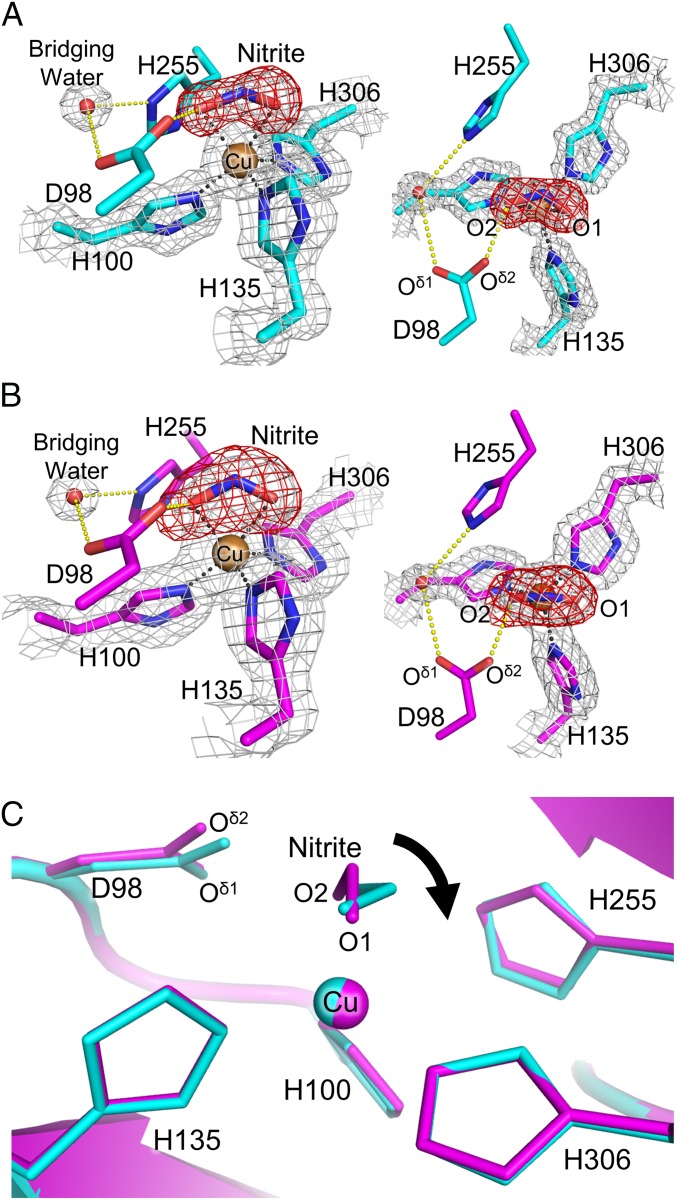
The cryogenic SRX NC structure was refined to 1.30-Å resolution (SI Appendix, Table S4), which is higher resolution than those of previous AfNiR NC structures (22, 46). T2Cu in the SRX NC structure showed ligand NO2− with 95% (molecule A) or 50% (molecules B and C) occupancy (SI Appendix, Fig. 1A, and SI Appendix, Fig. S4A). In molecules B and C, water with 50% occupancy was modeled near the O1 atom of NO2− (SI Appendix, Fig. S4A). The low NO2− occupancy and the presence of water indicates reduction of NO2− (21). The binding direction of NO2− was different from that observed in the high-resolution AfNiR NC structure (22) but similar to those in other CuNiR NC structures (SI Appendix, Fig. S4B). Ambiguity in assignment of nitrite binding modes in SRX structures may come from photoreduction of NO2−. The distances from the O1 and O2 atom to T2Cu were 2.07 ± 0.05 and 2.18 ± 0.03 Å, respectively, and the N atom was 2.16 ± 0.06 Å from T2Cu. The angle formed by the O1–N–O2 plane and the O1–T2Cu–O2 plane was 69 ± 2° (SI Appendix, Table S5). These values were similar to previously reported ones (7, 22, 23) (SI Appendix, Table S6) and showed the near face-on mode of NO2−. The Cu site geometries in the SRX NC structures are summarized in SI Appendix, Table S5.
Fig. 1.
NO2− binding in NC structures. (A) T2Cu site in the SRX NC structure (molecule A). The sigma-A–weighted 2Fo–Fc (1.5 σ) and omit Fo–Fc (6.5 σ) maps are shown as gray and red meshes, respectively. H-bonds (yellow) and coordination bonds (black) are represented by dashed lines. C, N, O, and Cu atoms are colored cyan, blue, red, and brown, respectively. (B) T2Cu site in the SFX NC structure (molecule A). The sigma-A–weighted 2Fo–Fc (1.0 σ) and omit Fo–Fc (4.5 σ) maps are shown as gray and red meshes, respectively. H-bonds and coordination bonds are represented as in A. C, N, O, and Cu atoms are colored magenta, blue, red and brown, respectively. (C) Comparison between the SFX NC (magenta) and SRX NC (cyan) structures.
SFX NC Structure.
To visualize the nondamaged NC structure, we performed SFX (SI Appendix, Fig. S5). Phase determination was performed with the single-wavelength anomalous diffraction (SAD) method using Cu as a phasing element (SI Appendix, Fig. S6). The protocol was the same as recent sulfur SAD phasing with SFX data (48) (SI Appendix). The SFX NC structure was refined to 1.60-Å resolution (SI Appendix, Table S7). The T1Cu site showed no significant difference between the SFX NC and SRX NC structures (SI Appendix, Table S5). Furthermore, both the SFX and SRX data showed that the T1Cu geometry was not dependent on NO2− binding (SI Appendix, Tables S3 and S5). Right above all T2Cu atoms in the SFX NC structure were asymmetric triangle-shaped electron densities, which could accommodate a bent triatomic molecule (SI Appendix, Fig. 1B, and SI Appendix, Fig. S7). We assigned NO2− with full occupancy because this model showed the best agreement with electron density (SI Appendix, Fig. S8). The distances from the O1 and O2 atom to T2Cu were 2.14 ± 0.05 and 2.00 ± 0.07 Å, respectively, and the N atom is 2.28 ± 0.02 Å from T2Cu. The angles between the O1–N–O2 plane and the O1–T2Cu–O2 plane in the SFX NC structure were 9° (molecule A), 39° (molecule B), and 23° (molecule C), showing a more vertical binding mode than the SRX NC structure (Fig. 1C and SI Appendix, Table S5).
Binding Mode of NO2−.
The vertical binding mode is found in many biomimetic model complexes of Cu(II)–NO2− (7, 26–29) and supported by computational chemistry (29, 49). However, synchrotron CuNiR structures have shown the near face-on modes (7, 22, 23). We then determined an SRX NC structure at 293 K (SRX NCRT, SI Appendix, Table S4 and Fig. S9A) to see whether experimental temperature will have an effect on NO2− binding modes. The angles between the O1–N–O2 plane and the O1–T2Cu–O2 plane in the SRX NCRT structure were 55° (molecule A), 33° (molecule B), and 50° (molecule C) (SI Appendix, Table S5); that is, the NO2− binding mode in the SFX NC structure was more vertical than in the SRX NCRT structure (SI Appendix, Fig. S9B and 1. Supplementary Discussion). This result is consistent with a previous study in which an SRX NC structure of CuNiR from Geobacillus thermodenitrificans (GtNiR) determined at 320 K showed NO2− assuming a near face-on mode (SI Appendix, Fig. S4B) (36). Therefore, it is most probable that the conformational change of NO2− from vertical to near face-on is induced by photoreduction. AfNiR NC structure determined at cryogenic temperature with an in-house X-ray source (46) is noteworthy because it shows relatively vertical binding modes of NO2− (SI Appendix, Table S6). Because the dose rate delivered by the in-house source is significantly lower than that of the synchrotron, the in-house cryogenic structure also implies that the near face-on mode corresponds to the binding mode when T2Cu is photoreduced. The difference of the NO2− coordination modes between Cu(I) and Cu(II) is not surprising, because model complexes of Cu(I)–NO2− generally show an N-coordination (7, 28, 50–52), not the O-coordination observed in Cu(II)–NO2−. Our present data, however, did not show a rearrangement from the O-coordination to the N-coordination, which was expected by model complexes and computational chemistry (28, 50–53).
Rotation of the Imidazole Ring of His255.
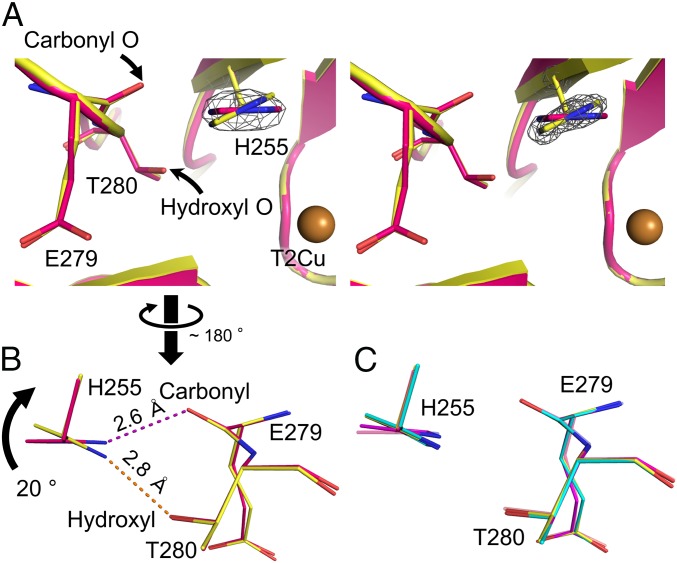
The Nδ1 atom of enzymatically important His255 can form a hydrogen bond (H-bond) with the carbonyl O atom of Glu279 and/or the hydroxyl O atom of Thr280, and this Glu–Thr pair is conserved in CuNiRs (SI Appendix, Fig. S10). Compared with the imidazole ring of His255 in the SFX RS structure, the imidazole ring in the SRX RS structure rotated about 20° and hence the H-bond partners of His255 were switched (Fig. 2 A and B and SI Appendix, Fig. S11). Similar rotation was observed in the SRX RSRT structure (SI Appendix, Fig. S12), although it was less obvious. This is presumably because the activation energy for the reverse rotation is not so high compared with the thermal energy at RT. The imidazole ring of His255 in the SRX NC structure significantly rotated as was observed in the SRX RS structure (Fig. 2C). Conversely, the imidazole ring in the SFX NC structure only showed slight rotation (Fig. 2C), indicating that NO2− binding was not the main cause for His255 rotation. The degree of His255 rotation in molecule A of the SRX NCRT structure was slightly larger than that in the SFX NC structure, although the difference was not significant in other monomers (SI Appendix, Fig. S13). Because NO2− binding itself causes slight rotation of His255, it was difficult to distinguish the effect of NO2− binding from other effects on His255 at RT, where the rotation is less obvious than at cryogenic temperature. We also solved an SRX structure in the chloride-bound form (SRX RSCL, SI Appendix, Table S8 and Fig. S14A). His255 in this structure was in the rotated form (SI Appendix, Fig. S14B), indicating that the differences of ligands are not the main reason of the rotation. Besides, the chloride ion in the SRX RSCL structure was shifted ∼1.0 Å toward the center of the catalytic site, probably because rotated His255 provided a wider space above T2Cu (SI Appendix, Fig. S14C). Because pH of a buffer at cryogenic temperature is significantly higher than at RT (54), deprotonation of His255 may be promoted at cryogenic temperature and may cause the structural change. However, the imidazole ring of His255 in the SRX RSRT structure was more rotated than that in the SFX RS structure (SI Appendix, Fig. S12). Moreover, we recently showed that the imidazole ring of His244 in GtNiR, which corresponds to His255 in AfNiR, rotates as a result of photoreduction, but not the difference of temperatures (55). Therefore, cryogenic temperature would not be the only factor for the rotation and the reduction of Cu may also cause it, as was predicted previously (15).
Fig. 2.
Conformational change of His255 (molecule A). (A) The sigma-A–weighted 2Fo–Fc maps (3.0 σ, gray) around His255 in the SFX RS structure (Left) and the SRX RS structure (Right). (B) Switching of H-bond partners. The SFX RS and SRX RS structures are shown in pink and yellow, respectively. Dashed lines represent H-bonds. (C) Comparison of the His255 conformation. The SRX RS, SFX NC, and SRX NC structures are shown in yellow, magenta, and cyan, respectively. The SFX RS structure is shown in light pink.
Using mutated AfNiR, we further proved that the rotated state of His255 is a transient conformation important for the CuNiR activity. The activity of the T280V and T280S mutants was, respectively, 20% and 29% of the WT activity. Because the T280V mutant lacks the hydroxyl O atom that can form an H-bond with His255, the rotation of His255 is inhibited in this mutant. Although the T280S mutant maintains a hydroxyl group in the side chain, it rotates more flexibly than that of Thr, which means that His255 is not always able to make an H-bond with Ser280. Therefore, the T280S mutant showed activity lower than that of WT but higher than that of T280V. Indeed, some natural CuNiRs containing Ser instead of Thr show lower activities than AfNiR (56, 57), and crystal structures of such Ser-containing CuNiRs demonstrate that Ser does not always form an H-bond with catalytic His under certain conditions (56, 57).
PCET and Nitrite Reduction Mechanisms.
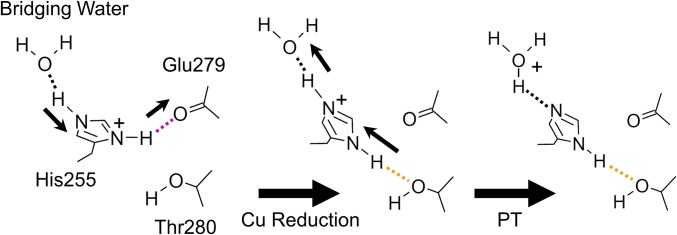
Apparently, His255 is not linked to either the T1Cu site or the T2Cu site. However, the side chain of Glu279 of the Glu–Thr pair is connected to His100 via an H-bond (SI Appendix, Fig. S15). His100 is not only a T2Cu ligand but also a terminal residue of a sensor loop, through which intramolecular ET between T1Cu and T2Cu is adjusted (9). Besides, His100 has a van der Waals and/or a π–π interaction with His255 (SI Appendix, Fig. S16). These observations suggest that structural change of His255 is involved in a redox-coupled reaction, although the precise mechanism by which His255 perceives electronic states of the Cu centers is unknown. Crystallographic (11, 12) and computational (53) studies support indirect PT from His255 to NO2− via bridging water after T2Cu reduction. The switching of the H-bond partners of His255 may facilitate this PT reaction. Because the hydroxyl O atom of Thr280 is less negatively charged than the carbonyl O atom of Gln279, the Nδ1 atom of His255 forms a longer and weaker H-bond with Thr280 (Fig. 2B). As a result, the H atom is more attracted to the Nδ1 atom and a proton on the Nε2 atom moves to bridging water (Fig. 3). Furthermore, the N atom of NO2− becomes closer to His255 when NO2− changes its conformation from vertical to near face-on (Fig. 1C), meaning that due to steric hindrance (<3.5 Å) near face-on NO2− might inhibit the reverse rotation of His255 and hence reverse PT. The catalytic activity of CuNiR dramatically drops below pH 5.0 (10, 13). This phenomenon has been explained by decreased intramolecular ET rate at low pH (15). Our model may provide another explanation: the imidazole ring rotation of His255 is difficult at low pH. Because the pH of our crystallization condition was ∼4.0, the unrotated state of His255 observed in the SFX structures should be the natural structure in the crystal. However, cryogenic temperature with increasing pH could assist the rotation of His255, which can explain why the observed rotation at cryogenic temperature was larger than that at RT.
Fig. 3.
Proposed mechanism of efficient PT driven by the rotation of His255. Dashed lines represent H-bonds. Strong and weak H-bonds involved in PCET are colored as in Fig. 2B. Thin black arrows illustrate the directions to which H atoms are attracted.
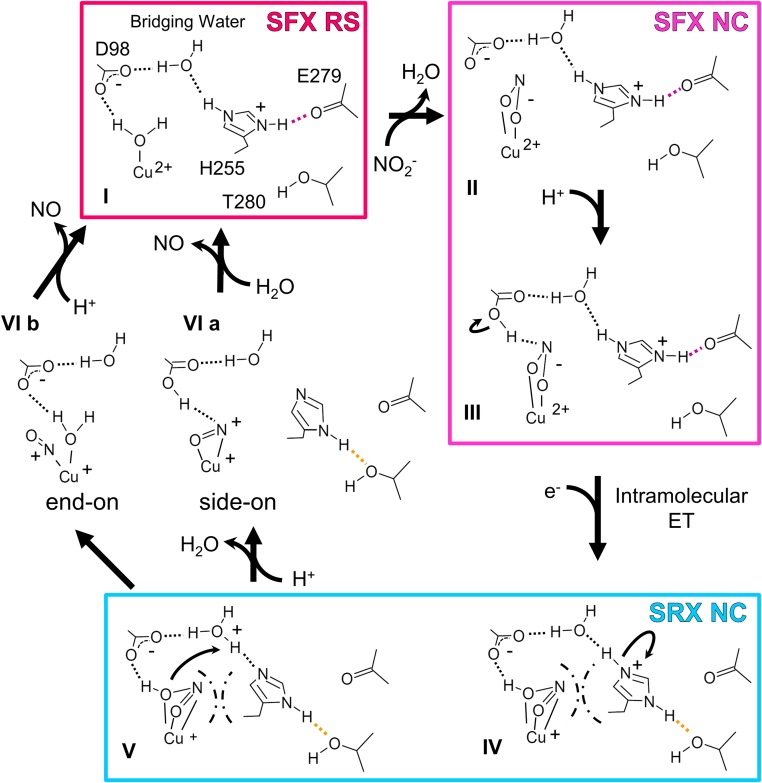
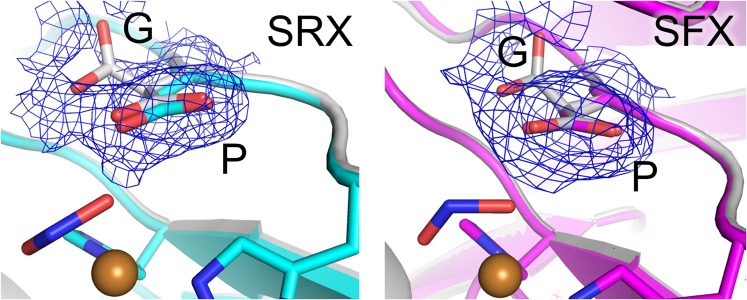
Fig. 4 describes the updated nitrite reduction mechanism. FTIR analysis with carbon monoxide (58) showed that Asp98 is deprotonated in the RS and just after binding of an external ligand (I and II). Because Asp98 is located at the end of the proton channel leading to bulk solvent (SI Appendix, Fig. S1) (17, 23), a proton may be provided through this residue (III). Intramolecular ET causes the structural changes described above (III → IV). The conformational change of NO2− makes the angle of N–O2–H to be about 120°, which may facilitate protonation of NO2−. Also, PT from His255 to bridging water occurs (IV → V). Atomic-resolution NC structures (23) revealed two conformations of Asp98, namely the gatekeeper (G) and proximal (P) conformations, indicating the catalytic importance of the movement of Asp98 (53); however, the G conformation is prohibited by steric hindrance in some CuNiRs (59). In our NC structures, Asp98 showed only a P conformation despite NO2− binding (Fig. 5), which has been thought to increase the population of the G state (23). Because the SFX data reflect the intact conformational population at RT (44), the G state reported previously may be generated by radiation damages and/or cryogenic manipulations. It is, however, noteworthy that our SRX RS structure showed a dual conformation of Asp98 due to slight movement (SI Appendix, Fig. S17). This movement may make the H-bond between the Oδ2 atom of Asp98 and HNO2 strong, which may accelerate the second PT from bridging water to the substrate (V). As was demonstrated by a density functional theory calculation (49), there is another possible reaction course, where NO2− is protonated only once by Asp98 before NO release. His255 can also function in this case as a switch to initiate a PT relay (SI Appendix, Fig. S18). A conundrum remained: Which Cu–NO species is produced in the enzymatic cycle (VI)? Whereas side-on NO is stabilized in crystal structures (22, 23, 25), spectroscopic and computational studies indicate that end-on NO is a physiological intermediate (37–40). Although visualization of an end-on NO species with short lifetime has been difficult, time-resolved SFX may enable it (60, 61). PCET is a fundamental phenomenon in living systems and expected to be applied to biomimetic electronic devices and enzyme-based green catalysts. Our study shows that SFX may contribute to studies toward the designing of such molecules.
Fig. 4.
Updated reaction mechanism of nitrite reduction. Dashed lines represent H-bonds. Strong and weak H-bonds involved in PCET are colored as in Fig. 2B. Chain lines mean steric hindrance between the near face-on substrate and His255.
Fig. 5.
Conformations of Asp98. The sigma-A–weighted 2Fo–Fc maps (0.2 σ) are shown as blue meshes. The SRX NC and SFX NC structure are shown in cyan and magenta, respectively. The structure showing G and P conformations of Asp98 (PDB ID code 2BWI) (21) is colored white.
Materials and Methods
Complete materials and methods used in this study are described in SI Appendix.
Sample Preparation and Activity Assay.
AfNiR with a C-terminal 6×His-tag was expressed in Escherichia coli BL21 (DE3) and purified by a Ni affinity column. After removing the His tag by thrombin, the sample was passed through an Ni affinity column to remove undigested proteins. Further purification was performed with an anion exchange column. Macrocrystals for SRX were prepared by the hanging-drop vapor-diffusion method. Crystals were grown at 20 °C in a solution composed of 100 mM sodium acetate (pH 4.1) and 7% PEG 4000. Nanoseed solution for micocrystals was prepared by sonicating the macrocrystals with a UD-211 ultrasonicator (Tomy Seiko Co.). The resulting solution was slightly centrifuged and the upper solution was collected and used as seeds. Microcrystals for SFX were prepared in a 15-mL centrifuge tube containing 500 μL of the protein solution (50 mg/mL) and precipitant solution [100 mM sodium acetate (pH 4.0), 12% PEG 4000, and 20 μL of the nanoseed solution]. The centrifuge tube was placed on the RT-50 rotator (Titec) at a speed of 30 rpm for 4 d at 20 °C to obtain microcrystals. The microcrystal solution was filtered through a 30-μm CellTrics filter (Chiyoda Science Co.) before the SFX experiments. The mutant proteins (T280V and T280S) were purified with the same protocol as the WT enzyme. The activity assay was performed at 25 °C as described elsewhere (57) with several modifications.
SRX Structure Determination.
For the SRX NC structure, a crystal was soaked in the reservoir solution containing 30% (vol/vol) glycerol and 60 mM NaNO2 for 15 min. For the SRX NCRT structure, a crystal was soaked in the reservoir solution containing 60 mM NaNO2 for 15 min. For the RS and RSCL structures, crystals were soaked in the reservoir solution containing 30% (vol/vol) glycerol. Diffraction data were collected at beamlines BL26B1, BL26B2, and BL44XU at SPring-8. The datasets were processed using HKL2000 (62). The phases were determined by molecular replacement (MR) using Phaser (63) with an AfNiR trimer (PDB ID code 1SJM) (22) as a search model. Manual model building was performed using Coot (64). The program Refmac5 (65) from the CCP4 suite (66) was used for structural refinement. The final models were checked for stereochemical quality using MolProbity (67).
SFX Structure Determination.
To prepare the NC, 1.2 M NaNO2 in the precipitant solution was added to the microcrystal sample in a 1.5-mL tube to give a final concentration of 60 mM. After incubation for 5 min, the sample was mixed with the grease matrix and packed in an injector syringe before data collection as described previously (68). To avoid the self-dismutation of NO2−, the totaled 18 samples of NC microcrystals were prepared at time of use, and data collection for each sample was completed within 50 min after addition of sodium nitrite. For the RS structure, microcrystals were mixed with the grease matrix and packed in an injector syringe before data collection. The diffraction patterns were collected with XFEL radiation at BL3 (EH4) of the SPring-8 Angstrom Compact Free-Electron Laser (69). The data were processed with CrystFEL (70). Indexing was performed by DirAx (71). The indexed diffraction images were merged using CrystFEL. The phase for the SFX RS data were determined by MR using Phaser with AfNiR (PDB ID code 1SJM) as a search model. The phase of the SFX NC data was determined by SAD with SHELX (72). Manual model building was performed using Coot. The program Refmac5 was used for structural refinement. The final modes were checked using MolProbity.
Supplementary Material
Acknowledgments
We thank beamline staffs at the SPring-8 (proposal 2014B6947) and the SPring-8 Angstrom Compact Free-Electron Laser (SACLA) for their support and A. Arrieta for her support in the activity assay. The serial femtosecond crystallography experiments were performed at BL3 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (Proposals 2014B8050, 2015A8026, 2015A8048, 2015A8049, and 2015B8047). This work was supported by the X-ray Free-Electron Laser Priority Strategy Program (Ministry of Education, Culture, Sports, Science, and Technology, MEXT), by a Grant-in-Aid for Scientific Research on Innovative Areas (MEXT), and by Japan Society for the Promotion of Science KAKENHI Grant 15K18487. We are grateful for support from the SACLA HPC system and the Mini-K super computer system. Experiments at BL26B1 and BL26B2 of SPring-8 were supported by the Platform for Drug Discovery, Informatics, and Structural Life Science (Proposals 2014B1146 and 2015B1146).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4YSC (SFX RS), 4YSE (SRX RS), 5D4H (SRX NC), 5D4I (SFX NC), 5D4J (SRX RSCL), 5F7B (SRX RSRT), 5F7A (SRX NCRT); and Coherent X-ray Imaging Data Bank ID: 34].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517770113/-/DCSupplemental.
References
- 1.Galloway JN, et al. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science. 2008;320(5878):889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 2.Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451(7176):293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 3.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tavares P, Pereira AS, Moura JJ, Moura I. Metalloenzymes of the denitrification pathway. J Inorg Biochem. 2006;100(12):2087–2100. doi: 10.1016/j.jinorgbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Fülöp V, Moir JWB, Ferguson SJ, Hajdu J. The anatomy of a bifunctional enzyme: Structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell. 1995;81(3):369–377. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- 6.Williams PA, et al. Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme. Nature. 1997;389(6649):406–412. doi: 10.1038/38775. [DOI] [PubMed] [Google Scholar]
- 7.Merkle AC, Lehnert N. Binding and activation of nitrite and nitric oxide by copper nitrite reductase and corresponding model complexes. Dalton Trans. 2012;41(12):3355–3368. doi: 10.1039/c1dt11049g. [DOI] [PubMed] [Google Scholar]
- 8.Godden JW, et al. The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science. 1991;253(5018):438–442. doi: 10.1126/science.1862344. [DOI] [PubMed] [Google Scholar]
- 9.Strange RW, et al. Structural and kinetic evidence for an ordered mechanism of copper nitrite reductase. J Mol Biol. 1999;287(5):1001–1009. doi: 10.1006/jmbi.1999.2648. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka K, Furusawa H, Takagi K, Yamaguchi K, Suzuki S. Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J Biochem. 2000;127(2):345–350. doi: 10.1093/oxfordjournals.jbchem.a022613. [DOI] [PubMed] [Google Scholar]
- 11.Boulanger MJ, Kukimoto M, Nishiyama M, Horinouchi S, Murphy ME. Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J Biol Chem. 2000;275(31):23957–23964. doi: 10.1074/jbc.M001859200. [DOI] [PubMed] [Google Scholar]
- 12.Boulanger MJ, Murphy MEP. Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: Structural model of a transient catalytic intermediate. Biochemistry. 2001;40(31):9132–9141. doi: 10.1021/bi0107400. [DOI] [PubMed] [Google Scholar]
- 13.Wijma HJ, Jeuken LJ, Verbeet MP, Armstrong FA, Canters GW. A random-sequential mechanism for nitrite binding and active site reduction in copper-containing nitrite reductase. J Biol Chem. 2006;281(24):16340–16346. doi: 10.1074/jbc.M601610200. [DOI] [PubMed] [Google Scholar]
- 14.Wijma HJ, Jeuken LJC, Verbeet MP, Armstrong FA, Canters GW. Protein film voltammetry of copper-containing nitrite reductase reveals reversible inactivation. J Am Chem Soc. 2007;129(27):8557–8565. doi: 10.1021/ja071274q. [DOI] [PubMed] [Google Scholar]
- 15. Kobayashi K, Tagawa S, Deligeer, Suzuki S (1999) The pH-dependent changes of intramolecular electron transfer on copper-containing nitrite reductase. J Biochem 126(2):408–412. [DOI] [PubMed]
- 16.Olesen K, et al. Spectroscopic, kinetic, and electrochemical characterization of heterologously expressed wild-type and mutant forms of copper-containing nitrite reductase from Rhodobacter sphaeroides 2.4.3. Biochemistry. 1998;37(17):6086–6094. doi: 10.1021/bi971603z. [DOI] [PubMed] [Google Scholar]
- 17.Leferink NG, et al. Proton-coupled electron transfer in the catalytic cycle of Alcaligenes xylosoxidans copper-dependent nitrite reductase. Biochemistry. 2011;50(19):4121–4131. doi: 10.1021/bi200246f. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S, et al. Demonstration of proton-coupled electron transfer in the copper-containing nitrite reductases. J Biol Chem. 2009;284(38):25973–25983. doi: 10.1074/jbc.M109.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson F, et al. Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: Changes in the interactions of the type 2 copper. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 9):1190–1198. doi: 10.1107/S0907444905017488. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson F, et al. pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J Biol Chem. 2007;282(9):6347–6355. doi: 10.1074/jbc.M605746200. [DOI] [PubMed] [Google Scholar]
- 21.Hough MA, Antonyuk SV, Strange RW, Eady RR, Hasnain SS. Crystallography with online optical and X-ray absorption spectroscopies demonstrates an ordered mechanism in copper nitrite reductase. J Mol Biol. 2008;378(2):353–361. doi: 10.1016/j.jmb.2008.01.097. [DOI] [PubMed] [Google Scholar]
- 22.Tocheva EI, Rosell FI, Mauk AG, Murphy ME. Side-on copper-nitrosyl coordination by nitrite reductase. Science. 2004;304(5672):867–870. doi: 10.1126/science.1095109. [DOI] [PubMed] [Google Scholar]
- 23.Antonyuk SV, Strange RW, Sawers G, Eady RR, Hasnain SS. Atomic resolution structures of resting-state, substrate- and product-complexed Cu-nitrite reductase provide insight into catalytic mechanism. Proc Natl Acad Sci USA. 2005;102(34):12041–12046. doi: 10.1073/pnas.0504207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tocheva EI, Eltis LD, Murphy MEP. Conserved active site residues limit inhibition of a copper-containing nitrite reductase by small molecules. Biochemistry. 2008;47(15):4452–4460. doi: 10.1021/bi7020537. [DOI] [PubMed] [Google Scholar]
- 25.Tocheva EI, Rosell FI, Mauk AG, Murphy MEP. Stable copper-nitrosyl formation by nitrite reductase in either oxidation state. Biochemistry. 2007;46(43):12366–12374. doi: 10.1021/bi701205j. [DOI] [PubMed] [Google Scholar]
- 26.Ruggiero CE, Carrier SM, Tolman WB. Reductive disproportionation of NO mediated by copper complexes: Modeling N,O generation by copper proteins and heterogeneous catalyst. Angew Chem Int Ed Engl. 1994;33(8):895–897. [Google Scholar]
- 27.Casella L, Carugo O, Gullotti M, Doldi S, Frassoni M. Synthesis, structure, and reactivity of model complexes of copper nitrite reductase. Inorg Chem. 1996;35(5):1101–1113. doi: 10.1021/ic950392o. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama H, Yamaguchi K, Sugimoto M, Suzuki S. CuI and CuII complexes containing nitrite and tridentate aromatic amine ligand as models for the substrate-binding type-2 Cu site of nitrite reductase. Eur J Inorg Chem. 2005;8:1435–1441. [Google Scholar]
- 29.Lehnert N, et al. Synthesis and spectroscopic characterization of copper(II)-nitrito complexes with hydrotris(pyrazolyl)borate and related coligands. Inorg Chem. 2007;46(10):3916–3933. doi: 10.1021/ic0619355. [DOI] [PubMed] [Google Scholar]
- 30.Schlichting I, et al. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287(5458):1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 31.Berglund GI, et al. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 2002;417(6887):463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 32.Antonyuk SV, Hough MA. Monitoring and validating active site redox states in protein crystals. Biochim Biophys Acta. 2011;1814(6):778–784. doi: 10.1016/j.bbapap.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462(7273):669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108(39):16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keedy DA, et al. Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR. Structure. 2014;22(6):899–910. doi: 10.1016/j.str.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda Y, Inoue T. High-temperature and high-resolution crystallography of thermostable copper nitrite reductase. Chem Commun (Camb) 2015;51(30):6532–6535. doi: 10.1039/c4cc09553g. [DOI] [PubMed] [Google Scholar]
- 37.Merkle AC, Lehnert N. The side-on copper(I) nitrosyl geometry in copper nitrite reductase is due to steric interactions with isoleucine-257. Inorg Chem. 2009;48(24):11504–11506. doi: 10.1021/ic9018376. [DOI] [PubMed] [Google Scholar]
- 38.Usov OM, Sun Y, Grigoryants VM, Shapleigh JP, Scholes CP. EPR-ENDOR of the Cu(I)NO complex of nitrite reductase. J Am Chem Soc. 2006;128(40):13102–13111. doi: 10.1021/ja056166n. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh S, et al. Resolution of the spectroscopy versus crystallography issue for NO intermediates of nitrite reductase from Rhodobacter sphaeroides. J Am Chem Soc. 2007;129(34):10310–10311. doi: 10.1021/ja072841c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujisawa K, et al. Structural and spectroscopic characterization of mononuclear copper(I) nitrosyl complexes: end-on versus side-on coordination of NO to copper(I) J Am Chem Soc. 2008;130(4):1205–1213. doi: 10.1021/ja075071d. [DOI] [PubMed] [Google Scholar]
- 41.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470(7332):73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kern J, et al. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proc Natl Acad Sci USA. 2012;109(25):9721–9726. doi: 10.1073/pnas.1204598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson LC, et al. Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nat Commun. 2013;4:2911. doi: 10.1038/ncomms3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342(6165):1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kakutani T, Watanabe H, Arima K, Beppu T. Purification and properties of a copper-containing nitrite reductase from a denitrifying bacterium, Alcaligenes faecalis strain S-6. J Biochem. 1981;89(2):453–461. doi: 10.1093/oxfordjournals.jbchem.a133220. [DOI] [PubMed] [Google Scholar]
- 46.Murphy MEP, Turley S, Adman ET. Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis. Mechanistic implications. J Biol Chem. 1997;272(45):28455–28460. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- 47.Solomon EI, Szilagyi RK, DeBeer George S, Basumallick L. Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem Rev. 2004;104(2):419–458. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- 48.Nakane T, et al. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 12):2519–2525. doi: 10.1107/S139900471501857X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh S, Dey A, Sun Y, Scholes CP, Solomon EI. Spectroscopic and computational studies of nitrite reductase: Proton induced electron transfer and backbonding contributions to reactivity. J Am Chem Soc. 2009;131(1):277–288. doi: 10.1021/ja806873e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halfen JA, Tolman WB. Synthetic model of the substrate adduct to the reduced active site of copper nitrite reductase. J Am Chem Soc. 1994;116:5475–5476. [Google Scholar]
- 51.Halfen JA, et al. Synthetic modeling of nitrite binding and activation by reduced copper proteins. Characterization of copper(I)-nitrite complexes that evolve nitric oxide. J Am Chem Soc. 1996;118:763–776. [Google Scholar]
- 52.Kujime M, Izumi C, Tomura M, Hada M, Fujii H. Effect of a tridentate ligand on the structure, electronic structure, and reactivity of the copper(I) nitrite complex: Role of the conserved three-histidine ligand environment of the type-2 copper site in copper-containing nitrite reductases. J Am Chem Soc. 2008;130(19):6088–6098. doi: 10.1021/ja075575b. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Hodak M, Bernholc J. Enzymatic mechanism of copper-containing nitrite reductase. Biochemistry. 2015;54(5):1233–1242. doi: 10.1021/bi5007767. [DOI] [PubMed] [Google Scholar]
- 54.Douzou P, Hoa GHB, Petsko GA. Protein crystallography at sub-zero temperatures: Lysozyme-substrate complexes in cooled mixed solvents. J Mol Biol. 1975;96(3):367–380. doi: 10.1016/0022-2836(75)90166-7. [DOI] [PubMed] [Google Scholar]
- 55.Fukuda Y, et al. Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography. J Biochem. 2016:mvv133. doi: 10.1093/jb/mvv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boulanger MJ, Murphy MEP. Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: A new class of copper-containing nitrite reductases. J Mol Biol. 2002;315(5):1111–1127. doi: 10.1006/jmbi.2001.5251. [DOI] [PubMed] [Google Scholar]
- 57.Lawton TJ, Bowen KE, Sayavedra-Soto LA, Arp DJ, Rosenzweig AC. Characterization of a nitrite reductase involved in nitrifier denitrification. J Biol Chem. 2013;288(35):25575–25583. doi: 10.1074/jbc.M113.484543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Boulanger MJ, Mauk AG, Murphy MEP. Carbon monoxide binding to copper-containing nitrite reductase from Alcaligenes faecalis. J Phys Chem B. 2000;104:10738–10742. [Google Scholar]
- 59.Fukuda Y, et al. Structural insights into the function of a thermostable copper-containing nitrite reductase. J Biochem. 2014;155(2):123–135. doi: 10.1093/jb/mvt107. [DOI] [PubMed] [Google Scholar]
- 60.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346(6214):1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513(7517):261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 63.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat Methods. 2015;12(1):61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 69.Tono K, et al. Beamline, experimental stations and photon beam diagnostics for the hard x-ray free electron laser of SACLA. New J Phys. 2013;15(8):083035. [Google Scholar]
- 70.White TA, et al. CrystFEL: A software suite for snapshot serial crystallography. J Appl Cryst. 2012;45(2):335–341. [Google Scholar]
- 71.Duisenberg AJM. Indexing in single-crystal diffractometry with an obstinate list of reflections. J Appl Cryst. 1992;25:92–96. [Google Scholar]
- 72.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.