Significance
The discovery of a novel modification system that inserts 7-deazaguanine derivatives in DNA, modifications thought until now to occur only in RNA, is an excellent illustration of the power of biological evolution to alter the ultimate function not only of the distinct proteins but also of entire metabolic pathways. The extensive lateral transfer of the gene cluster responsible for this modification highlights its significance as a previously unrecognized foreign DNA defense system that bacteria and phages use to protect their genomes. The characterization of these DNA modification pathways also opens the door to novel tools to manipulate nucleic acids.
Keywords: DNA modification, restriction–modification, 7-deazaguanine, comparative genomics, queuosine
Abstract
The discovery of ∼20-kb gene clusters containing a family of paralogs of tRNA guanosine transglycosylase genes, called tgtA5, alongside 7-cyano-7-deazaguanine (preQ0) synthesis and DNA metabolism genes, led to the hypothesis that 7-deazaguanine derivatives are inserted in DNA. This was established by detecting 2’-deoxy-preQ0 and 2’-deoxy-7-amido-7-deazaguanosine in enzymatic hydrolysates of DNA extracted from the pathogenic, Gram-negative bacteria Salmonella enterica serovar Montevideo. These modifications were absent in the closely related S. enterica serovar Typhimurium LT2 and from a mutant of S. Montevideo, each lacking the gene cluster. This led us to rename the genes of the S. Montevideo cluster as dpdA-K for 7-deazapurine in DNA. Similar gene clusters were analyzed in ∼150 phylogenetically diverse bacteria, and the modifications were detected in DNA from other organisms containing these clusters, including Kineococcus radiotolerans, Comamonas testosteroni, and Sphingopyxis alaskensis. Comparative genomic analysis shows that, in Enterobacteriaceae, the cluster is a genomic island integrated at the leuX locus, and the phylogenetic analysis of the TgtA5 family is consistent with widespread horizontal gene transfer. Comparison of transformation efficiencies of modified or unmodified plasmids into isogenic S. Montevideo strains containing or lacking the cluster strongly suggests a restriction–modification role for the cluster in Enterobacteriaceae. Another preQ0 derivative, 2’-deoxy-7-formamidino-7-deazaguanosine, was found in the Escherichia coli bacteriophage 9g, as predicted from the presence of homologs of genes involved in the synthesis of the archaeosine tRNA modification. These results illustrate a deep and unexpected evolutionary connection between DNA and tRNA metabolism.
Hypermodifications of DNA requiring more than one synthetic enzyme are not as prevalent and chemically diverse as RNA hypermodifications, but around a dozen have been identified in DNA to date (1). The functions of most DNA hypermodifications are still not known, but some have roles in protection against restriction enzymes, whereas others affect thermal stability temperature, DNA packaging, or transcription regulation (2). For example, the hypermodified DNA base β-d-glucosyl-hydroxymethyluracil, or base J, is an epigenetic factor that regulates Pol II transcription initiation in kinetoplastids of trypanosomes (3). The recently discovered phosphorothioate (PT) modification of the DNA backbone in bacteria was found to perform different functions in different organisms (4–6). In Salmonella Cerro 87, PT occurs on each strand of a GAAC/GTTC motif as part of a restriction–modification (R–M) system, whereas in Vibrio cyclitrophicus FF75, which lacks PT restriction enzymes, PT occurs on one strand of CpsCA motifs, and the function remains unclear (6). In 2013, Iyer et al. described the computational prediction of 12 novel DNA hypermodification systems in phage and bacteria (7), demonstrating the potential diversity and complexity of modifications yet to be discovered.
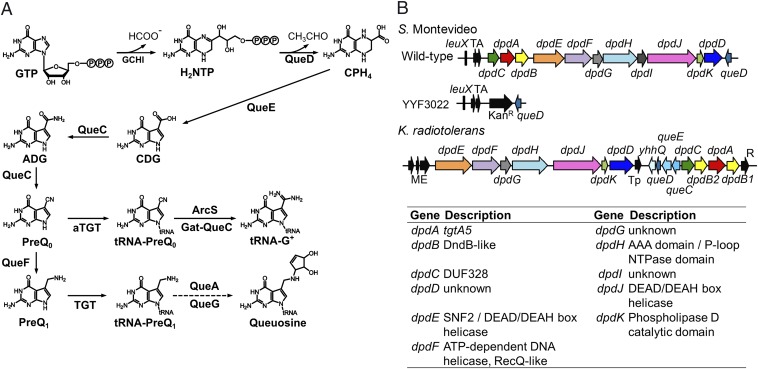
7-Cyano-7-deazaguanine (preQ0) is a common precursor of the widespread tRNA modifications queuosine (Q) and archaeosine (G+) (8) and of pyrrolopyrimidines such as toyocamycin or tubercidin (9). In both Archaea and Bacteria, preQ0 is synthesized from GTP in a pathway that has been fully characterized in the last 10 y (Fig. 1A). The first step catalyzed by GTP cyclohydrolase I (GCHI; FolE) is shared with the tetrahydrofolate synthesis pathway (10), and then three enzymes—6-carboxy-5,6,7,8-tetrahydropterin synthase (QueD), 7-carboxy-7-deazaguanine synthase (QueE), and 7-cyano-7-deazaguanine synthase (QueC)—lead to the formation of the preQ0 moiety (11, 12) (Fig. 1A).
Fig. 1.
Biosynthesis of 7-deazaguanine derivatives. (A) The biosynthetic pathways to queuosine and archaeosine in tRNA. ADG, 7-amido-7-deazaguanine; aTGT, archaeal TGT; CDG, 5-carboxydeazaguanine; CPH4, 6-carboxytetrahydropterin; G+, archaeosine; GCHI, GTP cyclohydrolase I (FolE); H2NTP, Dihydroneopterin phosphate; TGT, tRNA-Guanine transglycosylase. (B) Gene clusters of S. Montevideo (GCA_000238535.2; TgtA5 UniProt ID E7V8J4) and K. radiotolerans (NC_009664; TgtA5 UniProt ID A6WGA1) tgtA5/dpdA and S. Montevideo mutant strain YYF3022 (∆dpdC-dpdD::kan). Similar colors represent homologs. Red arrows indicate dpdA; black arrows represent genes predicted to be involved in HGT; gray arrows represent hypothetical proteins (hyp). int, integrase; ME, mobile element protein; R, resolvase; TA, toxin–antitoxin gene pair (ccdA, ccdB); Tp, transposase.
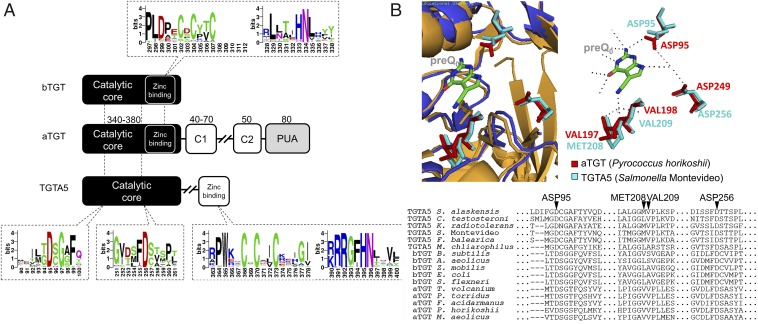
The synthesis of G+ and Q diverge after the formation of preQ0. In Bacteria, the tRNA guanosine (34) transglycosylase (bTGT; EC 2.4.2.29) enzyme that targets the G at position 34 of tRNAs with GUN anticodons (13) prefers the 7-aminomethyl-7-deazaguanine (preQ1) base that is derived from preQ0 in one step by NADPH-dependent enzyme preQ0 oxidoreductase (QueF) (14), but it can also use preQ0 when preQ1 is absent (14). Thus, preQ0 is reduced to preQ1 and inserted in tRNAs by bTGT (Fig. 1A). Two subsequent enzymatic steps, carried out by QueA and QueG, produce the final Q nucleoside (for recent review, see ref. 9). In Archaea, the tRNA guanosine (15) transglycosylase enzyme (aTGT; EC 2.4.2.48) is homologous to the bTGT enzyme and exchanges the G at position 15 with preQ0 in nearly all tRNAs (15, 16). The preQ0 is then modified to G+ by different types of amidotransferases [archaeosine synthase (ArcS), QueF-like, and glutamine amidotransferase class-II (GAT)-QueC] (17, 18) (Fig. 1A). Although the bTGT and aTGT recognize a guanosine at different positions of the tRNA and use different substrates, key residues involved in base exchange and in zinc binding are conserved (19) (Fig. 2A). In addition, signature residues involved in the differences in 7-deazaguanosine substrate recognition between the bTGT and aTGT enzymes have been identified (19) (Fig. 2B). The role of 7-deazaguanosine derivatives as precursors of modified bases in tRNAs and of secondary metabolites is well established (9), and the preQ0 molecule itself was recently found to have anticancer properties (20). These modified bases can also be detected in rRNA in vivo if labeled preQ1 is fed to Escherichia coli (21) or inserted in DNA in vitro with the bTGT enzyme (22), but the biological relevance of these last two observations is not clear.
Fig. 2.
Comparison of TGT and TgtA5 proteins. (A) Schematic representation of the domain architecture and arrangement of TgtA5 proteins and bacterial and archaeal TGT proteins (bTGT and aTGT, respectively). The numbering of the upper and lower logos refers to the S. Montevideo TGT and TgtA5 sequences, respectively. Sequence logos in dashed boxes show the two conserved Asp residues of TgtA5 and the zinc binding sites of bTGT (Top) and TgtA5 (Bottom). C1 and C2 represent C-terminal domains unique to aTGT (17, 19). (B) Model and alignments of proposed substrate-binding pocket of TgtA5. The aligned cartoon representation (Top) of the pockets of S. Montevideo TgtA5 and P. horikoshii aTGT (PDB ID code 1IT8) was produced by PyMol (version 1.3). The catalytic residues of aTGT, ASP95, VAL197, VAL198, and ASP249 (red) (18) and their TgtA5 counterparts ASP95, MET208, VAL209, and ASP256 (cyan) are indicated in stick models. Dashed lines among stick models indicate the catalytic residues interacting with preQ0. Sequence alignment (Bottom) of select aTGT, bTGT, and TgtA5 proteins was performed using MUSCLE (53). Dots indicate regions intentionally deleted for this figure. Dashes indicate gaps in the sequence alignment. UniProt IDs for proteins included in multiple alignment are as follows: S. Montevideo TgtA5, E7V8J4; F. balearica TgtA5, E1SVY3; S. alaskensis TgtA5, Q1GPS0; Comamonas testosteroni TgtA5, H1RRG1; K. radiotolerans TgtA5, A6WGA1; E. coli bTGT, P0A847; Z. mobilis bTGT, Q8GM47; Shigella flexneri bTGT, Q54177; Bacillus subtilis bTGT, L8AMH3; Aquifex aeolicus bTGT, O67331; P. horikoshii aTGT, O58843; Methanococcus aeolicus aTGT, A6UVD8; Thermoplasma volcanium aTGT, Q977Z3; Picrophilus torridus aTGT, Q6L1W3; Ferroplasma acidarmanus aTGT, S0AQ23.
In the computational analysis of DNA modification systems by Iyer et al., the presence of divergent tgt-like genes and preQ0 genes present in certain phage and bacteria clustering with ParB-like proteins and several families of helicases led the authors to hypothesize that these gene clusters encode a DNA modification system (7). Our own analysis of the distribution and physical clustering of tgt and preQ0 synthesis genes, described here, led us to a similar hypothesis that a preQ0 derivative would be found in DNA in specific bacteria and phages. This hypothesis was validated by the analysis of the DNA of organisms either possessing or lacking the gene cluster, leading to the discovery of complex modifications of DNA that had escaped identification to date and could be among the most chemically elaborate DNA modification systems found in nature thus far.
Results
A Bacterial TGT Variant, TgtA5, Must Be Involved in a PreQ0-Dependent Pathway Different from Q Synthesis.
Analysis of the distribution of all Q synthesis genes in bacteria was performed using the “dpd cluster” subsystem in the SEED database (23). Analysis of ∼12,000 bacterial genomes showed that some organisms, such as pathogenic strains of E. coli (strain E22) or Salmonella enterica subsp. enterica serovar Montevideo, contained two homologs of the tgt gene, whereas most other sequenced E. coli or Salmonella species contained only one (SI Appendix, Table S1). Synteny analysis revealed that one of the two tgt genes clustered with the Q synthesis gene queA and encoded the experimentally characterized bTGT enzyme (24) (SI Appendix, Figs. S1 and S2A), whereas the other, that we named tgtA5 (later changed to dpdA), was found in a different neighborhood context (Fig. 1B). Homologs of tgtA5 were found in nearly 284 complete prokaryotic genomes (SI Appendix, Table S1 and Fig. S10 A and B), and the TgtA5 proteins possess divergent features from the bTGT that inserts preQ1 at position 34 of tRNA (Fig. 2). Members of the TgtA5 family are larger proteins (average of ∼450 aa instead of ∼300 aa for bTGT), and only the core of TgtA5 shows significant similarity to the bTGT and aTGT enzymes (Fig. 2B). The key residues that catalyze the G exchange (Asp102 and Asp280 of Zymomonas mobilis bTGT and Asp95 and Asp249 of Pyrococcus horikoshii aTGT) (19), as well as the Zinc binding site (CXCXXCX22H motif), are conserved in TgtA5 (Fig. 2B). Analysis of the substrate binding pocket suggests that TgtA5 binds preQ0, like the aTGTs. The critical residues for preQ0 binding by aTGT are GVVPL[LM] at positions 196–201 of the P. horikoshii enzyme, differing from the bTGT preQ1 binding pocket residues GLAVGE at position 230–235 of the Z. mobilis enzyme (19). Alignments of TgtA5 sequences with bTGT and aTGT showed the binding pocket residues resembled aTGT more than bTGT (G[ML]VPL[KR] in TgtA5; Fig. 2B). Modeling of the S. Montevideo TgtA5 protein with the aTGT structure with preQ0 in the binding pocket demonstrated the similar placement of these binding pocket residues compared with the aTGT (Fig. 2B), supporting our hypothesis that TgtA5 binds preQ0. Finally, tgtA5 clusters with the preQ0 synthesis genes in 94% of analyzed genomes, but it does not cluster with queF, suggesting that the substrate is preQ0 or a derivative, and not preQ1. Sequence and genome context analyses predict that TgtA5 recognizes preQ0 as a substrate like the aTGT enzymes, but because TgtA5 proteins lack the tRNA binding PUA domain found in aTGTs (25), analyses strongly suggest that TgtA5 proteins do not target tRNAs (Fig. 2A). Also, tgtA5 genes are found in organisms that lack the canonical Q synthesis gene tgt, such as Kineococcus radiotolerans (SI Appendix, Table S1), and we confirmed that K. radiotolerans lacked Q and preQ0 in tRNA (SI Appendix, Fig. S1A). Finally, the tgt gene was deleted in the S. Montevideo strain that contained both a tgt and a tgtA5 gene, and tRNA extracted from the ∆tgt strain (YYF3020) lacked Q (SI Appendix, Figs. S1B and S2), confirming that TgtA5 is not involved in incorporation of Q in tRNA.
Strong physical clustering was observed between tgtA5 with homologs of a DndB-like protein that is involved with the PT modification of DNA in bacteria (5) (Fig. 1B and SI Appendix, Table S1). These dndB-like genes were present in 123 of 134 (92%) tgtA5 gene clusters analyzed, and in 21 clusters (15%), there were two distinct copies of this gene flanking the tgtA5 gene (Fig. 1B and SI Appendix, Table S1 and Fig. S4). According to HHPred analysis and previous work by Iyer et al. (7), DndB proteins contain a domain belonging to the superfamily of ParB nucleases involved in chromosome and plasmid partitioning (26, 27), suggesting a role in DNA recognition and binding. Studies have shown that DndB negatively regulates the PT modification (28–30) by binding to the promoter region and regulating expression of the DndBCDE operon (30). The S. Montevideo DndB-like protein (renamed DpdB) has 23% amino acid identity to the DndB protein of S. Cerro. Despite the low similarity, several residues are conserved among the PT-related DndB and TgtA5-clustered DndB-like proteins, including a QR doublet near the N terminus and a FXXXN motif near the middle of the sequence. Most striking, however, is the strictly conserved DGQQR motif in nearly all of the TgtA5-clustered DndB-like proteins, which differs by one residue from the DGQHR motif conserved among the DndB proteins involved in PT modification (SI Appendix, Fig. S3A). Although the PT-related DndB and the TgtA5-clustered DndB-like (DpdB) proteins share conserved motifs, they separate on a phylogenetic tree (SI Appendix, Fig. S3B), indicating that they comprise two subfamilies of the DndB-like family.
Strong physical clustering was also observed with tgtA5 and several other genes (Fig. 1 and SI Appendix, Figs. S4 and S5). Two genes of unknown function, which we call dpdC and dpdD, were present in 88% and 90% of the clusters analyzed, respectively. DpdC was predicted to encode a DUF328 domain-containing protein, and DpdD has little similarity to any known protein, although a small portion of the C terminus matched a DUF2325 domain (SI Appendix, Fig. S5 and Table S2). In 98% of the bacteria analyzed, the tgtA5 genes also clustered with several other putative DNA-binding enzymes, including a member of the DEAD/DEAH box helicase family, a SNF2-type helicase, and a RecQ-like Superfamily II DNA helicase (Fig. 1 and SI Appendix, Figs. S4 and S5 and Tables S1 and S2). This grouping allowed us to speculate that TgtA5 is involved in introducing preQ0-like modifications in DNA, a hypothesis previously proposed by Aravind and colleagues (7).
Nine of the analyzed genomes contained much larger TgtA5 proteins (∼700 aa). The TgtA5 domain of these proteins is similar to the rest of the TgtA5 family. The N-terminal half of the protein contains a DUF328 domain, like the one present in the DpdC of the S. Montevideo cluster. The genomic context of the longer tgtA5 genes includes preQ0 synthesis genes, similar to the other tgtA5 genes; however, one noticeable difference is the absence of some or all of the three genes conserved in the other clusters, the dndB-like dpdB, and dpdC and dpdD (see the Meiothermus chliarophilus DSM 9957 cluster in SI Appendix, Fig. S4 and others in SI Appendix, Fig. S6). Some of the long tgtA5 clusters encode a similar SNF2-type helicase, RecQ-like helicase, phospholipase-like domain-containing protein, and a DpdD, whereas others have other putative DNA-binding proteins and helicases, suggesting that the long TgtA5 protein is also involved in introducing a modification into DNA in these organisms.
The tgtA5 Cluster Is Responsible for the Insertion of PreQ0 and of 7-Amido-7-Deazaguanine in DNA.
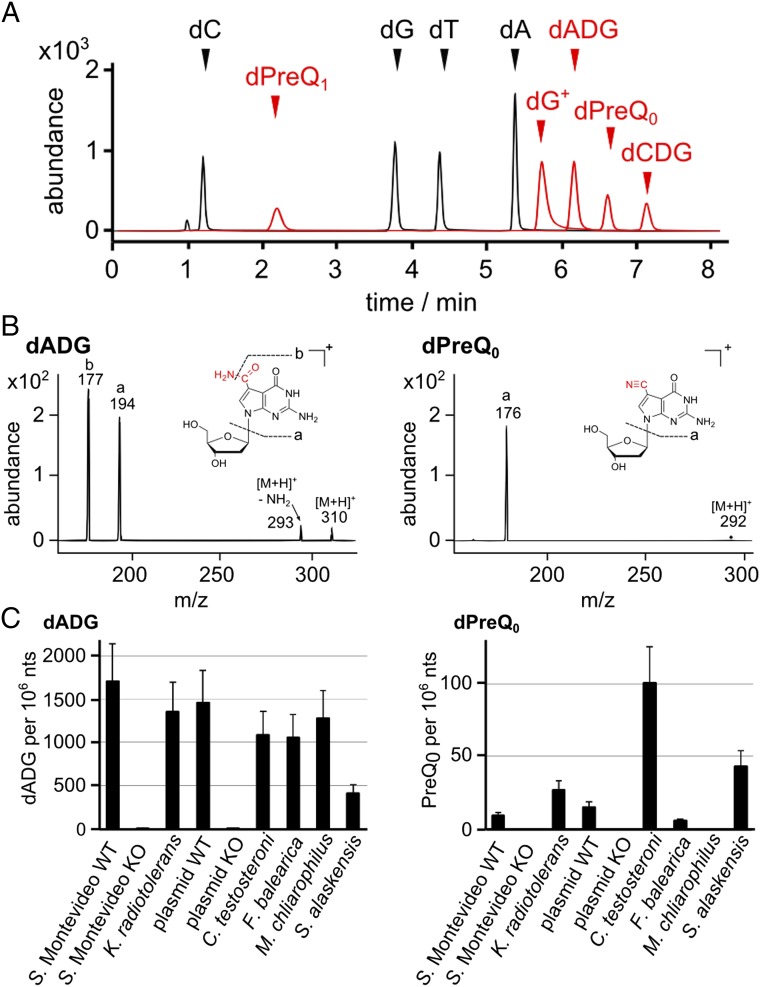
To test the hypothesis that 7-deazaguanine derivatives were inserted in the DNA of organisms that encode the tgtA5 cluster, a mass spectrometry-based approach was used to analyze DNA from two closely related Gram-negative bacteria possessing and lacking this gene cluster (S. Montevideo and S. Typhimurium LT2, respectively) and from the Gram-positive bacteria K. radiotolerans that also encodes the cluster (Fig. 1B). The strategy for discovering the 2’-deoxynucleosides was based on an initial presumption of the presence of 2’-deoxynucleosides containing any of the six 7-deazaguanine nucleobase structures formed in the tRNA queuosine biosynthetic pathway shown in Fig. 1A: 2’-deoxyCPH4, 2’-deoxyCDG, 2’-deoxyQ, 2’-deoxypreQ0, 2’-deoxypreQ1, and 2’-deoxyG+. A search for each candidate 2’-deoxynucleoside was conducted by neutral loss analysis mass spectrometry, in which product ions resulting from loss of a 2-deoxyribose during collision-induced dissociation could be traced back to the original 2’-deoxynucleoside eluting from the HPLC column at a specific retention time. In both S. Montevideo and K. radiotolerans, small amounts of putative 2’-deoxypreQ0 (dPreQ0) and a stronger signal for putative 2’-deoxyCDG (dCDG) were detected (SI Appendix, Fig. S7). Subsequent structural analysis revealed that the prediction of dCDG was incorrect and that the signal at m/z 311 was actually the M+1 isotopomer for a 2’-deoxy-7-amido-7 deazaguanosine (dADG; Fig. 3B). The identities of dPreQ0 and dADG were established by fragmentation analysis using high mass-accuracy quadrupole time-of-flight (QTOF) mass spectrometry (SI Appendix, Fig. S8) and by comparison with synthetic standards. Standards were also used to rule out detectable levels of dPreQ1 and 2’-deoxyArch (dG+) (SI Appendix, Fig. S8A). Using these standards, the optimal mass transitions (Fig. 3B and SI Appendix, Fig. S8 B–D) and retention times (SI Appendix, Fig. S9) of the modified 2’-deoxynucleosides were determined. Quantitative analysis by external calibration revealed ∼1,600 dADG modifications per 106 nt in S. Montevideo and ∼1,300 per 106 nt in K. radiotolerans (Fig. 3C). dPreQ0 levels were found to be significantly lower at 10 and 30 dPreQ0 per 106 nt, respectively (Fig. 3C). These results suggest that dADG is the main product of tgtA5 cluster, with dPreQ0 appearing as a side product.
Fig. 3.
Detection and quantification of 2′-deoxy-7-deazaguanosine derivatives by LC–MS/MS. (A) The LC–MS/MS analytical method is illustrated with an extracted ion chromatogram showing the HPLC retention of the various 7-deazaG–modified (red) and canonical (black) 2’-deoxynucleosides. Abundance denotes arbitrary units of signal intensity. (B) MS/MS fragmentation patterns for synthetic dADG and dPreQ0. Abundance denotes arbitrary units of signal intensity. (C) Detected quantities in DNA samples of various bacterial species displayed as modification per 106 nucleotides.
To confirm the role of the tgtA5 cluster in the insertion of dPreQ0 and dADG in DNA, a S. Montevideo derivative (YYF3022) with a 21-kb deletion eliminating nearly the entire cluster was constructed (Fig. 1B and SI Appendix, Fig. S2). Both the dPreQ0 and dADG modifications were absent in genomic DNA extracted from YYF3022 (Fig. 3C).
This discovery led us to rename the genes of the S. Montevideo cluster as dpdA-K (Fig. 1B), with dpd standing for 7-deazapurine in DNA.
The dpd Cluster Is Horizontally Transferred and Found in Genomic Islands.
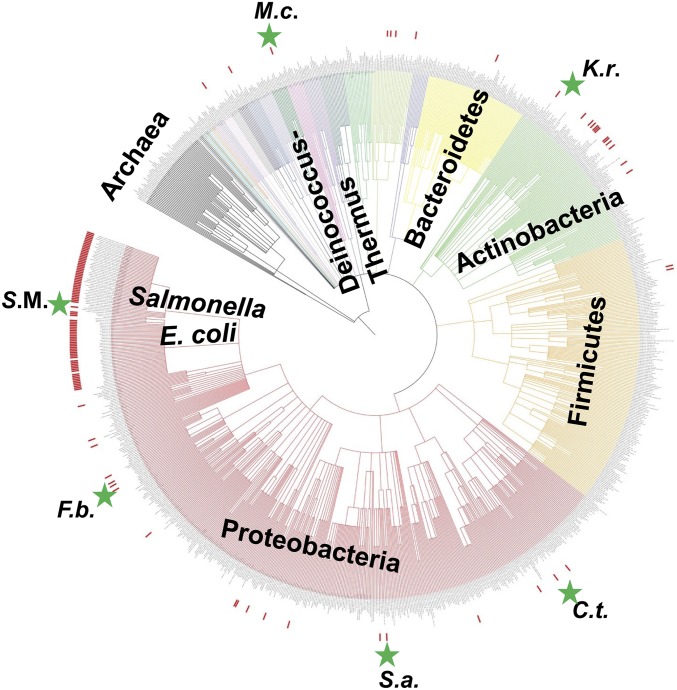
Analysis of the taxonomic distribution of the dpdA (tgtA5) gene (Fig. 4) showed that this gene is evenly distributed along the bacterial tree, suggesting either an ancestral origin in Bacteria accompanied by massive independent losses along the diversification of Bacteria or a more recent origin with propagation through horizontal gene transfers (HGTs). The phylogenetic analysis of the DpdA/TgtA5 homologs and the discrepancies observed between the topology of the resulting Bayesian and maximum likelihood DpdA/TgtA5 trees with the currently recognized systematics (compare Fig. 4 with SI Appendix, Fig. S10 A and B), such as the nonmonophyly of Gammaproteobacteria or Archaea or the robust grouping of Herbaspirillum massiliense (Betaproteobacteria) with Spirosoma spitsbergense (Bacteroidetes) and a Verrucomicrobia bacterium but not with other Betaproteobacteria (bootstrap value, 100%; posterior probability, 1.00; SI Appendix, Fig. S10 A and B, respectively), strongly favor the HGT hypothesis.
Fig. 4.
Taxonomic distribution of TgtA5. Taxonomic tree of ∼1,000 representative prokaryotes generated using iToL. Red bars indicate the presence of tgtA5 in species. Stars indicate organisms for which preQ0 and/or ADG were detected in DNA. C.t., C. testosteroni; F.b., F. balearica; K.r., K. radiotolerans; M.c., M. chliarophilus; S.a., S. alaskensis; S.M., S. Montevideo.
To confirm that the presence of dpdA is diagnostic of the presence of preQ0 and ADG in DNA, we analyzed the genomic DNA from a diverse set of organisms harboring the cluster (SI Appendix, Fig. S4 and Fig. 4). As shown in Fig. 3C, M. chliarophilus, Comamonas testosteroni, Sphingopyxis alaskensis, and Ferrimonas balearica all harbor dADG in DNA but in different quantities. Unlike the other strains analyzed, the DNA from M. chliarophilus contained only ADG and no detectable preQ0 (Fig. 3C). M. chliarophilus was the only strain tested with a long version of the dpdA and no dpdBCD genes (SI Appendix, Fig. S4).
In the S. Montevideo and E. coli strains that harbor the dpd cluster, it is inserted adjacent to the LeuX locus (SI Appendix, Fig. S11), a region that had been previously identified as highly variable (31). A pair of 19-bp direct repeats flanking the dpd cluster was identified (SI Appendix, Fig. S11), again indicative of a genomic island (32, 33). More recently, the sequencing of a large number of Salmonella strains confirmed this region as a novel variable island (SGI2) that contained different types of restriction systems, toxin/antitoxin modules, and mobile elements (34). Of the sequenced S. Montevideo strains analyzed, 92% contain the dpdA cluster at the SGI2 position.
Unlike the dpd islands of S. Montevideo and E. coli, neither tRNA genes nor direct repeats were identified in the region surrounding the K. radiotolerans dpd cluster. However, the region is flanked by mobile element proteins (e.g., a transposase-like protein and resolvase protein), and the IslandViewer software (35) identified this cluster as a genomic island. Additionally, the GC content of the ORFs is lower in this region compared with the rest of the genome (66% GC vs. 74% GC).
The dpd Cluster of S. Montevideo Encodes an R–M System.
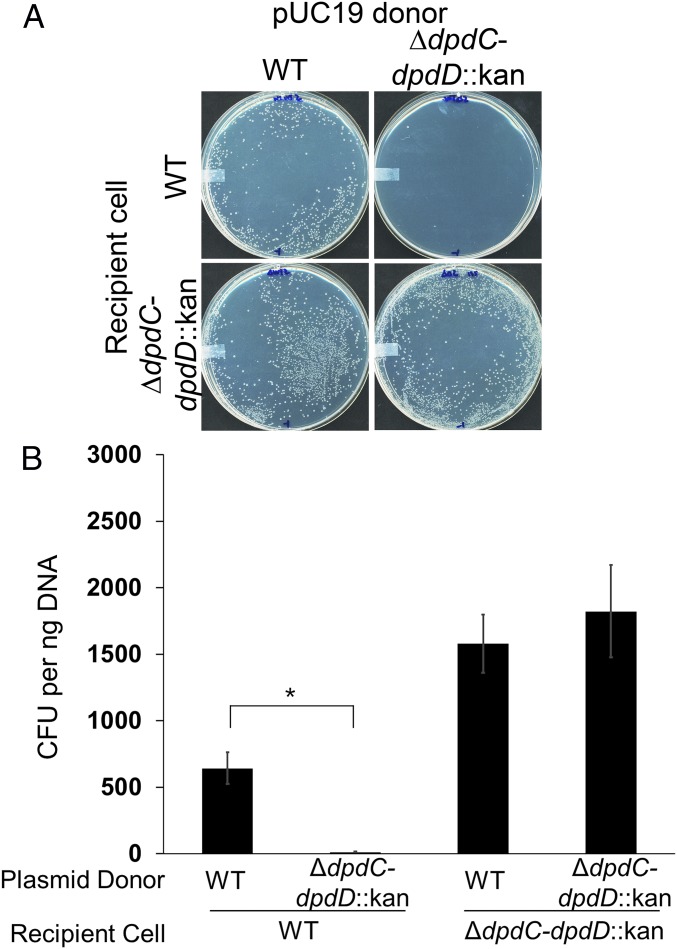
The discovery of a horizontally transferred DNA modification cluster logically suggested a potential role as a novel R–M system. To test this hypothesis, we compared the transformation efficiencies of the isogenic strains S. Montevideo WT and YYF3022 lacking the dpd cluster. pUC19 was propagated in each strain, and following extraction, 10 ng of the plasmid DNA was used to transform the WT and YYF3022 strains by electroporation. As shown in Fig. 5, pUC19 DNA extracted from YYF3022 transformed the WT strain with 100-fold less efficiency than pUC19 DNA extracted from the WT strain, indicating the unmodified plasmid is restricted in the WT host. The pUC19 extracted from YYF3022 transformed the YYF3022 strain with about 1,000-fold higher efficiency than the WT strain, suggesting that one or more of the genes in the dpd cluster was responsible for this restriction. No difference was seen with the plasmid extracted from the WT strain. Liquid chromatography (LC)–MS/MS analysis confirmed that dPreQ0 and dADG were present in the pUC19 extracted from S. Montevideo WT and not the mutant strain (Fig. 3C).
Fig. 5.
Transformation efficiency of modified and unmodified pUC19 DNA. (A) S. Montevideo WT and YYF3022 (ΔdpdC-dpdD::kan) transformed with 10 ng pUC19 extracted from either WT (modified) or ΔdpdC-dpdD::kan (unmodified) on LB agar plates containing ampicillin. (B) Transformation efficiencies of modified versus unmodified pUC19 in WT and ΔdpdC-dpdD::kan. Transformation efficiency per 1 ng DNA was calculated per 106 viable cfu. The average of three experiments is shown, with error bars representing SE (*P < 0.05, two-tailed Student’s t test).
PreQ0 Derivatives in Phage.
R–M systems and genomic islands are often transferred through phage transduction. Several examples of phage-encoded tgt-like genes and preQ0 genes have already been reported in the literature, including Mycobacteriophage Rosebush (36), Streptococcus phage Dp-1 (37), and the E. coli phage 9g (38) (Fig. 6A and SI Appendix, Fig. S12). In the characterization of phage 9g, Kulikov et al. (38) speculated that the restriction endonuclease-resistant nature of the phage DNA suggested it was heavily modified, and they proposed that tgt and preQ0 synthesis genes were involved in inserting Q into the DNA. To evaluate the prevalence of tgt paralogs and PreQ0 synthesis genes in phages, a similarity-based search was performed on all available phage genomes in the National Center for Biotechnology Information database. This revealed 36 bacteriophages and two archaeal viruses that encode a Tgt-like protein (SI Appendix, Table S5). Multiple sequence alignments of the phage Tgt-like proteins allowed the identification of catalytic residues (two conserved Asp residues) and of the preQ0-binding pocket (SI Appendix, Fig. S13) (19). The zinc-binding residues that are conserved in the aTGT, bTGT, and TgtA5 families were not found in the phage homologs; however, a His residue (H196 of phage 9g Tgt-like protein) is conserved specifically in the phage enzymes.
Fig. 6.
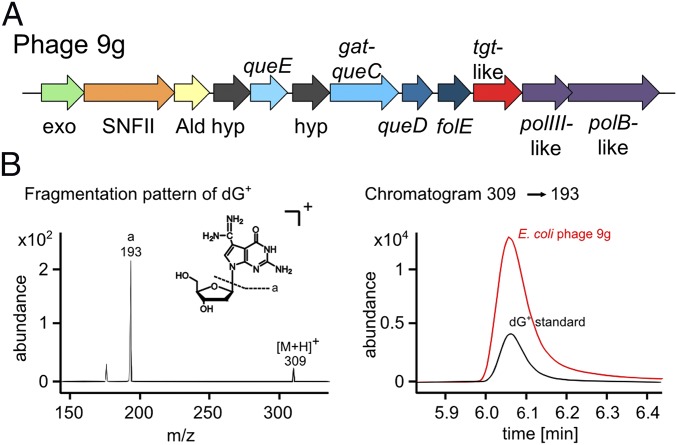
Phage 9g DNA is modified with dG+. (A) Gene cluster of phage 9g (NC_024146) tgt-like. Ald, aldolase; exo, Cas4-like exonuclease; hyp, hypothetical; polB-like, DNA polymerase B; polIII-like, DNA polymerase β-subunit; SNFII, superfamily II-like helicase. (B) MS/MS fragmentation pattern of synthetic dG+ (Left) used for subsequent detection of dG+ in E. coli phage 9g DNA (Right).
The preQ0 biosynthesis pathway genes (folE, queD, queE, and queC) were identified in 16 of Tgt-containing phages (SI Appendix, Table S5), three of which contained a homolog of Gat-QueC that is involved in the synthesis of archaeosine in some archaea (18). Two of the phages harbored a QueF homolog involved in preQ1 synthesis. Finally, one phage, phi13:1, encoded the three first genes of the preQ0 synthesis pathway (folE, queD, and queE) but no tgt homolog (SI Appendix, Table S5).
The genomic contexts of the phage preQ0/tgt clusters are different from those found in bacteria, however many of them encode DNA processing enzymes and could therefore insert 7-deazaguanosine derivatives in DNA. Some of these phages encode a homolog of ParB, an enzyme important for DNA binding and segregation (26), as previously pointed out by Aravind’s group (7) (e.g., Mycobacteriophage Rosebush). Other phages encode potential helicases and nucleases near the preQ0 cluster [e.g., Gp11 of 9g and Gp39 of JenKI contain SnfII-like domains (39), Gp11 of Dp-1 is a RecU-like protein (40), Gp40 of JenK1 encodes a putative exonuclease (41), and Gp10 of Dp-1 is a Cas4-like protein (42); SI Appendix, Fig. S12]. The presence of these nucleases is indicative of possible defense systems. The nature of the exact modification might differ with the specific phage, as the preQ0 pathway is found in Rosebush or BCD7, the preQ1 pathway in Dp-1, and the archaeosine pathway in 9g (SI Appendix, Table S5).
To test the hypothesis that some of these phages contained preQ0 derivatives, DNA from phage 9g, a phage predicted to insert archaeosine because of the presence of the gat-queC gene, was isolated and subjected to LC–MS/MS analysis as described above. As expected, 2’-deoxy-archaeosine (dG+) was found in the phage DNA (Fig. 6B). dG+ quantities were extremely high, allowing quantification by both MS/MS and UV analysis that revealed a conversion of dG to dG+ by 25% and 27%, respectively.
Discussion
The discovery of 7-deazaG derivatives in DNA of diverse bacteria and phages is a compelling example of the power of coupling in silico predictive approaches with bioanalytical validation (43, 44). These modifications would not have been identified if we had not purposely looked for them in very specific organisms. It is also another unexpected example of the crosstalk between RNA and DNA metabolism and of the strong evolutionary links found between RNA and DNA modifying enzymes (1). This crosstalk is advantageous in allowing plasticity in evolution of nucleic acid modification enzymes but also poses specificity problems that could lead to erroneous modification of a nontarget nucleic acid. It is poorly understood how bTGT enzymes, which, unlike aTGT proteins, lack any RNA-binding domains, recognize their RNA targets and also modify DNA in artificial situations. It will be interesting to understand if the TgtA5/DpdA family has evolved specific DNA-binding recognition or if they require the help of accessory proteins in the cluster.
We have clearly demonstrated that the DpdA-containing genomic islands are involved in inserting 7-deazapurine derivatives in DNA; however, the roles of the specific genes in the cluster and mechanism for modification are still to be elucidated. The well-characterized preQ0 synthesis genes, either encoded in the cluster or elsewhere on the chromosome, produce the ADG and preQ0. It was recently shown that ADG is an intermediate in the QueC-mediated reaction from CDG to preQ0 (45) (Fig. 1). ADG is the result of amidation of CDG, a reaction that occurs more rapidly compared with the subsequent dehydration to preQ0. DpdA is most likely involved in exchanging guanine in DNA for ADG or preQ0 in a base exchange reaction similar to bTGT and aTGT, but biochemical studies are required to test this hypothesis and determine if it can do it alone or if it requires other proteins encoded within the cluster. Because the majority of the detected modifications were dADG, it is likely that ADG is the preferred substrate of DpdA, and dPreQ0 may be present as the result of nonspecific insertion by DpdA. The presence of only dADG, and not dPreQ0, in the DNA of M. chliarophilus suggests that its DpdA has stricter substrate specificity. The DpdA of M. chliarophilus is a larger protein, with an additional 300 amino acids at the N terminus, and the substrate binding pocket of the longer DpdA proteins has a slightly different sequence motif (GGLAR vs. GGMVP of other DpdAs). The LA residues resemble the bTGT preQ1 binding pocket rather than the aTGT preQ0 pocket and may confer specificity for ADG.
The dpdB gene encodes a member of the DndB-like family. The DndB proteins appear to have a role in regulation of the PT modification of DNA. Deletion of dndB homologs in Streptomyces lividans and Salmonella Cerro led to increased PT modification (28, 29, 46). A recent study revealed that DptB (DndB homolog) of S. Cerro binds to the promoter region and negatively regulates the expression of the DptBCDE operon (30). It is possible that the DndB-like DpdB protein has a similar role in regulating the expression of the dpd modification genes.
The roles of DpdC and DpdD remain a mystery, and experimental studies are now underway. Very few hints as to a function have been derived from bioinformatics analyses. Although HHpred analysis identified a DUF328 domain present in DpdC, this domain has no known function. In a computational analysis of potential DNA modification clusters, Iyer et al. (7) predicted the DpdC protein to have an activity similar to ArcS, the amidotransferase that modifies preQ0 to archaeosine, based on long-range similarities. In our analysis of bacterial DNA, we did not detect archaeosine. Based on gene clustering, we predict that DpdC may be important in insertion of the modification, as it nearly always encoded adjacent to dpdA and/or dpdB. As for DpdD, only 90 residues of DpdD match a DUF2325 domain of unknown function, and we believe this is a completely novel protein family. The dpdD gene is present alongside the helicase-like genes and putative DNA-binding genes, leaving the possibility open that DpdD could be involved in the restriction system.
If these genomic islands do indeed encode R–M systems, as suggested by the reduced transformation efficiency of plasmids lacking the modification, the nucleases catalyzing cleavage of the unmodified DNA remain a mystery. Bioinformatic analyses of the genes in the clusters provided several candidates, although none stand out as a true endonuclease. We are currently working to elucidate the protein(s) involved in the restriction.
Finally, the discovery of archaeosine in DNA of phage 9g is another demonstration of the spectrum of DNA modifications that could occur in nature. It is unclear if the archaeosine modification of 9g is part of a restriction system. No restriction enzyme has been identified, although the SnfII-like protein encoded near the modification genes (Fig. 2) could be a potential candidate. Another role for this modification could be to provide resistance to restriction systems, essentially acting as an antirestriction system (47). The phage 9g DNA is resistant to most restriction enzymes tested (38), suggesting the presence of modified bases inhibits recognition or cleavage by these enzymes.
Further studies are needed to elucidate the roles of the 7-deazaguanine derivatives in bacterial and phage DNA, with potential functions varying among R–M systems, antirestriction systems, epigenetic marks, and unforeseen protective roles, as these modifications were found in organisms like K. radiotolerans that can resist radiation stress (48). We foresee that the molecular characterization of the enzymes involved in the synthesis, recognition, and cleavage of 7-deazaguanine derivatives in DNA could open the door to both biotechnological and antibacterial applications.
Materials and Methods
Bioinformatic Analyses.
Taxonomic distribution and physical clustering analysis of tgtA5 and preQ0 synthesis genes was performed on the public SEED server (pubseed.theseed.org/SubsysEditor.cgi) (23, 49). Results of the analysis are available in the dpd cluster subsystem and summarized in SI Appendix, Table S1. The taxonomic distribution of tgtA5 was then visualized using the Interactive Tree of Life (iToL, itol.embl.de) (50). Further details on all bioinformatic analyses can be found in SI Appendix, SI Materials and Methods.
Strains, Media, and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S3. S. Montevideo strains were routinely grown in LB (Tryptone 10 g/L, yeast extract 5 g/L, NaCl 5 g/L) at 37 °C. All other strains were grown in media and conditions as detailed in SI Appendix, SI Materials and Methods. S. Montevideo deletion constructs were made using the linear recombination method described by Datsenko and Wanner (51). Oligonucleotides used for deletion and confirmation of mutants are listed in SI Appendix, Table S4. Further details can be found in SI Appendix, SI Materials and Methods.
Plasmid Restriction Test.
Restriction of the plasmid pUC19 was tested as described in ref. 52 and is detailed in SI Appendix, SI Materials and Methods.
DNA Preparation.
Total DNA was extracted from bacteria and phage with phenol-chloroform followed by alcohol precipitation, as detailed in SI Appendix, SI Materials and Methods.
DNA Analysis.
DNA was enzymatically hydrolyzed to 2’-deoxynucleosides as described in SI Appendix, SI Materials and Methods. Modified 2’-deoxynucleosides were initially detected by LC–MS/MS, with subsequent structural corroboration of dPreQ0 and dADG by LC–QTOF. Quantification of dPreQ1, dPreQ0, dADG, dCDG, and dG+ was achieved by LC–MS/MS using external calibration curves. Details can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Rémi Zallot, Zdeněk Paris, and Juan Alfonzo for help with the determination of the presence of queuosine in tRNA; Sophie Alvarez for LC–MS/MS analyses of tRNA; Anne Oudard for sharing her local database of complete and ongoing genome projects; Max Teplitski for the S. Montevideo strain; and Paul Gulig and Kelly Rice for use of laboratory facilities. Mass spectrometric analyses were performed in the Bioanalytical Facilities Core of the MIT Center for Environmental Health Sciences, which is supported by National Institute for Environmental Health Sciences Grant ES002109. This work was supported by the National Institutes of Health Grant GM70641 (to V.d.C.-L.), Deutsche Forschungsgemeinschaft (S.M.K.), French National Agency for Research Grant ANR-10-BINF-01-01 and the Institut Universitaire de France (to C.B.-A.), Russian Scientific Foundation Grant RSF 15-15-00134 (to A.V.L.; this grant supported the work on phage 9g and preparation of its DNA and A.V.L.’s work on data analysis and writing), and the National Research Foundation of Singapore through its Singapore-MIT Alliance for Research and Technology (P.C.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518570113/-/DCSupplemental.
References
- 1.Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience; Austin, TX: 2009. pp. 1–18. [Google Scholar]
- 2.Warren RA. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 3.Ekanayake DK, et al. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol Cell Biol. 2011;31(8):1690–1700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA. 2011;108(7):2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3(11):709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 6.Cao B, et al. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Mol Microbiol. 2014;93(4):776–785. doi: 10.1111/mmi.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer LM, Zhang D, Burroughs AM, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41(16):7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46(1):69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 9.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips G, et al. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: A new role for GTP cyclohydrolase I. J Bacteriol. 2008;190(24):7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J Biol Chem. 2004;279(8):6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 12.McCarty RM, Somogyi A, Lin G, Jacobsen NE, Bandarian V. The deazapurine biosynthetic pathway revealed: In vitro enzymatic synthesis of PreQ0 from guanosine 5′-triphosphate in four steps. Biochemistry. 2009;48(18):3847–3852. doi: 10.1021/bi900400e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada N, Nishimura S. Isolation and characterization of a guanine insertion enzyme, a specific tRNA transglycosylase, from Escherichia coli. J Biol Chem. 1979;254(8):3061–3066. [PubMed] [Google Scholar]
- 14.Van Lanen SG, et al. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc Natl Acad Sci USA. 2005;102(12):4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai Y, Fox DT, Lacy JA, Van Lanen SG, Iwata-Reuyl D. Hypermodification of tRNA in Thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J Biol Chem. 2000;275(37):28731–28738. doi: 10.1074/jbc.M002174200. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M, et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem. 1997;272(32):20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- 17.Phillips G, et al. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J Biol Chem. 2010;285(17):12706–12713. doi: 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips G, et al. Diversity of archaeosine synthesis in crenarchaeota. ACS Chem Biol. 2012;7(2):300–305. doi: 10.1021/cb200361w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stengl B, Reuter K, Klebe G. Mechanism and substrate specificity of tRNA-guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism. ChemBioChem. 2005;6(11):1926–1939. doi: 10.1002/cbic.200500063. [DOI] [PubMed] [Google Scholar]
- 20.Xu D, et al. PreQ0 base, an unusual metabolite with anti-cancer activity from Streptomyces qinglanensis 172205. Anticancer Agents Med Chem. 2015;15(3):285–290. doi: 10.2174/1871520614666141027144653. [DOI] [PubMed] [Google Scholar]
- 21.Brooks AF, Vélez-Martínez CS, Showalter HD, Garcia GA. Investigating the prevalence of queuine in Escherichia coli RNA via incorporation of the tritium-labeled precursor, preQ1. Biochem Biophys Res Commun. 2012;425(1):83–88. doi: 10.1016/j.bbrc.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 22.Nonekowski ST, Kung FL, Garcia GA. The Escherichia coli tRNA-guanine transglycosylase can recognize and modify DNA. J Biol Chem. 2002;277(9):7178–7182. doi: 10.1074/jbc.M111077200. [DOI] [PubMed] [Google Scholar]
- 23.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J Biol Chem. 1982;257(11):6544–6550. [PubMed] [Google Scholar]
- 25.Sabina J, Söll D. The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation. J Biol Chem. 2006;281(11):6993–7001. doi: 10.1074/jbc.M512841200. [DOI] [PubMed] [Google Scholar]
- 26.Surtees JA, Funnell BE. Plasmid and chromosome traffic control: How ParA and ParB drive partition. Curr Top Dev Biol. 2003;56:145–180. doi: 10.1016/s0070-2153(03)01010-x. [DOI] [PubMed] [Google Scholar]
- 27.Grohmann E, Stanzer T, Schwab H. The ParB protein encoded by the RP4 par region is a Ca2+-dependent nuclease linearizing circular DNA substrates. Microbiology. 1997;143(Pt 12):3889–3898. doi: 10.1099/00221287-143-12-3889. [DOI] [PubMed] [Google Scholar]
- 28.Xu T, et al. DNA phosphorothioation in Streptomyces lividans: Mutational analysis of the dnd locus. BMC Microbiol. 2009;9:41. doi: 10.1186/1471-2180-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang J, et al. DNA modification by sulfur: Analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res. 2007;35(9):2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He W, et al. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci Rep. 2015;5:12368. doi: 10.1038/srep12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop AL, et al. Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNA(leuX) J Bacteriol. 2005;187(7):2469–2482. doi: 10.1128/JB.187.7.2469-2482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilde C, et al. Delineation of the recombination sites necessary for integration of pathogenicity islands II and III into the Escherichia coli 536 chromosome. Mol Microbiol. 2008;68(1):139–151. doi: 10.1111/j.1365-2958.2008.06145.x. [DOI] [PubMed] [Google Scholar]
- 33.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 34.Moreno Switt AI, et al. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One. 2012;7(7):e41247. doi: 10.1371/journal.pone.0041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhillon BK, Chiu TA, Laird MR, Langille MG, Brinkman FS. IslandViewer update: Improved genomic island discovery and visualization. Nucleic Acids Res. 2013;41(Web Server issue):W129–W132. doi: 10.1093/nar/gkt394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedulla ML, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113(2):171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 37.Sabri M, et al. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent phage Dp-1. J Bacteriol. 2011;193(2):551–562. doi: 10.1128/JB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulikov EE, et al. Genomic sequencing and biological characteristics of a novel Escherichia coli bacteriophage 9g, a putative representative of a new Siphoviridae genus. Viruses. 2014;6(12):5077–5092. doi: 10.3390/v6125077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorbalenya AE, Koonin EV. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991;291(2):277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- 40.McGregor N, et al. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure. 2005;13(9):1341–1351. doi: 10.1016/j.str.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian K, Rutvisuttinunt W, Scott W, Myers RS. The enzymatic basis of processivity in lambda exonuclease. Nucleic Acids Res. 2003;31(6):1585–1596. doi: 10.1093/nar/gkg266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Kasciukovic T, White MF. The CRISPR associated protein Cas4 is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012;7(10):e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405(6788):823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 44.El Yacoubi B, de Crécy-Lagard V. Integrative data-mining tools to link gene and function. Methods Mol Biol. 2014;1101:43–66. doi: 10.1007/978-1-62703-721-1_4. [DOI] [PubMed] [Google Scholar]
- 45.Nelp MT, Bandarian V. A single enzyme transforms a carboxylic acid into a nitrile through an amide intermediate. Angew Chem Int Ed Engl. 2015;54(36):10627–10629. doi: 10.1002/anie.201504505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Q, et al. Regulation of DNA phosphorothioate modifications by the transcriptional regulator DptB in Salmonella. Mol Microbiol. 2015;97(6):1186–1194. doi: 10.1111/mmi.13096. [DOI] [PubMed] [Google Scholar]
- 47.Stern A, Sorek R. The phage-host arms race: Shaping the evolution of microbes. BioEssays. 2011;33(1):43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagwell CE, et al. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLoS One. 2008;3(12):e3878. doi: 10.1371/journal.pone.0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I, Bork P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38(20):7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.