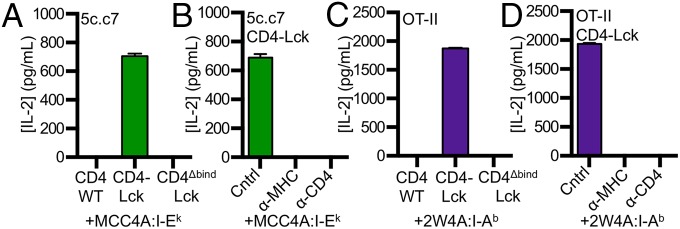
Significance
Most receptors co-evolve with specific ligands, whereas antibody–antigen pairing reflects interactions to a novel ligand (antigen) selected from a library of random antibodies. αβT-cell receptors (TCRs) lie somewhere in between. They are encoded by rearranged gene segments and screened for their ability to bind novel peptide fragments derived from foreign pathogens. But, unlike antibodies, they are restricted to recognizing peptides within allelic variants of related ligand scaffolds—MHCI and MHCII. It remains debated whether TCRs have an intrinsic specificity for MHC, like other receptor–ligand pairs, or if their recognition mirrors antibody–antigen interactions. This study presents functional evidence for TCRs interacting with MHC in an allele-independent and peptide sequence-independent manner, thus providing evidence for TCR-intrinsic recognition of MHC.
Keywords: TCR, MHC, restriction, CD4, Lck
Abstract
How T cells become restricted to binding antigenic peptides within class I or class II major histocompatibility complex molecules (pMHCI or pMHCII, respectively) via clonotypic T-cell receptors (TCRs) remains debated. During development, if TCR–pMHC interactions exceed an affinity threshold, a signal is generated that positively selects the thymocyte to become a mature CD4+ or CD8+ T cell that can recognize foreign peptides within MHCII or MHCI, respectively. But whether TCRs possess an intrinsic, subthreshold specificity for MHC that facilitates sampling of the peptides within MHC during positive selection or T-cell activation is undefined. Here we asked if increasing the frequency of lymphocyte-specific protein tyrosine kinase (Lck)-associated CD4 molecules in T-cell hybridomas would allow for the detection of subthreshold TCR–MHC interactions. The reactivity of 10 distinct TCRs was assessed in response to selecting and nonselecting MHCII bearing cognate, null, or “shaved” peptides with alanine substitutions at known TCR contact residues: Three of the TCRs were selected on MHCII and have defined peptide specificity, two were selected on MHCI and have a known pMHC specificity, and five were generated in vitro without defined selecting or cognate pMHC. Our central finding is that IL-2 was made when each TCR interacted with selecting or nonselecting MHCII presenting shaved peptides. These responses were abrogated by anti-CD4 antibodies and mutagenesis of CD4. They were also inhibited by anti-MHC antibodies that block TCR–MHCII interactions. We interpret these data as functional evidence for TCR-intrinsic specificity for MHCII.
Positive and negative selection limit the αβT-cell repertoire to cells expressing clonotypic T-cell receptors (TCRs) that distinguish the antigenicity of peptides embedded within class I and class II major histocompatibility complex molecules (pMHCI or pMHCII, respectively) based on their source of origin (i.e., self or foreign) (1–4). Approximately 7.5% of CD4+CD8+ double-positive (DP) thymocytes express TCRs that interact with self-pMHC above an affinity threshold required for positive selection, whereas 7.5% cross a higher affinity threshold that mediates negative selection and the remaining TCRs fail to direct positive selection (5). The rules that restrict TCR recognition of antigenic peptides within MHCI or MHCII are unresolved.
Two models have been proposed to explain MHC restriction. One posits that restriction is imposed by CD4 or CD8 during thymocyte development to eliminate TCRs that recognize non-MHC ligands (2, 6). Here, the CD4- and CD8-associated Src kinase, p56Lck [lymphocyte-specific protein tyrosine kinase (Lck)], is sequestered away from the immunoreceptor tyrosine-based activation motifs (ITAMs) of the TCR-associated CD3δε, CD3γε, and CD3ζζ signaling modules. Positively selecting signals are then generated in thymocytes expressing TCRs that bind MHCII or MHCI together with CD4 or CD8, respectively, as this localizes Lck to the ITAMs. Those thymocytes expressing TCRs that do not bind MHCI or MHCII would fail to localize Lck to the ITAMs and die. In the second model, germ line-encoded complementary determining regions (CDR) 1 and 2 allow each clonotypic TCR to bind distinct classes and alleles of MHC molecules via unique yet specific recognition codons that impose a canonical docking polarity and MHC restriction (1, 3, 4, 7, 8). Although it is not obvious that these models are mutually exclusive, the key distinction is that in the first model the randomly generated preselection TCR repertoire would contain TCRs that do and do not bind pMHC, whereas in the second model most if not all TCRs would have a specificity for MHC that is germ line-encoded, regardless of the class or allele of MHC.
The canonical docking polarity of TCRs on MHCI or MHCII observed in crystal structures, and the CDR1 and CDR2 contacts therein, provides evidence for germ line-encoded TCR–MHC interactions for positively selected TCRs (1, 3, 4, 7, 8). But this is taken as supporting either model, as germ line-encoded contacts are likely to be required to allow the formation of a TCR–CD3–pMHC–CD4/CD8 macrocomplex that situates the CD3 ITAMs and Lck in a functionally mandated orientation (1–4, 6, 9, 10). Structural insights from positively selected TCRs thus do not allow the basis of MHC restriction to be cleanly addressed, and functional data that support either model have been reported (11–15).
An open question that can shed light on the similarities and differences between the two models is whether TCRs participate in subthreshold scanning of MHC (4, 16). Scanning would allow a TCR to dock on MHC and survey its contents for peptides that increase the duration of TCR–pMHC interactions, via contacts with clonotypic CDR3s, and allow the formation of a TCR–CD3–pMHC–CD4/CD8 macrocomplex that generates signals (4, 10). In the co-receptor imposed model, a diverse preselection repertoire would contain TCRs with no intrinsic capacity to bind MHC, TCRs that interact with pMHC by atypical modalities, and TCRs that interact with a composite pMHC surface in a canonical modality in a lock-and-key manner akin to antibody–antigen recognition (2, 6). Once selected, this last group of TCRs would be predicted to scan composite pMHC with shapes (i.e., topology and chemical characteristics) related to the selecting pMHC—presumably the same MHC, or similar allelic variant, presenting related peptides. In the germ line-encoded recognition model, TCR scanning of MHC via recognition codons would be intrinsic to most if not all TCRs, regardless of the class of MHC, allelic variants, or the peptide sequence therein (4). At present, functional evidence for TCR scanning of MHC is lacking, regardless of whether it is MHC class-, allele-, and peptide sequence-dependent.
Recently, the frequency of Lck-associated CD4 molecules was proposed to influence if a TCR–pMHC interaction is of sufficient duration to direct a specific cell fate decision, such as negative selection (17). We thus hypothesized that genetically increasing the frequency of CD4–Lck association should allow for the detection of subthreshold TCR–pMHC interactions that are normally of insufficient duration to elicit a functional response. Here we show that T-cell hybridomas expressing 10 distinct TCRs along with a CD4–Lck fusion make IL-2 in response to APCs expressing selecting or nonselecting MHCII, regardless of the sequence of the presented peptide. These responses were independent of positive selection on MHCII, as TCRs that were positively selected on MHCI, or generated in vitro and thus not thymically selected, yielded similar responses. These data provide functional evidence for subthreshold TCR scanning of MHCII that is independent of the class of MHC, the allele, or the peptide sequence therein.
Results
Enhanced Responsiveness to Cognate pMHCII Stimulation.
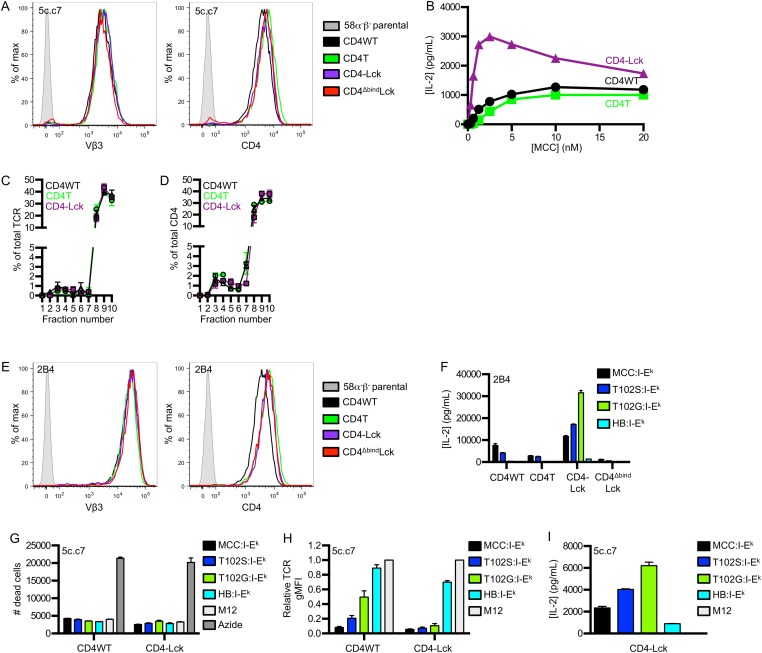
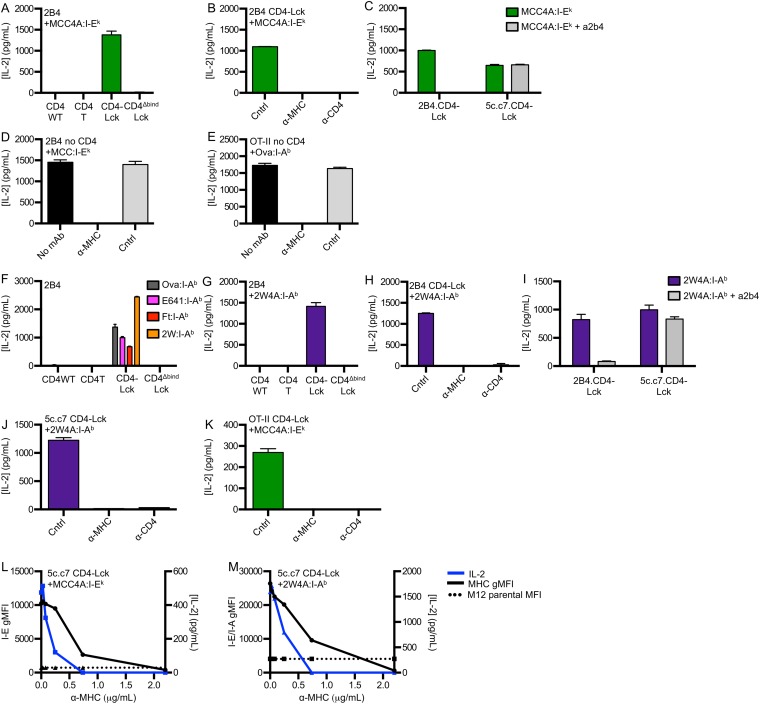
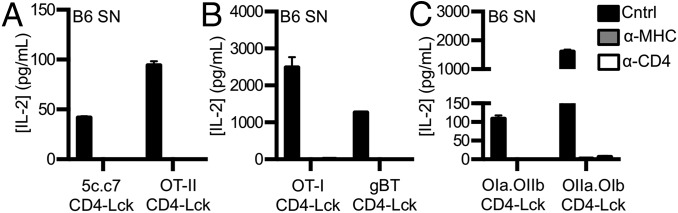
To confirm that fusing CD4 to Lck increases T-cell hybridoma sensitivity, as previously shown (18), we generated 58α−β− T-cell hybridoma lines expressing the 5c.c7 TCR, specific for a moth cytochrome c peptide (MCC 88–103) in I-Ek, along with wild-type CD4 (CD4WT), a C-terminally truncated CD4 (CD4T) that lacks the Lck-binding clasp domain, or a CD4–Lck fusion protein (18–20). Similar levels of TCR and CD4 were expressed on all cell lines (Fig. S1A). We then measured IL-2 production in response to varying MCC concentrations presented by Chinese hamster ovary cells ectopically expressing I-Ek (CHO I-Ek) (Fig. S1B) (21). The CD4–Lck cells produced more IL-2 than the CD4WT or CD4T cells, even at low MCC concentrations. The CD4T cells, in which Lck cannot interact with CD4 via the clasp (22), had lower responses than the CD4WT cells. These data confirm that fusing CD4 to Lck enhances T-cell hybridoma responses to agonist pMHC (18).
Fig. S1.
Fusing CD4 to Lck enhances responsiveness to cognate and null class II pMHC stimulation. 58α−β− hybridomas were transduced with the 5c.c7 or 2B4 TCR and either CD4WT, CD4T, CD4–Lck, or CD4ΔbindLck. (A) Surface expression for TCR and CD4 was assessed by flow cytometry as labeled. (B) IL-2 secretion from 58α−β− T-cell hybridomas after 16 h of coculture with CHO cells ectopically expressing I-Ek (CHO I-Ek) pulsed with MCC peptide at the indicated concentrations. (C and D) Membrane fractionation was performed on 58α−β− cells expressing the 5c.c7 TCR and CD4WT, CD4T, or CD4–Lck to analyze the percent of total (C) TCR or (D) CD4 in each fraction. Data were analyzed as in Fig. 1. (E) Surface expression of TCR and CD4 on 2B4 58α−β− cell lines was assessed by flow cytometry as labeled. (F) IL-2 secretion was detected after 16 h of co-culture of 58α−β− hybridomas expressing the 2B4 TCR and the indicated CD4 molecule with M12 cells expressing the indicated tethered peptide:I-Ek molecule. Data are mean ± SEM of triplicate wells and are representative of two independent experiments. (G–I) 5c.c7 CD4WT or CD4–Lck cells were cultured with M12 cells expressing the indicated tethered peptide:I-Ek molecule for 16 h and assessed for (G) cell death, (H) TCR down-regulation, or (I) IL-2 production. Cell death was determined by setting a dead cell gate in forward and side scatter on cells that had been treated with 1% sodium azide and then calculating the number of cells in this gate with count beads. The dead cell gate was confirmed using a live-dead cell stain. Data in G and I are mean ± SEM of triplicate wells and are representative of two independent experiments. Data in H are mean ± SEM of two experiments and are normalized to the parental M12 cell condition.
Increased Lck Localization in DSMs with the TCR.
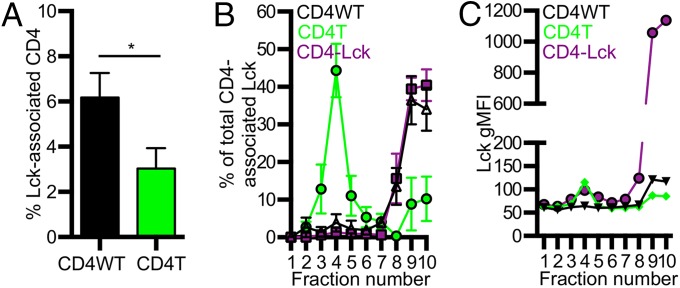
We next compared the amount of Lck associated with CD4 in the CD4WT and CD4T cells relative to the CD4–Lck cells (100% Lck association) to relate differences in responsiveness between these cells to Lck association with CD4. Immunoprecipitation of whole-cell lysates (WCLs) from CD4WT cells revealed that ∼6% of the CD4 signal was associated with Lck relative to the CD4–Lck fusion after detection with a flow-based flourophore-linked immunosorbant assay (FFLISA) (Fig. 1A), similarly to what has been shown for DP thymocytes (17). Interestingly, immunoprecipitation from CD4T WCL indicated that ∼3% of the CD4 signal was associated with Lck relative to the fusion even though CD4T lacks the clasp that mediates association with Lck. This is likely to be due to co-association in detergent-resistant membrane domains (DRMs) rather than direct interactions (23).
Fig. 1.
Fusing CD4 to Lck increases Lck in DSMs. (A) Percent of Lck-associated CD4 in WCL from CD4WT and CD4T cell lines relative to CD4–Lck lines (100%) (mean ± SEM from four experiments). (B) Membrane fractionation of 58α−β− cells expressing the 5c.c7 TCR plus CD4WT, CD4T, or CD4–Lck. gMFI −background (Bkg) within each fraction is shown as a percent of the total gMFI (–Bkg) for each cell line (mean ± SEM for three experiments). Background subtraction was performed with the gMFI of fraction 1 from each cell line. (C) Raw CD4-associated Lck signal (gMFI from each fraction). Data are representative of three experiments. *P < 0.05, Mann–Whitney.
Given these findings, we assessed the membrane compartmentalization of CD4 and CD4-associated Lck molecules relative to the TCR in each cell line to determine if responsiveness corresponded to the concentration of Lck and TCR in the same membrane fraction. The TCR is reported to localize to detergent-soluble membrane domains (DSMs) after sucrose fractionation, whereas Lck and Lck-associated CD4 localize to DRMs; however, the CD4–Lck fusion lacks a myristoylation site reported to impact Lck localization to DRMs and a palmitoylation site in CD4 that may also impact DRM localization (23, 24). We used FFLISA to measure TCR, CD4, and Lck in sucrose gradient DRM fractions 2–6 and DSM fractions 7–10. The TCR primarily localized to DSMs for all cells (Fig. S1C). Interestingly, CD4 was mostly detected in DSMs for all cells, with a smaller percentage being localized to DRMs (Fig. S1D).
We also assessed the amount of Lck that co-precipitated with CD4 in each fraction. Most CD4-associated Lck signals for CD4T cells were present in the DRM fractions (Fig. 1B). This suggests that CD4’s Lck-independent function might be partly attributed to CD4T molecules colocalizing with Lck in specific membrane domains in the absence of direct interactions. CD4T could thus recruit Lck to the TCR–CD3 complex even if with less efficiency than CD4WT. For both the CD4WT and CD4–Lck lysates, the CD4-associated Lck signal was most abundant in the DSMs rather than the DRMs (Fig. 1B). The total signal intensity for CD4-associated Lck was higher for the CD4–Lck cells compared with the CD4WT cells, indicating that there is higher colocalization of TCR and Lck in DSM fractions for the CD4–Lck cells (Fig. 1C). Thus, a higher frequency of CD4-associated Lck would be available for recruitment to TCR–pMHC interactions. One prediction of these results is that the CD4–Lck cells should be more sensitive to lower affinity TCR–pMHC interactions if activation depends on the frequency of Lck-associated CD4 molecules encountered during the course of TCR–pMHC interactions (17).
Fusing CD4 to Lck Reveals Responses to Null pMHCII.
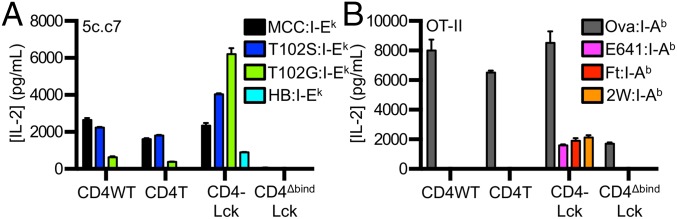
To test this prediction, we examined responses to TCR–pMHC interactions of different affinities using altered peptide ligands (APLs) of MCC for cells expressing the 5c.c7 or 2B4 TCRs. M12 cells expressing I-Ek tethered to an agonist (MCC), weak agonist (T102S), antagonist (T102G), or null peptide from mouse hemoglobin d allele peptide (Hb 64–76) were used as APCs (19, 25). Importantly, 5c.c7 and 2B4 interactions with T102G:I-Ek are too weak to measure by surface plasmon resonance, whereas both interact with a shorter half-life with T102S:I-Ek than MCC:I-Ek (16, 20, 26, 27). The 5c.c7 CD4WT cells produced more IL-2 in response to MCC than to T102S or T102G and none in response to HB (Fig. 2A) (20, 26). The CD4T cells followed a similar, albeit reduced, pattern of IL-2 production (Fig. 2A). Although it was surprising that a detectible response was observed for the CD4WT and CD4T cells to T102G, we suspect this is due to the high expression level of this single pMHC species.
Fig. 2.
Fusing CD4 to Lck reveals responses to null pMHCII. (A) 58α−β− cells expressing the 5c.c7 TCR plus the indicated CD4 molecules were co-cultured with M12 cells expressing the indicated tethered peptide:I-Ek. (B) 58α−β− cells expressing the OT-II TCR and the indicated CD4 molecule were co-cultured with M12 cells expressing the indicated tethered peptide:I-Ab constructs. Data are representative of two experiments (mean ± SEM of triplicate wells). IL-2 was measured by ELISA at 16 h.
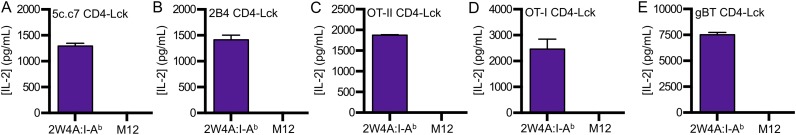
By comparison, the CD4–Lck cells produced similar amounts of IL-2 in response to MCC as the CD4WT cells, more upon stimulation with T102S, and even more to T102G. Surprisingly, they also produced IL-2 in response to the null HB peptide (Fig. 2A). The same results were obtained using the 2B4 TCR, which binds MCC in I-Ek with higher affinity than the 5c.c7 TCR (Fig. S1 E and F) (20). These results are consistent with an increased frequency of Lck-associated CD4 molecules amplifying signals from shorter duration TCR–pMHC interactions (17). In support of this, the inverse hierarchy of responses to APLs was not due to activation-induced cell death upon stimulation with MCC compared with T102S and T102G (Fig. S1G). Rather, we found that CD4WT cells down-regulated TCR proportionally to the potency of the pMHC (i.e., MCC > T102S > T102G > HB), whereas the CD4–Lck cells down-regulated TCR levels equally in response to MCC, T102S, T102G, and even slightly in response to HB relative to M12 cells not bearing a tethered pMHC (Fig. S1H). These results suggest that the increased IL-2 production by CD4–Lck cells in response to T102S, T102G, and HB cells compared with the CD4WT cells is due to increased TCR triggering and endocytosis.
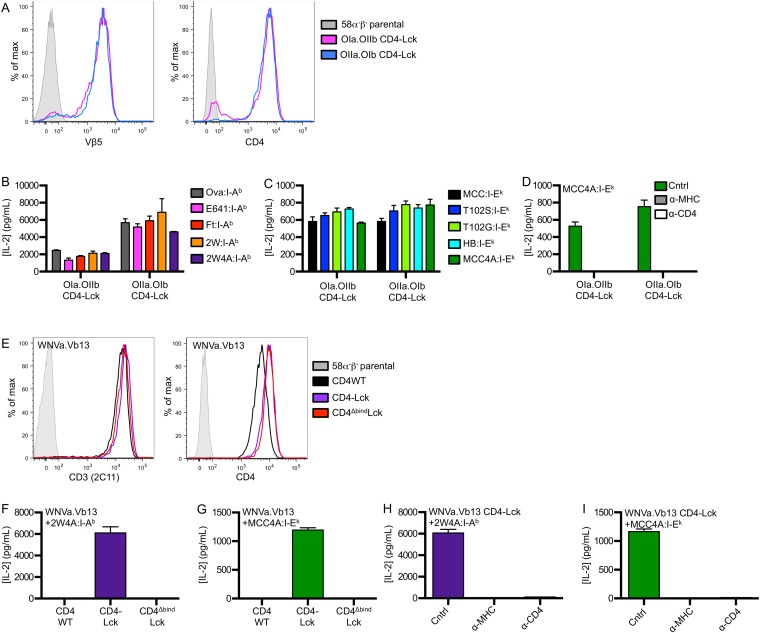
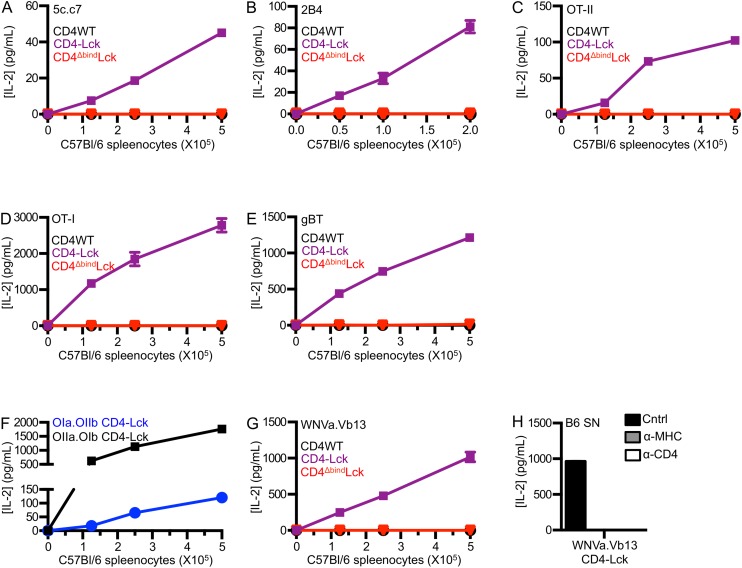
As an additional control, we generated cells expressing a CD4–Lck fusion mutated at the CD4 D1 domain (CD4ΔbindLck) to ensure that the response profiles reported above depended on CD4 binding to MHCII rather than increased Lck levels in the cell (19). The CD4ΔbindLck cells did not produce IL-2 in response to any of the peptides tested, so the responsiveness of the CD4–Lck fusion is dependent on binding to MHCII and not due to overexpression of Lck (Fig. 2A and Fig. S1F). Furthermore, superantigens (SAg) do not appear to be a factor in the HB response, as the CD4WT, CD4T, and CD4ΔbindLck cells failed to respond to HB and the CD4–Lck cells failed to respond to the parental M12 (Fig. S1I).
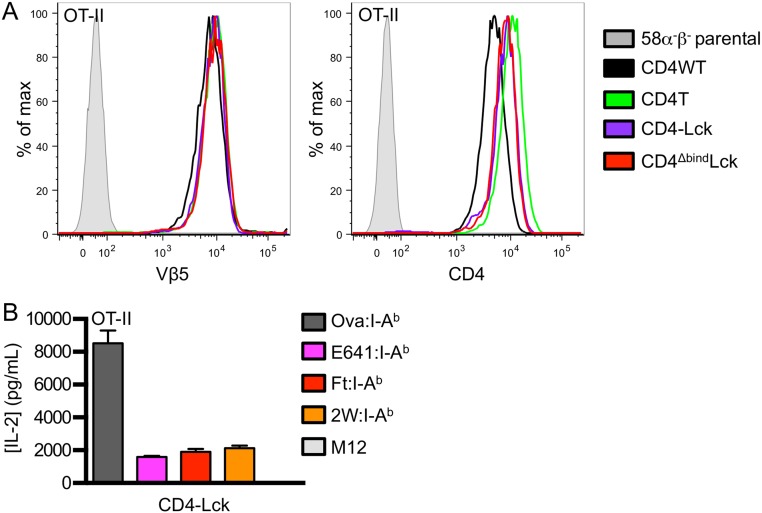
To evaluate if the CD4–Lck fusion would impart responsiveness to null pMHC for cells expressing a distinct TCR restricted to a different MHCII molecule, we generated cell lines expressing the OT-II TCR specific for an ovalbumin-derived peptide (Ova 326–338) in I-Ab along with the different CD4 constructs (Fig. S2A) (28). M12 cells were generated expressing I-Ab tethered to the Ova peptide or to three distinct peptides bound to I-Ab: the E641 peptide derived from the West Nile Virus Envelope protein (641–655), a peptide from the Francisella tularensis lipoprotein Tul4 (Ft 86–99), or the 2W peptide from I-Eα (52–68) (29–31). Although the OT-II CD4WT and CD4T cells responded to Ova, they failed to respond to the E641, Ft, or 2W:I-Ab pMHC complexes. The OT-II CD4–Lck cells produced similar amounts of IL-2 in response to Ova as the CD4WT or CD4T cells (Fig. 2B) but also responded to the three null peptides, whereas OT-II CD4ΔbindLck only responded weakly to Ova (Fig. 2B).
Fig. S2.
OT-II CD4–Lck cells do not respond to parental M12 cells. (A) Surface expression of TCR and CD4 on OT-II 58α−β− cell lines was assessed by flow cytometry as labeled. (B) IL-2 secretion was measured 16 h after OT-II CD4–Lck cells were cultured with M12 cells expressing the indicated peptide:I-Ab molecule or parental M12 cells. Data are mean ± SEM of triplicate wells and are representative of two independent experiments.
Altogether, these data suggest that fusing CD4 to Lck makes T-cell hybridomas sensitive to subthreshold TCR–MHC interactions that may reflect scanning of the MHC allele for which the TCR was positively selected. The failure of the CD4WT, CD4T, and CD4ΔbindLck OT-II cells to respond to the null peptides, and the absence of a response by the CD4–Lck cells to the parental M12 (Fig. S2B), suggests that SAgs are not a contributing factor.
Peptide Sequence-Independent Scanning of MHCII.
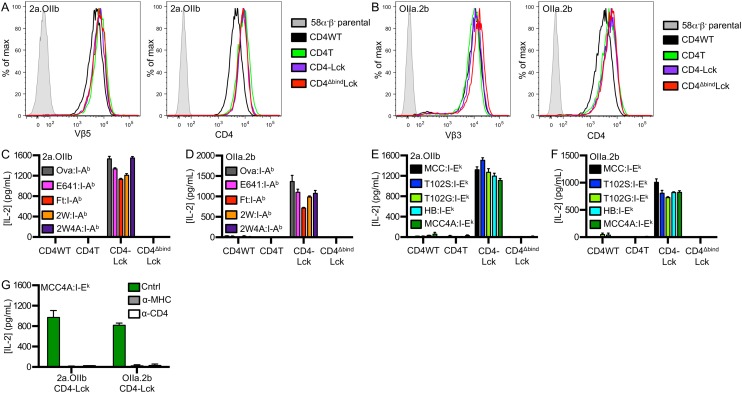
To determine if hybridomas expressing CD4–Lck can respond to MHCII bearing peptides that lack known TCR contact residues, we generated M12 APCs presenting a “shaved” variant of MCC in I-Ek in which the TCR contact residues were mutated to alanines (MCC4A). Cells expressing the 5c.c7 TCR and CD4–Lck, but not CD4WT or CD4ΔbindLck, produced similar amounts of IL-2 in response to MCC4A as they did to HB APCs (Figs. 2A and 3A). Antibodies that block CD4 and I-Ek abrogated this response, whereas those that block CD8 did not (Fig. 3B). Similar results were obtained with the 2B4 TCR when blocked with a clonotypic antibody against the 2B4 TCR (a2b4) (Fig. S3 A–C). Together with the CD4ΔbindLck results, these data indicate this peptide sequence-independent response is dependent on both CD4–MHC and TCR–MHC interactions.
Fig. 3.
Peptide sequence-independent TCR interactions with MHCII. 58α−β− cells expressing (A and B) 5c.c7 or (C and D) OT-II TCRs plus the indicated CD4 molecules were cultured with M12 cells expressing the tethered pMHC (A and B) MCC4A:I-Ek or (C and D) 2W4A:I-Ab. (B and D) Cells were cultured with control antibody (cntrl, anti-CD8), anti-MHCII, or anti-CD4 as described in Materials and Methods. Data are mean ± SEM of triplicate wells and are representative of two experiments. IL-2 was measured by ELISA at 16 h.
Fig. S3.
Detection of peptide sequence-independent TCR interactions with class II MHC. (A–C) IL-2 secretion was detected after 16 h of co-culture of 58α−β− hybridomas expressing the 2B4 TCR and the indicated CD4 molecule with M12 cells expressing the indicated peptide:I-Ek molecule. (B and C) CD4–Lck cells were cultured with MCC4A:I-Ek and either (B) control antibody (Cntrl, anti-CD8), anti-MHC, or anti-CD4 or (C) the clonotypic a2b4 antibody. (D and E) IL-2 was measured 16 h after co-culture of 58α−β− hybridomas expressing the (D) 2B4 or (E) OT-II TCR without CD4 with M12 cells expressing the indicated pMHC molecule and blocked with anti-MHC mAb or Cntrl mAb (anti-CD8). (F–I) 2B4 cells were cultured with M12 cells expressing the indicated tethered peptide:I-Ab molecule and either (H) control antibody (Cntrl, anti-CD8), anti-MHC, or anti-CD4 or (I) the clonotypic a2b4 antibody. (J) 5c.c7 or (K) OT-II CD4–Lck cells were cultured with M12 cells expressing the indicated tethered pMHCII molecule in the presence of control antibody (Cntrl, anti-CD8), anti-MHC, or anti-CD4. (L and M) 5c.c7 CD4–Lck cells were cultured with M12 cells expressing the indicated tethered pMHC molecule and blocked with a titration of either (L) anti–I-E (14-4-4) mAb or (M) anti–I-E/I-A (M5) mAb. M12 cells were then stained with the same mAb to detect gMFI of MHC, and IL-2 was detected by ELISA, as labeled. The dotted line indicates the gMFI of endogenous MHCII on M12 parental cells. Data are mean ± SEM of triplicate wells and are representative of two independent experiments for A–C and F–K or one experiment for D, E, L, and M.
To confirm that TCR–peptide contacts were not required, we tested OT-II CD4–Lck cell responses to pMHC in which TCR contact residues were mutated to reduce or eliminate CDR3 contributions. Here, we generated M12 APCs expressing a shaved variant of the 2W peptide in which we changed the residues that protrude from the MHC toward the TCR to alanines (2W4A) (32). The OT-II CD4–Lck cells, but not the OT-II CD4WT or CD4ΔbindLck cells, responded to APCs presenting the 2W4A peptide (Fig. 3C), and this peptide sequence-independent response was abolished by antibodies blocking MHC class II (M5) or CD4 (GK1.5) (Fig. 3D). The anti-MHCII antibodies also blocked IL-2 production in response to cognate peptide by hybridomas expressing the 2B4 or OT-II TCR without CD4, confirming that they block TCR–MHC interactions (Fig. S3 D and E).
Peptide Sequence-Independent Scanning of Nonselecting MHCII.
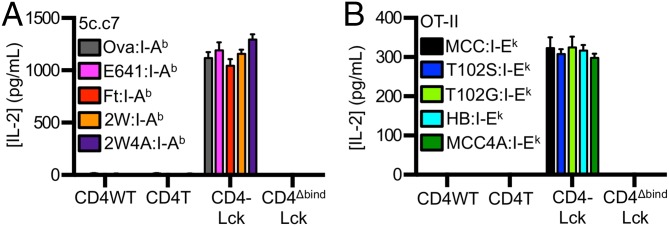
We next tested the ability of CD4–Lck cells to respond to a nonselecting MHCII. The I-Ek–restricted 5c.c7 and 2B4 CD4–Lck cells produced IL-2 in response to the Ova, E641, Ft, or 2W peptides in the nonselecting I-Ab, whereas the CD4WT, CD4T, or CD4ΔbindLck did not (Fig. 4A and Fig. S3F). Similarly, the I-Ab–restricted OT-II CD4–Lck cells responded to the MCC, T102S, T102G, and HB peptides in the nonselecting I-Ek (Fig. 4B), but the CD4WT, CD4T, or CD4ΔbindLck cells did not. Thus, the CD4–Lck fusion allowed for detection of peptide sequence-independent and allele-independent TCR interactions with MHC.
Fig. 4.
Peptide-independent TCR interactions with nonselecting MHCII. 58α−β− cells expressing (A) 5c.c7 or (B) OT-II TCRs plus the indicated CD4 molecules were co-cultured with M12 cells expressing tethered pMHCII as labeled. Experiments were performed and are labeled as in Fig. 3. Data are representative of two experiments.
To further evaluate if these responses reflected TCR interactions that were independent of peptide sequence, we measured the responses of 5c.c7, 2B4, and OT-II CD4–Lck cells to nonselecting pMHC complexes presenting the shaved peptides. The 5c.c7 and 2B4 CD4–Lck cells produced IL-2 in response to 2W4A:I-Ab (Fig. 4A and Fig. S3G), and the OT-II CD4–Lck cells responded to MCC4A:I-Ek (Fig. 4B). These responses depended on TCR–MHC and CD4–MHC interactions (Fig. S3 G–K) as well as the high ligand density of the shaved pMHC (Fig. S3 L and M).
We interpret these data as evidence that TCRs that have undergone positive selection on a particular MHCII can specifically interact with a different MHCII regardless of the sequence of the presented peptide. Because the three TCRs tested were selected on MHCII, the data do not address if a TCR can scan MHCII if it was selected on MHCI.
MHCI-Selected TCRs Can Scan MHCII.
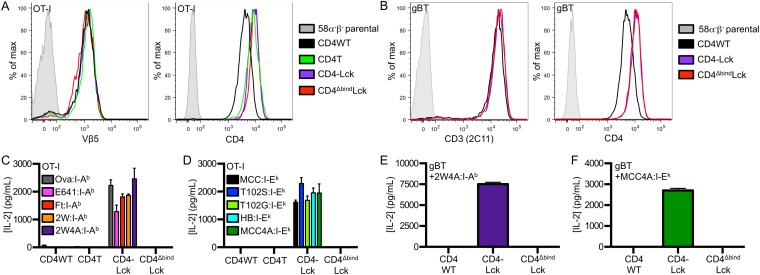
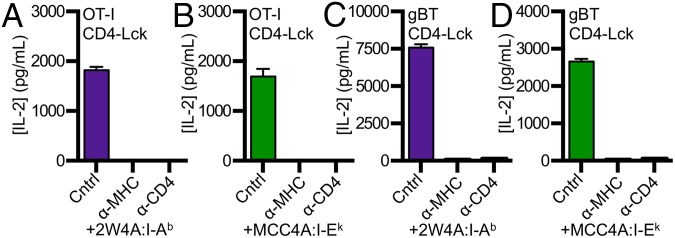
To address if TCRs that are positively selected on MHCI can scan MHCII, we generated cell lines expressing the well-defined OT-I TCR (Fig. S4A), which recognizes an ovalbumin-derived peptide (aa 257–264) in the context of H2-Kb (33), and cell lines expressing the gBT TCR, which recognizes the HSV-1 glycoprotein B peptide (aa 498–505) in H2-Kb (Fig. S4B) (34). OT-I CD4–Lck cells produced IL-2 in response to M12 cells expressing the Ova, E641, Ft, 2W, or 2W4A peptides in I-Ab as well as to M12 cells expressing the MCC, T102S, T102G, HB, or MCC4A peptides in I-Ek (Fig. S4 C and D), whereas the OT-I CD4WT, CD4T, or CD4ΔbindLck did not. Additionally, the response of OT-I CD4–Lck lines to 2W4A:I-Ab and MCC4A:I-Ek were blocked by anti-MHC and anti-CD4 (Fig. 5 A and B). Similarly, gBT CD4–Lck cells produced IL-2 in response to M12 cells expressing either 2W4A:I-Ab or MCC4A:I-Ek in a TCR–MHC- and CD4–MHC-dependent manner (Fig. 5 C and D), whereas cells expressing the gBT TCR with CD4WT or CD4ΔbindLck did not (Fig. S4 E and F). Importantly, CD4–Lck cells expressing the OT-I or gBT TCR, as well as the 5c.c7, 2B4, or OT-II TCRs, did not respond to parental M12 cells lacking high densities of ectopic pMHC (Fig. S5 A–E). We interpret these data as evidence for an inherent TCR specificity for MHCII, regardless of the class of the positively selecting MHC, the allelic variant, or the sequence of the presented peptide.
Fig. S4.
Detection of MHCII interactions with MHCI-restricted TCRs. (A and B) Surface expression of TCR or CD3 and CD4 on (A) OT-I or (B) gBT 58α−β− cell lines was assessed by flow cytometry as labeled. (C and D) OT-I CD4–Lck cells respond to M12 cells expressing the indicated tethered (C) peptide:I-Ab or (D) peptide:I-Ek molecule. (E and F) Hybridomas expressing the gBT TCR and the indicated CD4 molecule were cultured with M12 cells expressing the tethered pMHC (E) 2W4A:I-Ab or (F) MCC4A:I-Ek. Data are mean ± SEM of triplicate wells and are representative of two experiments.
Fig. 5.
Scanning of MHCII by MHCI-restricted TCRs. Anti-MHCII and anti-CD4 mAbs block OT-I CD4–Lck cell responses to (A) 2W4A:I-Ab and (B) MCC4A:I-Ek. gBT CD4–Lck cell responses to (C) 2W4A:I-Ab and (D) MCC4A:I-Ek were blocked with anti-MHCII and anti-CD4 mAbs. Experiments were performed and are labeled as in Fig. 3. Data are representative of two experiments.
Fig. S5.
58α−β− CD4–Lck cells do not respond to parental M12 cells. IL-2 secretion was detected after parental M12 cells or M12 cells expressing 2W4A:I-Ab were cultured with 58α−β− hybridomas expressing CD4–Lck and the (A) 5c.c7 TCR, (B) 2B4 TCR, (C) OT-II TCR, (D) OT-I TCR, or (E) gBT TCR. Data are mean ± SEM of triplicate wells and are representative of two experiments.
In Vitro-Generated TCRs Scan MHCII.
We next created cell lines expressing in vitro-generated TCRs to ask if TCRs that have not undergone thymic selection exhibit characteristics of MHC scanning. Accordingly, we coexpressed the 2B4 alpha and OT-II beta (2a.OIIb) or OT-II alpha and 2B4 beta (OIIa.2b) subunits to make two unique TCRs that mimic random pairing of TCRα and TCRβ subunits in the preselection repertoire (Fig. S6 A and B).
Fig. S6.
In vitro-generated 2a.OIIb and OIIa.2b TCRs display peptide-independent class II MHC specificity. Cell surface expression of TCR and CD4 were assessed by flow cytometry of 58α−β− cells expressing in vitro-generated TCRs made by pairing either (A) the TCR alpha subunit from the 2B4 TCR with the TCR beta subunit of the OT-II TCR (2a.OIIb) or (B) the TCR alpha subunit from the OT-II TCR with the TCR beta subunit of the 2B4 TCR (OIIa.2b). IL-2 secretion was detected by ELISA following 16 h of culture of 58α−β− cells expressing (C and E) 2a.OIIb or (D and F) OIIa.2b and the indicated CD4 molecule with M12 cells expressing the indicated peptide:MHC molecule. (G) IL-2 responses were blocked by addition of anti-MHC or anti-CD4 antibody (Cntrl, anti-CD8). Data are mean ± SEM of triplicate wells from one experiment.
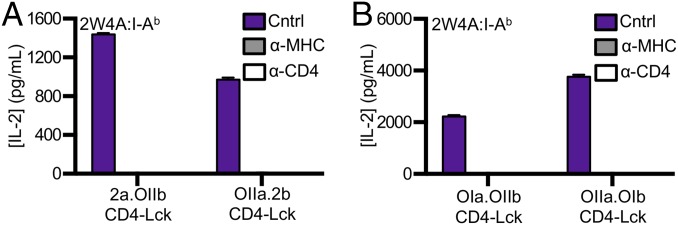
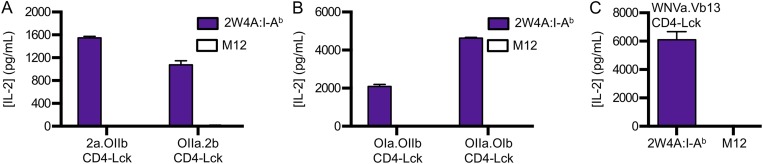
Both the 2a.OIIb and OIIa.2b CD4–Lck cells produced IL-2 in response to Ova, E641, Ft, and 2W in I-Ab (Fig. S6 C and D), as well as to MCC, T102S, T102G, and HB in I-Ek (Fig. S6 E and F), whereas the CD4WT, CD4T, or the CD4ΔbindLck cells did not. Importantly, the CD4–Lck cells also responded to 2W4A and MCC4A, suggesting that the responses occurred irrespective of CDR3 interactions with TCR contact residues on the peptide (Fig. 6A and Fig. S6 C–G). These responses depended on TCR–MHC and CD4–MHC interactions, as they could be abrogated by antibody blockade and the CD4Δbind mutation (Fig. 6A and Fig. S6 C–G).
Fig. 6.
Scanning of MHCII by in vitro-generated TCRs. 58α−β− cells expressing CD4–Lck plus the (A) 2a.OIIb or OIIa.2b or the (B) OIa.OIIb or OIIa.OIb in vitro-generated TCRs were cultured with M12 cells expressing 2W4A:I-Ab in the presence of control, anti-MHCII, or anti-CD4 mAbs as labeled. Experiments were performed as in Fig. 3. Data are representative of two experiments.
Because the subunits used to generate the 2a.OIIb and OIIa.2b TCRs originated from those that had undergone positive selection on MHCII, albeit on different alleles, we also generated cells expressing in vitro-generated TCRs comprised of the OT-I TCRα paired with the OT-II TCRβ (OIa.OIIb) or vice versa (OIIa.OIb) (Fig. S7A). Again, the CD4–Lck cells produced IL-2 in response to M12 cells expressing each of the five peptides presented in I-Ab as well as to the five peptides presented in I-Ek (Fig. S7 B and C). Anti-MHC and anti-CD4 abrogated the responses of both lines to 2W4A:I-Ab and MCC4A:I-Ek (Fig. 6B and Fig. S7D). Additionally, we generated a TCR expressing the TCRα from an MHCI-restricted TCR that recognizes a West Nile Virus Ns4b epitope (aa 2488–2496) in H2-Db (35) paired with a TCRβ of unknown specificity that uses TRBV14 (Vβ13) (WNVa.Vb13) (Fig. S7E) to extend our findings to another TCRα + TCRβ pairing. CD4–Lck cells expressing the WNVa.Vb13 TCR produced IL-2 in response to 2W4A:I-Ab and MCC4A:I-Ek, whereas cells expressing this TCR along with either CD4WT or CD4ΔbindLck did not (Fig. S7 F and G). Again, the response of CD4–Lck cells was blocked by anti-MHC or anti-CD4 (Fig. S7 H and I). Also, none of the CD4–Lck cells expressing in vitro-generated TCRs produced IL-2 in response to parental M12 cells lacking ectopically expressed pMHC (Fig. S8 A–C). Therefore, TCRs generated in vitro by random TCRα and TCRβ pairing, as would occur in the preselection repertoire, nevertheless specifically interact with MHCII in an allele-independent and peptide sequence-independent manner.
Fig. S7.
In vitro-generated OIa.OIIb, OIIa.OIb, and WNVa.Vb13 TCRs display peptide-independent class II MHC specificity. (A) Cell surface expression of TCR and CD4 were assessed by flow cytometry of 58α−β− cells expressing in vitro-generated TCRs made by expressing CD4–Lck with either the TCR alpha subunit from the OT-I TCR with the TCR beta subunit of the OT-II TCR (OIa.OIIb) or the TCR alpha subunit from the OT-II TCR with the TCR beta subunit of the OT-I TCR (OIIa.OIb). IL-2 secretion was detected by ELISA following 16 h of culture of 58α−β− cells expressing OIa.OIIb or OIIa.OIb and CD4–Lck with M12 cells expressing (B) the indicated peptide:I-Ab molecule or (C) the indicated peptide:I-Ek molecule. (D) IL-2 responses were blocked by addition of anti-MHC or anti-CD4 antibody (Cntrl, anti-CD8). (E) Cell surface expression of CD3 and CD4 were assessed by flow cytometry of cells expressing the in vitro-generated WNVa.Vb13 TCR and the indicated CD4 molecule. IL-2 secretion was detected by ELISA following 16 h of culture of 58α−β− cells expressing WNVa.Vb13 and the indicated CD4 molecule with (F) 2W4A:I-Ab or (G) MCC4A:I-Ek. (H and I) IL-2 responses were blocked by addition of anti-MHC or anti-CD4 antibody (Cntrl, anti-CD8). Data are mean ± SEM of triplicate wells and are representative of two experiments.
Fig. S8.
In vitro-generated TCR CD4–Lck 58α−β− cells do not respond to parental M12 cells. IL-2 secretion was detected after parental M12 cells or M12 cells expressing 2W4A:I-Ab were cultured with 58α−β− hybridomas expressing CD4–Lck and (A) the 2a.OIIb or OIIa.2b TCR, (B) the OIa.OIIb or OIIa.OIb TCR, or (C) the WNVa.Vb13 TCR. Data are mean ± SEM of triplicate wells and are representative of two experiments.
TCR Scanning of MHCII on Spleenocytes.
Finally, as the experiments above used cell lines expressing high levels of a single pMHCII species as APCs, we asked if CD4–Lck cells expressing the previously tested TCRs could respond to T-cell–depleted spleenocytes (SNs) from C57BL/6 mice. CD4–Lck cells expressing the 5c.c7, OT-II, OT-I, gBT, OIa.OIIb, or OIIa.OIb TCR all responded in a TCR–MHC and CD4–MHC interaction-dependent manner (Fig. 7 A–C and Fig. S9 A–F), as did those expressing the 2B4 or WNVa.Vb13 TCRs (Fig. S9 G and H). These data indicate that the CD4–Lck fusion reveals TCR interactions with MHCII on normal APCs presenting a diverse repertoire of peptides.
Fig. 7.
TCR scanning of MHCII on SNs. 58α−β− cells expressing CD4–Lck and the (A) 5c.c7 or OT-II TCR, (B) OT-I or gBT TCR, or (C) OIa.OIIb or OIIa.OIb TCR were cultured with 5 × 105 T cell-depleted C57BL/6 SNs and blocked with anti-MHCII and anti-CD4 mAbs. Data are mean ± SEM of triplicate wells and representative of at least two experiments.
Fig. S9.
TCR scanning of MHCII on T-depleted spleenocytes (SN). 58α−β− cells expressing the indicated CD4 molecule and the (A) 5c.c7 TCR, (B) 2B4 TCR, (C) OT-II TCR, (D) OT-I TCR, (E) gBT TCR, (F) OIa.OIIb or OIIa.OIb TCR, or (G) WNVa.Vb13 TCR were cultured for 16 h with a titration of T cell-depleted C57BL/6 SNs and IL-2 was measured. (H) IL-2 from 58α−β− cells expressing the WNVa.Vb13 TCR and CD4–Lck after co-culture with 5 × 105 T cell-depleted C57BL/6 SNs and blockade with anti-MHCII and anti-CD4 mAbs (Cntrl, anti-CD8). Data are mean ± SEM of triplicate wells and representative of at least two independent experiments.
Discussion
Unlike systems where single receptor–ligand interactions coevolve, or antibody–antigen recognition where a library of antibodies encoded by rearranged gene segments are screened for CDR1, 2, and 3 combinations that interact with a previously unencountered antigen, αβT cells use clonotypic TCRs encoded by rearranged gene segments to discriminate self from foreign peptides embedded within allelic variants of related ligand scaffolds—MHCI and MHCII. Such recognition poses interesting conceptual and experimental challenges for understanding how every T cell in the repertoire has arrived at a middle ground where TCRs are both MHC restricted and yet specific for one or a few related peptides out of the universe of potential peptides.
The two dominant models posited to explain MHC restriction make distinct predictions about TCR scanning of MHC. In the co-receptor imposed selection model, lock-and-key TCR–pMHC interactions should limit scanning to related shapes of the positively selecting MHC class, allele, and peptides. In contrast, the germ line-encoded recognition model predicts scanning occurs regardless of the class of MHC, allelic variants, or the peptide sequence. Because the TCRs used here can mediate positive selection or are composed of subunits that mediated positive selection in their original pairing, the responses observed with these TCRs in CD4–Lck cells can be evaluated within the context of these predictions.
The data are most consistent with subthreshold scanning that is independent of the MHC class, allele, and peptide sequence upon which the TCR was selected. For example, CD4–Lck cells expressing the 5c.c7 and 2B4 TCRs responded to a shaved peptide in I-Ab as well as SNs from C57Bl/6 mice. Because 5c.c7 and 2B4 Tg thymocytes do not positively select on the C57BL/6 (H-2b) background (36, 37), the data suggest that these TCRs can scan I-Ab but fail to encounter peptides that push TCR–pMHC interactions above the positively selecting threshold. The MHCI (H2-Kb)-restricted OT-I and gBT TCRs (33, 34), which direct CD8 lineage commitment in C57BL/6 mice, also interact with MHCII in an allele- and peptide sequence-independent fashion. This suggests that these TCRs can scan I-Ab in C57BL/6 mice but do not encounter peptides that push them above the threshold for positive selection of thymocytes to the CD4 lineage. These results are distinct from those showing that overexpression of CD8 in OT-I Tg thymocytes allows for the detection of specific self-peptides in the restricting MHCI (H2-Kb) (33). Finally, the five in vitro-generated TCRs, used here to mimic random pairing of TCRα and TCRβ in the preselection repertoire (15), are significant. If TCR restriction to pMHC mimics antibody–antigen recognition, such that scanning is limited to related pMHC shapes, then each TCRα and TCRβ pairing would create unique shape complementarity and new restrictions on the pMHC that can be scanned. However, each in vitro-generated TCR, including those with subunits from MHCI-selected TCRs, interacted with MHCII in an allele- and peptide sequence-independent manner. Altogether 10 distinct TCRs with distinct characteristics displayed functional evidence of allele-, peptide sequence-, and in two cases (i.e., OT-I and gBT), MHC class-independent scanning of MHCII. An exhaustive survey of all potential TCRα and TCRβ combinations with MHC is beyond the scope of this study, and some TCRs may not display intrinsic specificity for some MHC molecules. Nevertheless, the simplest interpretation of our data is that there exists an intrinsic, germ line-encoded recognition of MHCII by the TCR.
That MHC scanning was revealed by fusing Lck to CD4 is also important. For example, cells bearing the 5c.c7 or 2B4 TCRs responded to a low-affinity antagonist pMHC (T102G:I-Ek) proportionally to the extent of Lck association with CD4 within the cells (CD4–Lck > CD4WT > CD4T). Although CD4T cannot associate with Lck via the intracellular clasp, we detected small amounts of association within DRMs that suggest that CD4T may be able to recruit Lck to a TCR–CD3 complex at some low frequency and this may partially explain its Lck-independent function (38). Also of note, the CD4ΔbindLck cells that have 100% association with Lck but are mutated at their MHCII binding site (19) did not respond to T102G. These cells should have normal levels of free Lck, similar to the CD4T cells that were generated from the same parental cell line, which would not be sequestered away from the TCR–CD3 complex by a co-receptor. Nevertheless, these cells barely responded to agonist pMHC. Thus, the extent of CD4 association with Lck and its ability to interact with MHCII was integral to determining if the duration of TCR occupancy was converted to signaling. These data are consistent with a model in which occupied TCR–CD3 complexes scan co-receptors for those associated with active Lck, and these combination of events set signaling thresholds (17).
Altogether, the evidence for TCR scanning of MHC presented here is consistent with the hypothesis that the propensity of αβTCRs to recognize peptide presented by MHCI or MHCII is germ line-encoded and thus intrinsic to the TCR. The duration of TCR interactions with pMHCI or pMHCII, due to clonotypic CDR3 interactions with the peptide, and the extent of CD8 or CD4 association with active Lck would then determine if a clonotypic TCR exceeds a signaling threshold for positive selection to the CD8 or CD4 lineage, respectively. Co-receptor scanning by the TCR would thus impose MHCI or MHCII restriction through recruitment of active Lck to the TCR–CD3 complex to mediate positive selection (17). A positively selected TCR would then be specifically restricted to recognizing antigenic peptides within the context of the selecting class and allele of MHC, regardless of its intrinsic ability to scan any MHC.
Materials and Methods
Constructs.
MSCV-based retroviral expression vectors were used here (19). All proteins are described by amino acid number beginning at the start methionine. The 5c.c7 and 2B4 TCR constructs were previously described (19, 39). The OT-II, OT-I, and gBT TCRs were PCR-amplified from genomic DNA of transgenic mice (28, 34, 40). The WNVa TCRα chain (WNVa) was amplified from genomic DNA from WNV-NS4B transgenic mice and uses TRAV14-1 (Vα2) and TRAJ16. The last four amino acids of the V region, nucleotide additions, and first four amino acids of the J region are CAAS-ATSS. The Vb13 TCRβ chain was amplified from splenic cDNA from a C57BL/6 mouse and uses the TRBV14 (Vβ13) and TRBJ2-1 gene segment with the last four amino acids of the V region, nucleotide additions and D region, and first four amino acids of the J region as follows: CASS-YRV-AEQF. Full-length CD3δ, ε, γ, and ζ were encoded on a poly-cistronic construct as previously described (39, 41). CD4T (aa 1–421) was described elsewhere (19), and CD4–Lck encodes CD4T (aa 1–421) fused via a linker (AAAS) to Lck (aa 2–509). The CD4Δbind mutant was previously described (19). Constructs encoding T102G:I-Ek or the shaved MCC variant, MCC4A:I-Ek (ANERADLIAYLKQATK to ANERADAIAALAQAAK), were generated similarly to the previously described MCC:I-Ek, T102S:I-Ek, and HB:I-Ek constructs by fusing T102G or MCC4A to the N terminus of the I-Ek beta subunit via a short flexible linker (19). Those encoding Ova:I-Ab, E641:I-Ab, Ft:I-Ab, 2W:I-Ab, or the shaved 2W variant, 2W4A:I-Ab (EAWGALANWAVDSA to EAAAAAANAAVDSA), were generated in a similar manner by fusing Ova 326–338, E641 641–655, Ft 86–99, or 2W 52–68 to the N terminus of the full-length I-Ab beta subunit via a short linker. An independent vector encoded the I-Ab alpha subunit.
Cell Lines and Flow Cytometry.
The 58α−β− and M12 cell lines were generated as previously described (19) and the CHO cells expressing I-Ek (CHO I-Ek) as described elsewhere (21). Surface expression of TCRβ Vβ3 (mAb clone KJ25 PE), TCRβ Vβ5 (mAb clone MR9-4 FITC), CD3ε (mAb clone 145–2C11 PE-Cy7), and CD4 (mAb clone GK1.5 e450) expression on 58α−β− cells was assessed by flow cytometry. Cell death was determined by setting a dead cell gate in forward and side scatter on cells that had been treated with 1% sodium azide and then calculating the number of cells in this gate with count beads. TCR down-regulation was calculated with anti-Vβ3 by determining the geometric mean fluorescence intensity (gMFI) of TCR for the 58α−β− cells in each sample. Relative gMFI was calculated by normalizing the TCR gMFI of each sample to cells cultured with parental M12 cells.
Membrane Fractionation and FFLISA.
Membrane fractionation was performed similarly to Hur et al. (42). We lysed 4 × 107 58α−β− cells in 1 mL ice-cold 1% Triton X-100 in TNE (25 mM Tris, 150 mM sodium chloride, 5 mM EDTA) for 10 min followed by homogenization with a dounce homogenizer. Lysates were brought to 2.5 mL in an ultracentrifuge tube with lysis buffer and mixed with 2.5 mL 80% (wt/vol) sucrose in TNE. These were overlayed with 5 mL of 30% (wt/vol) sucrose in TNE followed by 3 mL of 5% (wt/vol) sucrose. After centrifugation for 18 h at 36,000 rpm at 4 °C using an SW40Ti rotor, fractions were collected from the top and the DRM band was visible at the interface between the 5% and 30% sucrose layers (fractions 3–4).
FFLISA was performed by immunoprecipitating TCR or CD4 from WCLs or membrane fractions using streptavidin-conjugated microspheres (Polysciences) coated with biotinylated anti-TCRβ (clone H57) or anti-CD4 (mAb clone RM4-4). Beads were washed with 0.1% lysis buffer in TNE followed by incubation with antibodies against either TCR (Vα11, mAb clone RR8-1 APC), CD4 (mAb clone GK1.5 PE), or Lck (mAb clone 3A5 PE) for 1 h at 4 °C and then washed. Protein was detected by flow cytometry.
Functional Assays.
We cultured 1 × 105 CHO I-Ek cells with 5 × 104 58α−β− cells and MCC 88–103 peptide in 96-well flat-bottom plates. Alternatively, 1 × 105 M12 cells expressing tethered pMHC were co-cultured with 5 × 104 58α−β− cells in 96-well round-bottom plates. For cultures with SNs, 5 × 104 58α−β− cells were cultured with either a titration of T-cell–depleted SNs from 8- to 12-wk-old male C57BL/6 mice or 5 × 105 SNs. Where indicated, anti–I-Ek (clone 14.4.4), anti–I-Ab/I-Ek (clone M5), anti-CD4 (clone GK1.5), anti-CD8 (clone 53.6.7), or the anti-2B4 TCR (clone a2b4) were added at 20 μg/mL. Supernatants were collected and assayed for IL-2 after 16 h of co-culture at 37 °C. Anti-mouse IL-2 (clone JES6-1A12, Biolegend) was used as a capture antibody, and biotin anti-mouse IL-2 (clone JES6-5H4, Biolegend) was used as the secondary antibody. Streptavidin-HRP and TMB substrate (Biolegend) were used for detection. The limit of detection for IL-2 was 4.4 pg/mL.
Acknowledgments
We thank Thomas Serwold, Lawren Wu, Lonnie Lybarger, Caleb R. Glassman, Mark S. Lee, Matthew Bronnimann, and Jeffrey Frelinger for thoughtful discussions and critical feedback, as well as members of the M.S.K., Nikolich, Frelinger, Wu, and Schenten laboratories. We thank Mark M. Davis for valuable reagents. Mark S. Lee, Hemant B. Badgandi, John Ryniawec, and Jessica Seng contributed technical assistance. The University of Arizona Cancer Center/Arizona Research Laboratories Cytometry Core and Cancer Center Support Grant CCSG-CA 023074 supported flow cytometry. M.S.K. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. This work was also supported by the University of Arizona College of Medicine (M.S.K.), the Bio5 Institute (M.S.K.), and NIH/National Institute of Allergy and Infectious Diseases Grant R01AI101053 (to M.S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 2809.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518499113/-/DCSupplemental.
References
- 1.Yin L, Scott-Browne J, Kappler JW, Gapin L, Marrack P. T cells and their eons-old obsession with MHC. Immunol Rev. 2012;250(1):49–60. doi: 10.1111/imr.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Laethem F, Tikhonova AN, Singer A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 2012;33(9):437–441. doi: 10.1016/j.it.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 2012;33(9):429–436. doi: 10.1016/j.it.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10(2):143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald BD, Bunker JJ, Erickson SA, Oh-Hora M, Bendelac A. Crossreactive αβ T cell receptors are the predominant targets of thymocyte negative selection. Immunity. 2015;43(5):859–869. doi: 10.1016/j.immuni.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27(5):735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Dai S, et al. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28(3):324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8(9):975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 9.Rangarajan S, Mariuzza RA. T cell receptor bias for MHC: Co-evolution or co-receptors? Cell Mol Life Sci. 2014;71(16):3059–3068. doi: 10.1007/s00018-014-1600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhns MS, Badgandi HB. Piecing together the family portrait of TCR-CD3 complexes. Immunol Rev. 2012;250(1):120–143. doi: 10.1111/imr.12000. [DOI] [PubMed] [Google Scholar]
- 11.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88(5):627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 12.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122(2):247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Merkenschlager M, et al. How many thymocytes audition for selection? J Exp Med. 1997;186(7):1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Laethem F, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154(6):1326–1341. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackman M, et al. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47(3):349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- 16.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418(6897):552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 17.Stepanek O, et al. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell. 2014;159(2):333–345. doi: 10.1016/j.cell.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Littman DR. A kinase-independent function of Lck in potentiating antigen-specific T cell activation. Cell. 1993;74(4):633–643. doi: 10.1016/0092-8674(93)90511-n. [DOI] [PubMed] [Google Scholar]
- 19.Parrish HL, et al. A transmembrane domain GGxxG motif in CD4 contributes to its Lck-independent function but does not mediate CD4 dimerization. PLoS One. 2015;10(7):e0132333. doi: 10.1371/journal.pone.0132333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell EW, et al. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) J Immunol. 2011;186(10):5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wettstein DA, Boniface JJ, Reay PA, Schild H, Davis MM. Expression of a class II major histocompatibility complex (MHC) heterodimer in a lipid-linked form with enhanced peptide/soluble MHC complex formation at low pH. J Exp Med. 1991;174(1):219–228. doi: 10.1084/jem.174.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner JM, et al. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60(5):755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 23.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Membrane compartmentation is required for efficient T cell activation. Immunity. 1998;8(6):723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 24.Zlatkine P, Mehul B, Magee AI. Retargeting of cytosolic proteins to the plasma membrane by the Lck protein tyrosine kinase dual acylation motif. J Cell Sci. 1997;110(Pt 5):673–679. doi: 10.1242/jcs.110.5.673. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz JD, et al. Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity. 1996;5(2):125–135. doi: 10.1016/s1074-7613(00)80489-6. [DOI] [PubMed] [Google Scholar]
- 26.Corse E, Gottschalk RA, Krogsgaard M, Allison JP. Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 2010;8(9):e1000481. doi: 10.1371/journal.pbio.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463(7283):963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76(1):34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 29.Rees W, et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci USA. 1999;96(17):9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brien JD, Uhrlaub JL, Nikolich-Zugich J. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol. 2008;181(12):8568–8575. doi: 10.4049/jimmunol.181.12.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentino MD, et al. Identification of a dominant CD4 T cell epitope in the membrane lipoprotein Tul4 from Francisella tularensis LVS. Mol Immunol. 2009;46(8-9):1830–1838. doi: 10.1016/j.molimm.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J Immunol. 2010;185(8):4705–4713. doi: 10.4049/jimmunol.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogquist KA, et al. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 1997;6(4):389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 34.Mueller SN, Heath W, McLain JD, Carbone FR, Jones CM. Characterization of two TCR transgenic mouse lines specific for herpes simplex virus. Immunol Cell Biol. 2002;80(2):156–163. doi: 10.1046/j.1440-1711.2002.01071.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, et al. A novel T-cell receptor mimic defines dendritic cells that present an immunodominant West Nile virus epitope in mice. Eur J Immunol. 2014;44(7):1936–1946. doi: 10.1002/eji.201444450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg LJ, et al. Antigen/MHC-specific T cells are preferentially exported from the thymus in the presence of their MHC ligand. Cell. 1989;58(6):1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 37.Canelles M, Park ML, Schwartz OM, Fowlkes BJ. The influence of the thymic environment on the CD4-versus-CD8 T lineage decision. Nat Immunol. 2003;4(8):756–764. doi: 10.1038/ni953. [DOI] [PubMed] [Google Scholar]
- 38.Killeen N, Littman DR. Helper T-cell development in the absence of CD4-p56lck association. Nature. 1993;364(6439):729–732. doi: 10.1038/364729a0. [DOI] [PubMed] [Google Scholar]
- 39.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26(3):357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 41.Holst J, et al. Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat Immunol. 2008;9(6):658–666. doi: 10.1038/ni.1611. [DOI] [PubMed] [Google Scholar]
- 42.Hur EM, et al. LIME, a novel transmembrane adaptor protein, associates with p56lck and mediates T cell activation. J Exp Med. 2003;198(10):1463–1473. doi: 10.1084/jem.20030232. [DOI] [PMC free article] [PubMed] [Google Scholar]