Significance
Photoreceptor proteins dynamically control many critical physiological processes in response to light across the whole phylogenetic order, including the regulation of circadian rhythms and photosynthesis. We created a comprehensive catalog of the protein architectures and biochemical functions of a ubiquitous class of natural photoreceptors, the light–oxygen–voltage sensitive (LOV) class of flavoproteins, including >4,000 new candidate LOVs, which nearly triples the sequence diversity known to date. Establishing the functional and structural diversity of LOVs will (i) shed light on how organisms adapt to environmental changes, (ii) elucidate the structure–function principles by which common photosensory inputs are transmitted into a multitude of cell signaling events, and (iii) beget novel “optogenetic” tools for light-driven physiological perturbation of cells expressing natural or engineered photoreceptors.
Keywords: photoreceptors, LOV, flavoproteins, optogenetics
Abstract
Light–oxygen–voltage sensitive (LOV) flavoproteins are ubiquitous photoreceptors that mediate responses to environmental cues. Photosensory inputs are transduced into signaling outputs via structural rearrangements in sensor domains that consequently modulate the activity of an effector domain or multidomain clusters. Establishing the diversity in effector function and sensor–effector topology will inform what signaling mechanisms govern light-responsive behaviors across multiple kingdoms of life and how these signals are transduced. Here, we report the bioinformatics identification of over 6,700 candidate LOV domains (including over 4,000 previously unidentified sequences from plants and protists), and insights from their annotations for ontological function and structural arrangements. Motif analysis identified the sensors from ∼42 million ORFs, with strong statistical separation from other flavoproteins and non-LOV members of the structurally related Per-aryl hydrocarbon receptor nuclear translocator (ARNT)-Sim family. Conserved-domain analysis determined putative light-regulated function and multidomain topologies. We found that for certain effectors, sensor–effector linker length is discretized based on both phylogeny and the preservation of α-helical heptad repeats within an extended coiled-coil linker structure. This finding suggests that preserving sensor–effector orientation is a key determinant of linker length, in addition to ancestry, in LOV signaling structure–function. We found a surprisingly high prevalence of effectors with functions previously thought to be rare among LOV proteins, such as regulators of G protein signaling, and discovered several previously unidentified effectors, such as lipases. This work highlights the value of applying genomic and transcriptomic technologies to diverse organisms to capture the structural and functional variation in photosensory proteins that are vastly important in adaptation, photobiology, and optogenetics.
The light–oxygen–voltage sensitive (LOV) domain subset of the Per-aryl hydrocarbon receptor nuclear translocator (ARNT)-Sim (PAS) superfamily is a ubiquitous photoreceptor class that enables organisms across multiple kingdoms to sense blue light (1–5). LOV photoreceptors consist of modular sensor and effector domains whose interactions are commonly mediated by an α-helical linker between the two (6). Blue light absorption initiates the reversible formation of a flavin-cysteinyl adduct in the LOV sensor hydrophobic core, triggering a conformational change in the overall protein tertiary structure that ultimately transduces the photosensory input into biochemical signaling outputs (4–7). These signaling events—often mediated by clusters of conserved protein domains that are indirectly light-regulated downstream of the primary effector—exert diverse physiological effects that underlie circadian rhythms (8), virulence (9), phototropism (10), and stress responses (11), across species in varied ecological settings. LOV proteins are also invaluable optogenetic tools for light-gated physiological perturbation of genetically targeted cells, either as natural proteins or engineered variants (12–16). Their modular design is advantageous for engineering chimeras between LOV sensors with effectors of choice, enabling strategies for dynamic gain-of-function of arbitrary proteins in cells. Thus, elucidating the diversity in the repertoire of effector functions, as well as the diversity in multidomain structural arrangements of LOV sensors and effectors, will respectively deepen collective understanding of what cellular adaptation processes are dynamically regulated by light and how these highly varied signals are transduced by the modular protein architecture in response to a common blue-light stimulus. More broadly, because PAS proteins share conserved signal transmission mechanisms in response to various sensory inputs (17) that include light (e.g., LOV, phytochrome), ligands (e.g., Cache domains, PDC domains) (18), and oxygen (e.g., HIF proteins) (19), new insights into LOV structure–function will enhance the overall understanding of the PAS superfamily of sensory proteins.
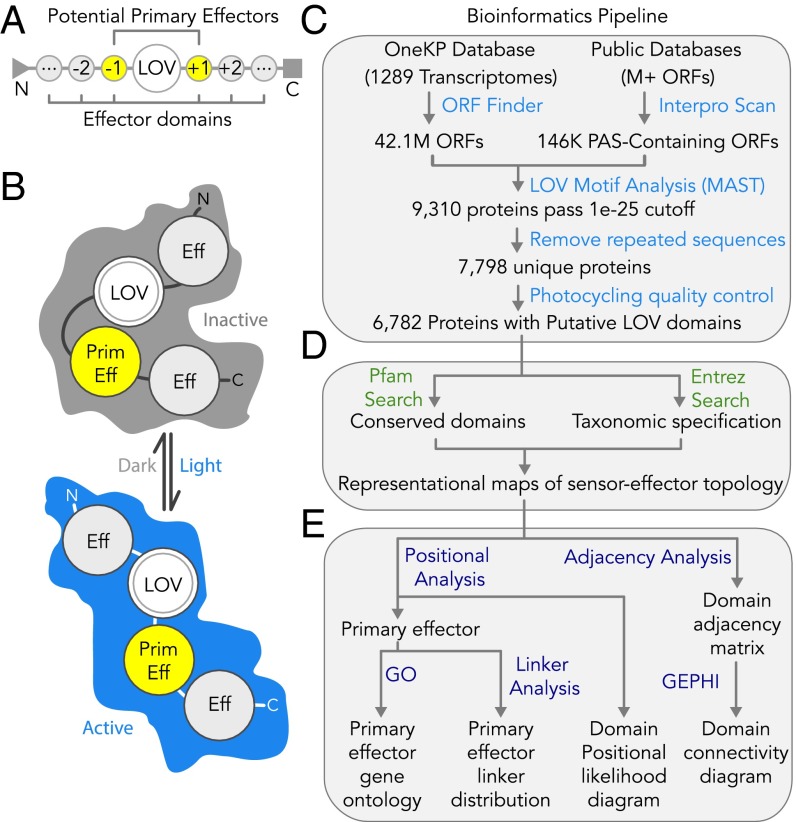
The modular sensor–effector topology of LOV proteins facilitates automated bioinformatics strategies in discovery and annotation. Because the conserved domains are encoded in discrete stretches of DNA, their identities and linear arrangements can be parsed within a single ORF. Here, we report the development of a fully automated bioinformatics pipeline written in Python (Fig. 1) that (i) identifies LOV sensors through motif analysis (20), (ii) identifies conserved domains in the up/downstream neighboring regions of the ORF via searches against the Pfam and Interpro databases (21–23), (iii) annotates predicted effector functions in computer-readable maps of LOV multidomain structures, and (iv) maps the functional and topological distributions across archaea, bacteria, fungi, protists (which hereon include algae), and land plants. Building on insights from previous BLAST-based analyses of published sequences (2–5, 24–26), we implemented an approach that would enhance the detection of LOV sensors from recently sequenced organisms (including ones reported here) that may not resemble well-studied LOV proteins.
Fig. 1.
Automated bioinformatics pipeline to identify LOV proteins and analyze their functional and structural diversity. (A) Multidomain topology of an LOV photosensor (or tandem sensors) fused to neighboring N- and/or C-terminal effectors (negative and positive positions, respectively). (B) Transduction of photosensory inputs into signaling outputs through light-gated structural rearrangements between sensor and neighboring effector(s). (C–E) Automated cataloging of LOV proteins via Python scripts. (C) Motif-based sensor identification from OneKP and PAS InterPro databases, followed by quality control measures and a check for the conserved cysteine required for photocycling and signal transmission. (D) Annotation of up/downstream conserved domains within the protein cluster by Pfam and InterPro database queries and taxonomic specification of organism of protein origin by Entrez query. (E) Analysis of functional and structural diversity from the resultant computer readable maps, for nearest effector GO, sensor–effector linker length, and multidomain positional likelihood and connectivity.
We identified 6,782 LOV proteins from ∼42 million ORFs (>5,700 organisms spanning two databases, Interpro and OneKP, the latter a recently generated collection of nearly 1,300 land plant and algal transcriptomes from >1,000 unique organisms) (27). The contributions here nearly triple the number of LOV sequences known and were chiefly derived from OneKP (4,163 from OneKP newly identified here vs. 2,619 from Interpro, consistent with a recent report) (2). We find that when effectors are grouped by function irrespective of relative position to each other or the sensor, LOV proteins are described by 119 “functional clusters” of associated domains that describe the extent to which LOV domain-based signaling is adaptable to complex physiological outputs. Maps of linker sequence length between the sensor and most proximal effector reveal discretized banding, possibly supporting the notion that linker structure is often modular (28). Additionally, we find an increased prevalence of effector functions [as determined by gene ontology (GO)] previously thought to be rare among LOV proteins, particularly those potentially implicated in G protein signaling, small-molecule biosynthesis, and catabolism. These rare functions were found in recently sequenced dikarya, heterokonts, and species diverging early in the evolutionary lineage of green algae, highlighting the importance of sequencing diverse organisms to capture the functional space of photosensory proteins. This comprehensive discovery, analysis, and cataloging of LOV domain diversity will inform how light regulates organismal behavior, beget new optogenetic tools or protein-based photocatalysts, and create a foundation for uncovering new insights into LOV photoreceptor structure–function and rational engineering principles through comparative structural genomics.
Results
Automated LOV Identification by de Novo Motif Analysis.
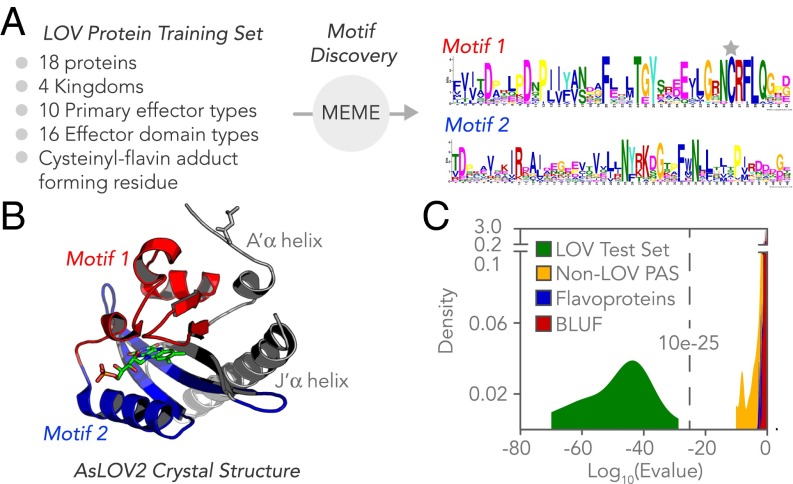
The pipeline (Fig. 1) identifies LOV domains by calculating a match score for candidate sequences to custom-developed LOV flavin-binding motifs, represented by position-weighted matrices that ascribe weights to various positions within a sequence pattern according to how strongly those positions are conserved. Because isolating motifs that relate to flavin binding and photocycling de-emphasizes the highly variable sequence contributions of the effectors also found within the ORF, a motif-based search created a clear stringency cutoff for defining the obligate LOV sensor domain. Conserved motifs were identified using the Multiple Em for Motif Elicitation (MEME) tool (20), based on 18 well-characterized LOV proteins that were selected to reflect a breadth in structural and functional diversity among known sensors (Fig. 2A and Dataset S1). Two highly conserved motifs emerged, of 43 and 48 amino acids in length, which mapped to the flavin-binding pocket when projected onto the 3D structure of AsLOV2 from Avena sativa (Fig. 2B) (29). Several submotifs had particularly high information contents, including a GX(N/D)C(R/H)(F/I)L(Q/A) submotif containing the key cysteine that forms the cysteinyl-flavin adduct during the LOV photocycle. Additionally, mutations to conserved residues in FXXXT(G/E)Y and N(Y/F)XXX(G/D)XX(F/L)XN submotifs are also known to impair blue-light sensation (30). It should be noted that although a covalent adduct can theoretically form between a flavin and noncysteine residue, the key cysteine is considered obligate here to maintain consistency with the best characterized form of the LOV photocycle.
Fig. 2.
Motif-based identification of LOV proteins and discrimination from related non-LOV proteins. (A) Sequence logos for motifs 1 and 2, identified by the MEME tool for a training set of 18 LOV proteins validated to photocycle, with the cysteine that forms the cysteinyl-flavin adduct during the photocycle marked with a gray star and (B) mapped onto the crystal structure of LOV2 from A. sativa (Protein Data Bank ID code 2V0U). The motifs encompass the flavin-binding pocket but not the linker region or the A’-alpha and J-alpha helices (shown in gray). (C) Histogram showing the likelihood (log10 of e-value) that motifs 1 and 2 are present in a given domain shows clear discrimination between known LOV sensors and closely related protein classes of non-LOV PAS proteins, BLUF domains, and other flavoproteins. When searching for the motifs in known test set LOV domains that were also in the training set, we applied a leave-one-out cross-validation scheme, in which the two sensor motifs were regenerated for the training LOV dataset minus one LOV photoreceptor, and the sensor motifs were then searched for with the MAST tool on the remaining LOV photoreceptor. The MEME training dataset proteins were selected to span a range of physiological functions, organisms of origin, and ecological niches and have been previously validated to photocycle. Training and test sets are provided in Dataset S1.
Importantly, the analysis readily distinguishes a LOV domain from its most closely related protein domains, which include non-LOV PAS domains (including PYP, “photoactive yellow protein”) and other flavoproteins, including BLUF domain photoreceptors (“Blue-Light Using FAD”) (18, 26, 31–33) (Fig. 2C and Dataset S1). The Motif Analysis & Search Tool (MAST) (20) was used to estimate the probability that both motifs were jointly present in a candidate protein, and a very clear distinction in e-values of the known LOV domains that comprised a “test set” (Dataset S1) versus related non-LOV proteins was found (see Methods). Given the large statistical separation between closely related proteins, we applied the automated query to two databases that would likely encompass the totality of potential LOV candidates: PAS-containing proteins cataloged in Interpro on structural grounds and OneKP on photobiology grounds. In total, 6,782 LOV-encoding sequences were discovered in both databases from analyzing ∼42 million ORFS from >5,700 organisms from archaea, bacteria, fungi, protists, and land plants.
Diversity of Nearest Neighboring Effectors.
The upstream and downstream sensor-neighboring regions were aligned to hidden Markov models of >14,000 conserved domain types from the Pfam database to identify (i) the primary or nearest neighboring effectors presumed to be directly modulated by the sensor, (ii) all conserved domains present in the protein-encoding region that are likely involved in the overall photosensory signaling pathway (abbreviations in Dataset S2), and (iii) the linker sequence length between sensor and primary effector. When no predicted Pfam effectors were found within 125 amino acids of the LOV sensor (roughly the size of a conserved domain but still within known sensor–effector linker length range), the candidate was triaged to an additional Interpro conserved domain search. When the nearest neighbor was another LOV sensor, similar to the tandem repeat architecture observed in LOV proteins from plants and algae (34), and also common to other mediators of protein–protein interactions (35, 36), the repeat was first collapsed into a single pseudodomain called tandem LOV, and then the linker lengths and effector positions were recalculated from the termini of the tandem. Tandem LOVs were found only in land plants and protists (1,756 total, 37% of land plant LOVs, 31% of protist LOVs) and never annotated as primary effectors. It should be noted that the interaction partner and/or most proximal effector to the LOV sensor in the tertiary protein structure might differ from the nearest neighbor in the linear polypeptide sequence. However, primary effectors and LOV signaling roles are routinely inferred from the domains with the shortest sequence linker polypeptides to the sensor, and thus, the definition applied here is reasonable for a dataset of nearly 42 million ORFs. From here on, the linker length refers to the number of residues, unless specified as physical distance.
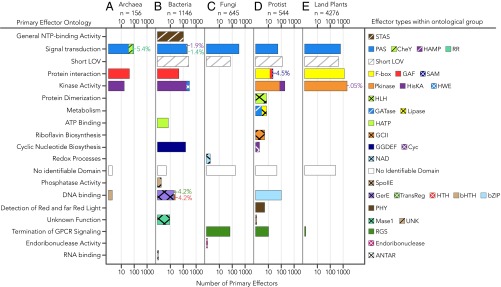
We identified 33 different primary effector types that are grouped according to their GO (Fig. 3). Five primary effector categories accounted for 83.1% of the LOV proteins in the sample set: protein kinase (serine/threonine kinase), F-box, Short LOVs (with terminal peptide extensions, similar to the fungal LOV domain VIVID) (7, 37–39), histidine kinase (HisKA), and PAS domains that may serve to integrate multiple environmental inputs with light (40). Nearly 1/10th of the sample set (7.2%) had no conserved domain matches in Pfam or Interpro despite extensions of 125–1,000+ residues that are much longer than those of short LOVs. This architecture is observed in candidates from both InterPro and OneKP, and hence it is unlikely to be attributable to de novo sequence assembly artifacts (see SI Text 1 and Fig. S1 for quality control assessments and direct comparisons between genome- vs. transcriptome-derived reads of matching genes), although one must always keep open to the possibility of truncations introduced by variation at the level of raw read in CG-rich regions. It is possible that these LOV with no identifiable conserved domains mediate protein interactions analogous to short LOVs. For example, in the well-described VIVID protein, light alters both LOV homodimerization interactions and consequent interaction with the White Collar complex to form a heterodimer that competes with the activated White Collar homodimer (41–43). It is also possible that the sensor-flanking regions are enzymatic or binding domains that have yet to be classified as conserved domains.
Fig. 3.
Diversity in primary effector identity and ontological function. Primary effectors are separated by (A) archaea, (B) bacteria, (C) fungi, (D) protists, and (E) land plants. Effectors are defined as the nearest conserved domain to sensors with respect to primary structure. Tandem LOVs are collapsed and treated as a single sensor domain, with possible effector domains N-terminal to the first LOV domain and C-terminal to the second LOV domain in the sequence. Bar plots indicate the number of effector domains of a given GO (assigned by Pfam and Interpro) for a given kingdom on a log10 scale. Bars are colored and hatched according to the fractional number (linear scale) and type of effector domains found with a given ontology. The percent relative distribution is provided for primary effectors that are not readily distinguishable by the eye. The order of domains in each figure legend corresponds to the priority with which bars were stacked, such that leftmost domains are stacked first and rightmost domains are stacked last. The total number of LOV proteins found in each kingdom is provided as n. Full names of effector abbreviations are provided in Dataset S2. Fifteen candidate sequences of uncertain taxonomic origin (Incertae sedis) are omitted.
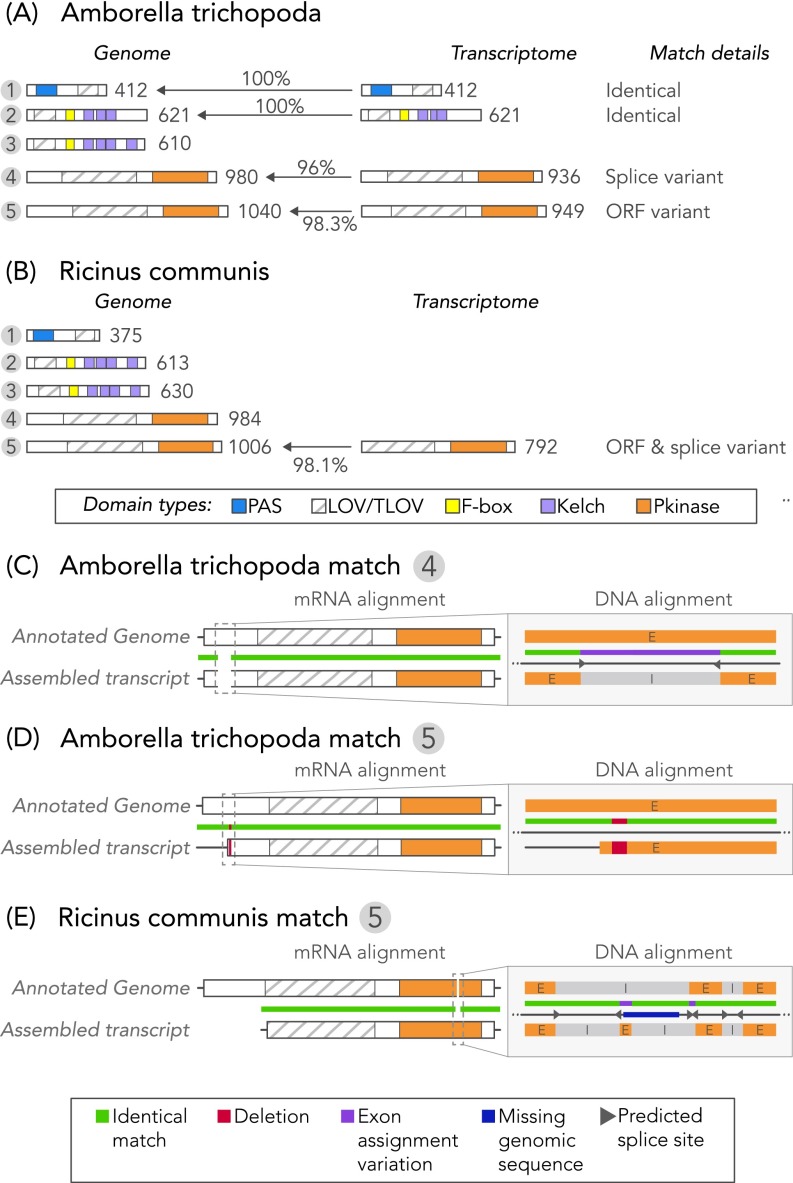
Fig. S1.
Comparisons between candidate gene sequence matches derived from the genome and transcriptome of the same organism. De novo transcriptome assembly quality does not affect reported LOV photoreceptor domain topologies. (A and B) Comparison of candidate sequence annotations for organisms represented in OneKP that also possess draft genomes of similar size to predicted genome sizes, (A) A. trichopoda and (B) R. communis, each with five unique genome-derived LOVs. Transcriptome-derived sequences had a genome-derived match based on domain combinations and order. Because only highly expressed transcripts are successfully assembled, half the genome-predicted candidates were found in the transcriptomes. Percentages represent protein sequence homology between matches. (C–E) Observed differences were attributable to assignments of splice site (A. trichopoda match #4) or attributable to raw sequence read of either the genome (R. communis match #5) or transcriptome (A. trichopoda match #5).
Several primary effector domains have not been previously described as LOV effectors to the best of our knowledge: GTP cyclohydrolase type II (five proteins from glaucophytes and chlorophytes), lipase (three proteins from chlorophytes), and glutamine amidotransferase (GATase, four proteins from chlorophytes) were all found more than once. We also found evidence that effectors previously thought to be rare may in fact be common—namely, 77 different LOV-RGS or regulators of G protein signaling primarily from fungi (dikarya) and protists (heterokonts) (3, 24, 44–46). Whereas a few LOV-RGS were previously identified by conserved domain analysis, the newfound abundance of LOV-RGS proteins was similar to more commonly studied LOV proteins that contain BZIP, STAS (sulfate transporter and anti-sigma factor), HTH (helix–turn–helix), and HLH (helix–loop–helix) domains. LOV proteins with recently described functions were derived from recent sequencing collaborations (OneKP and the Fungal Genome Initiative) that greatly expanded the breadth of organismal representation, begging the question of whether evolutionary diversity, sheer number of LOV photoreceptor gene sequences available, or number of organisms queried is the primary determinant of observed LOV diversity. As detailed further in the following section, evolutionary diversity within a kingdom, and neither sample size nor number of organisms queried, determines the observed diversity and complexity of LOV architectures within the kingdom.
Position and Connectivity of Multieffector Clusters.
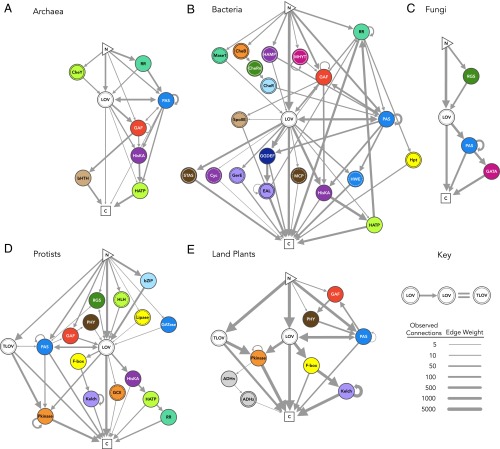
Fig. 4 shows the distribution of conserved domain positions relative to the sensor. Although both N- and C-terminal effectors are widely observed (negative and positive position number vs. sensor, respectively), different effector types preferentially locate to either N- or C-terminal to the sensor, with PAS, GAF, and RR as notable exceptions (although a preference is still largely maintained on a per kingdom basis) (Fig. 4). To illustrate which domains commonly associate in multidomain structures, an adjacency analysis (47) was visualized in the Gephi platform for networked systems (48) (Fig. 5). Many architectural aspects are conserved (e.g., LOV/PAS, short LOV, and LOV with no identifiable conserved domains), whereas others are highly kingdom-dependent. For example, tandem LOVs and serine/threonine protein kinases were only observed in land plants and protists, possibly as a two-sensor mechanism to tune the sensitivity of the system as suggested for phototropins (49, 50).
Fig. 4.
Effector position distribution within multidomain LOV proteins. Linear maps of multidomain polypeptides are separated by (A) archaea, (B) bacteria, (C) fungi, (D) protists, and (E) land plants. The x-axis represents domain position relative to a single or tandem LOV sensor. Sensors are assigned the zero positions, and conserved effector domains are numbered in increasing value toward the termini (negative N-terminal, positive C-terminal). Bar height (log10 scale) represents the total number of domains of any type observed at a given relative position. Fraction of each stacked bar (linear scale) that is uniquely colored and hatched corresponds directly to the fraction of domains at the given position of a specific domain type. Domains that constitute <10% of the fraction of any position for any kingdom are placed in “Other.” The order of domains in the figure legend corresponds to the priority with which bars were stacked, such that LOV domains are stacked first and the Other category is stacked last. Full names of effector abbreviations are provided in Dataset S2. Fifteen candidate sequences of uncertain taxonomic origin (I. sedis) are omitted.
Fig. 5.
Network maps of conserved domain connectivity. Connectivity networks are separated by (A) archaea, (B) bacteria, (C) fungi, (D) protists, and (E) land plants. Nodes represent sensor or effector domains. Nodes are colored and hatched according to effector domain type, where a solid ring inside the node indicates a single hatch and a dashed ring inside the node represents a crosshatch (to be consistent with all other figures). Edges between nodes represent a fusion of two domains (here, limited to connections observed ≥3 times for a kingdom), where edge weight corresponds to observed frequency of the connection on log2 scale. Networks originate at the N terminus, and arrows indicate the relative position of each domain in the polypeptide that culminates at the C terminus. Arrows that begin and end at the same node denote repeated effectors, with the exception of consecutive LOV sensors, which were grouped into tandem LOVs. Note that all pathways must pass through the LOV sensor in the diagrams. Full names of effector abbreviations are provided in Dataset S2. Fifteen candidate sequences of uncertain taxonomic origin (I. sedis) are omitted.
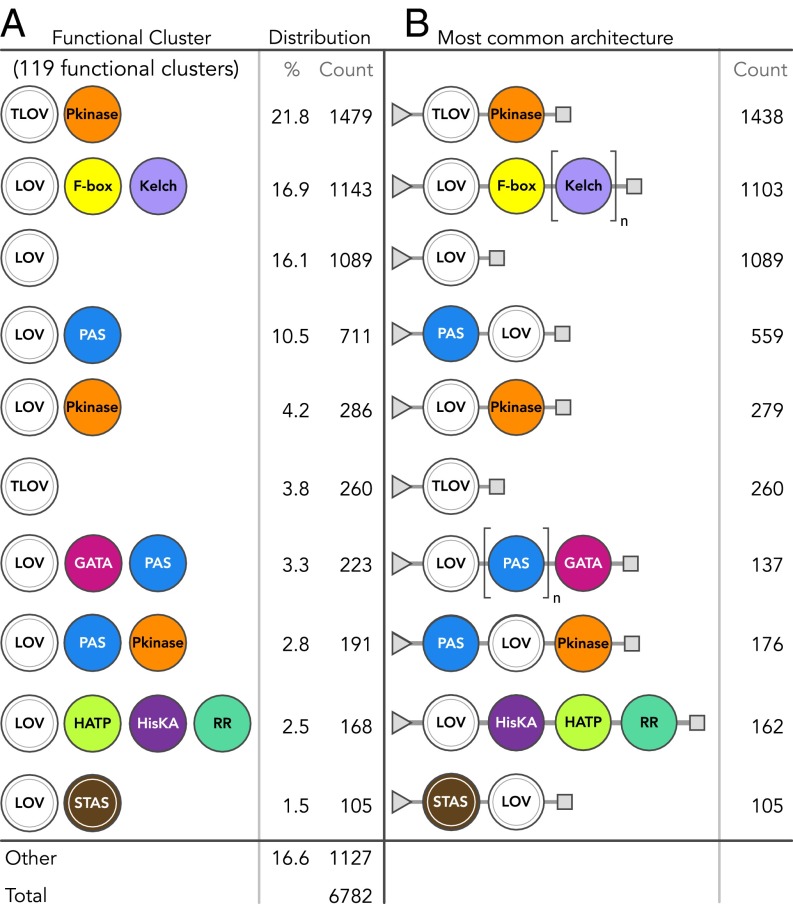
The position and connectivity information in these structural topology maps provide conserved associations and ordering between effector domains, from which multistep signaling pathways and native physiological roles may be inferred. For example, clear associations are seen between HisKA, histidine kinase-like ATPases (H-ATPases), and response regulators (RRs) across multiple kingdoms that implicate an evolutionarily conserved two-component signaling pathway (51, 52). Obligate associations can likewise be inferred. For example, LOV-associated Kelch repeats are always preceded by F-boxes even though β-propeller–forming Kelch repeats (53) do not require them. We classified these topologies into 119 functional clusters of associated domains, regardless of order or domain stoichiometry (Dataset S3), with Fig. 6 providing the 10 most prevalent clusters and their respective most common architecture. The 119 functional clusters reduce the overall protein architectural space and may facilitate physiological inferences by examining classes of domain associations instead of individual instances.
Fig. 6.
Grouping of conserved domains commonly associated in LOV proteins into functional clusters. (A) Ten most prevalent functional clusters of LOV proteins, where domains are grouped by composition, but independent of domain order and repeats. Frequency of occurrence is for each type of grouped domains or clusters, not individual domains. (B) Most common protein architecture for highly prevalent clusters (triangles, N terminus; squares, C terminus). Domains surrounded by brackets are commonly repeated, found n times total. Full names of effector abbreviations are provided in Dataset S2.
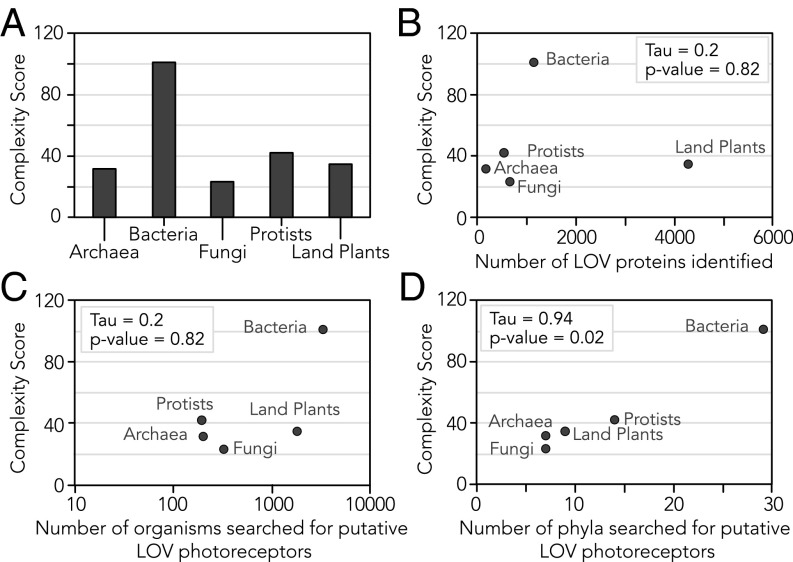
A computed complexity quotient, which quantifies domain architectural complexity as a function of both the number of domains and variety of domain types for a given set of proteins (47), shows that complexity across kingdoms varies widely, where bacteria exhibited the maximal overall architectural diversity (Fig. 7A). There is a clear trend that LOV complexity is proportional to evolutionary diversity (as estimated by the number of phyla searched for photoreceptors per kingdom) but not the sample size of LOV candidates or organisms searched for photoreceptors per kingdom (Fig. 7 B–D). Fungi interestingly lack architectural diversity with few conserved domains that are directly enzymatic (Figs. 5C and 7A) and instead rely on binding mediators such as peptide flanks (short LOVs) and zinc fingers. However, as previously discussed with VIVID, such binding domains can orchestrate multicomponent and multistep signaling pathways that are themselves complex, even if the domain architectures of fungal LOVs are “simple.”
Fig. 7.
Architectural complexity correlates with evolutionary diversity. (A) Computed complexity quotient for each kingdom quantifies domain architectural complexity as the product of the average number of effector domains per LOV photoreceptor in the kingdom and the total number of different effector types observed across the kingdom. (B–D) Complexity quotients for each kingdom plotted versus (B) the total number of putative LOV sequences identified in the kingdom, (C) the total number of organisms searched for LOV in the kingdom, and (D) the total number of phyla searched for LOV in the kingdom. Kendall’s rank correlation tau coefficients and their accompanying P values are shown on each scatterplot. A strong correlation between the number of phyla searched and the complexity of the resulting LOV photoreceptors suggests that evolutionary diversity is a greater predictor of complexity than sample size.
Discretization in Sensor–Effector Linker Length.
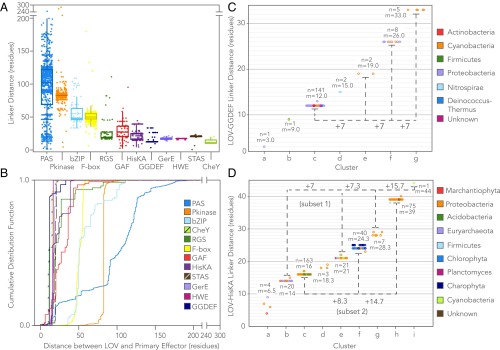
Linker sequence length was dependent on the primary effector type, with some effectors exhibiting highly discretized bands in linker length distribution (Fig. 8). Although some degree of effector-specific discretization is to be expected from common ancestry, the observed banding may also reflect key structure–function requirements for signal transmission. For example, the YF1 HisKA, a chimeric LOV engineered by substituting the cognate STAS effector from YtvA with a HisKA, exhibits cyclical light/dark effector behavior consistent with linker dependence on heptad periodicity; YF1 variants that differ in linker length by multiples of 7 retain light-inducible activity, whereas those with nonheptad additions or deletions exhibit reversed or no functionality (see figure 4 of ref. 28). The reported finding suggests that sensor–effector orientation is more critical than interdomain physical distance for natural or preformed dimers with extended coiled-coil linkers. In corollary to this insight from an engineered LOV, we conducted a structural genomics analysis to determine whether the distribution of linkers across wild-type LOV reflected a similar heptad repeat suggestive of extended coiled-coil linker regions (bands of linkers defined algorithmically by k-means clustering).
Fig. 8.
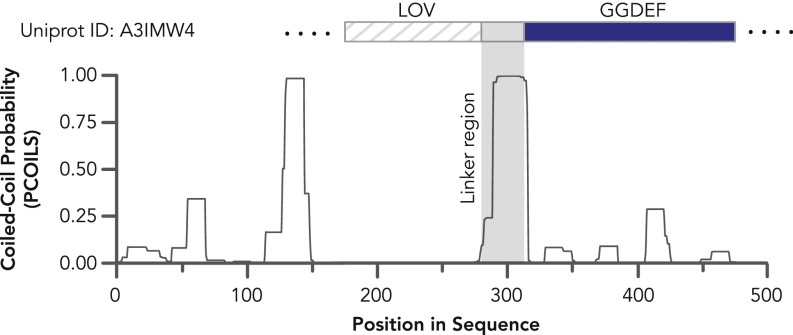
Effector-specific discretization in sensor–effector linker length. (A) Overlaid scatter- and box-and-whisker plots of the linker length between LOV or tandem LOV sensors and their nearest effector domains, shown for effectors observed >10 times (box, first to third quartile; internal band, median). (B) Cumulative linker length distributions for effector-specific linker length between LOV or tandem LOV sensors and their nearest effector domains. (C and D) Heptad periodicity observed for linker regions that adopt extended coiled-coil structures. Bands were defined by k-means clustering, where a Bayesian Information Criterion was used to optimally choose the number of clusters, k. The number of linkers in a given cluster (n) and cluster mean (m) are labeled on each cluster directly. Dotted lines grouping heptad repeats are provided to guide the eye, shown for (C) LOV-GGDEF and (D) LOV-HisKA. LOV-STAS proteins are omitted because only one linker band is observed. Tight banding observed in C and D is indicative of heptad repeats, potentially reflecting structural optimization of sensor–effector orientation and the capability to transmit photosensory structural changes over variable physical distances through an extended coiled-coil linker. Colors in C and D indicate phylum-level taxonomic origin of the LOV.
LOV-GGDEF proteins showed the clearest evidence of a heptad repeat dependence (Fig. 8C). In fact, linkers of up to three heptad repeats are found in nature, and thus, these proteins exhibit a surprising level of tolerance for variable sensor–effector physical distances of up to 32.4 Å, assuming the segment is linear and parallel (although it should be noted that coiled-coils and their dimers can be antiparallel). Although the crystal structures of LOV-GGDEF proteins have not yet been described, the crystal structure of a stimuli-responsive di-guanylate cyclase with a GGDEF-containing receiver WspR from Pseudomonas (54, 55) resembles the solved coiled-coil structure of the engineered YF1 LOV-HisKA (56). LOV-GGDEF linker regions have a remarkably high predicted probability of being coiled-coils based on PCOILS analysis (57) [Probability(Linker Region) > 0.9, Fig. S2]. Taken together with the heptad-cyclical phosphorylation seen with YF1, these convergent results suggest that LOV-GGDEF linkers form coiled-coils that constrain sensor–effector orientation and can transmit the signal over variable sensor–effector physical distances. This heptad repeat banding pattern is also in remarkable agreement with similar bioinformatics analyses of PAS-GGDEF linkers (see figure 5 of ref. 17).
Fig. S2.
Linker regions of LOV-GGDEF proteins are extended coiled-coil linkers. A representative trace of coiled-coiled probability as a function of sequence position is shown based on PCOILS prediction analysis with an analytical window size of 14. Sensor and primary effector domains are overlaid for clarity. The average coiled-coil probability of the linker region shown is nearly unity.
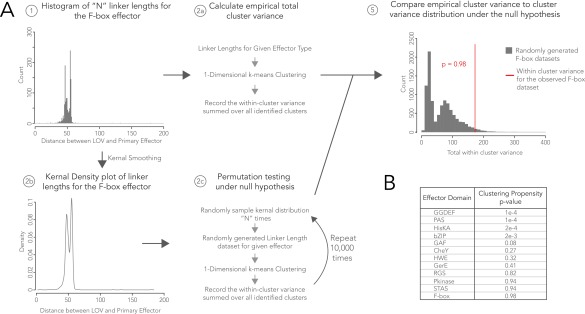
As expected in Fig. 8, YtvA-like LOV-STAS linkers were discretized but effectively only in one band. Naturally occurring LOV-HisKAs also exhibited banding in the linker region, although the heptad trend was not as strong as observed with LOV-GGDEF (Fig. 8D). This is consistent with findings that although some LOV-HisKAs follow a “tilting/rotation” model in which light induces modest structural changes, some members are known to be monomeric or stably oligomeric in the dark (4, 58–62) and thus are possibly less constrained with respect to sensor–effector orientation. The observed spread per linker band may reflect subpopulations of LOV-HisKAs. For example, multiple of the bands have an m + 1 population (where m = k-means cluster) suggestive of opposite transitions in light/dark activity, as seen with the engineered YF1 protein (28). Likewise, LOV-HisKA linkers appear to group into two populations of heptad repeats that are offset by two residues, in a (7n + 2) trend similar to non-LOV PAS-HisKAs (see figure 5 of ref. 28). It should be noted that the heptad repeat is not a perfect multiple of 7 but rather 7.2 residues. Notably, monomeric LOV-HisKAs have recently been described (62) with similar helical linkers separating LOV and HisKA domains, and our data may help suggest sequence preferences that direct these coiled-coil elements to favor interactions in cis with their own sensor domains versus in trans to another coiled-coil. Resampling analysis supports the finding that the discretization in linker length between sensor and GGDEF or HisKA effectors over a large range of lengths is not random (Fig. S3).
Fig. S3.
Resampling analysis to test statistical significance of linker length clustering. (A) Overview of resampling methodology, shown with linker lengths for the F-box effector domain as an example. (B) P values calculate the likelihood that a randomly resampled dataset will form clusters with as small or smaller variance as the observed linker length dataset, where P values < 0.05 are considered significant. GGDEF and HisKA linker length distributions exhibit linker length clustering more tightly than what would be expected by random chance (P < 0.001) for a linker length distribution with the corresponding range.
In contrast, LOVs that undergo larger conformational changes and “unfold” in response to light into monomeric or dimeric forms do not show demonstrable heptad banding. Existing photochemical and structural analyses show that, by and large, these structures do not form stable dimers in the dark [bZIPs such as aureochrome (63), zinc fingers such as White Collar (64–66), HTH proteins such as EL222 (67), and short LOVs such as VIVID (7, 37–39)] or are oriented in antiparallel fashion inconsistent with a parallel extended coiled-coil model, such as phototropins (68) and F-box/Kelch repeats like FKF1 (69, 70). Thus, the observed trend of linker length discretization by effector type and phylum of origin (Fig. 8) reflects that the structural conservation is due to both the functional consequences of preserving sensor–effector orientation and a shared evolutionary ancestry.
SI Text 1
SI Methods.
Transcriptome quality control.
The quality of the assembled contigs from de novo transcriptomes was assessed using TransRate (www.stevekellylab.com/software/transrate) using the program default settings. The program estimates the probability that a contig is both structurally complete and derived from a single transcript (as opposed to a hybrid of two or more) by (i) mapping paired raw transcriptome reads to it, (ii) computing the proportion of those pairs that consist of two reads oriented inwardly and completely contained within the assembled transcript, and (iii) estimating the probability that the read coverage is best modeled by a single Dirichlet distribution. Contigs that did not properly associate with mapped reads of uniform coverage (probability of uniform coverage < 0.05) were removed from the bioinformatics pipeline (1 sequence or 0.015% of the total dataset).
The quality of the assembled contigs from de novo transcriptomes was additionally assessed via genome–transcriptome comparisons. Fully sequenced genomes and corresponding proteomes of LOV-containing species were accessed from NCBI and Uniprot for Amborella trichopoda and Ricinus communis strains and then passed through the bioinformatics pipeline in Fig. 1. These two organisms were chosen as ones with well-annotated genomes because their draft genome sizes were >90% the hypothetical size (87, 88). For each species, the LOV candidates from the two sources were aligned with BLAST. Homologous sequences were manually examined for equivalent domain architectures, and the putative gene product matches are schematically compared in Fig. S1. Splice site prediction in the plant transcripts was performed using the SplicePort server and a score threshold of –0.85, which corresponds to a sensitivity of 97.7–98.9% and a false positive rate of 7.0–8.3% (89).
Redundancy analysis.
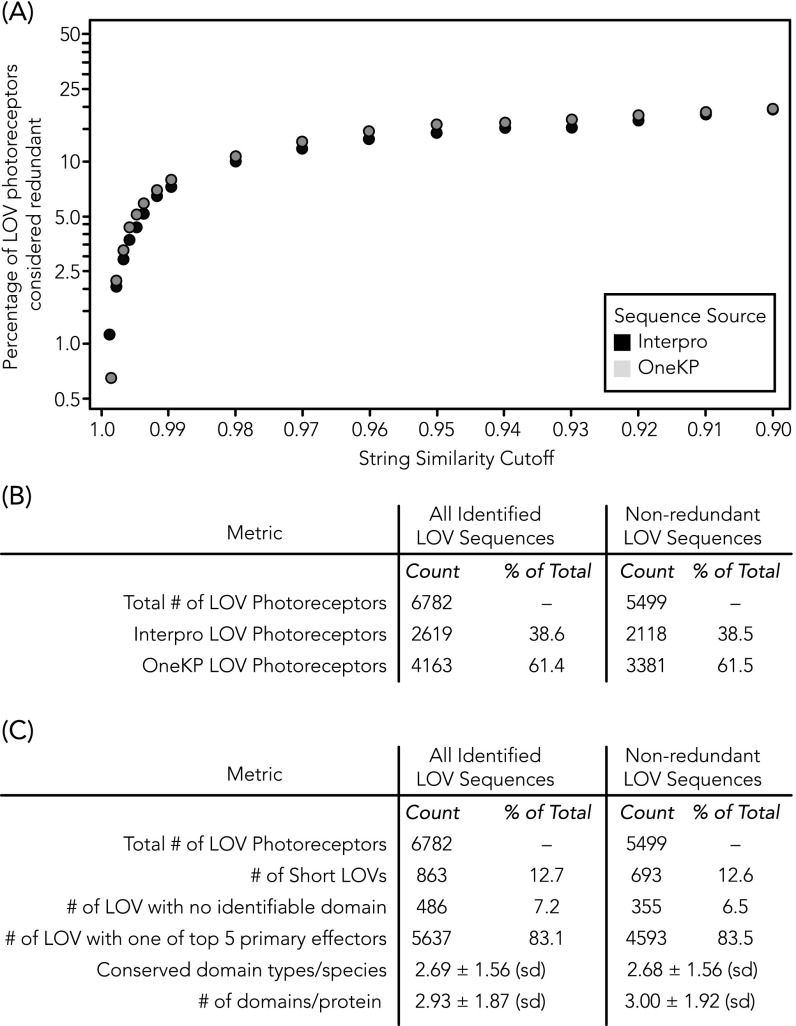
To evaluate the degree to which in-frame point mutations, deletions, and additions contribute to double counting otherwise identical photoreceptor sequences, all reported LOV proteins identified for a given organism were clustered with the CD-HIT tool (www.bioinformatics.org/cd-hit/) at varying thresholds (81, 82). Pairs with similarity scores greater than the allowable similarity threshold were considered theoretically redundant and collapsed into a single consensus sequence (Fig. S4). The longest sequence in a given group of matched sequences was considered to be the “parent sequence” and shorter sequences to be “child sequences.” The fully annotated database of parent sequences only for a similarity threshold of 0.9 (CD-HIT default setting) is available as a spreadsheet tab in Dataset S4, with child sequences listed as an additional column.
Fig. S4.
Relative redundancy is the same across InterPro and OneKP databases. (A) Percentage of LOV photoreceptor sequences from each database that would be considered redundant and collapsed into a single sequence according to a given similarity threshold. Similar percentages of redundant LOV photoreceptors were derived from each source for all similarity thresholds tested. (B) Clustering of redundant sequences into consensus parent sequences reduces the dataset size by ∼20%, similarly for both OneKP and InterPro. (C) Key finding metrics are similar between the nonclustered dataset (i.e., all candidates) and the clustered parent-only dataset, including the prevalence of the five most commonly found conserved domain effectors, as well as domain architectures susceptible to truncation artifacts in redundant sequences (short LOV and LOV with no identifiable conserved domain effector).
Resampling analysis.
A resampling analysis tested if the discretized linker length pattern characterized by tight clustering observed for GGDEF and HisKA effectors over a large range of linker lengths was beyond what would be expected by random chance. The analysis proceeded as follows: (i) Observed linker length distributions of size n were k-means clustered in one dimension and the total variance across all clusters found was recorded; (ii, a) a Gaussian kernel smoothing function was applied to the observed linker length distribution to estimate the underlying probability density function; (ii, b) the estimated probability density function was drawn from n times to compile a “randomized” linker length dataset; (ii, c) k-means clustering in one dimension was run for the randomized dataset and the total variance across all clusters found was recorded; (ii, d) steps ii, b and c were repeated 10,000 times to build the null distribution, which was the range of total variances expected if the linker length dataset was drawn at random from the same underlying probability density function as the observed data; and (iii) the observed total variance was compared with the null distribution. One-tailed P values were calculated as the fraction of variances in the null distribution smaller than the true variance.
SI Results.
Fig. S1 shows comparisons between putatively matching draft genome-derived and de novo transcriptome-derived candidate sequences from A. trichopoda and R. communis. Five candidate genes of identical functional cluster repertoires were identified in the draft genome for both organisms. All transcriptome-derived sequences had a clear genomic match, but given that only highly expressed transcripts are successfully assembled, only five LOV photoreceptor transcripts were identified in total between the two species (Fig. S1 A and B). Two of the transcripts were identical in sequence and length to the genome-predicted counterpart. A. trichopoda match #4 differed in predicted versus actual splice sites; the apparent “deletion” in the transcript versus the genome-predicted product was exactly flanked by independently predicted splice sites (Fig. S1C). A. trichopoda match #5 differed because of a frame shift, with either a two-base deletion in the transcript contig (position 295) or two-base addition in the genome read; the sequences were 100% homologous upstream and downstream of the site (Fig. S1D). Without isolating the protein from the native organism, the following remain possible origins of the frame shift: (i) the genome or transcriptome assemblies, (ii) genome or transcriptome sequencing errors (Illumina sequencers were used in both cases), or (iii) natural sequence variation from different samples of the same organism. Lastly, differences in R. communis match #5 were attributable to start codon variation as well as missing genomic sequences (denoted as “?” in the draft genome) (88) (Fig. S1E).
None of the matching pairs showed any differences in functional cluster architecture, linker length, and sequence between any conserved domains, or the C terminus. In summary, we conclude that the de novo transcriptome assemblies here are in strong agreement with the predicted proteomes of next-generation sequencing-derived draft genomes and that differences are reasonable within the limits of natural variation (splice variants, sample specificity, tissue specificity, etc.) and accuracy of predicted start codon and intron/exon during draft genome assemblies.
Discussion
Expanded Functional Diversity from Broadly Surveying Organismal Diversity.
The analysis balanced various factors—namely, throughput and broad representation of organisms. For example, the motif-based analysis revealed that LOV protein regions that form the flavin-binding pocket and mediate photocycling are highly conserved, whereas those that interface with and transmit signals to effector domains, such as the A′-alpha and J-alpha helices, are not (Fig. 2B). Limiting the length of the query sequence to motifs implicated only in flavin binding and light sensing augmented computational throughput and reduced the likelihood that a potential LOV candidate would be excluded on the basis of an unusual mechanism for effector domain regulation. Although other position-weighted approaches exist like PSI-BLAST, which compares sequences against the National Center for Biotechnology Information (NCBI) database (71), they would not have allowed for a self-consistently generated dataset because ∼60% of the raw data analyzed here resided in other databases or are not yet available in annotated forms. Likewise, because LOV is not yet a domain class of its own in InterPro, which also lacks much of the dataset studied here, custom analyses were necessary to annotate the functional and topological diversity in full breadth.
Although most physiological roles deduced from ontological functions and multidomain topologies were consistent with previous descriptions in signaling, transcriptional regulation, and cytoskeletal movement, rare effectors were often putatively involved in biosynthesis of molecules beyond cyclic nucleotides, such as lipase or glutamine amidotransferase primary effectors (Dataset S3). Previously unidentified effector functions were all found in early-diverging green algae that were only recently sequenced by OneKP, which highlights the value of broadly sampling organismal diversity in addition to optimizing algorithms. The expanded range of physiological roles shows how adaptable LOV sensors regulate both evolutionarily conserved processes and specialized organism-specific functions. Importantly and as previously stated, the inherent LOV diversity found in nature is correlated with the evolutionary diversity within a kingdom (Fig. 7). As species become more evolutionarily diverse, so do their LOV proteins.
LOV proteins with primary effectors of new and/or rare ontological functions may push the protein class into new signaling physiological roles. For example, LOV-RGS proteins are the likely photoreceptors that govern steering in brown algal negative phototaxis, based on previously reported microspectrophotometry, proteomics, and immunofluorescence imaging studies (45, 72). A putative role for LOV-RGS in fungi is less apparent despite their surprising prevalence. Deletion of the Magnaporthe oryzae MoRGS5 (73), which previously was described as a PAS/RGS but whose sensor is identified here as a LOV, causes no observable phenotypic difference. It is possible that LOV-RGS proteins were shared between brown/golden algae and pathogenic fungi by horizontal gene transfer in light of the fact that fungal LOVs possess few effectors like RGS that are directly enzymatic. The physiological roles and evolutionary history of LOV-RGS and the many other functional clusters reported here warrant future studies by photophysical and structural characterization, genomics, and organismal physiology.
Bioinformatics-Guided Engineering of Optogenetic Tools.
Natural and engineered photosensory proteins, when heterologously or ectopically expressed in genetically targeted cells, are powerful optogenetic tools to control cellular physiology and transcriptional circuits (12). The previously unidentified effectors and expansion in sequence diversity of rare effectors reported here are important for several reasons. First, natural LOVs with effector functions such as lipase activity and RGS-based tuning of GPCR activity may be highly useful in cell signaling. Second, screening phylogenetic diversity is a valuable strategy for enhanced performance and trafficking in optogenetic tool development. For example, the natural LOV-HTH EL222 (16) is a high-performance optogenetic transcription factor with light/dark ratios in transcriptional activity of >100-fold. Likewise, diversity-driven discovery has begotten numerous electrogenic rhodopsins widely used for controlling excitable cells, including the first optogenetic tools to elicit behavioral changes in primates (74–76) and achieve noninvasive inhibition in rodents (77), as well as spectrally diverse channelrhodopsins for truly orthogonal activation of two colocalized cell populations with two colors of light (78).
The inherent modularity of LOV proteins makes them invaluable in creating chimeric optogenetic tools by swapping natural effectors with arbitrary proteins to confer photosensitivity to the latter. Although most structure–function studies on LOV proteins to date focus on flavin photocycling, the structure–function of the linker region, which is arguably more critical for engineering high-performance chimeras, is more varied and less established. Most reported chimeras are constructed from one of three LOV proteins whose sensor–linker interactions have been described by structural NMR (AsLOV2) (6) or by crystallography (VIVID, YtvA) (56, 58, 79). Further establishing principles of how optical signals are transmitted between sensor and effector through the linker region enhances our ability to rationally engineer novel and improved protein-based tools. Through sequence analysis of natural LOV photoreceptors, which complements previous structural analyses of an engineered chimeric HisKA (28), we find further convergent evidence that linker length in parallel extended coiled-coils reflects not only evolutionary history (as evident by conservation of banding across phyla in Fig. 8 C and D) but also a mechanistic optimization of sensor–effector orientation with a surprising tolerance for variable sensor–effector physical distances. This finding suggests that when photosensitizing an effector-of-choice, the signal transmission mechanism through the linker is a critical element in choosing appropriate LOV sensors for the chimera. Future bioinformatics or structural analyses that establish new photoswitching models will greatly advance optogenetic tools and consequent scientific discoveries from their application.
Resource for LOV Photoreceptor Comparative Genomics.
Beyond the insights reported here, we have created a valuable resource that contains an enormous wealth of LOV gene sequences that nearly triples the number of sequences known to date and catalogs the functional and architectural diversity of LOV photoreceptors. The entire dataset is provided in text-searchable format (Dataset S4), which contains the (i) sequence of each putative LOV protein identified, (ii) flavin-binding motif, (iii) primary effector and ontological function, (iv) sequence and position of all conserved domains, (v) multidomain cluster architectures, (vi) linker length, (vii) taxonomy of organism of origin, and (viii) presence of likely integral membrane proteins (TMHMM Server v. 2.0) (80). Combinations of these entry fields may be queried in an automated manner in Python (SI Text 2 provides instructions and Dataset S5 provides a sample script for automated data extraction). To evaluate the degree to which natural variation could affect the counting of candidates (i.e., redundancies from in-frame point mutations, splice variations, deletions, and additions of the same gene), all reported LOV sequences identified for a given organism were clustered with the “CD-HIT” tool (81, 82). Sequences derived from OneKP or InterPro were equally likely to be labeled redundant, and no physiologically relevant changes were introduced by collapsing the redundant set to the longest consensus sequence (Fig. S4 and Dataset S4, with “redundant sequences” in an additional column). These resources may beget numerous new insights by facilitating rapid comparative analyses of highly specific features (e.g., all HisKA s with a given linker length range, all LOV domains from a specific phylum, etc.), thereby offering great proliferative benefit to the overall understanding of LOV photobiology of photosynthetic and nonphotosynthetic organisms.
In summary, our study highlights the value of genomic surveys of broad ranges of organisms and ecological niches for establishing sensory protein diversity. By customizing the bioinformatics analysis to thoroughly capture that diversity, we created an annotated dataset of >6,700 LOV proteins for exploring LOV structure–function through comparative structural genomics, understanding the expanded photosensory signaling roles of newfound proteins, and inventing optogenetic reagents for light-driven control of physiology in targeted cells.
SI Text 2
Dataset S4 is a resource that consists of putative LOV photoreceptors that have been annotated and analyzed. It consists of protein sequences sourced from two databases: Interpro (see www.ebi.ac.uk/interpro/) and OneKP (https://sites.google.com/a/ualberta.ca/onekp/). The tab labeled “All_sequences” contains the >6,700 LOV photoreceptors identified by the bioinformatics pipeline. The tab labeled “Non-redundant” contains only consensus sequences identified through a clustering analysis (see SI Methods).
If you have problems, questions, ideas, or suggestions, please contact us at chowlab.seas.upenn.edu.
Spreadsheet Fields.
Database source.
The database source specifies from where the protein sequence was sourced.
Sequence ID.
The sequence ID specifies a unique ID number attached to each protein sequence analyzed. For Interpro protein sequences, this number may be used as a search term to pull up further information in Uniprot (www.uniprot.org). For OneKP protein sequences, this number reflects a unique identifier generated during next-generation sequencing and transcriptome assembly.
GenBank ID.
The corresponding GenBank ID for each protein sequence is listed. It may be used as a search term to pull up the sequence entry in the GenBank database (www.ncbi.nlm.nih.gov/genbank/). GenBank accession numbers marked with a hash sign (#) correspond to assembled contigs with multiple reading frames; this may represent a frame-shift mutation at the raw sequencing read level or be a consequence of natural variation.
Primary structure.
The primary structure is the sequence of amino acids that comprise the ORF within which a putative LOV photoreceptor was identified. See Methods for the ORF definition.
GXNCRFLQ.
The consensus flavin-adduct LOV protein motif is GXNCRFLQ. This field identifies the actual putative flavin-adduct binding sequence for each photoreceptor identified, which may differ from the consensus sequence to a variable degree.
Protein length.
The protein length is the length of the putative photoreceptor in the amino acids.
Domain structure.
This is a text-based representation of the domain structure of each LOV photoreceptor analyzed and is presented in the form “Domain Type: (starting location, stopping location).” All domains are listed in order from the amino (N) to carboxy (C) terminus, with arrows between domains indicating an amino to carboxy direction. Domain names are generated directly from either the pFAM or Interpro conserved domain databases. For further information about each domain type, please search these conserved domain databases with the domain type name given.
Functional cluster.
The functional cluster is the domain composition for a LOV photoreceptor, independent of domain order and repeats. Domains are listed in alphabetical order. Full names of effector abbreviations are provided in Dataset S2.
Primary effector.
The primary effector is the nearest neighboring effector domain to the LOV sensor in the linear polypeptide sequence. Full names of effector abbreviations are provided in Dataset S2.
Primary effector gene ontology.
The primary effector gene ontology is the corresponding GO for the primary effector domain, as described by pFAM and Interpro database GO assignments.
Linker length.
This is defined as the number of amino acid residues that separate the primary effector domain from the LOV sensor in the linear polypeptide sequence.
Number of predicted transmembrane helices.
Each sequence was run through the TMHMM2.0 transmembrane helix prediction program, and the number of predicted transmembrane helices is reported.
Transmembrane helix topology.
The transmembrane helix topology is the ropology of predicted transmembrane helices from the TMHMM2.0 program where both orientation and position are listed. “i” indicates a loop on the inside, and “o” indicates a loop on the outside. A topology of “i10-32o50-72i” would correspond to a set of two helices where the first is predicted to start on the inside and go from residues 10–32 and end outside the membrane. The second predicted helix would continue outside and begin at residue 50. It would continue to residue 72 and end on the inside (80).
Kingdom.
LOV photoreceptors were assigned to one of six kingdoms: bacteria, fungi, protists, land plants, archaea, or unknown.
Phylum.
The phylum was assigned to a LOV photoreceptor via an automated search of the NCBI Taxonomy browser. Missing NCBI taxonomy entries were supplemented according to Algaebase and the ITIS.
Class.
The class was assigned to a LOV photoreceptor via an automated search of the NCBI taxonomy browser. Missing NCBI taxonomy entries were supplemented according to Algaebase and the ITIS.
Family.
The family was assigned to a LOV photoreceptor via an automated search of the NCBI taxonomy browser. Missing NCBI taxonomy entries were supplemented according to Algaebase and the ITIS.
Genus.
For LOV photoreceptors identified from the Interpro database, the genus was assigned to a LOV photoreceptor via an automated search of the NCBI taxonomy browser. Missing NCBI taxonomy entries were supplemented according to Algaebase and the ITIS. For LOV photoreceptors from the OneKP database, information about the genus was supplied with each tissue sample to be sequenced.
Species.
For LOV photoreceptors identified from the Interpro database, the species was assigned to a LOV photoreceptor via an automated search of the NCBI taxonomy browser. Missing NCBI taxonomy entries were supplemented according to Algaebase and the ITIS. For LOV photoreceptors from the OneKP database, information about the species was supplied with each tissue sample to be sequenced.
Child transcript variant sequences.
All reported LOV proteins identified for a given organism were clustered with the CD-HIT tool at the default threshold (see SI Methods) Pairs with similarity scores greater than the allowable similarity threshold were considered theoretically redundant and collapsed into a single consensus sequence (Fig. S4). The longest sequence in a given group of matched sequences was considered to be the parent sequence and shorter sequences to be child sequences. In the “nonredundant” tab of Dataset S4, an extra column lists the child sequences for each consensus parent sequence.
Scripts.
There is a script available for common search needs. The script is written in the python language. Python must be installed for this script to properly run. Dataset S4 must also be located in the same folder as the python script for proper function. The script allows the user to perform a defined query and offers output either to a print screen or in an excel spreadsheet format. See the script in Dataset S5. Instructions for using the script are provided within the script and are distinguished from surrounding commands by the # symbol. The script searches only the “All_sequences” tab in Dataset S4.
Methods
De Novo Motif Prediction for LOV Sensor Domains.
Sequence patterns were identified by motif analysis with the MEME Suite (20) in 18 LOV domains known to photocycle (Fig. 2). MEME tool parameters were set to find two motifs of ≤50 amino acids that must be present in all query sequences. Identified motifs were exported as .xml files.
LOV Photoreceptor Identification.
OneKP database (27) ORFs were required to begin with a start codon (ATG) and end with a stop codon and were predicted with EMBOSS-6.6.0 using the standard codon table #0 and a minimum ORF length of 100 amino acids (83). The predicted protein list was pooled with protein sequences deposited in the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) protein database with PAS domains (IPR000014) that were identified with Interpro (22, 23). A Python script removed duplicate proteins or exact subsets of longer proteins on a per-species basis to ensure uniqueness of each candidate, which was then searched for the sensor motifs with the MAST tool (20) with e-value threshold ≤1e-25. Residues enclosed by a predicted PAS fold were identified by Pfam (21), and then a Python script labeled proteins as bona fide LOV sensors if both motifs (i) aligned with a P value ≤1e-15, (ii) were separated by <75 residues, and (iii) were bound within a PAS fold and (iv) a cysteine residue was present in the flavin-binding site of motif 1.
Comparator datasets for validating the ability to distinguish between LOV and structurally related non-LOV proteins were (i) ligand binding PAS-fold proteins, taken from figure 3 of ref. 18 (listed in Dataset S1), (ii) flavin-binding BLUF photoreceptors in InterPro collection IPR007024, (iii) flavoproteins from InterPro collection IPR00382, and (iv) a test set of known LOV domains selected from figure 2 of ref. 84 (listed in Dataset S1). MAST searched for the joint presence of the two sensor motifs in each candidate protein and reported an e-value defined as the expected number of sequences in a random database of the same size that would match the group of motifs as well as the sequence does. When searching for the motifs in known test set LOV domains also contained in the training set, we applied a “leave-one-out cross-validation” scheme, where the two sensor motifs were regenerated for the training set minus one LOV photoreceptor, and the sensor motifs were then searched for with MAST on the remaining LOV photoreceptor.
LOV Photoreceptor Annotation.
Potential effectors were searched against the Pfam HMM database with the UNIX command-line HMMER v3.1b1 tool (85), with an e-value ≤1e-3. A Python script parsed the results to generate protein maps that specify where predicted effector and sensor domains are located along the candidate polypeptide sequence. In cases when possible effectors overlapped in polypeptide sequence, the conserved domain with the lowest e-value associated with identification by Pfam was chosen. If the nearest effector was another LOV sensor reflective of the tandem LOV architecture, the tandem LOV was collapsed into a single tandem LOV pseudodomain, and the annotations continued as they would for a single LOV. Maps were labeled with taxonomic information from kingdom to species, using the entrez command line tool to search the NCBI taxonomy database. Missing NCBI taxonomy entries were supplemented according to Algaebase and the Integrated Taxonomic Information System (ITIS). If no Pfam effector domains were assigned within 125 amino acids of a putative LOV sensor domain, the region was subjected to an additional Interpro conserved domains search.
Linker bands were defined by k-means clustering in one dimension where a Bayesian Information Criterion was used to optimally choose the number of clusters, k, according to default settings of the Ckmeans.1d.dp package for the R statistical programming language (86). Cumulative linker length distributions were generated with the ecdf function in R.
Domain Connectivity Analysis.
To establish domain connectivity, Python scripts analyzed annotated LOV proteins and executed a domain adjacency analysis (47) by scanning through LOV maps comprised of n distinct effector and sensor domain types to produce an n × n matrix, where the off-diagonal entry aij is the number of times domain type i is followed by domain type j, and the diagonal entry aii is equal to the number of times domain type i is adjacent to itself. The resultant domain adjacency matrix was visualized with the network software program Gephi (48), such that (i) domains are nodes, (ii) edges are connections between domains, and (iii) line thickness is proportional to log2 of aij or aii. To perform a domain-positional analysis, Python scripts analyzed annotated LOV maps and determined the position of each domain relative to the LOV or tandem LOV sensor, which was assigned position 0. N- and C-terminal domains were assigned negative and positive values, respectively. The primary effector was defined as the domain with the shortest linker length (in polypeptide sequence) to the LOV or tandem LOV sensor.
Dataset Release.
The fully annotated database is available as Dataset S4 and can be manually text-searched or examined by automated data extraction (instructions provided in SI Text 2, and sample Python code provided in Dataset S5 which makes use of the “xlrd” and “xlwt” Python packages to import/export spreadsheets).
Note Added in Proof.
While this paper was in production, GenBank BLASTx analysis showed that 18% of newly identified OneKP-derived LOV candidates have multiple hits to a single existing protein sequence in the NCBI database after translating in multiple frames. This result may be indicative of natural variation between organisms or a frame-shift mutation introduced at the raw sequencing read level. This BLASTx result is consistent with findings in Fig. S1, which assesses agreement between matching candidates derived from OneKP transcriptomes to literature-reported genome predictions of the same organism (one of the five matches varied by a possible frame shift). We have marked the corresponding GenBank entries with a hash sign (#) in Dataset S4. We thank GenBank for conducting the analysis on our behalf.
Supplementary Material
Acknowledgments
The authors thank Ben Voight, Danielle Bassett, Arjun Raj, Daniel Schmidt, and all members of the B.Y.C. laboratory for helpful discussions. The authors also thank Stuart Levine and Huiming Ding of the MIT Broad Institute for early technical support and the Hibberd laboratory for access to the TransRate software. S.T.G. is supported by the National Science Foundation (NSF) Graduate Research Fellowship Program. B.Y.C. was funded by NSF Biophotonics (CBET 126497), the W. W. Smith Charitable Trust for the Heart, the Brain Research Foundation Fay Frank Program, the Penn Medicine Neuroscience Center, and the NIH/National Institute on Drug Abuse Grant 1R21 DA040434-01. B.Y.C. and E.S.B. were funded by Defense Advanced Research Projects Agency Living Foundries HR0011-12-C-0068. The 1000 Plants (1KP) initiative, led by G.K.-S.W., is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures, Innovates Centres of Research Excellence, Musea Ventures, BGI-Shenzhen, and China National Genebank. E.S.B. was funded by the MIT Media Lab, Office of the Assistant Secretary of Defense for Research and Engineering, Harvard/MIT Joint Grants in Basic Neuroscience, NSF (especially CBET 1053233 and EFRI 0835878), NIH (especially 1DP2OD002002, 1R01NS067199, 1R01DA029639, 1R01GM104948, 1RC1MH088182, and 1R01NS075421), the Wallace H. Coulter Foundation, Alfred P. Sloan Foundation, Human Frontiers Science Program, New York Stem Cell Foundation Robertson Neuroscience Investigator Award, Institution of Engineering and Technology A. F. Harvey Prize, and Skolkovo Institute of Science and Technology. K.H.G. was funded by NIH Grant R01 GM106239 and Cancer Prevention Research Institute of Texas Grant RP130312.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU698078–KU702192).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509428113/-/DCSupplemental.
References
- 1.Zoltowski BD, Gardner KH. Tripping the light fantastic: Blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50(1):4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losi A, Mandalari C, Gärtner W. From plant infectivity to growth patterns: The role of blue-light sensing in the prokaryotic world. Plants. 2014;3(1):70–94. doi: 10.3390/plants3010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauss U, et al. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J Bacteriol. 2009;191(23):7234–7242. doi: 10.1128/JB.00923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9(10):713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42(1):2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 6.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 7.Zoltowski BD, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316(5827):1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426(6964):302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 9.Swartz TE, et al. Blue-light-activated histidine kinases: Two-component sensors in bacteria. Science. 2007;523(2001):1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 10.Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7(4):473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avila-Pérez M, Hellingwerf KJ, Kort R. Blue light activates the sigmaB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188(17):6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Möglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem Photobiol Sci. 2010;9(10):1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 13.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105(31):10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lungu OI, et al. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19(4):507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9(3):266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 16.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10(3):196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17(10):1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erbel PJA, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100(26):15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL, et al. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server Issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell A, et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43(Database issue):D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak GP, Losi A, Gärtner W. Metagenome-based screening reveals worldwide distribution of LOV-domain proteins. Photochem Photobiol. 2012;88(1):107–118. doi: 10.1111/j.1751-1097.2011.01024.x. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa M, et al. Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta. 2009;230(3):543–552. doi: 10.1007/s00425-009-0967-6. [DOI] [PubMed] [Google Scholar]
- 26.Losi A, Gärtner W. The evolution of flavin-binding photoreceptors: An ancient chromophore serving trendy blue-light sensors. Annu Rev Plant Biol. 2012;63(1):49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- 27.Matasci N, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3(1):17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385(5):1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46(49):14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 30.Zayner JP, Antoniou C, French AR, Hause RJ, Jr, Sosnick TR. Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophys J. 2013;105(4):1027–1036. doi: 10.1016/j.bpj.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5(3):533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopal S, Moffat K. Crystal structure of a photoactive yellow protein from a sensor histidine kinase: Conformational variability and signal transduction. Proc Natl Acad Sci USA. 2003;100(4):1649–1654. doi: 10.1073/pnas.0336353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamoto Y, Kataoka M. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem Photobiol. 2007;83(1):40–49. doi: 10.1562/2006-02-28-IR-827. [DOI] [PubMed] [Google Scholar]
- 34.Kasahara M, et al. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002;129(2):762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajava AV. Tandem repeats in proteins: From sequence to structure. J Struct Biol. 2012;179(3):279–288. doi: 10.1016/j.jsb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Di Domenico T, et al. RepeatsDB: A database of tandem repeat protein structures. Nucleic Acids Res. 2014;42(Database issue):D352–D357. doi: 10.1093/nar/gkt1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104(3):453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 38.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22(18):4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47(27):7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Möglich A, Ayers RA, Moffat K. Addition at the molecular level: Signal integration in designed Per-ARNT-Sim receptor proteins. J Mol Biol. 2010;400(3):477–486. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 41.Hunt SM, Thompson S, Elvin M, Heintzen C. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci USA. 2010;107(38):16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci USA. 2010;107(38):16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malzahn E, Ciprianidis S, Káldi K, Schafmeier T, Brunner M. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell. 2010;142(5):762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Idnurm A, Verma S, Corrochano LM. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet Biol. 2010;47(11):881–892. doi: 10.1016/j.fgb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu G, Nagasato C, Oka S, Cock JM, Motomura T. Proteomics analysis of heterogeneous flagella in brown algae (stramenopiles) Protist. 2014;165(5):662–675. doi: 10.1016/j.protis.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 46.de Mendoza A, Sebé-Pedrós A, Ruiz-Trillo I. The evolution of the GPCR signaling system in eukaryotes: Modularity, conservation, and the transition to metazoan multicellularity. Genome Biol Evol. 2014;6(3):606–619. doi: 10.1093/gbe/evu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: New insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007;61:453–475. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- 48.Bastian M, Heymann S, Jacomy M. 2009. Gephi: An open source software for exploring and manipulating networks. Third International AAAI Conference on Weblogs and Social Media:361-362. [DOI] [PMC free article] [PubMed]
- 49.Okajima K, et al. Light-induced conformational changes of LOV1 (light oxygen voltage-sensing domain 1) and LOV2 relative to the kinase domain and regulation of kinase activity in Chlamydomonas phototropin. J Biol Chem. 2014;289(1):413–422. doi: 10.1074/jbc.M113.515403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58(1):21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 51.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 52.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 53.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 2000;10(1):17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 54.De N, et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6(3):e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7(10):724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 56.Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21(7):1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 57.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 58.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373(1):112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Correa F, Ko W-H, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: Enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52(27):4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purcell EB, McDonald CA, Palfey BA, Crosson S. An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry. 2010;49(31):6761–6770. doi: 10.1021/bi1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104(46):18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera-Cancel G, Ko WH, Tomchick DR, Correa F, Gardner KH. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci USA. 2014;111(50):17839–17844. doi: 10.1073/pnas.1413983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hisatomi O, Nakatani Y, Takeuchi K, Takahashi F, Kataoka H. Blue light-induced dimerization of monomeric aureochrome-1 enhances its affinity for the target sequence. J Biol Chem. 2014;289(25):17379–17391. doi: 10.1074/jbc.M114.554618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ballario P, Talora C, Galli D, Linden H, Macino G. Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins. Mol Microbiol. 1998;29(3):719–729. doi: 10.1046/j.1365-2958.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- 65.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297(5582):815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 66.Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem. 2003;278(6):3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- 67.Rivera-Cancel G, Motta-Mena LB, Gardner KH. Identification of natural and artificial DNA substrates for light-activated LOV-HTH transcription factor EL222. Biochemistry. 2012;51(50):10024–10034. doi: 10.1021/bi301306t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katsura H, Zikihara K, Okajima K, Yoshihara S, Tokutomi S. Oligomeric structure of LOV domains in Arabidopsis phototropin. FEBS Lett. 2009;583(3):526–530. doi: 10.1016/j.febslet.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 69.Nakasako M, Matsuoka D, Zikihara K, Tokutomi S. Quaternary structure of LOV-domain containing polypeptide of Arabidopsis FKF1 protein. FEBS Lett. 2005;579(5):1067–1071. doi: 10.1016/j.febslet.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 70.Nakasone Y, Zikihara K, Tokutomi S, Terazima M. Kinetics of conformational changes of the FKF1-LOV domain upon photoexcitation. Biophys J. 2010;99(11):3831–3839. doi: 10.1016/j.bpj.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhagwat M, Aravind L. In: PSI-BLAST Tutorial. Comparative Genomics, Methods in Molecular Biology. Bergman N, editor. Vol 395. Totowa, NJ: Humana Press; 2008. pp. 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu G, et al. Ubiquitous distribution of helmchrome in phototactic swarmers of the stramenopiles. Protoplasma. 2015 doi: 10.1007/s00709-015-0857-7. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H, et al. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011;7(12):e1002450. doi: 10.1371/journal.ppat.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han X, et al. A high-light sensitivity optical neural silencer: Development and application to optogenetic control of non-human primate cortex. Front Syst Neurosci. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavanaugh J, et al. Optogenetic inactivation modifies monkey visuomotor behavior. Neuron. 2012;76(5):901–907. doi: 10.1016/j.neuron.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gerits A, et al. Optogenetically induced behavioral and functional network changes in primates. Curr Biol. 2012;22(18):1722–1726. doi: 10.1016/j.cub.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17(8):1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11(3):338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a light-activated LOV protein dimer that regulates transcription. Sci Signal. 2011;4(184):ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 81.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 83.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 84.Mandalari C, Losi A, Gärtner W. Distance-tree analysis, distribution and co-presence of bilin- and flavin-binding prokaryotic photoreceptors for visible light. Photochem Photobiol Sci. 2013;12(7):1144–1157. doi: 10.1039/c3pp25404f. [DOI] [PubMed] [Google Scholar]
- 85.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Song M. Ckmeans. 1d. dp: Optimal k-means clustering in one dimension by dynamic programming. R J. 2011;2:29–33. [PMC free article] [PubMed] [Google Scholar]
- 87.Project AG. Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342(6165):1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 88.Chan AP, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28(9):951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dogan RI, Getoor L, Wilbur WJ, Mount SM. SplicePort—An interactive splice-site analysis tool. Nucleic Acids Res. 2007;35(Web Server issue):W285–W291. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.