Significance
The cell envelope of Gram-negative bacteria contains two membranes and a cell wall located in the aqueous compartment between them. The outer membrane (OM) functions as a barrier that contributes to antibiotic resistance. We describe a dominant mutation in a gene for an OM lipoprotein that leads to cell death under starvation conditions in medium with limited cation concentrations. We show that death occurs not by rapid cell lysis but by a previously uncharacterized mechanism involving flow of material from the inner membrane to the OM that results in rupture of the inner membrane and the slow leakage of cytoplasmic contents. Our study highlights the vital need for balanced synthesis across the Gram-negative envelope and may empower the development of new therapeutics.
Keywords: outer membrane, lipid transport, lipopolysaccharide, lysophospholipids, single-cell imaging
Abstract
Gram-negative bacteria balance synthesis of the outer membrane (OM), cell wall, and cytoplasmic contents during growth via unknown mechanisms. Here, we show that a dominant mutation (designated mlaA*, maintenance of lipid asymmetry) that alters MlaA, a lipoprotein that removes phospholipids from the outer leaflet of the OM of Escherichia coli, increases OM permeability, lipopolysaccharide levels, drug sensitivity, and cell death in stationary phase. Surprisingly, single-cell imaging revealed that death occurs after protracted loss of OM material through vesiculation and blebbing at cell-division sites and compensatory shrinkage of the inner membrane, eventually resulting in rupture and slow leakage of cytoplasmic contents. The death of mlaA* cells was linked to fatty acid depletion and was not affected by membrane depolarization, suggesting that lipids flow from the inner membrane to the OM in an energy-independent manner. Suppressor analysis suggested that the dominant mlaA* mutation activates phospholipase A, resulting in increased levels of lipopolysaccharide and OM vesiculation that ultimately undermine the integrity of the cell envelope by depleting the inner membrane of phospholipids. This novel cell-death pathway suggests that balanced synthesis across both membranes is key to the mechanical integrity of the Gram-negative cell envelope.
The Gram-negative bacterial cell envelope is a remarkably complex structure with critical functions for cellular growth and viability. It protects the cell from rapidly changing and potentially harmful environments and must do so while also allowing the selective import of nutrients and export of waste (1). Structurally, the Gram-negative cell envelope consists of an inner membrane (IM) and an outer membrane (OM) that delimit an aqueous compartment known as the periplasm (1, 2). Within the periplasmic space is a mesh-like network of peptide-crosslinked glycan chains, known as the peptidoglycan cell wall (1, 3, 4). This structure shapes the cell and provides mechanical resistance to turgor pressure-driven expansion (3). After inoculation into fresh medium, cells use nutrients in the medium to carry out processes essential to growth. Once these nutrients are depleted, cells enter stationary phase, during which they undergo gross morphological and physiological changes and stop growing (5). Throughout these growth phases and during septum formation and cytokinesis, synthesis of the various layers of the cell envelope must remain coordinated.
The Escherichia coli OM is an asymmetric bilayer that contains phospholipids (PLs) in the inner leaflet and LPS in the outer leaflet (6). This structure functions as a robust, highly selective permeability barrier that protects the cell from harmful agents such as detergents, bile salts, and antibiotics (1). The effectiveness of the OM can be attributed to the hydrophobicity of and strong lateral interactions between LPS molecules (6); E. coli must properly synthesize and transport LPS to the outer leaflet of the OM to survive (7). Many proteins contribute to LPS biosynthesis and assembly (for a review, see refs. 8 and 9). By contrast with LPS, how lipids are transported to the OM is virtually unknown.
When LPS biosynthetic or transport proteins are compromised, PLs are flipped from the inner to the outer leaflet of the OM to accommodate the reduction in LPS abundance (10). In the outer leaflet, it is thought that PLs form rafts (11), creating patches in the membrane that are more susceptible to the influx of hydrophobic, toxic molecules. To prevent damage resulting from surface-exposed PLs in wild-type E. coli cells, several mechanisms destroy or remove these PLs from the outer leaflet. The OM β-barrel protein PagP is a palmitoyltransferase that removes a palmitate from the sn-1 position of a surface-exposed PL and transfers it to lipid A or phosphatidylglycerol (12, 13). Another OM β-barrel phospholipase, PldA, removes both sn-1 and sn-2 palmitate moieties from PLs and lyso-PLs (14).
The Mla (maintenance of lipid asymmetry) ABC transport system is a third mechanism for maintaining lipid asymmetry. Mla proteins are present in all compartments of the cell envelope and facilitate retrograde phospholipid transport from the OM back to the IM (15). MlaA is the lipoprotein component that interacts with OmpC in the OM (16) and is thought to remove PLs from the outer leaflet of the OM and shuttle them to MlaC, the soluble periplasmic component. MlaC delivers the PLs to the IM MlaFEDB complex, which is presumed to aid in the reintegration of PLs into the IM. Null mutations in any mla gene increase the permeability of the OM, rendering cells susceptible to detergent by an increase in surface-exposed PLs (15).
Here we show that a dominant mutation in mlaA disrupts the lipid balance of the OM by a mechanism that does not require the other mla gene products but does require active PldA. Cells carrying this mutation are sensitized to the transition to stationary phase in medium with low divalent cation concentrations. This transition triggers an unexpected cell-death trajectory in starving cells in which death is correlated with increased OM blebbing at sites of cell division concomitant with decreased cytoplasmic volume and IM surface area, suggesting that lipid flow from the IM to the OM compensates for lipid loss by OM blebs and vesicles. Thus, our data may provide insights into the long-standing question of how lipids are transported to the OM.
Results
A Gain-of-Function mlaA* Allele Increases OM Permeability.
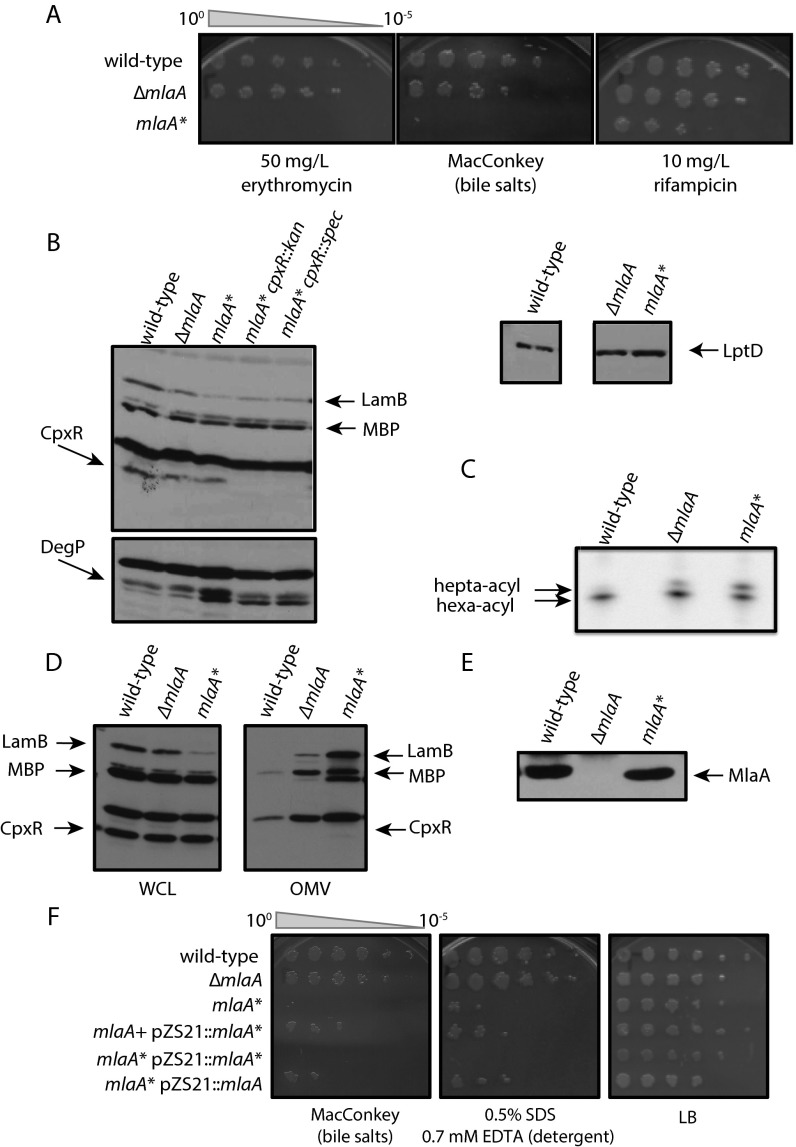
To date, only null mutants in the Mla pathway have been identified and characterized (15). These null mutants display a moderate OM permeability defect: PLs accumulate at the cell surface, and cells are more susceptible to detergent. In selecting for a bacitracin-resistant suppressor of an lptD mutant that exhibits defects in LPS assembly, we isolated a mutation in mlaA, mlaA*, that is a deletion resulting in the loss of the residues at positions 41 (N) and 42 (F) of the signal sequence-containing precursor protein. When mlaA* was moved into an otherwise wild-type background, the strain exhibited increased sensitivity to detergents relative to a ΔmlaA strain (Fig. 1A). Furthermore, mlaA* cells were sensitive to bile salts, erythromycin, and rifampicin, in contrast to ΔmlaA cells (Fig. S1A), suggesting that the mlaA* allele generally increases OM permeability. mlaA* also induces the σE and Cpx envelope stress responses (Fig. S1B), which are activated by OMP/LPS perturbations and by cues such as misfolded proteins in the IM and periplasm, high pH, and surface adhesion (17). To determine whether this increased sensitivity was related to an increase in PLs in the outer leaflet of the OM, we used a PagP assay to detect surface exposure of PLs (18, 19). mlaA* cells had 1.8 times more hepta-acylated lipid A than ΔmlaA cells and five times more than wild-type cells (Fig. S1C), indicating an increase in surface-exposed PLs. In addition, the mlaA* strain hypervesiculated in comparison with wild-type and ∆mlaA cells (Fig. S1D). None of these phenotypes can be explained by a change in protein levels, because MlaA* levels are comparable to MlaA levels in wild-type cells (Fig. S1E).
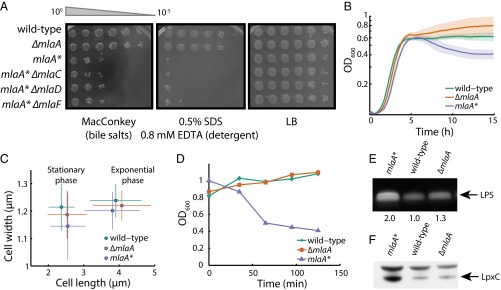
Fig. 1.
mlaA* increases OM permeability and causes cell death in stationary phase. (A) Efficiency of plating assays on MacConkey or LB agar plates with the indicated supplements. Ten-fold dilutions of cultures are indicated above the left plate. mlaA* cells exhibited increased sensitivity to bile salts and detergent. Introduction of ΔmlaC::kan, ΔmlaD::kan, or ΔmlaF::kan in an mlaA* background did not decrease the severity of the mlaA* phenotype. (B) OD600 of a culture of mlaA* cells decreased during entrance into stationary phase, unlike wild-type or ∆mlaA cells. Maximal growth rates were approximately the same for all three strains (0.88 ± 0.04/h, 0.86 ± 0.03/h, and 0.90 ± 0.04/h, respectively). Solid curves are mean values from six independent samples; shaded areas represent the standard deviation (SD) of the samples. (C) Single-cell microscopy measurements of cellular dimensions for exponential-phase and stationary-phase cells. Wild-type, ΔmlaA, and mlaA* cells had virtually identical distributions of cellular dimensions during exponential growth and in stationary phase, with stationary-phase cells being shorter. Data points represent mean ± SD across >200 cells. (D) In the spent medium transition assay (Materials and Methods), the OD of mlaA* cells decreased within 30 min after the switch from exponential growth to resuspension in supernatant from an overnight culture. (E) mlaA* cells have higher LPS levels, as measured by SDS/PAGE (Materials and Methods). (F) Whole-cell protein lysates were prepared, separated by SDS/PAGE, and analyzed by immunoblotting (Materials and Methods). mlaA* increases the LpxC protein level.
Fig. S1.
mlaA* phenotypes indicate perturbations to OM composition. (A) Efficiency of plating assay on LB agar plates with the indicated supplements. Ten-fold dilutions of cultures are indicated above the left plate. mlaA* cells exhibit increased sensitivity to bile salts, erythromycin, and rifampicin. (B) Whole-cell protein lysates were prepared, separated by SDS/PAGE, and analyzed by immunoblotting (Materials and Methods). mlaA* induces the σE and Cpx envelope stress responses. (C) 32PO4-radiolabeled lipid A samples were isolated and separated by TLC (Materials and Methods). mlaA* cells have more hepta-acylated lipid A than wild-type or ΔmlaA cells. (D) Whole-cell lysates (WCL) and OM vesicles (OMVs) were prepared, separated by SDS/PAGE, and analyzed by immunoblotting. mlaA* cells hypervesiculated in comparison with wild-type or ΔmlaA cells. (E) Whole-cell lysates were prepared, separated by SDS/PAGE, and analyzed by immunoblotting. MlaA* levels are comparable to levels of MlaA in wild-type cells. (F) The efficiency of plating assay on LB agar or MacConkey plates with the indicated supplements indicates that mlaA* is a gain-of-function mutation. Ten-fold dilutions of cultures are indicated above the left plate.
Introduction of ΔmlaC::kan, ΔmlaD::kan, or ΔmlaF::kan alleles into an mlaA* background neither increased nor decreased the severity of the mlaA* phenotype (Fig. 1A), indicating that the OM permeability phenotypes are not dependent on downstream Mla proteins. Because mlaA* did not phenocopy ΔmlaA, we cloned mlaA* with its native promoter and ribosome-binding site into a low-copy plasmid (pZS21) and performed diploid analysis in the presence of the wild-type protein. Whether mlaA* was present on the chromosome or plasmid-borne, we observed dominant effects with respect to sensitivity to detergents and bile salts (Fig. S1F), suggesting that mlaA* is a gain-of-function mutation. Taken together, our data indicate that the activity of the dominant MlaA* protein is epistatic to that of downstream Mla proteins and hence reflects an independent function of MlaA that induces envelope stress responses, perhaps because of the altered composition and properties of the OM.
mlaA* Causes Cell Death in Stationary Phase.
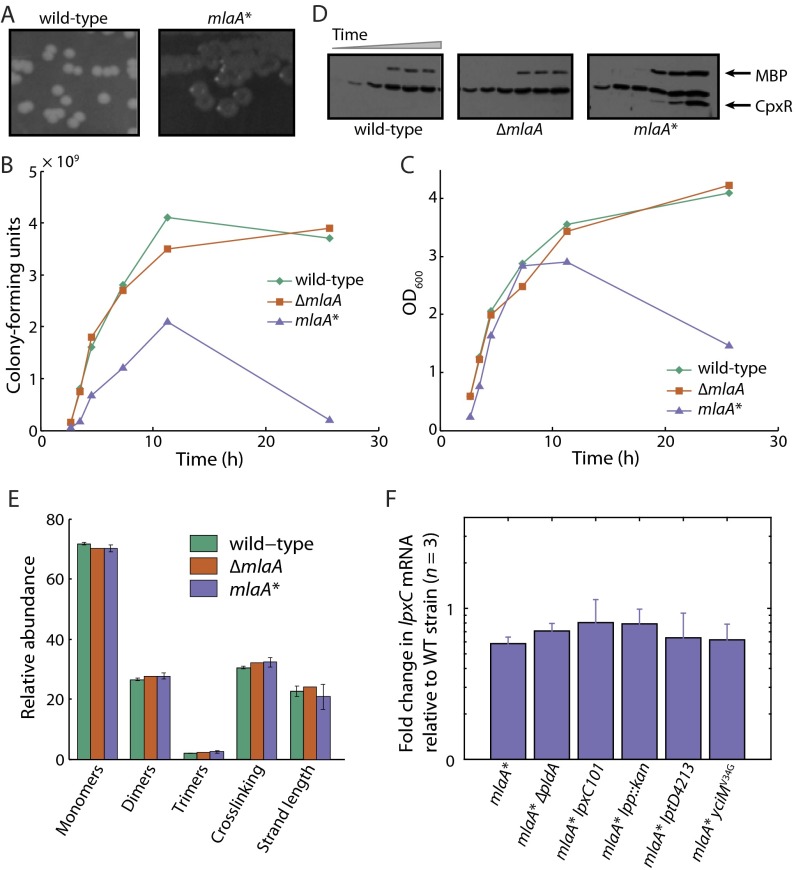
Working with the mlaA* strain in either liquid lysogeny broth (LB) or on plates suggested that the mutation causes a growth defect. Overnight cultures of mlaA* had visibly lower optical density (OD) than wild-type and ΔmlaA strains, and mlaA* cells formed colonies that had distinct depressed colony centers (Fig. S2A). To characterize this growth defect, we monitored the growth of the wild-type, ΔmlaA, and mlaA* strains by OD600. Although the maximal growth rates were the same for all three strains (Fig. 1B), there was a marked decrease in OD600 for mlaA* cells during entry to stationary phase (Fig. 1B), suggesting that death was occurring and cells had lost integrity. To corroborate these data with more direct evidence of death, we simultaneously quantified OD600 colony-forming units (to determine the number of viable cells) and the concentration of cytoplasmic proteins such as the envelope stress response protein CpxR in samples of the culture supernatant taken over the entire growth curve. The coincident decrease in OD600 and the number of viable mlaA* cells (Fig. S2 B and C) and, at later time points, the accumulation of CpxR in the culture supernatants (Fig. S2D) indicate substantial cell death. We did not observe these trends in wild-type or ΔmlaA cells (Fig. S2 B–D). Death was not caused by obvious differences in cell morphology: Populations of wild-type, ΔmlaA, and mlaA* cells had virtually identical distributions of cellular dimensions both during exponential growth and in stationary phase (Fig. 1C).
Fig. S2.
mlaA* causes partial cell death in stationary phase, and cell death is related to LPS levels. (A) mlaA* cells form colonies with distinct depressed colony centers. (B–D) Growth assays with simultaneous quantification of cells by enumerating colony-forming units and immunoblotting for CpxR protein levels in the culture supernatant. Time points shown for each strain on each graph correspond to the six lanes on the immunoblot. The number of viable mlaA* cells decreased upon entrance into stationary phase. At later time points, the cytoplasmic stress response protein CpxR and the periplasmic maltose-binding protein (MBP) accumulated in the culture supernatant. (E) Digested wild-type, ΔmlaA, and mlaA* sacculi had quantitatively similar muropeptide abundances as measured by UPLC. Monomers refer to uncrosslinked muropeptides; dimers and trimers refer to double- and triple-crosslinked muropeptides, respectively. Crosslinking is the number of crosslinked muropeptides divided by the total number of disaccharides, and strand length is the length of the glycan strands as measured by the presence of terminating anhydro-muropeptides (55). Bars represent mean ± SD for three independent samples. (F) Quantitative RT-PCR of lpxC mRNA levels, normalized to the wild-type level. lpxC mRNA levels remain unchanged in mlaA* cells and in suppressors of the mlaA* phenotype.
To interrogate the dynamics of cell death at the single-cell level, we required a method of rapidly inducing the transition of exponentially growing cells into stationary phase. To do so, we pelleted exponentially growing cultures and resuspended the cells in spent LB from an overnight culture of wild-type cells (Materials and Methods). Because the nutrients already had been depleted from this medium, growth was severely limited, and the cells entered stationary phase. We observed very little increase in the OD600 of wild-type cultures after resuspension, but the OD600 of mlaA* cells decreased within 30 min (Fig. 1D). Thus, no matter how closely the shift to spent medium mimics the transition into stationary phase, our assay provides a means to investigate mlaA*-mediated cell death.
The mlaA* Cell Envelope Has Increased Levels of LPS but No Obvious Peptidoglycan Defects.
Lysis is generally thought to occur as cells burst because of defects in the cell wall, i.e., areas of weakened cell envelope that are unable to stretch to provide the necessary resistance to turgor pressure (20–22). In principle, however, cell death can occur after the slow leakage of critical cytoplasmic contents from a cell envelope that is largely intact. To test whether MlaA* altered the biochemical composition of the peptidoglycan in a manner that compromises mechanical integrity, we isolated intact sacculi and analyzed muropeptide abundances using ultra-performance liquid chromatography (UPLC). Wild-type, ΔmlaA, and mlaA* chromatograms were quantitatively similar (Fig. S2E), indicating that the peptidoglycan does not vary among these samples, at least at the level of composition.
Because no obvious peptidoglycan defects were associated with the mlaA* strain, we wondered whether there were any differences in the composition of the OM. When we assessed LPS levels in whole-cell lysates, we observed a marked increase in LPS levels in mlaA* compared with wild-type and ΔmlaA (Fig. 1E). Increased LPS levels correlated with an increase in the protein levels of the enzyme that catalyzes the first committed step in LPS biosynthesis, LpxC (Fig. 1F). The mlaA* mutation does not alter lpxC transcription as measured by quantitative RT-PCR (qRT-PCR) (Fig. S2F). LpxC is known to be regulated by FtsH-mediated proteolysis (23), and thus we suspect that the mlaA* mutation stabilizes the LpxC protein (likely indirectly), resulting in increased synthesis and assembly of LPS in this mutant background.
The Death Trajectory of mlaA* Cells in Spent Medium Implicates Loss of Membrane Integrity.
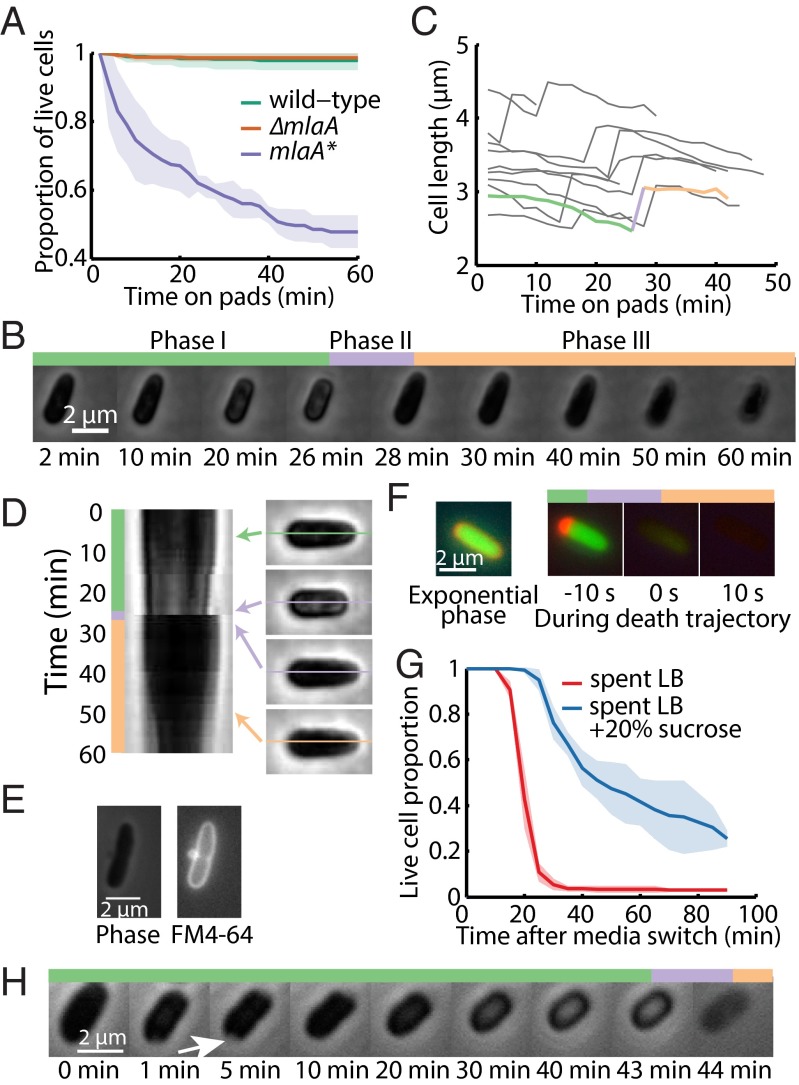
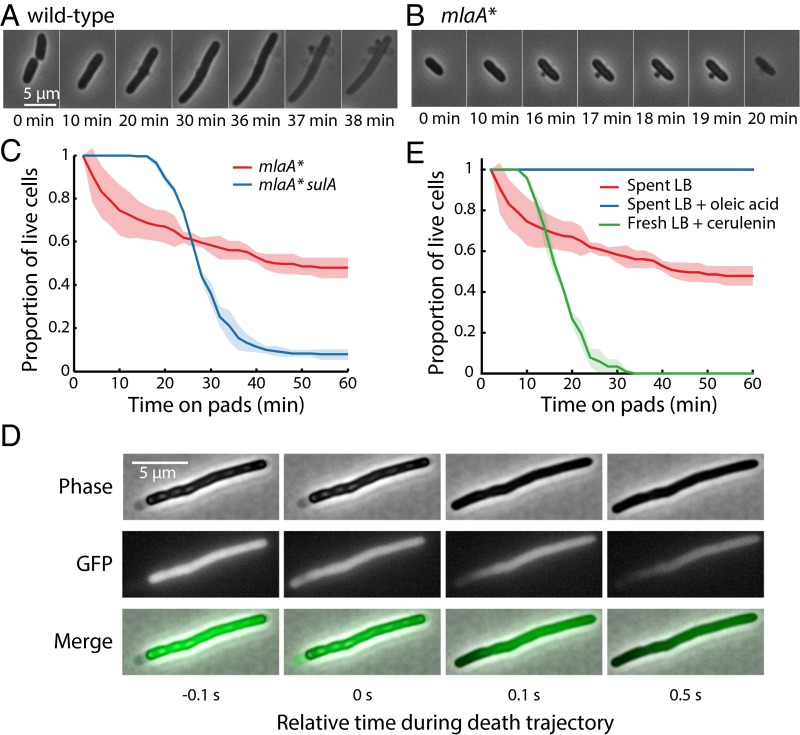
To probe the dynamics and cause(s) of cell death, we transferred exponentially growing mlaA* cells onto agarose pads made from spent LB (Materials and Methods) and imaged the cells using time-lapse microscopy. Wild-type and ∆mlaA cells exhibited a marked increase in the frequency of cell division, producing shorter and hence rounder cells after 1 h of growth in spent medium; death events were rare (Fig. 2A). In contrast, a substantial fraction of mlaA* cells died within 10 min of the transition, although ∼50% of the mlaA* cells remained alive after 1 h (Fig. 2A), similar to our bulk measurements using liquid cultures (Fig. 1D). All the cells that died shared a novel death trajectory that included three phases (Fig. 2B and Movie S1). In phase I, cells shifted from exponential elongation to a mode of gradual shortening for 10–20 min. In phase II, they experienced a rapid “pop” in which they dramatically expanded in length within 2 min. Finally, in phase III, they slowly decreased in length. We could identify these phases robustly by evaluating the dynamics of cell length (Fig. 2B); the time of death was determined based on the popping event in phase II (Fig. 2C), enabling us to quantify death as a function of time (Fig. 2A).
Fig. 2.
Single-cell growth trajectories implicate an increase in cytoplasmic density and OM blebbing at the septum in cell death. (A) Growth on agarose pads made with spent LB medium led to a dead-cell fraction over 1 h similar to that observed in liquid LB (Fig. 1D). Dead cells were identified by the three phases discussed below, and death was defined by the rapid popping event. Solid curves are mean values from three independent experiments; shaded areas represent SDs. Each measurement involved >80 cells. (B) For cells that died, cell length displayed a characteristic trajectory of three phases involving gradual shortening (phase I), a rapid popping event (phase II), and steady shrinking coupled to a decrease in phase contrast toward the background level, indicating a slow loss of cytoplasmic contents (phase III). (C) Dynamics of cell length reflected the three phases of death, with the cell in B denoted by the thick curve; colors correspond to the three phases in B. (D) Kymograph of cell midline for the cell in B. In phase I, part of the cell gradually became bright; this bright region disappeared once the cell entered phase II at ∼30 min. (E) FM 4-64 labeling of the OM (Right) revealed a midcell bleb that is typical of the death trajectory and is not clearly visible under phase-contrast microscopy (Left). (F, Left) Exponentially growing mlaA* cells had an approximately uniform distribution of periplasmic mCherry surrounding the cytoplasmic GFP signal. (Right) During the death trajectory upon transition into spent LB, mCherry initially accumulated at one pole in the periplasm, indicating cytoplasmic shrinkage away from the OM. At the time of the popping event, both fluorescence signals leaked out of the cytoplasm, and the fluorescence intensity decreased to background levels within 10 s of popping. (G) The addition of sucrose to spent medium delayed cell death by tens of minutes. (The popping event following cell shrinkage was used as a signature of cell death.) The spent LB control also was performed in a microfluidic flow cell, without the hyperosmotic shock. Solid curves are mean values from three fields of view, and shaded areas represent SDs. Each measurement involved >50 cells. (H) After a hyperosmotic shock in a microfluidic flow cell caused by transfer from fresh LB to spent LB supplemented with 20% sucrose at t = 0 min, mlaA* cells plasmolyzed, with visible detachment of the cytoplasm from the cell wall and OM forming a concave region (arrow). The IM then shrank in area and resolved the concave region. The cells eventually underwent a death trajectory similar to that in B.
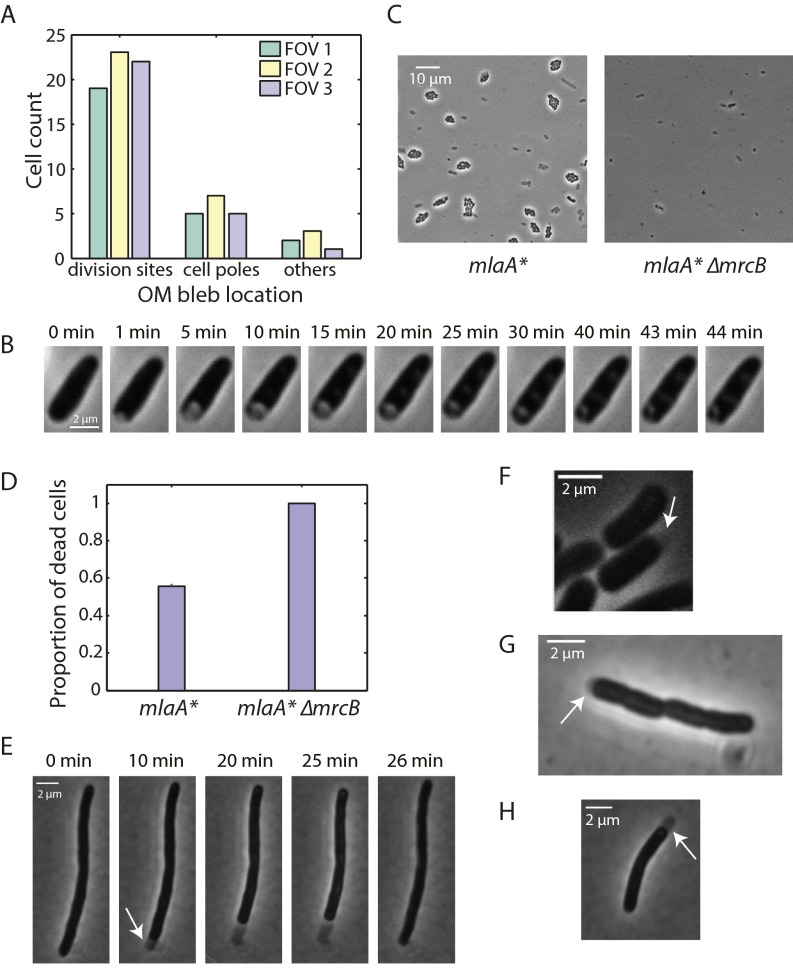
During phase I, we noted that the shrinking was associated with the cell interior becoming phase bright (Fig. 2 B and D), suggesting that the cytoplasm was compressed to a higher density as the cell volume decreased and turgor pressure increased. The cell reverted back to a phase-dark state once the cell experienced phase II (Fig. 2 B and D), as would be expected if the increase in turgor pressure caused by the decreasing volume during phase I was relieved. To explore further the changes in the OM during cell death, we stained mlaA* cells with the OM dye FM 4-64 and imaged the death trajectory upon the transition to spent medium. When FM 4-64–stained mlaA* cells were exposed to spent medium, most of them exhibited OM blebbing at midcell before death occurred (Fig. 2E). Unlike β-lactam–induced blebs, which contain cytoplasmic contents (24), the blebs in mlaA* cells were not visible in phase-contrast images (Fig. 2E), suggesting that they are composed primarily of OM and periplasmic contents. Midcell blebs usually occurred at sites with obvious constriction (Fig. S3A), indicating that the OM is less stably connected to the cell wall at sites of cell division. During phase III, cells gradually lost phase contrast over tens of minutes, but even after 1 h of imaging they still were more phase dark (Fig. 2B) than the “ghosts,” which had lost most cellular contents.
Fig. S3.
mlaA*-mediated stationary-phase death is related to cell division and IM shrinkage. (A) mlaA* cells stained with FM 4-64 exhibited blebbing when placed on agarose pads made with spent medium. The frequency of blebbing events was higher at division sites and cell poles than at other locations. Frequencies were measured in three fields of view (FOVs). (B) After a hyperosmotic shock in a microfluidic flow cell induced by transfer from fresh LB to spent LB supplemented with 20% sucrose at t = 0 min, wild-type cells first plasmolyzed, with visible detachment of the cytoplasm from the cell wall and OM that formed a concave region. The concave region gradually resolved because of the expansion of cytoplasm until the entire cell outline was again phase dark. (C) Exponentially growing mlaA* and mlaA* ∆mrcB cells were spotted onto agarose pads containing spent LB. The pads were incubated for 2.5 h and then were imaged. Dead cells appeared as gray ghosts, and live cells grew into microcolonies. (Left) Approximately half of the mlaA* cells were dead after 2.5 h. (Right) Nearly all mlaA* ∆mrcB cells died. (D) The proportions of cells that underwent cell death in the spent medium transition assay in fields of view similar to those in C were calculated by counting the number of dead cells and microcolonies. The proportion of cell death is defined as the number of dead cells divided by the sum of dead cells and microcolonies. Bars represent mean ± SD for three fields of view. (E) SulA-induced mlaA* cells were placed onto an agarose pad containing spent LB. The cells exhibited a death trajectory similar to that of nonfilamentous cells as shown in Fig. 2B. In phase I, the IM detached from the cell wall and OM, leaving a visible periplasmic region (arrow). In phase II (25–26 min), the cytoplasmic region expanded to fill the whole region delimited by the cell wall and OM. (F) IM detachment also occurred during phase I of the death trajectory of nonfilamentous mlaA* cells. The periplasmic region is harder to observe because the cells are shorter. (G and H) IM detachment still occurred with the addition of the proton ionophore CCCP to spent LB in the transition assay, in both nonfilamentous mlaA* (G) and SulA-induced mlaA* cells (H).
To test the integrity of the cell envelope further, we constructed an mlaA* strain expressing both cytoplasmic GFP and periplasmic mCherry and analyzed the localization of both fluorescent proteins along cell-death trajectories. In phase I, both compartments were intact, the fluorescence signals were well separated, and the periplasmic region was larger and brighter than in exponential phase (Fig. 2F). When cells entered phase II, most fluorescence signal dissipated within 10 s, indicating that both membranes in the envelope became permeable to the diffusion of proteins and other small molecules (Fig. 2F).
To elucidate the cause of cell shrinkage during phase I, we grew exponential phase mlaA* cells in a microfluidic flow cell (Materials and Methods) and switched from fresh LB to sucrose-supplemented spent LB. The addition of sucrose did not prevent cell death, but a prolonged period before cell death was observed, and the rate of cell death decreased (Fig. 2G). When sucrose was added, plasmolysis occurred rapidly, and the cytoplasm contents shrank and detached from the cell wall and OM, exposing a visible region of concavity in the phase signal (Fig. 2H, white arrow). The cytoplasm remained roughly the same size, but the concave region gradually resolved (Fig. 2H), suggesting loss of membrane material and resulting contraction of the cytoplasm. This shrinkage is in contrast to the response of wild-type cells to hyperosmotic shock in which the cytoplasm expands back to the size dictated by the cell wall (Fig. S3B). After this shrinkage, mlaA* cells continued shortening, and the cell interior became phase bright (Fig. 2H), similar to phase I behavior in our experiments without sucrose (Fig. 2B). Finally, cells lost their integrity and faded away after death (Fig. 2H). Based on these results and our FM 4-64 data demonstrating OM blebbing (Fig. 2E), we conclude that, when exposed to spent medium, mlaA* cells begin to lose membrane faster than it can be replaced, causing the cell to shrink throughout phase I. Such shrinkage leads to an increase in turgor pressure, eventually causing the cell to rupture because of mechanical failure (phase II), and the cell contents gradually leak out (phase III). The addition of sucrose reduces the turgor pressure, permitting a longer period of shrinking, and delays death (Fig. 2G); nonetheless, the continuous loss of OM eventually results in mechanical destabilization and cell death (Fig. 2H).
mlaA*-Mediated Death Is Linked to Cell Division.
In spent medium, mlaA* cells exhibited OM blebbing, predominantly from division sites or cell poles (Fig. S3A), as a prelude to death. Moreover, a previous chemical screen revealed that ∆mlaA cells display increased sensitivity to cefsulodin (25), a β-lactam antibiotic that inhibits the transpeptidation activity of the bifunctional enzymes PBP1A and PBP1B, the latter of which contributes to growth of the septal wall during division (26). However, aside from a small increase in lag time, mlaA* cells do not exhibit any growth-rate or morphological phenotypes before stationary phase (Fig. 1 B and C). Thus, we postulated that, because the transition into stationary phase decouples cell division and cell mass increase (27), frequent divisions exacerbated the slight defects in mlaA* cells during the coordination of septal cell-wall and OM synthesis. This notion suggested that other perturbations to septal peptidoglycan synthesis also may amplify mlaA* phenotypes. In agreement with this idea, we found that in the mlaA* background the loss of PBP1B amplified the mlaA* phenotype, with death occurring in virtually all cells (Fig. S3 C and D).
To provide further evidence for defective coordination between septal cell-wall and OM synthesis, we placed exponentially growing wild-type and mlaA* cells in a microfluidic flow cell and monitored the changes in cell growth after treatment with 10 µg/mL cephalexin, a β-lactam that inhibits the septal transpeptidase PBP3. As expected, wild-type cells elongated for several doublings without dividing, eventually dying because of bulging defects followed by rapid mechanical failure (Fig. 3A); the death trajectories of these cells were typical of β-lactam treatment and did not resemble that of mlaA* cells in spent medium. In contrast, mlaA* cells rapidly exhibited blebs near midcell or at the poles and died within 20 min of cephalexin treatment (Fig. 3B). Thus, perturbations to septal cell-wall synthesis are synergistic with mlaA* in causing cell death.
Fig. 3.
Cell death in mlaA* cells is linked to cell division and fatty acid limitation. (A) Under 10 µg/mL cephalexin treatment, wild-type cells elongated for several doublings without dividing, eventually lysing because of bulging defects followed by rapid mechanical failure. (B) mlaA* cells rapidly developed OM blebs near midcell or at the poles and died within 20 min of cephalexin treatment. (C) Induction of sulA caused cell filamentation and delayed cell death in mlaA* cells for ∼20 min. (D) Phase-contrast and cytoplasmic GFP fluorescence images of a sulA-induced mlaA* cell during a short interval encompassing the popping event in the cell-death trajectory. Initially, the cytoplasm and IM shrank away from the cell wall, exposing periplasmic space near the left pole, and GFP signal was confined to the cytoplasm. At the popping event (t = 0 s), GFP filled the entire region encompassed by the cell envelope. After this time, GFP exited the cell, leaving a gradient of fluorescence increasing toward the right pole. (E) Cerulenin treatment of exponentially growing mlaA* cells on fresh LB pads caused cells to die, with a death trajectory at the single-cell level similar to that of untreated cells shifted onto spent LB pads. The addition of 1 mg/mL oleic acid in spent LB completely suppressed cell death.
The links between mlaA* cell death and division suggest that blocking cell division should rescue mlaA*-mediated death to some extent. To test this possibility, we transformed a plasmid with inducible sulA into mlaA* cells. SulA is a cell-division inhibitor that blocks FtsZ ring assembly and thus prevents septum formation (28). SulA induction caused mlaA* cells to form long filaments in exponential phase, and, upon transitioning into spent LB, these cells’ dying was delayed for 20 min compared with nonfilamentous mlaA* cells (Fig. 3C and Fig. S3E). Thus, the loss of membrane material from septal OM blebs likely accelerates cell death. The timing of the popping event was more synchronized across the population of sulA-expressing cells, leading to a sharper transition in the proportion of live cells than in non–sulA-expressing cells (Fig. 3C). mlaA* cells lost OM material through both midcell blebs and OM vesicles, whereas sulA-expressing cells exhibited only OM vesicles and hence would be expected to lose OM at a rate proportional to the total membrane area. Based on this reasoning, we expected that sulA-expressing cells would die at approximately the same time, when OM loss after the transition to spent medium reaches a common threshold. By contrast, in non–sulA-expressing cells, midcell blebs are predominantly responsible for membrane loss (Fig. S3A) and thus can accelerate cell death in some cells, leading to a broader range of cell death timing.
During the transition to spent medium, SulA-induced mlaA* cells also experienced dramatic detachment of the IM from the OM and the cell wall at one pole during phase I shrinkage (white arrow in Fig. S3E). Upon closer examination, we found that such detachment also occurred in mlaA* cells in the absence of SulA induction (Fig. S3F) but was less apparent because of the shorter cell lengths. The detachment still occurred with the addition of the proton ionophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which depletes the proton-motive force, in both mlaA* (Fig. S3G) and SulA-induced mlaA* cells (Fig. S3H). To resolve which membrane, IM or OM, was compromised first upon cell death, we rapidly imaged cytoplasmic GFP with a temporal resolution of 10 frames/s. In SulA-induced cells, GFP molecules were confined within the IM during most of phase I; approximately 0.1 s before phase II, the IM ruptured, and GFP filled the periplasm, still confined by the OM. In phase II, GFP quickly diffused out of the periplasm through the pole, and the whole cell gradually lost fluorescence intensity (Fig. 3D). Because lipids in mlaA* cells are lost through OM vesicles (Fig. S1D) and blebs (Fig. 2E and Fig. S3A), and because the OM cannot shrink because of the underlying rigid cell wall, our data suggest that lipids flow from the IM to the OM to compensate for the lipid loss in the OM. Such lipid flow would account for the IM shrinkage we observe and eventually would cause the IM to rupture.
mlaA*-Mediated Death Is Linked to Fatty Acid Depletion.
Cells carrying the mlaA* mutation consume fatty acids faster than wild-type cells because of the increased synthesis of LPS and the lipid loss through OM vesicles. Upon transition to spent medium, the slow shrinkage in phase I indicates that cells are losing membrane faster than new fatty acids can be synthesized, and this imbalance ultimately leads to cell death. Therefore, we hypothesized that providing fatty acids to starving mlaA* cells could block cell death. To test this hypothesis, we imaged the transition of mlaA* cells on agarose pads with spent LB supplemented with 1 mg/mL oleic acid (29). As expected, cells stopped growing, but no shrinkage was observed, and all cells remained alive after 1 h (Fig. 3E), indicating that fatty acid depletion causes cell death. To test whether mlaA* cells were generally sensitized to decreases in fatty acid production, we imaged exponentially growing cells treated with 100 µg/mL cerulenin, a specific inhibitor of fatty acid biosynthesis (30). Cerulenin treatment did not cause any cell death in wild-type or ∆mlaA cells, but mlaA* cells underwent the same death trajectory as untreated cells in spent LB (Fig. 3E), consistent with our measurements of increased LPS and OM vesicle production in mlaA* cells relative to wild-type cells during exponential growth. Taken together, these experiments demonstrate that mlaA* cells are generally fatty acid-limited, likely because of increased LPS synthesis and lipid flow from the IM that compensates for the loss of material from the OM.
Suppressor Analysis Implicates Increased Levels of LPS as a Major Cause of Cell Death.
To understand better the processes affected by MlaA*, we used a random, selection-based approach to isolate genetic suppressors of MlaA*-mediated death. We selected for suppressors on maltose MacConkey agar and on LB agar supplemented with 50 µg/mL erythromycin. Resistant colonies arose quite frequently (10−6) on both media, so we biased toward picking suppressors with wild-type colony morphology to select mutants that suppress both the OM permeability and death phenotypes. On both media, we mainly isolated insertions or deletions in the mlaA gene that simply destroyed the dominant mlaA* allele.
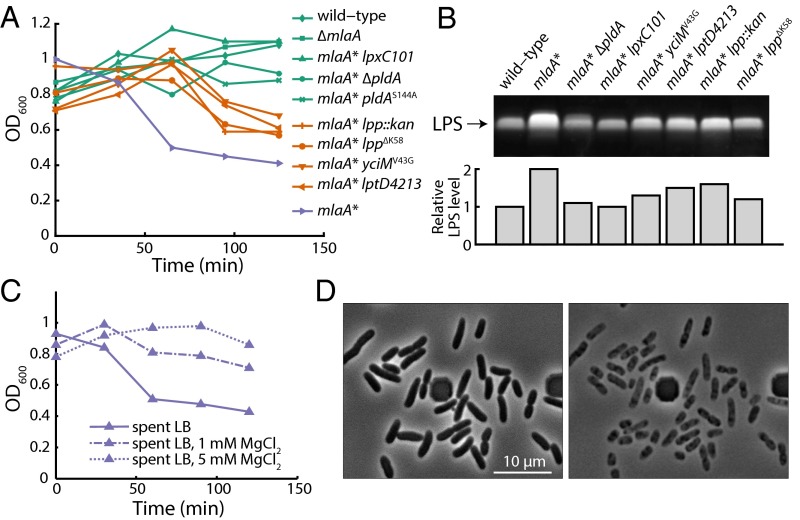
We identified three different extragenic suppressors on maltose MacConkey agar. Two of the suppressors mapped to lpp, and the other mapped to yciM. Lpp is the major lipoprotein in E. coli and is one of the most abundant proteins in the cell (31, 32); some Lpp molecules are covalently attached to peptidoglycan (32, 33). One suppressor contained a large insertion in lpp, resulting in a null mutation. The other suppressor, lppY76C, had been isolated previously and shown to decrease the amount of Lpp bound to peptidoglycan (34). To determine whether these suppressors act by decreasing connections between the OM and the cell wall, we introduced an lpp::kan-null allele, as well as the lppΔK58 allele which completely abolishes linkage of Lpp to the cell wall (34), into mlaA*. The lpp::kan and lppΔK58 alleles suppressed MacConkey sensitivity, but all the lpp alleles and the yciM mutation suppressed cell death only partially (Fig. 4A).
Fig. 4.
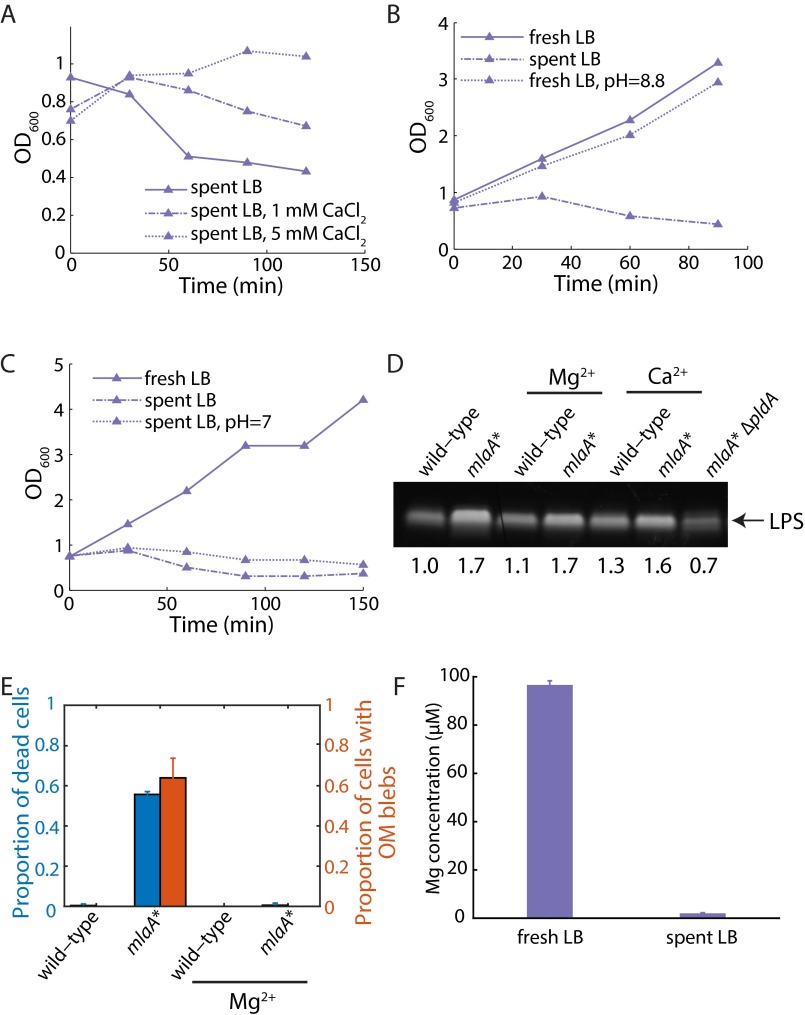
Genetic suppressors of cell death reverse the increases in LPS levels in mlaA* cells, whereas divalent cations stabilize the OM and suppress death. (A) The spent-medium transition growth assay demonstrated that mlaA* suppressors prevent death to differing degrees. (B) LPS levels correlated with the degree of suppression of mlaA*-mediated death during the transition to spent LB. (C) During a spent medium transition assay (Materials and Methods), supplementation with MgCl2 suppressed death in a concentration-dependent manner. (D) After the shift to spent LB in a microfluidic flow cell with constant flow, nearly every mlaA* cell died, with a death trajectory similar to that on pads shown in Fig. 2B. The dark square-like structures are part of the microfluidic device.
yciM encodes a gene that recently has been implicated in negatively regulating LpxC, which catalyzes the second step in LPS biosynthesis (35, 36), and lpp-null mutations have been isolated as suppressors of yciM loss-of-function mutations. Therefore, we wondered whether the yciM and lpp mutations suppressed death in an mlaA* background by reducing LPS levels, thereby stabilizing the OM. Indeed, these mutations do reduce LPS levels (Fig. 4B), but, as is consistent with the lpp and yciMV43G mutations being weak suppressors of cell death, their effects on LPS levels are modest.
To confirm that reductions in LPS levels suppress cell death, we singly introduced mutations that decrease LPS synthesis (lpxC101) (37) or transport to the OM (lptD4213) (38) into an mlaA* background and found that all mutations suppressed the death phenotype (Fig. 4A). The strongest suppressor of cell death is lpxC101 (Fig. 4A); this mutation reduced LPS levels to wild-type levels (Fig. 4B). In all cases, we conclude that reductions in LPS levels correlate with suppressor strength and that the increased LPS levels in the mlaA* background ultimately lead to the cell-death phenotype by increasing fatty acid consumption and perhaps by destabilizing the OM and stimulating the formation of OM vesicles.
Depletion of Extracellular Mg2+ Is Partially Responsible for mlaA* Cell Death in Stationary Phase.
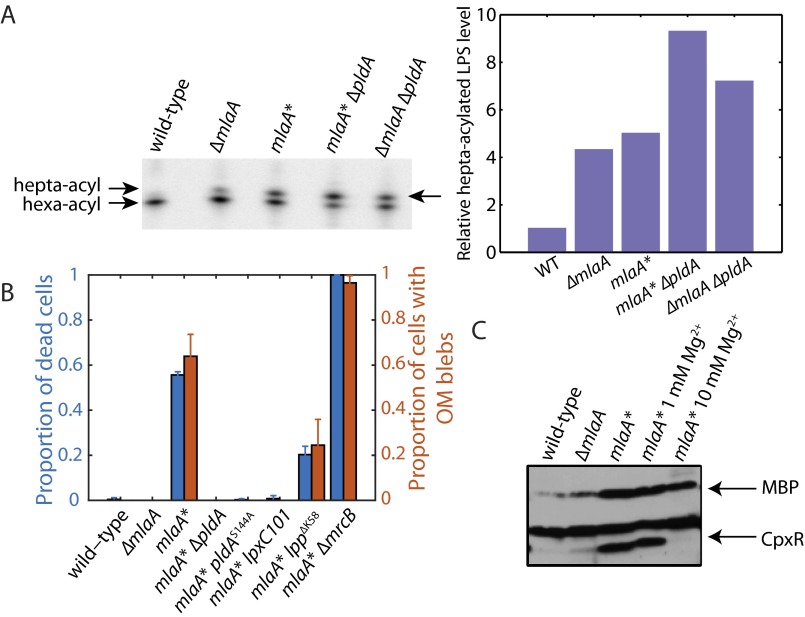
We wondered if the increased LPS levels in the mlaA* background destabilized the OM and contributed to cell death by promoting vesiculation and lipid loss. Mg2+ levels could be an especially important issue for mlaA* cells, because Mg2+ stabilizes the OM by associating with negatively charged LPS molecules and anionic PLs (6), and Mg2+ is known to be present at low levels in LB (39). Accordingly, we supplemented spent medium with increasing concentrations of Mg2+ and assayed for death. Interestingly, supplementation with 1 mM MgCl2 partially suppressed death, and 5 mM MgCl2 fully suppressed death (Fig. 4C). mlaA* strains grown overnight in the presence of excess Mg2+ did not accumulate the cytoplasmic protein CpxR in the culture supernatant; such accumulation would have been indicative of cell rupture (Fig. S4C).
Fig. S4.
Suppressors of mlaA*-mediated death reduce LPS levels and suppress OM blebs. Chemical suppression of mlaA*-mediated stationary-phase death indicates restoration of OM mechanical stability. (A) 32PO4-radiolabeled lipid A samples were isolated and separated by TLC (Materials and Methods). pldA::kan in an mlaA* background has increased levels of hepta-acylated LPS, indicative of an increase in the levels of PLs in the outer leaflet of the OM. (Left) Gel image. (Right) Relative hepta-acylated LPS levels quantified from the gel. (B) Transition assay into spent medium was performed on mlaA* suppressor strains stained with FM 4-64. The wild-type, ∆mlaA, mlaA* lpxC101, mlaA* ∆pldA, and mlaA* pldAS144A strains did not show any cell death; the mlaA* lpp∆K58 strains and the mlaA* ∆mrcB strains showed less and more death than mlaA*, respectively. The degree of OM blebbing in each case correlated with cell-death proportion. Bars represent mean ± SD in three fields of view. (C) Whole-cell protein lysates were prepared, separated by SDS/PAGE, and analyzed by immunoblotting (Materials and Methods). mlaA* strains grown overnight in the presence of excess magnesium do not accumulate CpxR in the culture supernatant.
Because Mg2+ also is required for the function of a variety of intracellular enzymes (39) and cytoskeletal proteins such as MreB (40), we asked whether other divalent cations could suppress stationary-phase death. Unlike Mg2+ (41), intracellular Ca2+ levels are low (42), but extracellular Ca2+ is known to stabilize the OM (43). The effects of CaCl2 on death suppression in mlaA* cells were nearly identical to those of Mg2+: 1 mM and 5 mM CaCl2 partially and fully suppressed death, respectively (Fig. S5A). We confirmed that adding Ca2+ or Mg2+ does not reduce LPS levels (Fig. S5D). Nonetheless, the addition of Mg2+ does suppress the formation of midcell blebs (Fig. S5E), indicating that divalent cations suppress death by physically stabilizing the OM and thereby preventing loss of material. Moreover, pH changes associated with the transition to stationary phase did not contribute to cell death (Fig. S5 B and C). Thus, excess divalent cations are necessary and sufficient to prevent stationary-phase death in mlaA* cells, as is consistent with the suggestion that increased levels of LPS destabilize the OM in spent LB.
Fig. S5.
Divalent cations are sufficient to suppress mlaA*-mediated cell death by mechanically stabilizing the OM. (A) The effects of CaCl2 on the suppression of death of mlaA* cells during the spent medium transition assay are similar to those of MgCl2 (Fig. 4C). (B) Growth in fresh LB with pH increased to 8.8 does not induce death of mlaA* cells. (C) Shifting the pH of spent LB to 7 does not suppress the death of mlaA* cells in the spent medium transition assay. (D) The addition of Ca2+ or Mg2+ does not reduce LPS levels in mlaA* cells. (E) The transition assay into Mg2+-supplemented spent medium was performed on mlaA* cells stained with the membrane dye FM –4-64. OM blebbing and cell death in mlaA* cells were both suppressed with 5 mM Mg2+. No blebs were observed in 136 of 136 mlaA* cells in medium supplemented with Mg2+. Bars represent mean ± SD in three fields of view. (F) Magnesium levels in LB and spent LB, measured by coupled plasma optical emission spectrometry. Fresh LB contains ∼100 µM Mg. The Mg level drops to ∼2 µM in spent LB. Bars represent mean ± SD in three independent samples.
In our single-cell imaging on agarose pads (Fig. 2A) and our bulk OD600 measurements in liquid broth (Fig. 1 B and D), ∼50% of the cells survived the transition into spent medium. These cells do not contain suppressor mutations; when they were subcultured, they underwent stationary-phase death in this second round of growth. What enables these cells to survive? We hypothesized that in both cases, dead cells release molecules that stabilize the OM of the surviving cells. To test this hypothesis, we grew cells in a microfluidic flow cell (Materials and Methods) in which dead-cell contents were washed away rapidly and cells were kept in a chemostat-like environment. After the shift to spent medium, nearly every mlaA* cell died, with a death trajectory similar to that seen on pads (Fig. 4D). Therefore, the deaths of mlaA* cells in stationary phase likely exert altruistic effects by releasing molecules that protect other cells.
To identify whether the dead cells released enough Mg2+ to protect other cells, we directly measured the concentration of elemental Mg in LB and spent LB using inductively coupled plasma optical emission spectrometry. Fresh LB contained ∼100 µM Mg, and the Mg level dropped to ∼2 µM in spent LB (Fig. S5F), suggesting that the contents released by dead mlaA* cells in spent LB can result in a significant increase in Mg2+ concentration that stabilizes other cells. However, such an increase could not restore the Mg2+ level any higher than the starting level of 100 µM and thus would not be sufficient to rescue the remaining cells fully. Therefore, it is likely the release of several different molecules, such as divalent cations and fatty acids and other nutrients, from dead mlaA* cells stabilizes the envelope of other cells.
MlaA* Mediates Cell Death by Activating PldA.
Because our genetic evidence suggests that OM composition can generally modulate the propensity for death in mlaA* cells, and because both MlaA and PldA function to remove PLs from the outer leaflet of the OM (15), we hypothesized that the phenotypes conferred by MlaA* would be exacerbated by the loss of PldA. Strikingly, however, when we introduced pldA::kan or pldAS144A, a mutation that alters the active site of PldA (44), into mlaA*, we observed complete suppression of death (Fig. 4A). This suppression was specific to PldA, because death was not suppressed by inactivating pagP (12). Knocking out PldA function is as potent a suppressor of mlaA*-mediated death as lpxC101 (Fig. 4A). As expected, deletion of pldA in an mlaA* background increased both OM permeability and the accumulation of PLs in the outer leaflet relative to mlaA* alone (Fig. S4A). Thus, pldA::kan does not remedy the defect in OM symmetry and PL trafficking caused by mlaA*, separating the permeability and death phenotypes of mlaA*. Rather, like lpxC101, the pldA-null mutations suppress the death phenotype by reducing LPS levels to nearly wild-type levels (Fig. 4B).
We labeled several of the suppressed strains with FM 4-64 and imaged their transition from exponential phase into spent medium. As expected, wild-type, ∆mlaA, mlaA* lpxC101, mlaA* ∆pldA, and mlaA* pldAS144A did not show any cell death, whereas mlaA* lpp∆K58 and mlaA* ∆mrcB showed less and more death than mlaA*, respectively (Fig. S4B). The degree of OM blebbing in each case was predicted by the degree of suppression; in particular, no visible blebs were visible on mlaA* lpxC101, mlaA* ∆pldA, or mlaA* pldAS144A cells (Fig. S4B). These results suggest that MlaA* activates PldA and that this increase in PldA activity causes the increased levels of LPS that are responsible for the cell death observed.
Discussion
In this study we have described the isolation and characterization of a gain-of-function mutation in mlaA that specifies the OM lipoprotein component of the retrograde Mla PL transport pathway. Unlike the null mutation, mlaA* renders the OM severely defective, accumulating even more PLs in the outer leaflet as indicated by an increase in the levels of hepta-acylated LPS (Fig. S1C). This accumulation results in more dramatic OM permeability defects (Fig. 1A and Fig. S1A), activation of the σE and Cpx stress responses (Fig. S1B), and increased OM vesiculation (Fig. S1D). The phenotypes conferred by this gain-of-function mutation are dominant over the wild-type allele (Fig. S1F) and are independent of downstream mla gene products (Fig. 1A). Perhaps the most striking phenotype of mlaA* cells is death in stationary phase (Fig. 1B). Single-cell analysis reveals a previously unidentified, lengthy death trajectory for mlaA* cells (Fig. 2 B and C) that begins with the loss of OM membrane material and eventually ends by rupture of the IM (Fig. 2F and Fig. 3D).
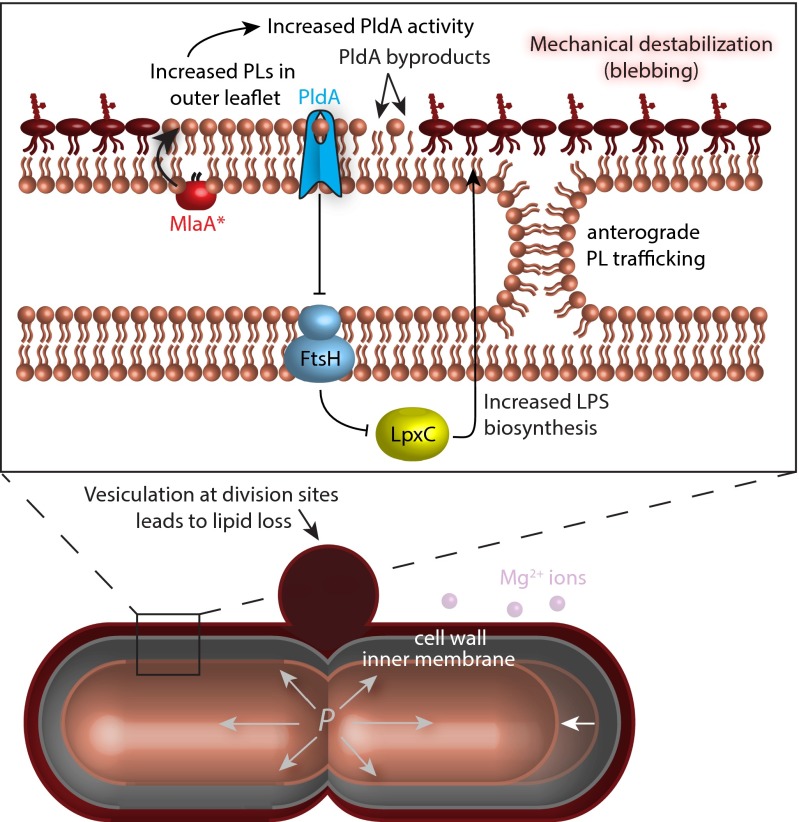
Underlying this novel cell-death pathway are the increased levels of LPS triggered by mlaA*. Moreover, death requires a special set of circumstances that occur as cells enter stationary phase in LB: (i) an increased frequency of cell division, (ii) severely limited concentrations of divalent cations, and (iii) depletion of the carbon sources necessary for fatty acid synthesis. Together with the increased levels of LPS, these conditions destabilize the OM, causing midcell blebs at division sites (Fig. 2E) and the production of OM vesicles (Fig. S1D). Because the area of the OM is dictated by the rigid peptidoglycan cell wall, lipids lost from the OM by vesiculation must be replaced by lipids from the IM. This transfer of lipids causes the IM to shrink, decreasing the volume of the cytoplasm and increasing its density. Eventually, this cytoplasmic concentration leads to mechanical rupture of the IM and ensuing cell death (Fig. 5).
Fig. 5.
Model of MlaA*-mediated death. The mlaA* allele increases LPS levels, and we hypothesize that this increase is caused by the cell’s response to the activation of PldA caused by MlaA* transferring PLs from the inner to the outer leaflet (the reverse of its wild-type activity). Because of this change in composition, OM material is lost through vesiculation at cell-division sites when cations are depleted. Because the area of the OM is constrained to be larger than the rigid cell wall, this loss is compensated by lipid flow to the OM from the IM in an energy-independent manner, possibly through sites of membrane hemifusion. Starved cells cannot synthesize fatty acids, and consequently the IM shrinks away from the cell wall (white arrow) and eventually ruptures because of the increase in turgor pressure. Cells die through slow leakage of cytoplasmic contents out of the envelope rather than through rapid lysis.
Multiple lines of evidence support our explanation of this remarkable cell-death pathway. First, genetic analysis demonstrates that mutations that decrease LPS levels suppress cell death, counteracting the increased LPS biosynthesis observed in mlaA* cells. Weak suppressors of death, such as lpp, show a modest decrease in LPS levels, whereas potent suppressors, such as lpxC101 and ∆pldA, restore LPS levels to those in wild-type cells. Second, mlaA*-mediated death is delayed by the production of the cell division inhibitor SulA, which suppresses midcell blebs at division sites (Fig. 3C). Based on an estimate of the bleb radius (r) of ∼0.3–0.5 µm (Fig. 2E) and the average length (l) of exponentially growing mlaA* cells of ∼4 µm (Fig. 1C), the midcell blebs would give a fractional area loss of 2r/l∼0.15–0.25. This estimate is in reasonable agreement with the total membrane lost in sulA-expressing cells as measured by cell length (∆l/l, ∼0.15–0.2). Third, cell death can be suppressed by the addition of divalent cations to fortify the low cation levels in spent LB (Fig. 4C and Figs. S4C and S5A). Finally, increased synthesis of LPS and OM destabilization leading to OM vesicle production and midcell bleb formation forces the cell to replace OM material with lipids from the IM. However, starved cells are unable to replenish IM lipids that are lost, leading to cytoplasmic shrinkage. Preventing starvation by adding a carbon source such as glucose prevents cell death. Even more revealing, the addition of external fatty acids fully rescues mlaA* cells (Fig. 3E), even though this addition does not restore growth as glucose addition does. Moreover, the exquisite sensitivity of mlaA* cells to fatty acid levels is demonstrated by the fact that inhibiting fatty acid synthesis in exponential phase, where nutrient and cation concentrations are high, also initiates the mlaA* cell-death pathway (Fig. 3E). Ultimately, the loss of lipids from the IM breaks the balance across the layers of the cell envelope, resulting in unsustainable cytoplasmic densities.
We do not yet understand why the mlaA* mutation increases LPS levels. Genetic analysis indicates that the increase in LPS levels caused by mlaA* is dependent upon PldA. Indeed, pldA-null mutations restore LPS levels (Fig. 4B) and are strong suppressors of cell death (Fig. 4A). Equally strong suppression is observed with the pldA active-site mutation (pldAS144A), demonstrating that the loss of phospholipase activity (44), rather than some unknown activity of the protein, is responsible for suppression. It is important to note that lpxC101 and pldA-null mutations do not repair the OM permeability defects associated with mlaA* (Fig. S4A); in fact, the pldA-null mutation increases the levels of surface-exposed PLs and exacerbates the permeability defects (Fig. S4A). Although the functions of the Mla proteins and PldA are related, they act in separate pathways and eliminate PLs from the outer leaflet in different ways (14, 15). The Mla pathway completely removes PLs (15), whereas PldA destroys PLs by cleaving fatty-acid side chains from the glycerophosphodiester backbones of PLs and lyso-PLs (14). Although the exact mechanism linking PldA activity and LPS levels remains to be determined, we propose that E. coli cells interpret increases in the levels of PldA reaction products (free fatty acids and lyso-PLs) as a signal that more LPS is needed in the OM.
At the molecular level, the accumulation of more surface-exposed PLs in mlaA* cells than in ∆mlaA cells (Fig. S1C) indicates that MlaA* has a novel function that likely is related to its dominant phenotype. If MlaA* selectively left a particular type of PL in the OM but removed other PLs, deleting downstream Mla proteins would change the phenotype because the subset of PLs removed by MlaA* would accumulate and change the composition of the OM to that observed in ∆mla cells. If MlaA* removed PLs from the inner leaflet instead of the outer leaflet, the downstream Mla proteins still would affect the phenotype, again in contrast with our observations. Instead, if MlaA* removed PLs from the inner leaflet and put them in the outer leaflet (the reverse reaction of wild-type MlaA), all the phenotypes that we identified would be explained. Removing downstream Mla proteins would not affect the phenotype of mlaA* (Fig. 1A), and such a mutant would accumulate even more PLs in the outer leaflet than an mlaA-null mutant (Fig. S1C). We suggest that the increase in PLs in the outer leaflet activates PldA. This mechanism is consistent with our diploid analysis; we observed effects that were dominant, but not completely so (Fig. S1F), reflecting a race between MlaA and MlaA* for insertion and removal of surface-exposed PLs. The activation of PldA by the presence of PLs in the outer leaflet of the OM caused by the activity of MlaA* results in production of lyso-PLs and free fatty acids, which may be linked to OM vesiculation and the increased production of LPS.
Since the turn of the century, the cellular components required for the transport of integral OM proteins, lipoproteins, and LPS from their sites of synthesis in the cytoplasm or the IM have all been identified (1). However, the mechanism by which PLs are transported to the OM remains mysterious. Decades ago, it was shown that PLs, unlike any other OM component, can move backward from the OM to the IM; this retrograde movement occurs even with lipids not normally present in the OM, such as cholesterol (45). To account for these results, the authors proposed zones of adhesion between the IM and the OM. Although proteinaceous bridges across the periplasm such as those required for LPS assembly have been demonstrated, sites of direct membrane contact have not, and they remain controversial (1). Nonetheless, our data indicating substantial motion of lipids from the IM to the OM in mlaA* cells suggest the presence of zones of adhesion that are sites of hemifusion where the outer leaflet of the IM is fused with the inner leaflet of the OM. Hemifusion would permit diffusive flow of phospholipids, resulting in a rapid and substantial net flux to the OM when vesiculation occurs. This lipid flow persisted in the presence of the uncoupler CCCP (Fig. S3 G and H) or mutations that remove ATP synthase, suggesting that lipid movement from the IM to OM is passive and concentration dependent. Direct demonstration of hemifusions, which likely are transient because of the dynamics of growth, will be challenging and will require the development of new methodologies. Nevertheless, our study demonstrates the vital importance of homeostasis across the layers of the Gram-negative bacterial cell envelope and the utility of single-cell imaging and genetics for revealing the molecular and cellular mechanisms coupling these layers together.
Materials and Methods
Bacterial Strains and Growth Conditions.
All strains used in this study are isogenic derivatives of NR754, an araD+ revertant of E. coli MC4100 (Table S1) (46, 47). Strains were constructed by generalized P1 transduction or transformation (48). kan cassettes were excised using the Flp recombinase (49). Unless otherwise noted, strains were grown in LB medium or on LB agar at 37 °C. When necessary, LB was supplemented with 25 µg/mL ampicillin, 20 µg/mL chloramphenicol, 25 µg/mL kanamycin, or 25 µg/mL tetracycline.
Table S1.
Strains used in this study
| Strain | Relevant features | Reference/source |
| MC4100 | F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR thi | 46 |
| NR754 | MC4100 Ara+ | 47 |
| HC494 | NR754 ΔlptD pET2342::lptDD57V/A59P/P95L | This study |
| HC687 | NR754 mlaA* ΔyfdI | 49, 62, and this study |
| HC723 | HC687 cpxR::kan | 63 and this study |
| HC724 | HC687 cpxR::spc | 63 and this study |
| HC726 | HC687 pldA::kan | 62 and this study |
| HC727 | HC687 rpoS::kan | 62 and this study |
| HC735 | NR754 ΔyfdI | 62 and this study |
| HC736 | NR754 ΔmlaA ΔyfdI | 49, 62, and this study |
| HC741 | HC687 bolA::kan | 62 and this study |
| HC742 | HC735 bolA::kan | 62 and this study |
| HC747 | HC735 rpoS::kan | 62 and this study |
| HC748 | HC736 bolA::kan | 62 and this study |
| HC752 | HC736 pldA::kan | 62 and this study |
| HC753 | HC736 rpoS::kan | 62 and this study |
| HC756 | HC687 pldAS144A bla | 62 and this study |
| HC783 | HC687 lpp::kan | 62 and this study |
| HC810 | HC687 lppΔK58 ynhG::kan | 64 and this study |
| HC818 | HC687 rfaC::kan | 62 and this study |
| HC819 | HC687 rfaP::kan | 62 and this study |
| HC829 | HC687 lptD4213 carB::Tn10 | 65 and this study |
| HC833 | HC687 mrcB::kan | 62 and this study |
| HC849 | HC687 ompA::kan | 62 and this study |
| HC850 | HC687 pal::kan | 62 and this study |
| HC851 | HC687 tolB::kan | 62 and this study |
| HC853 | HC687 lpxL::kan | 62 and this study |
| HC854 | HC687 lpxM::kan | 62 and this study |
| HC855 | HC687 skp::kan | 62 and this study |
| HC858 | HC687 pagP::kan | 62 and this study |
| HC899 | HC687 yraP::kan | 62 and this study |
| HC900 | HC736 pZS21::mlaA | This study |
| HC919 | HC687 pZS21::mlaA | This study |
| HC920 | HC735 pZS21::mlaA* | This study |
| HC921 | HC687 pZS21::mlaA* | This study |
| HC922 | HC736 pZS21::mlaA* | This study |
| HC1024 | HC687 erfK::kan | 62 and this study |
| HC1025 | HC687 ybiS::kan | 62 and this study |
| HC1026 | HC687 ycbB::kan | 62 and this study |
| HC1027 | HC687 ycfS::kan | 62 and this study |
| HC1028 | HC687 ynhG::kan | 62 and this study |
| HC1029 | HC687 amiA::kan | 62 and this study |
| HC1030 | HC687 amiB::kan | 62 and this study |
| HC1032 | HC687 envC::kan | 62 and this study |
| HC1033 | HC687 nlpD::kan | 62 and this study |
| HC1046 | HC687 mipA::kan | 62 and this study |
| HC1247 | HC687 lapBV43G ycjM::Tn10 | 15 and this study |
| HC1390 | HC687 lpxC101 leuB::Tn10 | 37, 66, and this study |
| HC1396 | HC687 amiC::kan | 62 and this study |
| HC1413 | HC687 mlaC::kan | 62 and this study |
| HC1414 | HC687 mlaD::kan | 62 and this study |
| HC1415 | HC687 mlaF::kan | 62 and this study |
Suppressor Selection and Tn10 Mapping.
Spontaneous drug-resistant suppressors of lptDD57V/A59P/P95L were obtained by plating 100 μL of an overnight culture onto LB supplemented with 625 µg/mL bacitracin and overnight incubation at 37 °C. Suppressor frequency was calculated based on the ratio of the number of suppressor colonies and the number of cells plated at a given dilution. Suppressors were purified at 37 °C. Mutations were mapped using pools of random Tn10 insertions as described previously (15) and identified by DNA sequencing.
Plasmid Construction.
All plasmids used in this study are listed in Table S2. pZS21::mlaA was constructed using primer HC245 (5′-TTGTCTCGAGCAACTGACGGGAGTTACTCTG-3′) and primer HC333 (5′-TTGTTCTAGAGACTGAGAGAACCAAACGACG-3′) to amplify mlaA with its native promoter and ribosome-binding site from the chromosome with XhoI and XbaI sites (underlined), and the product was cloned into pZS21::bamD. Digestion with XhoI and XbaI was used to remove bamD from this plasmid and to insert mlaA under the control of its native promoter.
Table S2.
Plasmids used in this study
| Plasmid | Relevant features | Reference/source |
| pET2342:: lptDD57V/A59P/P95L | Site-directed mutagenesis to introduce lptDD57V/A59P/P95L into pET2342::lptD | 67 |
| pZS21::bamD | bamD cloned into pZS21::luc | 68 |
| pZS21::mlaA | mlaA cloned into pZS21::bamD | This study |
| pZS21::mlaA* | Site-directed mutagenesis to introduce mlaAΔN41ΔF42 (mlaA*) into pZS21::mlaA | This study |
Site-Directed Mutagenesis.
The mlaA* allele was introduced using site-directed mutagenesis into pZS21::mlaA. The Platinum Pfx polymerase (Invitrogen) was used to introduce the mutations with the following conditions for PCR: 95 °C for 5 min, followed by 18 cycles of 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 6 min, and one final extension at 68 °C for 10 min. Products were digested with DpnI (New England Biolabs) at 37 °C for 2.5 h and subsequently were used to transform electrocompetent DH5α cells. Transformations were plated on LB agar supplemented with 25 µg/mL kanamycin and were incubated at 37 °C overnight. Transformants were purified, plasmids were isolated, and mutations were confirmed by DNA sequencing.
Sensitivity Assays.
Efficiency of plating assays were used to assess sensitivity to bile salts, detergent, and antibiotics. Overnight cultures were serially diluted and replica-plated onto lactose MacConkey agar, LB agar supplemented with the indicated antibiotic or detergent concentrations, or LB agar alone. Plates were incubated overnight at 37 °C.
Immunoblot Analyses.
For whole-cell lysate samples, cultures were grown overnight and subcultured 1:500 into fresh LB. Exponential-phase cultures were harvested at OD600 ∼0.6–0.8, 1 mL of culture was pelleted (5,000 × g for 2 min at room temperature), and this pellet was resuspended in SDS/PAGE resuspension buffer in a volume equal to the OD600 divided by six. Samples were boiled for 10 min and resolved by 10% SDS/PAGE. Proteins were transferred to a nitrocellulose membrane, and immunoblotting was performed using the following dilutions of polyclonal rabbit antibodies: CpxR-MBP (1:5,000), DegP (1:20,000), LamB (1:30,000), LptD (1:7,500), LpxC (1:5,000), and MlaA (1:5,000). Donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase was used (1:15,000 for LpxC and 1:5,000 for others). ECL film (Amersham) and X-ray film (Denville) were used to develop the immunoblots. When indicated, densitometry analysis was performed using Fiji 1.49e (19).
To measure supernatant protein levels, at each time point, 3 mL of culture were pelleted, and the supernatant was passed through a 0.2-μm syringe filter to exclude whole cells. Two milliliters of this filtered supernatant were concentrated on an Amicon Ultra-15 3K Centrifugal Filter unit (Millipore) by centrifugation at 5,000 × g at room temperature for 15 min. Concentrated protein samples were resuspended from the filter in 100 μL SDS/PAGE sample buffer, boiled, and resolved by SDS/PAGE. Immunoblotting was performed as described above.
LPS Analyses.
LPS levels were analyzed as previously described (50). Briefly, 5 × 108 cells from overnight (16-h) cultures were collected by centrifugation, and cell pellets were resuspended in 100 μL of 1× LDS sample buffer (Invitrogen) + 4% β-mercaptoethanol. Samples were boiled for 10 min, allowed to cool, and treated with 125 ng/μL Proteinase K (New England Biolabs) at 55 °C for 16 h. Proteinase K was heat-inactivated at 100 °C for 5 min, and the lysates were separated by SDS/PAGE using 4–12% bis-Tris NuPAGE gradient gels (Invitrogen). Gels were stained with Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (Molecular Probes) according to the manufacturer’s instructions. LPS bands were visualized by UV transillumination, and relative band intensities were determined using the Quantity One imaging software (Bio-Rad).
OM Vesicle Preparation.
To isolate OM vesicles, cells were grown to midexponential phase (OD600 ∼0.6–0.8). A 1-mL sample was used as a whole-cell lysate control. Twenty milliliters of the sample with the lowest OD600 were pelleted (5,000 × g for 2 min at room temperature), and volumes of the other samples were adjusted according to the difference in OD600. Equal volumes of the culture supernatants were filtered through 0.2-μm filters to eliminate whole cells and large cellular debris. This filtered supernatant was subjected to a second round of filtration through an Amicon Ultra-15 100K centrifugal filter (Millipore) to isolate and concentrate OM vesicles. Samples were resuspended from the 100K filter in 50 μL of SDS/PAGE buffer, boiled for 10 min, and resolved by SDS/PAGE. Immunoblotting was performed as described above.
Growth Assays.
For spectrophotometer-based growth curves, overnight cultures grown at 37 °C were diluted 1:500 in 25 mL of LB and were grown in a 37 °C shaking water bath. OD600 was measured at the indicated time points.
For growth curves with more frequent OD600 readings, overnight cultures were back-diluted 1:200 into 200 µL LB and grown with shaking at 37 °C in a Tecan M200 plate reader. At 7.5-min intervals, the absorbance at wavelength 600 nm was measured. The natural logarithm of the OD was fit to the Gompertz equation (51) to determine a maximum specific growth rate. Each growth curve was fit individually, and the fitting parameters were averaged to generate a mean fit.
Quantification of Viable Cells.
To measure the number of viable cells, strains were serially diluted, and 100 μL of the 10−5, 10−6, and 10−7 dilutions were plated on LB agar plates. These plates were incubated overnight at 37 °C, and colonies were counted to determine the number of colony-forming units present at each dilution.
Spent Medium Death Assay.
NR754 cells were grown overnight in LB at 37 °C, after which the cells were pelleted and the supernatant was passed through a 0.2-μm filter to eliminate cells and large cellular debris. This filtered supernatant was used as spent medium. For death assays, overnight cultures grown at 37 °C were diluted in fresh LB and grown at 37 °C with shaking until late exponential phase (OD600∼0.8). For bulk measurements, these cultures were pelleted, resuspended in spent medium to induce the transition into stationary phase, and incubated at 37 °C while OD600 was monitored. Where indicated, the spent medium was supplemented with sterile MgCl2, CaCl2, or CCCP (Sigma).
Analysis of Radiolabeled Lipid A.
Overnight cultures grown at 37 °C were diluted 1:100 in 5 mL LB with 5 μCi/mL 32PO4 and were grown at 37 °C in a shaking water bath for 3 h. Lipid A abundance was analyzed with a mild acid-hydrolysis method (18, 52). Briefly, 32PO4-radiolabeled lipid A samples were dried under a stream of nitrogen gas. The following day, samples were resuspended in 100 μL chloroform:methanol (4:1) and spotted on a 20 × 20 cm SILGUR-25 TLC plate with UV254 coating (Macherey Nagel). Samples were normalized according to scintillation counts. The TLC solvent was chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol). TLC plates were dried and exposed to a phosphor screen for 16–18 h. The phosphor screen was read using a Typhoon 9410 Imager (GE Healthcare).
Purification of Sacculi and UPLC of Peptidoglycan Composition.
UPLC samples were prepared as previously described (53). Overnight cultures were diluted 1:100 in 250 mL LB and grown at 37 °C to an OD600 of 0.7. Cultures were centrifuged at 5,000 × g for 10 min at room temperature, and the resulting pellet was resuspended in 3 mL LB. Cell suspensions were lysed by boiling in SDS for 3 h. Lysed cell suspensions were ultracentrifuged at 400,000 × g at room temperature to purify sacculi, which then were digested with muramidase into muropeptides. Samples were pH-adjusted and injected into a Waters H Class UPLC system equipped with a BEH C18 1.7-µm column (Waters) and were eluted with sodium phosphate buffers. Peaks were quantified and identified as particular muropeptide species from their elution times based on a previous study (54), and the crosslinking density and average strand length of each strain were calculated from the muropeptide abundances as previously described (55).
RNA Isolation.
Cells were grown to midexponential phase (OD600 ∼0.6–0.8) in LB at 37 °C, and total RNA was extracted using an RNeasy Mini Kit (QIAGEN) with on-column DNase I treatment, according to the manufacturer’s instructions.
cDNA Synthesis and qRT-PCR.
One microgram of total RNA from experimental samples was converted to cDNA using random primers (Applied Biosystems) and the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s protocol. cDNAs were diluted 1:100, and 3.75 μL of each were used as a template for qRT-PCR using PerfeCTa SYBR Green FastMix (Quanta Biosciences; 20-μL reaction volumes). Primers specific for lpxC mRNA (KM1qPCR_lpxC_F: TACGTGTCTGGTCAACGAGC; KM2qPCR_lpxC_R: TTTTGGCGCAGTTCAACTCG) and rpoD mRNA (KM9qPCR_rpoD_F: GCACCGTTGAACTGTTGACC; KM10qPCR_rpoD_R: CCGGTGATCAGATCGGACAG) were designed using Primer3Plus (56) and were used at a final concentration of 250 nM. Thermal cycling was performed on a StepOnePlus RT-PCR system (Applied Biosystems) with cycling conditions consisting of 95 °C for 3 min, 40 repetitions of 95 °C (15 s), 60 °C (15 s), and 72 °C (30 s), followed by a melt-curve analysis. The resulting cycle threshold (Ct) values were recorded for lpxC mRNA templates and normalized to total rpoD mRNA. The relative amounts of lpxC mRNA in a strain with the wild-type mlaA allele (HC735) and the various mutants were compared using the comparative Ct (ΔΔCt) method.
Single-Cell Imaging.
Cells were imaged on a Nikon Eclipse Ti-E inverted fluorescence microscope with a 100× (NA 1.40) oil-immersion objective (Nikon Instruments). Images were collected on a DU885 electron-multiplying CCD camera (Andor Technology) or a Neo sCMOS camera (Andor Technology) using µManager version 1.4 (https://micro-manager.org/) (57). Cells were maintained at 37 °C during imaging with an active-control environmental chamber (Haison Technology).
For experiments conducted on agarose pads, 1 µL of cells was spotted onto a pad of 1% agarose in fresh LB or spent medium, with 1 µg/mL FM 4-64 (Invitrogen) as indicated. Flow-cell experiments were performed in ONIX B04A microfluidic chips (CellASIC). Exponentially growing rod-shaped cells were loaded into the imaging chamber, and media reservoirs were filled with fresh or spent LB. When indicated, 10 µg/mL cephalexin or 20% sucrose was added to the medium.
Image Analysis.
Time-lapse and static images were segmented using a custom MATLAB (MathWorks) software package (58). Cell width and length were calculated using the MicrobeTracker meshing algorithm (59).
SI Results
mlaA* Induces Extracytoplasmic Stress Responses and Hypervesiculates.
Based on the defects in OM permeability exhibited by mlaA* cells, we conjectured that the mutation may induce envelope-stress responses (17). The σE extracytoplasmic stress response is activated by perturbations to OM composition and LPS levels. The Cpx stress response is induced by many cues, including misfolded proteins in the IM and periplasm, high pH, and surface adhesion. These stress responses have some overlap in terms of the genes they regulate; for example, degP is induced by both σE and Cpx (17). Thus, we compared DegP levels in wild-type, ΔmlaA, and mlaA* strains; increased levels of full-length DegP and its proteolytically cleaved form indicate that one or both of the σE and Cpx responses is turned on (Fig. S1B). When we deleted the mlaA* strain for cpxR, the gene encoding the response regulator for the Cpx stress response (60), DegP levels decreased, but not to wild-type levels, suggesting that both stress responses are turned on in these cells (Fig. S1B). Additionally, there was a slight increase in the level of LptD and a decrease in the level of LamB, as expected because they are regulated by the σE stress response (Fig. S1B).
OM defects also can result in the shedding of OM vesicles (61). To measure OM vesicle production in mlaA* cells, we filtered culture supernatants to exclude whole cells and concentrated these samples on 100K centrifugal filters, which retain these vesicles. We then performed SDS/PAGE and immunoblotted for OM and periplasmic proteins, whose levels are indicative of OM vesiculation (Materials and Methods). The mlaA* strain hypervesiculated in comparison with wild-type and ΔmlaA cells (Fig. S1D), as is consistent with the OM permeability defects caused by mlaA*. Thus, the mlaA* allele induces envelope stress responses, perhaps through the altered composition and properties of the OM caused by hypervesiculation.
Increased pH Does Not Cause mlaA* Cell Lysis.
In addition to cellular characteristics, the extracellular environment changes over the course of growth as nutrients are removed from the medium and cells expel waste products into the medium (5). As a result, pH increases over the course of growth of E. coli cells; we measured a pH of 7 for fresh LB, whereas spent LB had a pH of 8.8. To test the effect of pH on lysis, we collected spent LB, reduced the pH to 7, and assayed for lysis. Although this spent LB had the same pH as fresh LB, mlaA* cells still lysed (Fig. S4C). Alternatively, in fresh LB in which the pH was increased to 8.8, mlaA* cells grew and did not exhibit any signs of lysis (Fig. S4D). Therefore, the change in pH as cells transition from exponential growth to stationary phase is not responsible for mlaA* cell lysis.
Supplementary Material
Acknowledgments
We thank Juliana Malinverni and Marcin Grabowicz for strains and helpful discussions and Enrique Rojas and members of the K.C.H. and T.J.S. laboratories for insightful discussions. This work was supported by National Institute of General Medical Sciences Grant GM34821 (to T.J.S.), National Science Foundation CAREER Award MCB-1149328 (to K.C.H.), National Institute of Health Director’s New Innovator Award DP2OD006466 (to K.C.H.), the National Science Foundation Graduate Research Fellowship Program under Grant DGE1148900 (to H.A.S.), and a Stanford Agilent Fellowship (to H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601375113/-/DCSupplemental.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos MP, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 3.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10(2):123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollmer W, Blanot D, De Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 5.Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34(4):476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Quinn PJ. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog Lipid Res. 2010;49(2):97–107. doi: 10.1016/j.plipres.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido H. Restoring permeability barrier function to the outer membrane. Chem Biol. 2005;12(5):507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol. 2006;1(6):385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 12.Bishop RE. Structural biology of membrane-intrinsic beta-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim Biophys Acta. 2008;1778(9):1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci USA. 2014;111(5):1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker N. Outer-membrane phospholipase A: Known structure, unknown biological function. Mol Microbiol. 2000;35(4):711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 15.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106(19):8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong ZS, Woo WF, Chng SS. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol. 2015;98(6):1133–1146. doi: 10.1111/mmi.13202. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz N, Silhavy TJ. Sensing external stress: Watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8(2):122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Jia W, et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279(43):44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 19.Schindelin J, et al. FIJI: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alemohammad MM, Knowles CJ. Osmotically induced volume and turbidity changes of Escherichia coli due to salts, sucrose and glycerol, with particular reference to the rapid permeation of glycerol into the cell. J Gen Microbiol. 1974;82(1):125–142. doi: 10.1099/00221287-82-1-125. [DOI] [PubMed] [Google Scholar]
- 21.Cayley DS, Guttman HJ, Record MT., Jr Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys J. 2000;78(4):1748–1764. doi: 10.1016/s0006-3495(00)76726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32(2):259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 23.Schäkermann M, Langklotz S, Narberhaus F. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J Bacteriol. 2013;195(9):1912–1919. doi: 10.1128/JB.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105(49):19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144(1):143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertsche U, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61(3):675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 27.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47(1):855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 28.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175(4):1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis: Complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284(43):29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg I, Walker JR, Bloch K. Inhibition of lipid synthesis in Escherichia coli cells by the antibiotic cerulenin. Antimicrob Agents Chemother. 1973;3(5):549–554. doi: 10.1128/aac.3.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun V, Bosch V. Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur J Biochem. 1972;28(1):51–69. doi: 10.1111/j.1432-1033.1972.tb01883.x. [DOI] [PubMed] [Google Scholar]
- 32.Inouye M, Shaw J, Shen C. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem. 1972;247(24):8154–8159. [PubMed] [Google Scholar]
- 33.Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur J Biochem. 1970;13(2):336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang WY, Wu HC. Alterations of the carboxyl-terminal amino acid residues of Escherichia coli lipoprotein affect the formation of murein-bound lipoprotein. J Biol Chem. 1992;267(27):19560–19564. [PubMed] [Google Scholar]
- 35.Mahalakshmi S, Sunayana MR, SaiSree L, Reddy M. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol. 2014;91(1):145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 36.Nicolaes V, et al. Insights into the function of YciM, a heat shock membrane protein required to maintain envelope integrity in Escherichia coli. J Bacteriol. 2014;196(2):300–309. doi: 10.1128/JB.00921-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169(12):5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122(3):491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lusk JE, Williams RJ, Kennedy EP. Magnesium and the growth of Escherichia coli. J Biol Chem. 1968;243(10):2618–2624. [PubMed] [Google Scholar]
- 40.van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413(6851):39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 41.Moncany ML, Kellenberger E. High magnesium content of Escherichia coli B. Experientia. 1981;37(8):846–847. doi: 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 42.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262(26):12570–12574. [PubMed] [Google Scholar]
- 43.Katowsky M, Sabisch A, Gutberlet T, Bradaczek H. Molecular modelling of bacterial deep rough mutant lipopolysaccharide of Escherichia coli. Eur J Biochem. 1991;197(3):707–716. doi: 10.1111/j.1432-1033.1991.tb15962.x. [DOI] [PubMed] [Google Scholar]
- 44.Horrevoets AJ, Verheij HM, de Haas GH. Inactivation of Escherichia coli outer-membrane phospholipase A by the affinity label hexadecanesulfonyl fluoride. Evidence for an active-site serine. Eur J Biochem. 1991;198(1):247–253. doi: 10.1111/j.1432-1033.1991.tb16008.x. [DOI] [PubMed] [Google Scholar]
- 45.Jones NC, Osborn MJ. Translocation of phospholipids between the outer and inner membranes of Salmonella Typhimurium. J Biol Chem. 1977;252(20):7405–7412. [PubMed] [Google Scholar]
- 46.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105(14):5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1984. [Google Scholar]
- 49.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 50.Stead CM, Zhao J, Raetz CR, Trent MS. Removal of the outer Kdo from Helicobacter pylori lipopolysaccharide and its impact on the bacterial surface. Mol Microbiol. 2010;78(4):837–852. doi: 10.1111/j.1365-2958.2010.07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwietering MH, Jongenburger I, Rombouts FM, van ’t Riet K. Modeling of the bacterial growth curve. Appl Environ Microbiol. 1990;56(6):1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desmarais SM, Cava F, de Pedro MA, Huang KC. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp. 2014;83(83):e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263(21):10088–10095. [PubMed] [Google Scholar]
- 55.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172(2):451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 56.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(Web Server issue) suppl 2:W71-4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer Control of Microscopes Using µManager. John Wiley & Sons, Inc., Hoboken, NJ; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ursell TS, et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA. 2014;111(11):E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80(3):612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong J, Iuchi S, Kwan HS, Lu Z, Lin EC. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene. 1993;136(1-2):227–230. doi: 10.1016/0378-1119(93)90469-j. [DOI] [PubMed] [Google Scholar]
- 61.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188(15):5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2(1):1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahoney TF, Silhavy TJ. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol. 2013;195(9):1869–1874. doi: 10.1128/JB.02197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol. 2011;79(5):1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121(2):307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 66.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45(5):1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 67.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103(31):11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol. 2012;194(5):1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.