Significance
The retrovirus human T-lymphotropic virus type 1 (HTLV-1) causes inflammatory and malignant diseases in humans. To maintain latency and avoid immune detection in vivo, HTLV-1 minimizes expression of genes on the plus-strand of the integrated provirus but allows constitutive expression of the minus-strand gene, which maintains clonal persistence. It is not understood how this gene expression is regulated. We show that CTCF, a master regulator of chromatin structure and gene expression, binds to HTLV-1, forms loops between the provirus and host genome, and alters expression of proviral and host genes. Because a typical HTLV-1–infected host carries >104 infected T-cell clones, each containing a provirus integrated in a different genomic site, CTCF binding gives HTLV-1 the potential to cause widespread abnormalities in the human genome.
Keywords: retrovirus, latency, epigenetics, HTLV-1, CTCF
Abstract
Human T-lymphotropic virus type 1 (HTLV-1) is a retrovirus that causes malignant and inflammatory diseases in ∼10% of infected people. A typical host has between 104 and 105 clones of HTLV-1–infected T lymphocytes, each clone distinguished by the genomic integration site of the single-copy HTLV-1 provirus. The HTLV-1 bZIP (HBZ) factor gene is constitutively expressed from the minus strand of the provirus, whereas plus-strand expression, required for viral propagation to uninfected cells, is suppressed or intermittent in vivo, allowing escape from host immune surveillance. It remains unknown what regulates this pattern of proviral transcription and latency. Here, we show that CTCF, a key regulator of chromatin structure and function, binds to the provirus at a sharp border in epigenetic modifications in the pX region of the HTLV-1 provirus in T cells naturally infected with HTLV-1. CTCF is a zinc-finger protein that binds to an insulator region in genomic DNA and plays a fundamental role in controlling higher order chromatin structure and gene expression in vertebrate cells. We show that CTCF bound to HTLV-1 acts as an enhancer blocker, regulates HTLV-1 mRNA splicing, and forms long-distance interactions with flanking host chromatin. CTCF-binding sites (CTCF-BSs) have been propagated throughout the genome by transposons in certain primate lineages, but CTCF binding has not previously been described in present-day exogenous retroviruses. The presence of an ectopic CTCF-BS introduced by the retrovirus in tens of thousands of genomic locations has the potential to cause widespread abnormalities in host cell chromatin structure and gene expression.
Retroviruses integrate a dsDNA copy of their genome, the provirus, into the genome of the cell they infect. Human T-lymphotropic virus type 1 (HTLV-1) is an exogenous retrovirus, widespread in the tropics. Most infected people are asymptomatic carriers, but ∼10% develop a malignant or inflammatory disease. Adult T-cell leukemia (ATL) is a leukemia of HTLV-1–infected CD4+ T cells. ATL cells frequently contain chromosomal abnormalities, and the disease is refractory to conventional chemotherapy. HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP) is a chronic inflammatory disease of the spinal cord. HTLV-1 spreads within the individual both by cell-to-cell transmission and by clonal proliferation of infected cells: HTLV-1 gene products induce proliferation and enhance survival of infected cells (1, 2). In addition to the viral genes that encode enzymes and structural proteins, HTLV-1 encodes several regulatory and accessory genes in the pX region, between the env gene and the 3′ long terminal repeat (LTR). The HBZ gene is constitutively expressed from the minus strand of the integrated provirus (3), whereas plus-strand expression, required for viral propagation to uninfected cells, is suppressed or intermittently expressed in vivo, allowing escape from host immune surveillance (2, 4). It is unknown how HTLV-1 maintains this chromatin state and strand-selective transcription.
One mechanism used by HTLV-1 to suppress transcription of the plus strand is methylation of the 5′ LTR, whereas there is little DNA methylation in the 3′ LTR (5). The DNA methylation is sharply reduced at the middle of the provirus (6). This observation raised the question of whether there is a regulatory mechanism that divides the methylated 5′ part from the unmethylated 3′ part of the provirus, perhaps to allow the constitutive expression of the HBZ gene that appears to be required for clonal persistence of HTLV-1 (3, 7).
A chromatin insulator is a DNA region that separates transcriptionally active and inactive regions by binding to certain proteins. The best-characterized insulator-binding protein in higher eukaryotes is CTCF, an 11 zinc-finger protein highly conserved from flies to humans (8), which binds to tens of thousands of sites in the human genome and regulates chromatin structure, transcriptional activation, repression, silencing, imprinting, and alternative splicing (9). We therefore set out to test the hypothesis that CTCF binds to the HTLV-1 provirus at an epigenetic border and regulates proviral transcription.
Results
Epigenetic Border in the HTLV-1 Provirus.
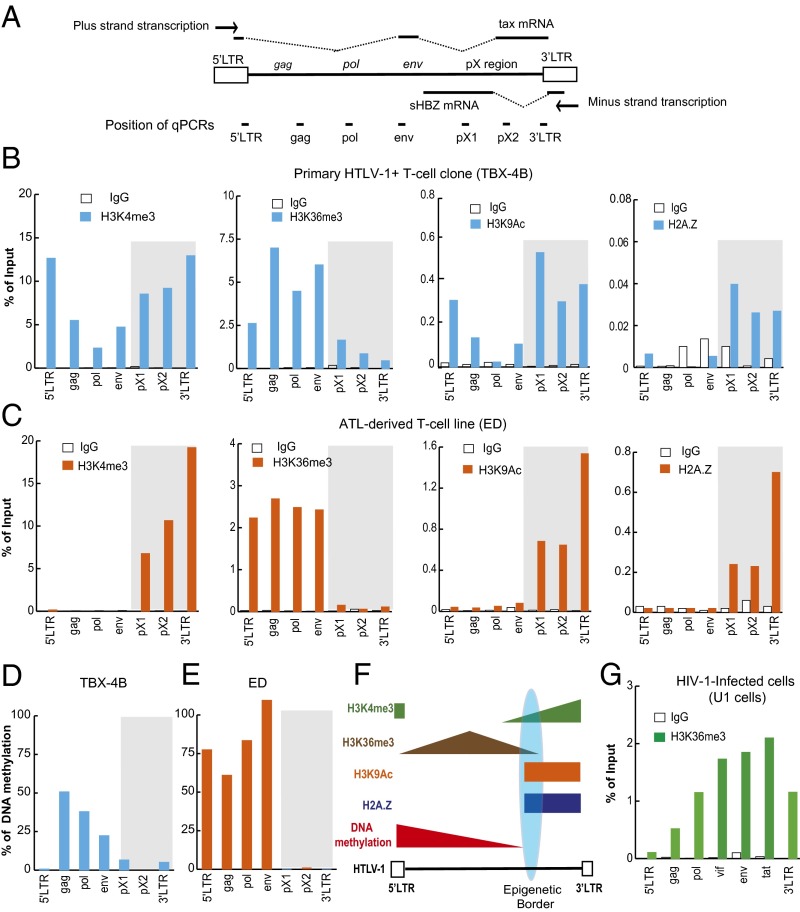
We used chromatin immunoprecipitation (ChIP) assays to identify epigenetic marks over the HTLV-1 provirus (Fig. 1A), including H3K36me3 and H3K9Ac, each associated with actively transcribed genes; H3K4me3, characteristic of enhancer and promoter regions; and H2A.Z, a histone variant frequently observed in promoter and enhancer regions (10). The results showed a sharp border in each of these epigenetic modifications in the pX region of the provirus in both a nonmalignant primary T-cell clone (Fig. 1B) and an ATL cell line (Fig. 1C). The epigenetic border was more sharply defined in the ATL cell line, in which the spliced HBZ transcript (sHBZ) is transcribed but plus-strand transcription is significantly suppressed (Fig. S1), than in nonmalignant T-cell clones, in which both tax, a plus-strand transcript, and sHBZ, a minus-strand transcript, are actively transcribed. The same pattern was also observed in three other HTLV-1–infected T-cell lines (Fig. S2 A–C) and in peripheral blood mononuclear cells (PBMCs) freshly isolated from patients with ATL (Fig. S3 A–F). In addition, a border was observed in DNA methylation, consistent with previous observations (6) (Fig. 1 D and E). The results are summarized schematically in Fig. 1F. The observed pattern of epigenetic modifications was consistent with the observation that transcription of the proviral minus-strand exceeds transcription of the plus-strand both ex vivo (4) and frequently in HTLV-1–infected clones in vitro (Fig. S1). In contrast, in the provirus of another human retrovirus, HIV-1, the mark H3K36me3 showed a progressive rise, with no border (Fig. 1G); this result was expected, because H3K36me3 is characteristic of actively transcribed gene bodies, increasing toward the 3′ end of the gene (10). These observations, together with previous reports (5, 6), suggested the presence of a regulatory mechanism that establishes the distinctive pattern of respective epigenetic modifications in the 5′ and 3′ portions of the HTLV-1 provirus.
Fig. 1.
Epigenetic border in the pX region of the HTLV-1 provirus. (A) HTLV-1 provirus showing the four main viral coding regions (gag, pol, env, and pX; above provirus), the regions analyzed by real-time quantitative PCR (qPCR) (primers shown in Table S1) in the ChIP assay (below provirus), and the viral transcripts tax and sHBZ. Distribution of H3K4me3, H3K36me3, H3K9Ac, and H2A.Z (B and C) and DNA methylation (D and E) over the HTLV-1 provirus in a primary nonmalignant clone (TBX-4B) (B and D) and an ATL cell line (ED) (C and E). The ChIP signal is shown as the percentage of input DNA. An IgG of unrelated specificity was used as a negative control. Representative results from three independent experiments are shown. (F) Schematic distribution of epigenetic modifications in the HTLV-1 provirus in relation to the CTCF-BS. (G) Distribution of H3K36me3 over the HIV-1 provirus in a latently infected cell line (U1).
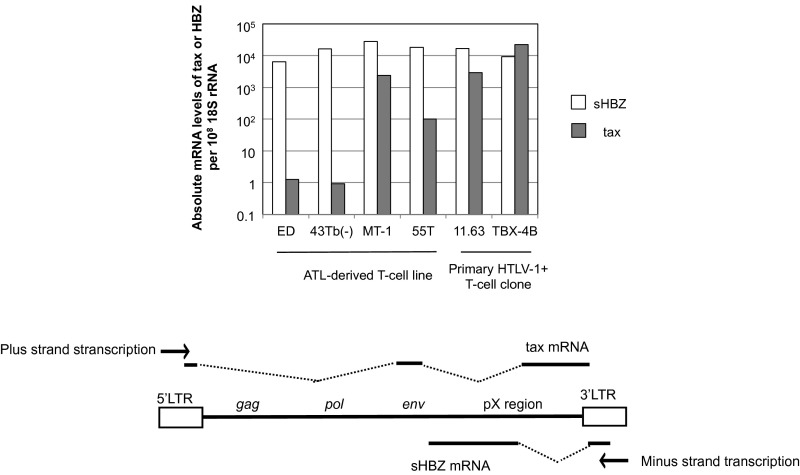
Fig. S1.
Quantification of transcription from the plus-strand and the minus-strand of the HTLV-1 provirus. Expression of tax and HBZ was quantified by qRT-PCR using standard plasmid DNA to calculate the copy number of the transcripts. The copy numbers of each transcript per 108 molecules of 18S rRNA are shown as means of triplicate values. A representative result from two independent experiments is shown. A schematic figure of the viral transcripts tax and sHBZ is shown below the graph.
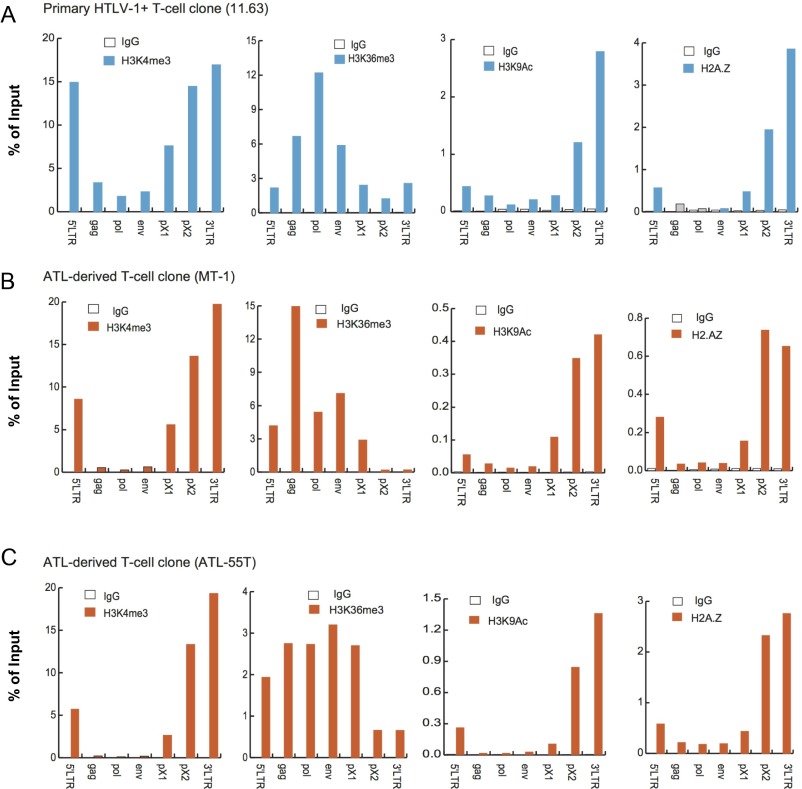
Fig. S2.
Epigenetic border at the proviral CTCF-BS in three additional HTLV-1–associated T-cell lines. ChIP signals of H3K4me3, H3K36me3, H3K9Ac, and H2A.Z over HTLV-1 provirus in a nonmalignant T-cell clone, 11.63 (A), and two ATL cell lines, MT-1 (B) and ATL-55T (C), are shown as the percentage of input DNA. Immunoprecipitated DNA was analyzed by real-time PCR using the primer sets shown in Fig. 1A. An IgG of unrelated specificity was used as a negative control. Representative results from three independent experiments are shown.
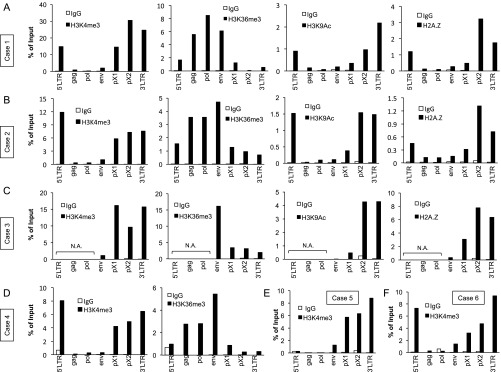
Fig. S3.
Epigenetic border at the proviral CTCF-BS in fresh PBMCs of patients with ATL. (A–F) Distribution of H3K4me3, H3K36me3, H3K9Ac, and H2A.Z over the HTLV-1 provirus in ATL cells was analyzed by ChIP assay. The results are shown as the percentage of input DNA. Immunoprecipitated DNA was analyzed by real-time PCR using the primer sets as shown in Fig. 1A. N.A., not amplified in qPCR assay of input DNA, indicating that the ATL cells had a defective provirus.
The Host Insulator-Binding Protein CTCF Binds to the HTLV-1 Provirus in Vivo.
Insulator elements demarcate the boundary between transcriptionally active (euchromatic) and inactive (heterochromatic) regions of the genome (11) and prevent enhancers from activating inappropriate promoters. We therefore tested the hypothesis that CTCF binds to HTLV-1 at the observed boundary in epigenetic modifications (Fig. 1). First, we searched for the consensus CTCF DNA-binding motif (9) (Fig. S4 A–C) in the HTLV-1 genome and identified several candidate binding sites (Fig. 2A); the site in pX showed the highest similarity to the consensus sequence. CTCF ChIP assays revealed CTCF binding in pX [viral CTCF-binding site (vCTCF-BS)], but not in the other proviral regions tested (Fig. 2 B and C); similar results were obtained in fresh PBMCs from patients with either HAM/TSP (Fig. 2D) or ATL (Fig. 2E), demonstrating that CTCF binds to the pX region of HTLV-1 in natural infection in vivo. In contrast, CTCF did not bind to the provirus of HIV-1 (Fig. 2F), consistent with the absence of a border in the epigenetic mark H3K36me3 within the HIV-1 provirus (Fig. 1G).
Fig. S4.
Calculation of the similarity score to the consensus CTCF-binding sequence. (A) Consensus sequence for CTCF binding reported by Kim et al. (9). (B) At each position in the motif, the respective nucleotide was weighted by the frequency of occurrence of that nucleotide in the consensus sequence. The score for that candidate site was then calculated by adding the weighting factors of each core nucleotide position from position 5 to position 18. (C) Score of the consensus motif (10.253) was taken as 100%.
Fig. 2.
Identification of CTCF (vCTCF)-BS in the HTLV-1 genome. (A) Similarity scores were calculated using the method shown in Fig. S4 to identify potential CTCF-BSs in the HTLV-1 genome (National Center for Biotechnology Information GenBank accession no. AB513134). The four positions with a score >80 (shown in red) were all in the plus-strand (sense). (B–E) CTCF specifically localized at the putative CTCF-BS of the pX region. Results of CTCF-ChIP assay in a nonmalignant HTLV-1–infected T-cell clone, 11.63 (B); an ATL cell line, ED (C); PBMCs from a patient with HAM/TSP (D); PBMCs from a patient with ATL (E); and an HIV-1–infected cell line (U1) (F). Representative results are shown from three (B and C) or two (D and E) independent experiments, respectively. TC-1, positive control region for CTCF binding in the human genome.
ChIP-Sequencing Analysis Showed a Sharp Peak of CTCF Binding in the Region of the Epigenetic Border.
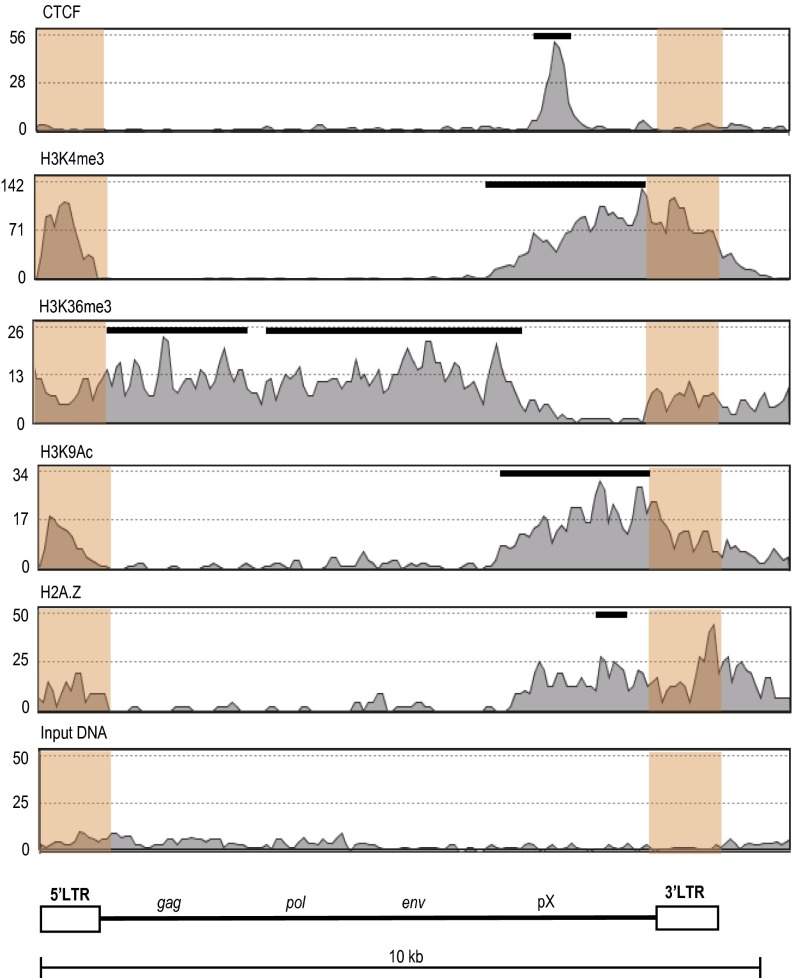
To define CTCF binding and the epigenetic border in the HTLV-1 provirus with greater precision, we performed ChIP-sequencing (ChIP-seq) analysis of a cell line (ED) derived from a patient with ATL, which contains a single-copy HTLV-1 provirus. Because the 5′ LTR and 3′ LTR are identical in sequence, sequence reads that lie within the LTR cannot be specifically mapped. LTR reads were therefore randomly mapped to either the 5′ or 3′ LTR. The results showed a single peak of CTCF binding in the pX region, consistent with the ChIP-quantitative PCR result (Fig. 3 and Fig. S5). The pattern of histone modifications and the histone variant H2A.Z also changed at the CTCF-binding region, extending a short distance beyond the binding site in each case, as observed by others (12).
Fig. 3.
Epigenetic landscape of HTLV-1 provirus analyzed by ChIP-seq. Enrichment of CTCF, H3K4me3, H3K36me3, H3K9Ac, and H2A.Z in ED cells is shown. These data are aggregated from two biological replicate experiments; each replicate showed a similar epigenetic profile. LTR sequences are shaded in orange. LTR reads were randomly mapped to the 5′ LTR or 3′ LTR, because their sequences are identical. Peaks of each signal were analyzed by the model-based analysis of ChIP-seq algorithm and are shown as black bars above the histogram. The significance of enrichment of each signal over input-DNA peaks was calculated with a cutoff P value of 10−5. The ChIP-seq profile of the flanking human genome is shown at a larger scale (∼100 kb) in Fig. S5.
Fig. S5.
Epigenetic landscape of the HTLV-1 provirus at a larger scale. This figure is generated by using the same ChIP-seq data as in Fig. 3. The HTLV-1 genome in ED cells is integrated into the host gene IFT81 with antisense orientation.
CTCF Binds Directly to HTLV-1 pX DNA in a Sequence-Dependent Manner.
CTCF exerts its pleiotropic functions by interacting with various cofactors (8, 13), suggesting that CTCF might bind indirectly to HTLV-1. To test this possibility, we performed an EMSA using recombinant CTCF and three synthetic oligonucleotides, each corresponding to a candidate CTCF-BS in the HTLV-1 genome, and one from the well-characterized CTCF-binding DNA region H19/DMR, the differentially methylated region of the H19 gene (14, 15) (Fig. S6A); each oligonucleotide contained a different variant of the consensus sequence for CTCF binding (Fig. S6B). The results showed specific binding of CTCF to vCTCF in the HTLV-1 pX region, but not to either the LTR or env (Fig. S6C). Competition assays confirmed these results (Fig. S6D). The introduction of six nucleotide substitutions in the consensus CTCF-binding sequence in vCTCF abrogated the ability of the oligonucleotide to compete with the WT pX sequence in binding to CTCF, showing that CTCF binds directly to HTLV-1 DNA in the pX region in a sequence-dependent manner (Fig. S6E). The consensus sequence for the CTCF-BS in pX is conserved both in the HTLV-1 sequences in the GenBank (Fig. S7A) and in all cases of ATL and asymptomatic carriers examined (Fig. S7B).
Fig. S6.
CTCF binds directly to the HTLV-1 provirus in vitro. (A) HTLV-1 provirus showing (below provirus) the position of each probe used in the EMSA for CTCF. (B) Consensus sequence for CTCF binding, as reported by Kim et al. (9), in each probe is shown in red. Nucleotide substitutions in the mutant probes are shown in bold black. We used H19/DMR, a well-studied insulator region (14, 15), as a positive control. (C) Direct binding of CTCF to the vCTCF sequence. Nonradioactively labeled duplex probes of ∼100 bp for each of the three viral candidate regions were incubated with recombinant CTCF and anti-CTCF Ab. CTCF–DNA complexes and supershifted complexes, detected with the anti-CTCF Ab, are indicated by solid and open arrowheads, respectively. (D) Specific CTCF binding to the pX region (vCTCF). Excess nonlabeled vCTCF and H19-DMR probes inhibited the binding of labeled H19-DMR to CTCF, but LTR and env probes did not inhibit the binding. (E) Six nucleotide substitutions in the core consensus sequence of the vCTCF-WT probe strongly reduced the binding to CTCF. Excess nonlabeled vCTCF-WT probes inhibited the binding of labeled vCTCF-WT to CTCF, but mutant probes (vCTCF-mut1 or vCTCF-mut2) did not inhibit the binding.
Fig. S7.
Conservation of the proviral CTCF-BS among various HTLV-1 strains (A) and in HTLV-1–infected cells [PBMCs of patients with ATL or asymptomatic carriers (AC) or ATL-derived cell lines] (B). A hyphen (-) denotes the same sequence as the consensus nucleotide. This vCTCF sequence has strong similarity to the human genome CTCF core motif consensus (Fig. S4; similarity score of 88%).
HTLV-1 pX Has Enhancer-Blocking Activity.
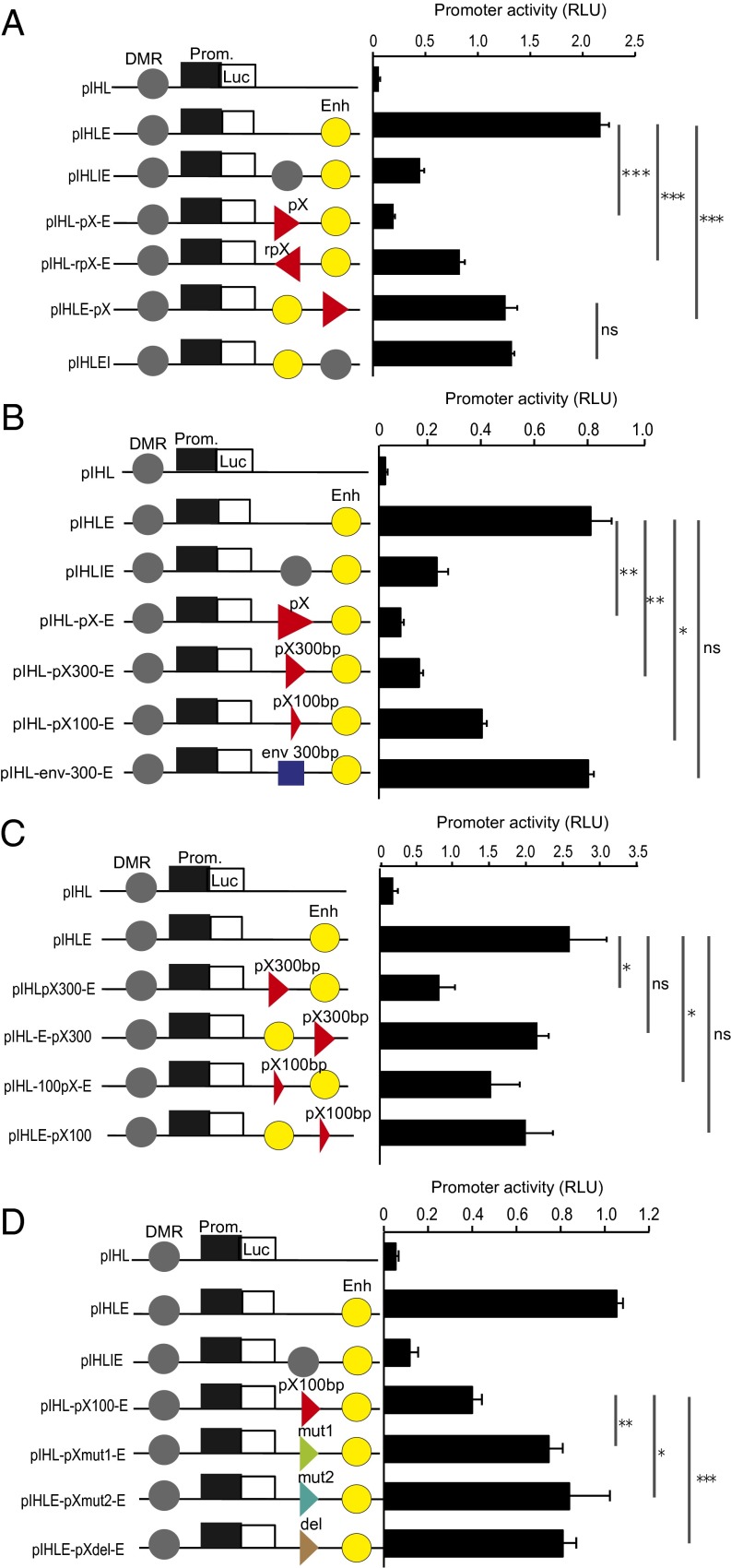
CTCF bound to DNA between an enhancer and a promoter can block the activation of the promoter by the enhancer. A 1-kb DNA fragment of pX containing the CTCF-BS showed strong enhancer-blocking (EB) activity, similar to the positive control, H19/DMR (Fig. 4A). EB function was observed with pX inserted in either orientation between the enhancer and the promoter, and it was reduced, but not eliminated, when the pX fragment was inserted outside the promoter-enhancer region, suggesting that pX contains a bidirectional insulator function (Fig. 4A). The EB activity was detected using 1-kb, 300-bp, or 100-bp fragments (Table S2) of the pX sequence, with each containing the CTCF-binding region (Fig. 4B); the progressively stronger EB effect observed with longer fragments suggests that additional host factors bind the provirus near the CTCF site. In contrast, there was no EB activity with the HTLV-1 env sequence, which does not bind CTCF. A degree of promoter suppression was observed even when the pX sequence was located beyond the promoter-enhancer (Fig. 4A). In these experiments, we used circular plasmids, in which the enhancer might affect the promoter either upstream or downstream. We therefore repeated the EB assay after linearizing the plasmids. The results showed that 300-bp or 100-bp pX fragments exerted EB activity only when inserted between the promoter and the enhancer (Fig. 4C). The EB activity of the pX region was significantly reduced by mutation of the CTCF-BS (Fig. S6B), indicating that CTCF plays an important role in the EB function (Fig. 4D).
Fig. 4.
HTLV-1 pX region exerts CTCF-dependent EB activity. (A) pIHL-pX-E and pIHL-rpX-E plasmids were constructed by inserting a fragment of ∼1,000 bp, containing the CTCF-BS from the pX region, inserted in either a sense or antisense orientation between the promoter and the enhancer in pIHLE reporter vector. The H19 DMR insulator was used as a positive control for insulator function. For pIHLE-pX, the pX fragment was inserted downstream of the enhancer in pIHLE reporter vector. Firefly luciferase activities were normalized to Renilla luciferase and are shown as relative light units (RLU). (B) Plasmids pIHL-pX300-E and pIHL-pX100-E were constructed by inserting fragments, containing the pX CTCF-BS, of ∼300 bp and ∼100 bp, respectively. The pIHL-env-300-E was constructed by inserting a 300-bp fragment from the HTLV-1 env region. (C) For pIHLE-pX300 and pIHLE-pX100, the pX fragment was inserted downstream of the enhancer. Plasmids linearized with the restriction enzyme MluI were used in the assay. (D) Plasmids pIHLpXmut1-E and pIHLpXmut2-E were generated by inserting a DNA fragment of 100 bp containing the CTCF-BS with the same nucleotide substitutions as shown in Fig. S6B. In pIHL-pXdel-E, the core CTCF-BS was deleted. DMR, H19 DMR insulator; Enh, SV40 enhancer; Luc, luciferase gene; Prom., promoter of H19 gene. Data shown are representative of three independent experiments (t test: *P < 0.05; **P < 0.01; ***P < 0.001). ns, not significant.
Table S2.
Primers used in the construction of reporter vectors
| Fragment | Sense | Antisense |
| pX | 5′-CCCTTGAGAATCGAGTCCT-3′ | 5′-AGTAGGGCGTGACGATGTAG-3′ |
| pX300 | 5′-CTGCTCTTCCTGCTTCCTCC-3′ | 5′-ACAAGGAGGAGGAGGAAGCT-3′ |
| pX100 | 5′-CCGATCACGATGCGTTTC-3′ | 5′-ACGGTTTGCTATCCTTAGAAGA-3′ |
CTCF Influences Proviral Transcription and RNA Splicing.
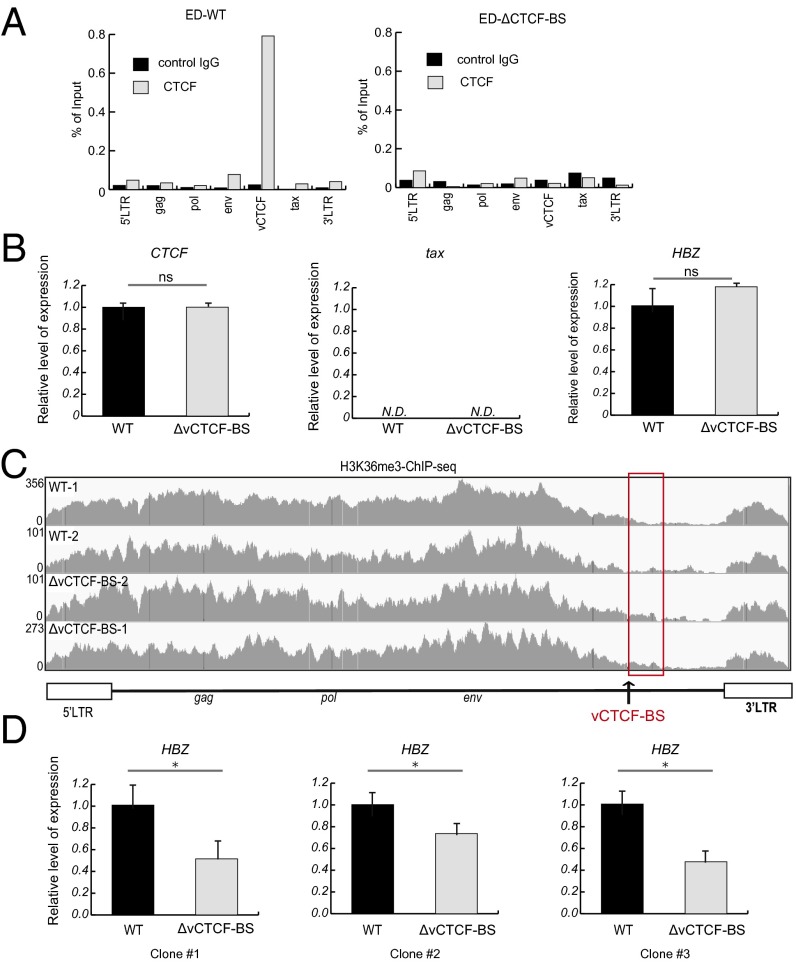
To investigate the effect of CTCF on proviral transcription, we transfected an ATL cell line (MT-1) with shRNA to knock down CTCF expression (Fig. S8 A and B). There was a slight suppressive effect of CTCF knockdown (KD) on the proviral transcriptome (Fig. S8C). Further analysis by quantitative RT-PCR (qRT-PCR) assay with transcript-specific primers (Fig. S8D) revealed significant repression of the HTLV-1 p30 gene (Fig. S8E). We further analyzed the effect of mutations in the viral CTCF-binding region by using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system in another ATL cell line, ED. We confirmed the loss of CTCF binding by mutating six key nucleotides (Fig. S6B; vCTCF-mut1) in the proviral core CTCF-BS (Fig. 5A). Initially, there was no significant change in expression of tax or HBZ (Fig. 5B), but a slight change was evident in the border of H3K36me3 (Fig. 5C). Because HBZ expression plays a critical role in the proliferation of ATL cells (3), ED cells with low HBZ expression may be counterselected during in vitro growth. To reduce the dependency of ED cell proliferation on HBZ, we cultured the cells in the presence of IL-2. After 4 wk, the loss of the vCTCF-BS (ΔvCTCF-BS) resulted in a significant decrease in sHBZ transcription (Fig. 5D). The tax gene was not reactivated; the provirus of ED cells appears to be epigenetically stabilized by DNA hypermethylation in the 5′ LTR (Fig. 1E). Together with the results shown in Figs. 1–5 and Figs. S1–S3 and S8, these data indicate that CTCF plays a role in the regulation of HTLV-1 proviral transcription and RNA splicing.
Fig. S8.
CTCF KD in an ATL cell line, MT-1. Plasmids expressing shRNA for CTCF (CTCF-1) or nonspecific shRNA plasmids as a negative control (NC) were transfected by electroporation into MT-1 cells. The efficiency of CTCF KD was evaluated by qRT-PCR (A) or Western blotting (B) 2 d after transfection. (C) HTLV-1 transcriptome of NC or CTCF KD MT-1 cells is shown. (D) Schematic figure of viral transcripts and primers used for qRT-PCR. (E) Representative qRT-PCR result of viral transcripts from HTLV-1 provirus. 18S rRNA was also analyzed as an internal control. Data shown are representative of three independent experiments (t test: *P < 0.05; ***P < 0.001). ns, not significant.
Fig. 5.
Effect of mutations in the vCTCF sequence on proviral transcription and a histone modification, H3K36me3. (A) Introduction of mutations in the vCTCF sequence (using CRISPR-Cas9) abolished the CTCF binding to the pX region. Representative CTCF-ChIP-quantitative PCR results of ED-WT and ED ΔvCTCF-BS cells are shown. (B) Representative qRT-PCR result of viral transcripts from HTLV-1 provirus. 18S rRNA was also quantified as an internal control. Data shown are representative of two independent experiments. Tax expression is inhibited in ED cells by dense DNA methylation in the 5′ LTR and by a premature stop codon in the tax gene. N.D., not detectable. (C) H3K36me3–ChIP-seq results from WT and ED ΔvCTCF-BS cells (two independent experiments in each case). Because the 5′ LTR and 3′ LTR are identical in sequence, the LTR reads were randomly mapped to either the 5′ LTR or the 3′ LTR. The black arrow indicates a vCTCF-BS. The red rectangle indicates the region immediately downstream of the CTCF-BS. (D) Representative qRT-PCR result of viral transcripts from HTLV-1 provirus. 18S rRNA was quantified as an internal control. Data shown are representative of two independent experiments on each of three independent sets of clones after long-term cultivation in the presence of IL-2 (100 IU/mL). Statistical significance was assessed by Student’s t test (*P < 0.05).
HTLV-1 Alters Local Higher Order Chromatin Structure and Gene Expression in the Host Genome.
To test the hypothesis that the HTLV-1 provirus forms CTCF-mediated chromatin interactions with the flanking host genome, we used the chromosome conformation capture (3C) assay to analyze chromatin from ED cells containing either WT HTLV-1 or the mutant HTLV-1 ΔvCTCF-BS, in which CTCF binding was abrogated (Fig. 5A). A strong CTCF ChIP-seq signal [cellular CTCF (cCTCF)] was observed in the host genome, 48 kb upstream of the proviral integration site (Fig. 6A). Quantitative 3C analysis demonstrated that the cCTCF sites made long-range interactions with the WT provirus significantly more frequently than with the ΔvCTCF-BS mutant (Fig. 6B). These CTCF sites are oriented toward the provirus, which is integrated in the genome in the negative orientation (Fig. 6A). The vCTCF-BS and host CTCF-BS are therefore oriented toward each other; convergent CTCF-BSs appear to be required for chromatin loop formation (16–18). We also examined the effect of the ΔvCTCF-BS on local host gene expression: TCHP, GIT2, and C12orf76 (Fig. 6C). Transcription of two host genes near an upstream CTCF-BS was affected by vCTCF-BS mutation (Fig. 6D). These results show that the integrated HTLV-1 provirus can form CTCF-dependent chromatin loops with the flanking host genome and may cause CTCF-dependent changes in host gene expression.
Fig. 6.
HTLV-1 alters local higher order chromatin structure and gene expression in the host genome. (A) CTCF ChIP-seq data reveal binding of CTCF to the HTLV-1 provirus and the flanking host genome across a 60-kb region in ED cells. The green and red arrowheads denote the orientation of the respective CTCF-BS (inferred from the DNA sequence) in the cellular genome (cCTCF-BS) and the provirus (vCTCF-BS). (B) Frequency of long-range chromatin interactions was assessed by quantitative 3C analyses on six independent libraries (three ED-WT and three ED-ΔvCTCF-BS), using primers within the NlaIII fragments containing the cCTCF-BS (green bar in A) and vCTCF-BS (red bar in A) and the TaqMan probe encompassing the ligation junction of two fragments. Results are mean ± SE. The mean interaction frequency in the mutant was significantly lower than in the WT (three biological replicates of each) (Welch two-sample t test: *P = 0.0186). (C) Schematic figure of host genes near the integration site of HTLV-1 in ED cells. CTCF-binding regions in ED cells are also shown as vertical bars below the figure. The red bar shows a CTCF-BS within the HTLV-1 provirus, and the green bar shows the interacting host CTCF site shown in A. (D) Representative result of qRT-PCR analysis. Expression levels of GIT2, TCHP, and C12orf76 genes were analyzed in ED-WT and ED-ΔvCTCF-BS cells after long-term cultivation in the presence of IL-2. Representative results from two independent experiments are shown. The level of expression of GIT2 and TCHP was significantly lower in the ΔvCTCF-BS line than in the WT (Student’s t test: **P < 0.01; *P < 0.05).
Discussion
We show that the chromatin insulator protein CTCF binds directly to the HTLV-1 provirus. CTCF-BSs have been propagated throughout the genome by transposons in certain nonhuman primate lineages (19), and CTCF has been shown to regulate latency programs in the herpesviruses Epstein-Barr virus (EBV) (20) and Kaposi's sarcoma herpesvirus (KSHV) (21). However, CTCF binding has not been described in present-day exogenous retroviruses. The diverse functions ascribed to CTCF include enhancement or repression of transcription, promoting or blocking communication between enhancers and promoters, acting as a barrier to the propagation of epigenetic changes, and regulation of mRNA splicing (22, 23). We show here that KD of CTCF changed the pattern of splicing of the p30 transcript, in which the CTCF-BS is present near the splicing junction (Fig. S8 D and E). The actions of CTCF at a given location depend on the local context, involving selective interactions between CTCF and both transcriptional cofactors and the primary DNA sequence (24). Homodimerization of CTCF bound to two genomic sites can cause chromatin looping (8), forming structural chromatin loops (∼1 Mb in size), known as topologically associating domains (TADs), which are largely shared between cell types, and smaller functional loops (sub-TADs). Sub-TADs, typically contained within TADs, appear to regulate transcription by bringing a distant enhancer near the respective promoter (8, 16, 18, 25). However, although CTCF plays a central role in establishing chromatin structure, epigenetic borders, and transcriptional activity, removal of CTCF does not invariably lead to changes in chromatin looping (26) and epigenetic marks flanking the border (12). Further, gene expression can be changed in cis at some distance from the abrogated CTCF-BS (12, 27). The epigenetic pattern and chromatin structure may be maintained in the absence of CTCF by cohesin or by other DNA-binding factors, such as transcription factors, other zinc finger proteins (e.g., ZNF143), or RNAs (8, 13). We show here that CTCF binding to HTLV-1 plays a role in EB (Fig. 4) and proviral gene expression (Fig. 5); however, abolition of CTCF binding did not cancel the effect completely (Figs. 4D and 5C), suggesting that other cofactors also contribute to these mechanisms.
Binding of CTCF to the HTLV-1 provirus may serve several functions that benefit the virus, among which we identify three possibilities that are not mutually exclusive. First, by forming a barrier to epigenetic modifications, CTCF may cause or perpetuate differential transcription of the 5′ and 3′ parts of the provirus, resulting in persistent expression of HBZ, which is required for clonal persistence (2, 7), while promoting escape from the strong host immune response to the plus-strand gene products Tax, Pol, and Gag (4) by temporary silencing of the plus-strand genes (3, 5, 6). The results reported here and the previous observation of partial DNA methylation in the provirus (3, 5, 6) suggest that CTCF establishes an epigenetic border in the provirus, which may be stably maintained thereafter by DNA methylation. Second, CTCF may block aberrant activation of the 5′ and 3′ viral promoters by the enhancer present in the 3′ LTR and 5′ LTR, respectively. Third, CTCF might tether the provirus to the nucleolar periphery (28), allowing cell cycle-dependent proviral silencing, and thereby promoting immune escape. This possibility is consistent with our observation (29) of selective survival in vivo of HTLV-1 proviruses integrated in acrocentric chromosomes, which are associated with the nucleolar periphery. The impact of the HTLV-1 provirus on higher order host chromatin structure and the consequent effects on gene expression are adventitious, because they depend on the proviral integration site: They are therefore unlikely to benefit the virus systematically. The consequences of CTCF binding will differ between clones of HTLV-1–infected cells, because the proviral integration site is unique to each clone. A chromatin loop that brings either the 5′ LTR or the 3′ LTR adjacent to a host gene may dysregulate cell growth and cause or contribute to malignant transformation of the cell (30, 31). HTLV-1–infected individuals typically possess >104 infected T-cell clones, each containing one provirus in a unique genomic location (32). HTLV-1 therefore has the potential to cause widespread abnormalities in chromatin structure and gene expression of the human genome by inserting ectopic CTCF-BSs. These abnormalities may, in turn, contribute to the HTLV-1–associated diseases ATL and HAM/TSP (33).
Materials and Methods
This study was conducted according to the principles expressed in the Declaration of Helsinki and was approved by the review boards of the respective institutions (National Research Ethics Service, UK; Ethics Committee of Kumamoto University Graduate School of Medicine, Japan). Written informed consent was obtained from each subject. Full details of the materials and methods used are provided in SI Materials and Methods, including cell culture, qRT-PCR, EMSA, ChIP assay, methylated DNA immunoprecipitation assay, EB assay, shRNA KD of CTCF, RNA-sequencing, CRISPR modification of the HTLV-1 genome, quantitative 3C, and identification of candidate CTCF-BSs in the HTLV-1 genome.
SI Materials and Methods
Cell Culture.
Six HTLV-1–infected cell lines were used in the present study. Four [(ED, ATL-43Tb(−), ATL-55T, and MT-1] are derived from patients with ATL, identified as clones by either a monoclonal integration site of the HTLV-1 provirus or a monoclonal recombination of the T-cell receptor gene. TBX-4B and 11.63 are CD4+ T-cell clones derived by limiting dilution of PBMCs from patients with nonmalignant HTLV-1 infection; each clone contains a single integrated HTLV-1 provirus (data not shown). The T-cell lines and HeLa cells were cultured in RPMI plus 10% (vol/vol) FCS and antibiotics (penicillin and streptomycin). Recombinant IL-2 was added in culture medium for TBX-4B and 11.63 (200 IU/mL; Peprotech) T-cell clones or CRISPR-modified ED cells (100 IU/mL; Peprotech). Samples were also analyzed from clinical cases, including one patient with HAM/TSP, 11 patients with ATL, and eight asymptomatic HTLV-1 carriers; each subject provided written informed consent. PBMCs were isolated from peripheral blood of the patients using Ficoll-Hypaque (GE Healthcare). This study was conducted according to the principles expressed in the Declaration of Helsinki and approved by the review boards of the respective institutions. The cell line U1, chronically infected with HIV-1 (34), was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH (U1/HIV-1).
qRT-PCR.
RNA was extracted with the RNeasy kit (Qiagen), and cDNA was generated using ReverTra Ace reagent (TOYOBO), which contains a mixture of random and oligo dT primers. The expression level of CTCF, TCHP, GIT2, C12orf76, tax, env, p30, p13, and HBZ was evaluated by quantitative PCR (qPCR) (StepOne Plus; ABI) relative to 18S rRNA as an internal control. The sequences of the primers are detailed in Tables S1 and S2. The level of expression was calculated by the ∆∆Ct cycle threshold method. For absolute quantification, we generated TA-cloning vectors (pGEM-T Easy; Promega) containing sequences from Tax, HBZ, or 18S rRNA RT-PCR, respectively.
Table S1.
Primers used in qRT-PCR, EMSA, ChIP, and MeDIP
| Experiment | Target | Sense | Antisense |
| qRT-PCR | 18S rRNA | 5′-GTAACCCGTTGAACCCCATT-3′ | 5′-CCATCCAATCGGTAGTAGCG-3′ |
| CTCF | 5′-CTTTGCAGCCACGGAGAG-3′ | 5′-TCTCCTTTCCTTTAATAAAAGTTTCG-3′ | |
| Tax | 5′-CCGGCGCTGCTCTCATCCCGGT-3′ | 5′-GGCCGAACATAGTCCCCCAGAG-3′ | |
| sHBZ | 5′-GGACGCAGTTCAGGAGGCAC-3′ | 5′-CCTCCAAGGATAATAGCCCG-3′ | |
| TCHP | 5′-GGGGACTCTTCGGAAACTCC-3′ | 5′-TTGGAGCTGCAGATGTCAGA-3′ | |
| GIT2 | 5′-GGGTCTTCTTGCACTCTGAC-3′ | 5′-GCACCGAAGATGTCATCAGG-3′ | |
| C12orf76 | 5′-CCGAATGGGCTTGTAGACAC-3′ | 5′-GGAAGTGGCCTCTCTGCT-3′ | |
| EMSA probe synthesis | LTR | 5′-GAGAAACAGAAGTCTGAAAAGGTCA-3′ | 5′-GACACGTCAGGGCCTAGC-3′ |
| Env | 5′-GCTAGTTCTGCCCAGTGGAT-3′ | 5′-TTGGTGGTCTTTTTCTTTGG-3′ | |
| pX | 5′-CCGATCACGATGCGTTTC-3′ | 5′-ACGGTTTGCTATCCTTAGAAGA-3′ | |
| ChIP HTLV-1 | 5′ LTR | 5′-GACAGCCCATCCTATAGCACTC-3′ | 5′-CTAGCGCTACGGGAAAAGATT-3′ |
| Gag | 5′-CAGAGGAAGATGCCCTCCTATT-3′ | 5′-GTCAACCTGGGCTTTAATTACG-3′ | |
| Pol | 5′-CAGCCCATTCGGCAAG-3′ | 5′-TGAGAGTAGTAGTAGGTCCTCATGG-3′ | |
| Env | 5′-GCTAGTTCTGCCCAGTGGAT-3′ | 5′-TTGGTGGTCTTTTTCTTTGG-3′ | |
| vCTCF | 5′-CCGATCACGATGCGTTTC-3′ | 5′-ACGGTTTGCTATCCTTAGAAGA-3′ | |
| tax | 5′-CTCCTTCCGTTCCACTCAAC-3′ | 5′-GTGGTAGGCCTTGGTTTGAA-3′ | |
| 3′ LTR | 5′-AATACACCAACATCCCCATTTC-3′ | 5′-GTTTTTCACTGGGAGGCTCTAA-3′ | |
| ChIP HIV-1 | 5′ LTR | 5′-TCAAGTAGTGTGTGCCCGTC-3′ | 5′-TCGCTTTCAGGTCCCTGTTC-3′ |
| Gag | 5′-GCAGAGAGGCAATTTTAGGAACC3′ | 5′-CCTTTTTCCTAGGGGCCCTG-3′ | |
| Pol | 5′-TCCCTTAGATGAAGACTTCAGGA-3′ | 5′-CCATCCCTGTGGAAGCACAT-3′ | |
| Pol2 | 5′-TCAGAAGACTGAGTTACAAGCAA-3′ | 5′-TGTGCTTGAATGATTCCTAATGCA-3′ | |
| vif | 5′-CCAGCAAAGCTCCTCTGGAA-3′ | 5′-GCCACACAATCATCACCTGC-3′ | |
| Env | 5′-CTAGAGCCCTGGAAGCATCC-3′ | 5′-TCCGCTTCTTCCTGCCATAG-3′ | |
| tat | 5′-TGTACACATGGAATTAGGCCAGT-3′ | 5′-TGGTTTTAGCATTGTCCGTGAAA-3′ | |
| nef | 5′-CAACTCACAGTCTGGGGCAT-3′ | 5′-TAGCATTCCAAGGCACAGCA-3′ | |
| 3′ LTR | 5′-GGGGACTGGAAGGGCTAATT-3′ | 5′-GGCCCTGGTGTGTAGTTCTG-3′ | |
| TC-1 | 5′-TCTCCAGCACTTCTTGCTCA-3′ | 5′-TGGGATGGCTAACCTGTTGT-3′ | |
| MeDIP HTLV-1 | 5′ LTR | 5′-GACAGCCCATCCTATAGCACTC-3′ | 5′-CTAGCGCTACGGGAAAAGATT-3′ |
| Gag | 5′-CCTATGTCAAGACCCAACTCACT-3′ | 5′-GGAGGTCTAATAGGAGGGCATCT-3′ | |
| Pol | 5′-CCCCTATGTCAAGACCCAAC-3′ | 5′-GCGGGGAGGTCTAATAGGAG-3′ | |
| Env | 5′-GCTAGTTCTGCCCAGTGGAT-3′ | 5′-TTGGTGGTCTTTTTCTTTGG-3′ | |
| vCTCF | 5′-CCGATCACGATGCGTTTC-3′ | 5′-ACGGTTTGCTATCCTTAGAAGA-3′ | |
| tax | 5′-CTCCTTCCGTTCCACTCAAC-3′ | 5′-GTGGTAGGCCTTGGTTTGAA-3′ | |
| 3′ LTR | 5′-AATACACCAACATCCCCATTTC-3′ | 5′-GTTTTTCACTGGGAGGCTCTAA-3′ |
EMSA.
CTCF EMSA was performed as described previously (25) with minor modifications. CTCF was synthesized in vitro using a coupled transcription/translation reaction, with the TnT T7 Quick Coupled Transcription/Translation System (Promega), according to the manufacturer’s instructions. The sequences of the probes were as follows: H19 DMR (5′-TGGCACGGAATTGGTTGTAGTTGTGGAATCGGAAGTGGCCGCGCGGCGGCAGTGCAGGCTCACACATCACAGCCCGAG CCCGCCCCAACT-3′), LTR (5′-GAGAAACAGAAGTCTGAAAAGTCAGGGCCCAGACTAAGGCTCTGACGTCTCCCCCCGGAGGGACAGCTCAGCACCGGCTCGGGCTAGGCCCTGACGTGTC-3′), Env (5′-GCTAGTTCTGCCCAGTGGATCCCGTGGAGACTCCTCAAGCGAGCT GCATGCCCAAGACCCGTCGGAGGCCCCGCCGATCCCAAAGAAAAAGACCACCAA-3′), and pX 5′-CCGATCACGATGCGTTTCCCCGCGAGGTGGCGCTTTCTCCCCTGGAGGGCCCCGTCGCAGCCGGCCGCGGCTTTCCTCTTCTAAGGATAGCAAACCGT-3′). Probes corresponding to the different HTLV-1 proviral sequences were prepared by amplifying the respective segments with the primers detailed in Table S2. For supershift assays, 2 μL of anti-CTCF Ab (612149; BD Biosciences) was included in the reaction mixture. For competition assays, an excess of nonlabeled probe containing specific mutations, as shown in Fig. S6B, was added to the reaction mixture.
ChIP Assay.
Cells were cross-linked with 1% formaldehyde at room temperature for 5 min. Nuclear cell lysates were sonicated with a Bioruptor USD-200 (Diagenode). The following Abs were used for the immunoprecipitation step: anti-CTCF (07-729; Millipore), anti-H2A.Z (ab4174; Abcam), anti-H3K4me3 (07-473; Millipore), anti-H3K9Ac (17-658; Millipore), anti-H3K36me3 (ab9050; Abcam), and rabbit polyclonal control IgG (ab46540; Abcam). DNA enrichment in the ChIP samples was determined by qPCR using Thunderbird SYBR mix (TOYOBO) and the StepOnePlus Real-Time PCR System (Applied Biosystems). The sequences of the primers targeting the different regions within the HTLV-1 and HIV proviruses are detailed in Tables S1 and S2. As a positive control for CTCF binding, we used TC-1 primers to amplify a well-characterized CTCF-binding region (35) in the human genome. Data are expressed as the percentage of input DNA (Tables S1 and S2). One representative result from three independent experiments is shown depending on the availability of the samples (Tables S1 and S2).
For ChIP-seq analysis, ChIP-DNAs of ED cells were prepared as described above. A total of between 2 ng and 1,000 ng of ChIP or input DNAs was used for library generation with a NEBNext Ultra DNA Library Prep Kit for Illumina and Multiplex Oligos for Illumina (New England Biolabs) according to the manufacturer’s instructions. The libraries were quantified by qPCR and diluted to the optimal concentration for cluster generation. Seventy-five base-pair single-end reads plus a 6-bp index read were acquired on the Illumina NextSeq 500. Each DNA sequence was mapped using the Burrows–Wheeler Alignment (BWA)-MEM algorithm (36) to the human reference genome hg19, modified by insertion of the HTLV-1 sequence (GenBank accession no. AB513134.1) into the integration site of ED cells. Peak detection was performed using the model-based analysis of the ChIP-seq algorithm (37) in Strand NGS software (Strand Life Sciences). CTCF binding or histone modification at each region was identified by the significant enrichment of each signal over input-DNA peaks at a P cutoff value of 10−5. Visualization of ChIP-seq data was done using Strand NGS software. We performed ChIP-seq analysis of CRISPR-modified ED cells according to the protocol described above, with minor modifications. To increase the sequencing efficiency of HTLV-1, we enriched HTLV-1 sequences with 120-bp biotinylated DNA probes covering the whole HTLV-1 sequence according to the manufacturer’s instructions (Integrated DNA Technology).
Methylated DNA Immunoprecipitation Assay.
The level of DNA methylation within the HTLV-1 provirus was quantified by methylated DNA immunoprecipitation (MeDIP) assay. The MeDIP assay was performed with MethylCollector Ultra (ActiveMotif) according to the manufacturer’s protocol. The HTLV-1 molecular clone pACH (38) was methylated in vitro by CpG methyltransferase (M.SssI; New England Biolabs). The MeDIP value of the methylated pACH was taken as 100% methylation. Relative DNA methylation was analyzed by qPCR, using the primers shown in Tables S1 and S2 in each respective region of the HTLV-1 provirus.
EB Assay with Luciferase Reporter Plasmids.
The vectors pIHL, pIHLE, pIHLEI, and pIHLIE were constructed as previously described (27). Reporter vectors were constructed, containing 1,000 bp (pX), 300 bp (pX300), and 100 bp (pX100) segments of the HTLV-1 pX region, respectively, using the primers detailed in Table S2 to amplify the specific fragments. Sequences containing the same mutations described for the competitor probes in the EMSA were also prepared using the pX100 reporter plasmid. A deletion mutant with a 20-bp deletion in the consensus sequence for CTCF binding was also generated using the pX100 reporter plasmid. The reporter vectors (0.025 pmol) were transfected into HeLa cells (5 × 104 cells) in a 24-well plate, using HilyMax reagent (Dojindo). In some experiments, the plasmids were linearized with the restriction enzyme MluI before transfection. Luciferase activity was analyzed 24 h later, using the Dual Luciferase Reporter Assay System (Promega). Graphs show the mean ± SD of triplicates of one of at least three independent experiments.
shRNA KD of CTCF.
An shRNA expression vector targeting CTCF (pSilencer CTCF-1) was constructed by using pSilencer-3.1-H1-puro vector (pSilencer-NC; Ambion) as previously described (27). The oligonucleotides used for pCTCF1 were as follows: 5′-GATCCGTGTCTAAAGAGGGCCTTGTTCAAGAGACAAGGCCCTCTTTAGACACTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGTGTCTAAAGAGGGCCTTGTCTCTTGAACAAGGCCCTCTTTAGACACG-3′. To transfect MT-1 cells, we used NEPA21 (Nepagene) according to the manufacturer’s instructions. One million cells were transfected with 5–10 μg of plasmid. After 24 h, transfected cells were selected with puromycin (2 μg/mL), and they were harvested 24 h and 48 h later; dead cells were removed by Ficoll separation before RNA extraction. We also confirmed the efficiency of CTCF KD by Western blotting as described previously, with minor modifications. Dead cells were removed by Ficoll separation before protein extraction.
RNA-Sequencing.
RNAs were extracted from MT-1 cells as described above. We used the SureSelect Strand-Specific RNA Library Kit (Agilent Technologies) to prepare libraries for high-throughput sequencing. Paired-end sequencing was performed on an Illumina HiSeq 1500 with a TrueSeq Rapid PE cluster kit (Illumina) and an SBS kit (200 cycles; Illumina). We obtained 49,570,644 and 54,813,994 reads from negative control and CTCF KD MT-1 cells, respectively. After removal of reads with low-sequencing quality, reads were mapped to the human reference genome (hg19) or HTLV-1 genome (GenBank accession no. AB513134.1) using TopHat, a spliced read mapper for RNA-sequencing (RNA-seq). Mapping rate visualization of RNA-seq was done using Strand NGS software.
Identification of CTCF Consensus Sequence in HTLV-1 Genome.
We first identified possible CTCF-BSs by using EMBOSS 6.3.1:fuzznuc (mobyle.pasteur.fr/cgi-bin/portal.py?#forms::fuzznuc) within the HTLV-1 genome (GenBank accession no. AB513134.1) as those sites with the same nucleotide as the consensus motif identified by Kim et al. (9) at positions 6, 11, 14, and 16. We then calculated a similarity score for each possible site based on the observed frequencies of 14 core nucleotides of the consensus CTCF-binding sequence (position 5 to position 18 inclusive) (details are provided in the legend for Fig. S4).
Establishment of ED Clones with a Mutated CTCF-BS.
To generate ED cells containing a mutated CTCF-BS, lentiCas9_gRNA retroviral vectors were prepared. The guide RNA (gRNA) oligonucleotides (5′-CACCGGAGAAAGCGCCACCTCGCG-3′ and 5′-AAACGCGCGAGTGGCGCTTTCTCC-3′) were annealed and cloned into the BsmBI site of lentiCRISPRv2 (no. 52961; Addgene). The resulting plasmid was transfected into 293T cells, together with pCMV-VSV-G (no. 8454; Addgene) and psPAX2 plasmids (no. 12260; Addgene), to obtain the lentiCas9_gRNA vectors in the culture supernatant. Five million ED cells were transfected by electroporation (Amaxa Nucleofector; Lonza) with 30 pmol of single-stranded template DNA (5′-CTTAGAAGAGGAAAGCCGCGGCCGGCTGCGACGGGGCCCTCCAGGGGAGAAAGCCCCGCCAAGAGGTGAAACGCATCGTGATCGGCAGCGACGGGCTGAGGAGAAGAGGAAGCGAAAAAA-3′) (Integrated DNA Technologies). Mutations within the CTCF-BS are indicated in bold. Three hours later, transfected cells were transduced with the lentiviral vector lentiCas9_gRNA by spinoculation (1,000 × g, 60 min) in the presence of 8 μg/mL polybrene (Sigma). Two days later, selection with 2 μg/mL puromycin was started. After 2 wk of selection, single clones were obtained by limiting dilution. The clones were maintained in RPMI 10% (vol/vol) FBS supplemented with antibiotics (penicillin and streptomycin). For long-term cultivation, recombinant IL-2 (100 IU/mL; Peprotech) was added to the culture medium.
Quantitative 3C Analysis.
Cells were cross-linked with 1% formaldehyde at room temperature for 10 min and then lysed with lysis buffer [10 mM Tris⋅HCl (pH 8.0), 10 mM NaCl, 0.2% Nonidet P-40, and Protease Inhibitor Mixture (Roche)]. Nuclei (5 × 106 cells) were digested with 1,000 units of NlaIII (New England Biolabs) overnight at 37 °C, and DNA was diluted (0.5 ng/μL) and ligated with 1,600 units of T4 DNA ligase (New England Biolabs) at 16 °C overnight. After reversing the cross-links at 65 °C overnight in the presence of proteinase K (100 μg/mL) followed by treatment with RNase A (40 μg/mL) at 37 °C for 1 h, the circular DNA was purified by phenol/chloroform extraction and ethanol precipitation and resuspended in 400 μL of 10 mM Tris⋅HCl (pH 7.5). The DNA (3C library) was repurified by an additional ethanol precipitation. Six 3C libraries were made from three independent cultures of ED cells with WT HTLV-1 and HTLV-3 with the CTCF-BS deletion mutant of HTLV-1. Real-time 3C qPCR was then performed in quadruplicate or triplicate on each library, using a QuantStudio 7 Flex Real-Time PCR System utilizing TaqMan Gene Expression Master Mix (Applied Biosystems). To assess the interaction between the viral CTCF-BS and cCTCF-BS, we primers used were vCTCF_1 (5′-CAAGGATAATAGCCCGTCCA-3′) and cCTCF_1 (5′-TGTCCAAGGCCAGACAAA-3′) and the probe A (5′-TCCGGGCATGGTGGTACTGCTA-3′). As controls, we assessed the interactions between the vCTCF-BS and two regions adjacent to the view point. For the interaction shown as b in Fig. 6A, the primers used were vCTCF 3C_2 (5′-CTCCTCCTCCTTGTCCTTTAAC-3′) and Control_B (5′-GCTCAGCTCTACAGTTCCTTATC-3′), and the probe used was probe B (5′-TCCGGGCATGGCCTGGAGGAA-3′). For the interaction shown as c in Fig. 6A, the primers used were vCTCF 3C_3 (5′-TAATAGCCCGTCCACCAATTC-3′) and Control_C (5′-TCTGTTCTGGGCAGCATAC-3′), and the probe used was probe C (5′-CCTCCGGGCATGAGGTGGACAAAGATA-3′). To normalize the qPCR, we amplified a 101-bp sequence from the provirus using the primers IC_1 (5′-CGCGGCTTTCCTCTTCTAA-3′) and IC_2 (5′-ATTGGTGGACGGGCTATTATC-3′) and the probe (5′-AAGCACAGCTTCCTCCTCCTCCTT-3′). Data analysis was done with LinRegPCR software (version 2014.5) The Welch two-sample t test was used to test whether the mean interaction frequency was significantly different between the WT and mutant samples (three biological replicates for each).
Acknowledgments
We thank Graham Taylor for clinical samples and Michiyo Tokunaga for technical support. We thank Nicholas Proudfoot, Douglas Higgs, Jim Hughes, Matthias Merkenschlager, and members of the C.R.M.B. laboratory and Y.S. laboratory for discussion. This work was supported by a senior investigator award from the Wellcome Trust UK (to C.R.M.B.), Leuka (to C.R.M.B.), Japan Society for the Promotion of Sciences Grant-in-Aid for Scientific Research scheme (KAKENHI) (Grant 26461428 to Y.S.), Takeda Science Foundation (to Y.S.), the Japan Science of Technology Agency (to Y.S.), Core Research for Evolutionary Science and Technology (to M.N.), and The Naito Foundation (to M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: ChIP-sequencing data reported in this paper have been deposited in the DNA Data Bank of Japan (accession no. DRA004162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423199113/-/DCSupplemental.
References
- 1.Igakura T, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299(5613):1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 2.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7(4):270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 3.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006;103(3):720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bangham CR, et al. The immune control of HTLV-1 infection: Selection forces and dynamics. Front Biosci (Landmark Ed) 2009;14:2889–2903. doi: 10.2741/3420. [DOI] [PubMed] [Google Scholar]
- 5.Koiwa T, et al. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J Virol. 2002;76(18):9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi Y, et al. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology. 2005;2:64. doi: 10.1186/1742-4690-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold J, et al. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107(10):3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong CT, Corces VG. CTCF: An architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128(6):1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347(6225):1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouwman BA, de Laat W. Getting the genome in shape: The formation of loops, domains and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 15.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 16.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163(7):1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148(1-2):335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tempera I, Lieberman PM. Epigenetic regulation of EBV persistence and oncogenesis. Semin Cancer Biol. 2014;26:22–29. doi: 10.1016/j.semcancer.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H, Cho H, Sung GH, Lieberman PM. CTCF regulates Kaposi’s sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming. J Virol. 2013;87(3):1789–1799. doi: 10.1128/JVI.02283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34(3):271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Reports. 2013;3(5):1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishiro T, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28(9):1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang R, et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23(5):733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13(2):291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 29.Cook LB, et al. The role of HTLV-1 clonality, proviral structure, and genomic integration site in adult T-cell leukemia/lymphoma. Blood. 2014;123(25):3925–3931. doi: 10.1182/blood-2014-02-553602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babaei S, Akhtar W, de Jong J, Reinders M, de Ridder J. 3D hotspots of recurrent retroviral insertions reveal long-range interactions with cancer genes. Nat Commun. 2015;6:6381. doi: 10.1038/ncomms7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laydon DJ, et al. Quantification of HTLV-1 clonality and TCR diversity. PLOS Comput Biol. 2014;10(6):e1003646. doi: 10.1371/journal.pcbi.1003646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Markus J, Bies J, Paul T, Wolff L. Three murine leukemia virus integration regions within 100 kilobases upstream of c-myb are proximal to the 5′ regulatory region of the gene through DNA looping. J Virol. 2012;86(19):10524–10532. doi: 10.1128/JVI.01077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238(4828):800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe T, et al. Higher-order chromatin regulation and differential gene expression in the human tumor necrosis factor/lymphotoxin locus in hepatocellular carcinoma cells. Mol Cell Biol. 2012;32(8):1529–1541. doi: 10.1128/MCB.06478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimata JT, Wong FH, Wang JJ, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204(2):656–664. doi: 10.1006/viro.1994.1581. [DOI] [PubMed] [Google Scholar]