Significance
Cytokinin regulates plant drought adaptation via a multistep component system consisting of histidine kinases, histidine phosphotransfer proteins, and type A and B response regulators (RRs). The functional dissection of individual members of cytokinin signaling and identification of their downstream targets in drought responses are of high importance to provide a complete picture of how cytokinin controls plant drought adaptation. Previous studies have identified functions of several histidine kinases, histidine phosphotransfer proteins, and type A RRs in drought responses of Arabidopsis; however, the roles of type B RRs remain elusive. This comprehensive functional analysis of three type B RRs provides further insight into how cytokinin signaling regulates plant drought adaptation through the proposed yin-yang strategy, enabling efficient application of cytokinin biology in stress tolerance-oriented plant biotechnology.
Keywords: cytokinin signaling, drought adaption, comparative transcriptome analysis, type B response regulators
Abstract
In this study, we used a loss-of-function approach to elucidate the functions of three Arabidopsis type B response regulators (ARRs)—namely ARR1, ARR10, and ARR12—in regulating the Arabidopsis plant responses to drought. The arr1,10,12 triple mutant showed a significant increase in drought tolerance versus WT plants, as indicated by its higher relative water content and survival rate on drying soil. This enhanced drought tolerance of arr1,10,12 plants can be attributed to enhanced cell membrane integrity, increased anthocyanin biosynthesis, abscisic acid (ABA) hypersensitivity, and reduced stomatal aperture, but not to altered stomatal density. Further drought-tolerance tests of lower-order double and single mutants indicated that ARR1, ARR10, and ARR12 negatively and redundantly control plant responses to drought, with ARR1 appearing to bear the most critical function among the three proteins. In agreement with these findings, a comparative genome-wide analysis of the leaves of arr1,10,12 and WT plants under both normal and dehydration conditions suggested a cytokinin (CK) signaling-mediated network controlling plant adaptation to drought via many dehydration/drought- and/or ABA-responsive genes that can provide osmotic adjustment and protection to cellular and membrane structures. Expression of all three ARR genes was repressed by dehydration and ABA treatments, inferring that plants down-regulate these genes as an adaptive mechanism to survive drought. Collectively, our results demonstrate that repression of CK response, and thus CK signaling, is one of the strategies plants use to cope with water deficit, providing novel insight for the design of drought-tolerant plants by genetic engineering.
Water deficit is one of the most detrimental environmental stresses commonly encountered by plants, resulting in a significant loss of crop yield worldwide. To deal with drought, plants have evolved various adaptive strategies during the process of evolution that help them survive. Various phytohormones, including abscisic acid (ABA) and cytokinin (CK), have been known to regulate the protective responses in plants to drought (1, 2). During water deficit, ABA accumulates to help plants survive by different mechanisms, such as promotion of stomatal closure, acceleration of leaf senescence, and induction of the biosynthesis of various protective metabolites. In contrast, CK is known to delay both stomatal closure and leaf senescence, and thus, to overcome drought, CK biosynthesis has been reported to be repressed at some critical stages in plants with the involvement of ABA (2–5). These findings suggest the existence of an interaction between ABA and CK, and perhaps other hormones, and that regulation of ABA and CK homeostases is critical for plant adaptation to drought (1, 6, 7).
Recently, genetic studies in Arabidopsis have provided evidence that CK acts as negative regulator of plant adaptation to drought through the CK phosphorelay system (4, 6, 7), which consists of three CK receptor Arabidopsis histidine kinases (AHKs), five authentic His-containing phosphotransfer proteins (AHPs), and 21 typical Arabidopsis response regulators (ARRs) (8). The ARRs are further classified into type A and type B ARRs, of which the 10 type A ARRs are CK-inducible, and the 11 type B ARRs are not CK-inducible. Type A ARRs, which act as negative feedback regulators of CK signaling, have only a receiver domain, whereas type B ARRs, which mediate the CK-regulated gene expression as myeloblastosis (MYB)-like transcription factors (TFs), have both receiver and output domains. Like many gene families in Arabidopsis, significant functional overlap has been observed within the AHK, AHP, type A ARR, and type B ARR families, as evidenced by results of comparative analyses of single and multiple mutants in CK responses (8).
Among the CK signaling elements, a number of studies have indicated that the AHK2 and AHK3, as well as the AHP2, AHP3, and AHP5, play a negative and redundant role in the adaptation of plants to drought in both ABA-dependent and -independent manners (9–11). However, only scant genetic evidence is available with regard to the functions of the ARRs in drought responses, perhaps because of the significantly higher number of ARR genes and the complexity resulted from their cross-talk (8, 12). Only a preliminary phenotyping study of three higher-order type A arr3,4,5,6, arr5,6,8,9, and arr3,4,5,6,8,9 mutants is available in the literature, which suggests that type A ARR3 and -4 might act as positive regulators, and ARR8 and -9 as negative regulators in plant responses to water stress stimulated by sorbitol (13). On the other hand, no reports are yet available with regard to the function of type B ARRs in plant responses to drought.
Given that dissection of the role of each component of the CK phosphorelay system in drought responses is an important and challenging task, in this study we used a loss-of-function approach to characterize detailed functions of three type B ARR members—namely ARR1, ARR10, and ARR12—in regulating drought responses. Various combinations of arr single, double, and triple mutants were explored to elucidate the functions of these three ARRs, as well as the mechanisms by which they modulate Arabidopsis plant adaptation to drought. Our results demonstrate that ARR1, ARR10, and ARR12 redundantly act as negative regulators of drought responses in both ABA-dependent and -independent pathways. We also identify an enhanced ABA response in the arr1,10,12 triple mutant, which is an evidence for cross-talk between ABA and CK signalings, and may be critical for its higher drought tolerance relative to WT. Furthermore, retention of leaf water content, maintenance of cell membrane stability, and enhancement of anthocyanin biosynthesis were found to contribute to the enhanced drought tolerance of the arr1,10,12 mutant. Comparative genome-wide analyses of the arr1,10,12 triple mutant and WT reveal several potential pathways, including the anthocyanin biosynthesis pathway, that are activated by mutations of ARR1, ARR10, and ARR12 under both normal and dehydration conditions, further strengthening our findings.
Results
arr1,10,12 Triple Mutant Displays Enhanced Tolerance to Drought.
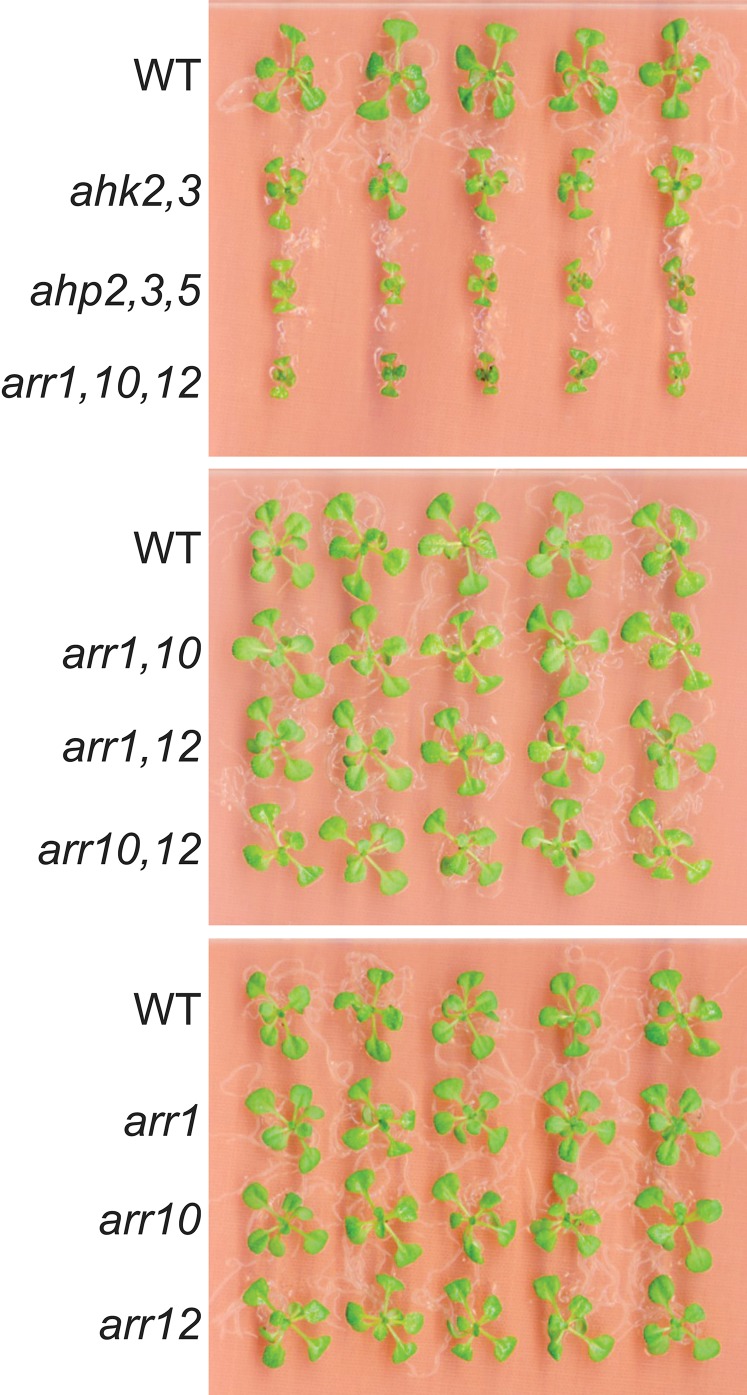
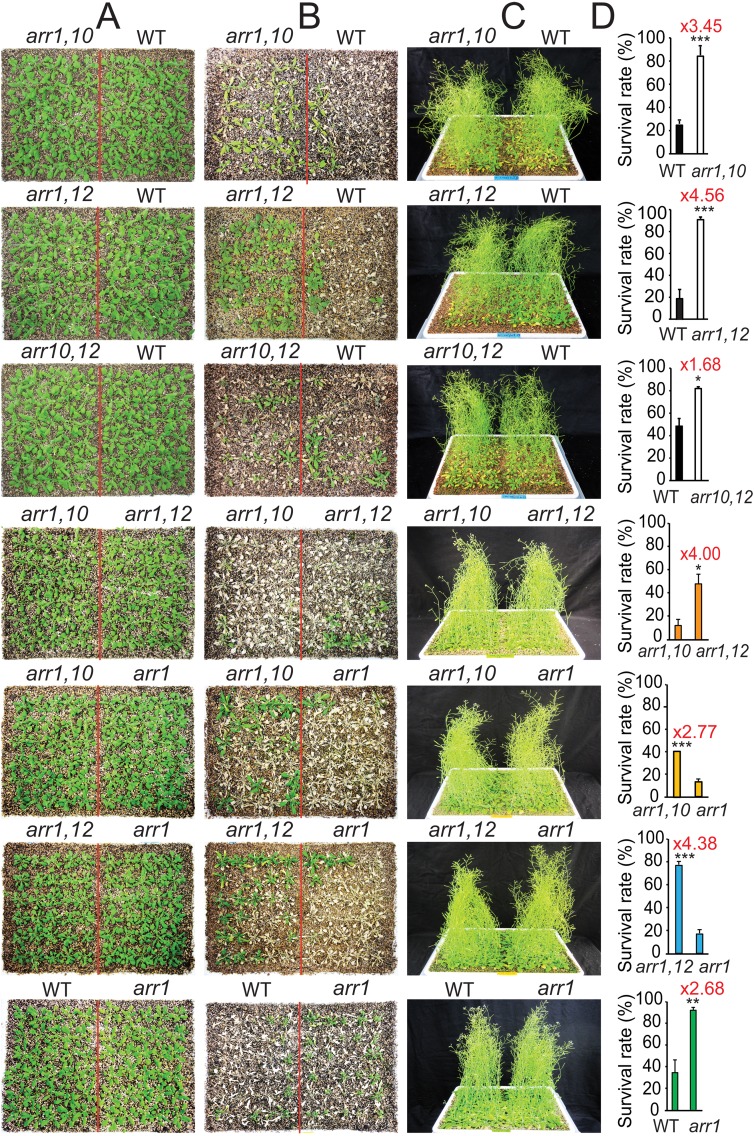
Previous reports studying the functions of AHKs and AHPs in drought responses demonstrated that among the various combinations of ahk and ahp mutations, the ahk2,3 double and ahp2,3,5 triple mutants showed the strongest tolerance to drought (9–11). Mutants harboring mutated alleles of these two ahk or three ahp genes displayed a noticeable shoot-growth retardation compared with other combinations that do not possess these combined ahk or ahp mutations, suggesting that reduced shoot stature might be, at least in part, an adaptive trait of CK signaling mutants to drought. Thus, to study the functions of other downstream components of the CK signaling, namely the type B ARRs, in drought responses, we screened for type B ARR genes whose mutation resulted in dwarfism. Among the 11 type B ARR genes, mutant combinations of arr1, arr10, and arr12 genes (arr1–3, 10–5, 12–1 alleles, referred to as arr1, arr10, and arr12 hereafter) exhibited similar shoot-growth retardation as ahk2,3 and ahp2,3,5 mutant plants (Fig. S1) (12). We therefore focused on elucidating the functions of the ARR1, ARR10, and ARR12 genes in plant adaptation to drought using a loss-of-function approach.
Fig. S1.
Growth phenotype of various Arabidopsis histidine kinase ahk2,3, Arabidopsis histidine phosphotransfer ahp2,3,5 mutants and all mutant combinations for Arabidopsis response regulators ARR1, ARR10, and ARR12 genes. Seeds of WT and mutant plants were sowed on GM plates that were incubated at 22 °C for 10 d. Representative plants were subsequently transferred to new GM plates and grown for 4 additional days, and photographs were taken.
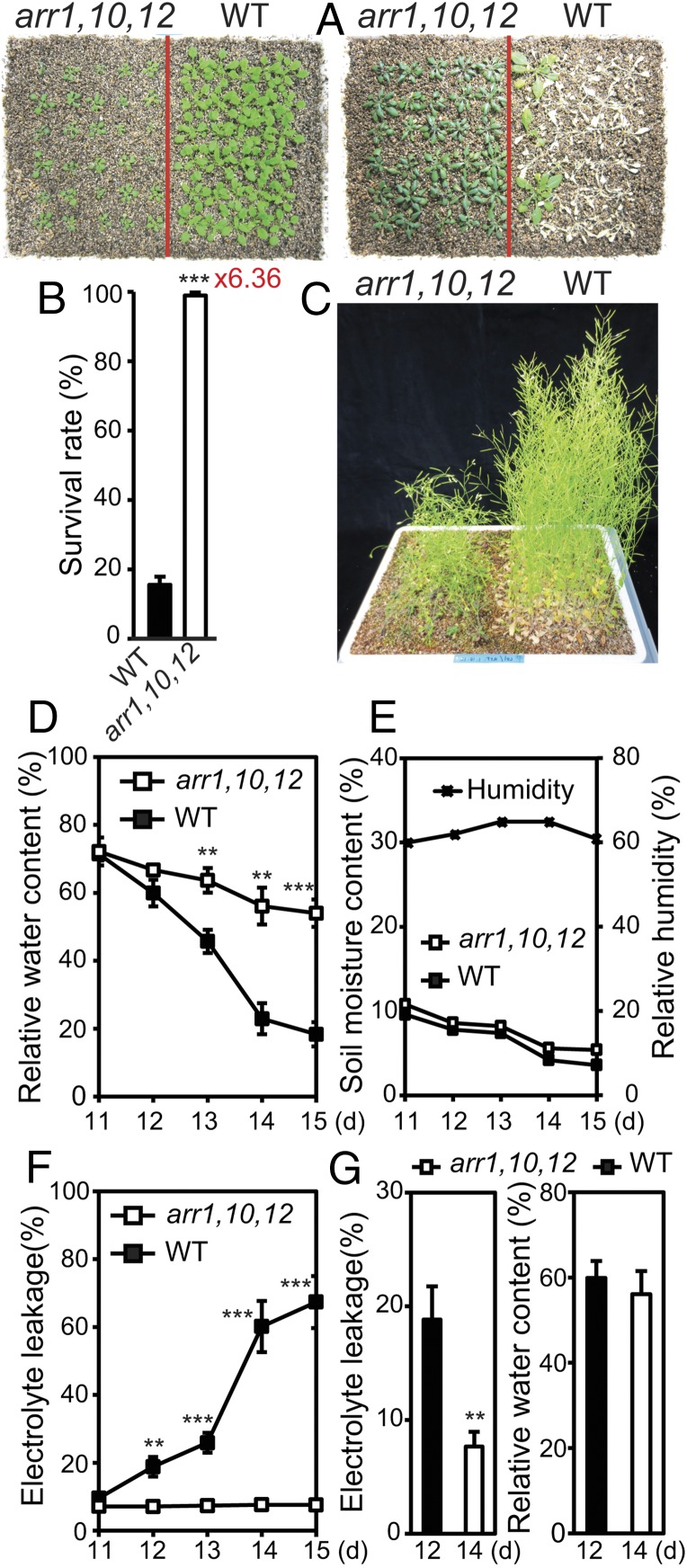
To examine the function of the ARR1, ARR10, and ARR12 genes in drought responses, first the drought tolerance of 3-wk-old arr1,10,12 mutant and WT plants were compared on drying soil in a long-term drought treatment using the same-tray method developed for comparing genotypes with different growth rates (Fig. 1 A–C) (4). A significantly high number of arr1,10,12 mutant plants survived the water deficit, whereas most of the WT plants died following their drought exposure (Fig. 1 A and B). Although the same-tray method applied ensures a valid comparison of genotypes with different growth rates (14), a more stringent method was also used to compare the drought tolerance of WT and arr1,10,12 plants. In this method, the two genotypes were grown in an alternate order, as shown in Fig. S2A. Results also indicated that arr1,10,12 showed higher drought tolerance than the WT (Fig. S2 B and C). For comparison, both the arr1,10,12 and WT plants grew healthily under well-watered conditions (Fig. 1C and Fig. S2D). Additionally, we found that arr1,10,12 plants retained higher relative water content (RWC) than WT at similar soil moisture content in the soil-drying experiment (Fig. 1 D and E). These results demonstrate that loss-of-function of ARR1, ARR10, and ARR12 genes enhances drought tolerance, suggesting that these three type B ARR proteins mediate Arabidopsis responses to drought as negative regulators.
Fig. 1.
Loss-of-function of ARR1, ARR10, and ARR12 results in enhanced drought tolerance. (A) Two-week-old arr1,10,12 and WT plants were transferred from GM plates to trays containing soil and grown for an additional week (Left). Three-week-old plants were exposed to a drought treatment by water withholding for 15 d and then rewatered for 3 d. Photographs were taken subsequent to rewatering and after the removal of inflorescences (Right). (B) Survival rates and SEs (error bars) were calculated from the results of three independent experiments (n = 30 plants per genotype). Red number next to the asterisks represents the fold-change over the WT. (C) For control, 2-wk-old arr1,10,12 and WT plants were transferred from GM plates to trays and grown under well-watered conditions in parallel with the drought tolerance assay. (D) RWC of the arr1,10,12 and WT plants grown and exposed to drought treatment as described in A. Error bars represent SEs (n = 5). (E) Soil relative moisture contents (n = 5) and relative humidity were monitored from day 11 until the end of drought tolerance test. (F) Electrolyte leakage of the arr1,10,12 and WT plants grown and exposed to drought treatment as described in A. Error bars represent SEs (n = 5). (G) Differential electrolyte leakage (Left) between arr1,10,12 and WT plants collected at a similar RWC (Right) during drought treatment. Error bars represent SEs (n = 5). Asterisks indicate significant differences as determined by a Student’s t test analysis (**P < 0.01; ***P < 0.001).
Fig. S2.
Comparison of the drought tolerance of the Arabidopsis response regulator arr1,10,12 and WT plants. (A) Two-week-old arr1,10,12 and WT plants were transferred to soil in an alternate order and grown for an additional week. (B) Three-week-old plants were exposed to a drought treatment, and plants were photographed 16 d after withholding of water after inflorescences were removed. (C) Photograph was taken after 3 d of rewatering. (D) The arr1,10,12 and WT plants were grown in an alternate order for 37 d in parallel under well-watered control conditions.
Enhanced Cell Membrane Integrity of arr1,10,12 Under Drought Stress.
An enhancement in cell membrane integrity and stability might contribute to the differential RWC observed between arr1,10,12 and WT (Fig. 1D) (14). Thus, to elucidate the mechanisms by which ARR1, ARR10, and ARR12 mediate drought responses, first we compared the degree of cell-membrane injury induced by drought in arr1,10,12 and WT plants by measuring electrolyte leakage from both genotypes at various time points in a soil-drying experiment. Data indicated that loss-of-function mutations in all three ARR genes resulted in a remarkably lower electrolyte leakage in the arr1,10,12 mutant in comparison with WT, especially on the days where significant differences in RWC of the two genotypes were recorded (Fig. 1 D–F). Even at the time point when the RWC of the two genotypes was almost equally reduced to below 60%, arr1,10,12 plants showed significantly lower level of ion leakage than did the WT plants (Fig. 1G). These findings suggest that loss-of-function of these three type B ARR genes enhances cell membrane integrity and stability as an adaptive mechanism, contributing to the improved drought tolerance of arr1,10,12 plants by retaining higher water content in the plants under drought.
Comparative Analyses of ABA Responsiveness and Stomatal Regulation in arr1,10,12 and WT Plants.
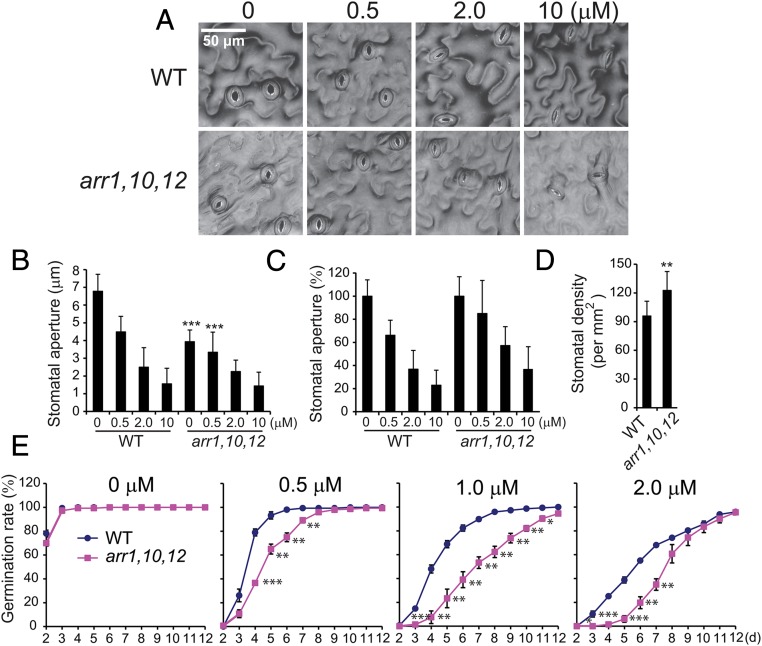
Another trait that might result in difference in RWC of the arr1,10,12 and WT is that related to stomatal regulation and ABA, namely ABA-mediated stomatal closure and/or stomatal density and/or ABA response (3). As shown in Fig. 2 A and B, the stomatal aperture in the arr1,10,12 mutant was significantly narrower than that in WT with or without ABA. However, both genotypes exhibited comparable stomata closing rates (after aperture normalization) in response to ABA (Fig. 2C). Furthermore, arr1,10,12 plants were observed to have higher—but not lower—stomatal density than WT plants (Fig. 2D). These results indicate that the ABA-independent impairment of stomatal opening by loss-of-function of the three ARR genes may reduce the water loss in the mutant, thereby contributing to enhanced drought tolerance of arr1,10,12 rather than alteration in ABA-mediated stomatal closure or stomatal density.
Fig. 2.
Comparison of stomatal aperture, stomatal density, and ABA responsiveness of arr1,10,12 and WT plants. (A) Representative guard cells of rosette leaves from 4-wk-old arr1,10,12 and WT plants in the presence or absence of ABA for 2 h. (B and C) Average stomatal aperture of rosette leaves from 4-wk-old arr1,10,12 and WT plants in the absence or presence of indicated ABA concentrations. Aperture data are expressed in micrometers (B) or in percentage of the aperture data obtained without ABA (C). Error bars represent SD values (n = 35). (D) Average stomatal density of rosette leaves (abaxial side) from 4-wk-old arr1,10,12 and WT plants. Error bars represent SDs (n = 8). (E) Percent germination of arr1,10,12 and WT seeds treated with different levels of exogenous ABA. Data represent the means and SEs of data pooled from three independent experiments (n = 3; 50 seeds were used per genotype per experiment). Asterisks indicate significant differences as determined by a Student’s t test analysis (*P < 0.05; **P < 0.01; ***P < 0.001).
Although the arr1,10,12 and WT plants responded similarly to ABA-mediated inhibition of stomatal opening, the arr1,10,12 mutant exhibited higher ABA responsiveness than WT in a germination assay (Fig. 2E). This result suggests that loss-of-function of these three ARR genes may also enhance the general ABA response, which induces expression of genes that function in cellular dehydration tolerance, thereby enhancing the survival of arr1,10,12 plants under drought.
Enhanced Anthocyanin Biosynthesis in arr1,10,12 Plants Under Drought.
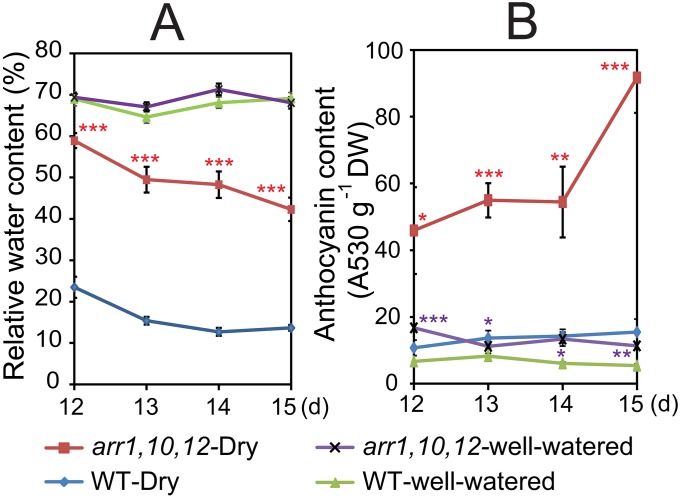
We observed that arr1,10,12 developed a darker leaf coloration than WT in our drought-tolerance assays (Fig. 1A and Fig. S2). Thus, we suspected that arr1,10,12 might have higher anthocyanin content than WT, which could also improve the performance of arr1,10,12 under drought because anthocyanins protect plants against various environmental stresses, including drought (15). To test this hypothesis, we measured anthocyanin contents in the aerial parts of arr1,10,12 and WT plants at various time points after exposing plants to a drought treatment on soil. As shown in Fig. S3, the arr1,10,12 plants possessed significantly higher levels of anthocyanins than WT under both drought and well-watered control conditions. These data support the notion that enhancement of anthocyanin production in the arr1,10,12 mutant could be interpreted as an adaptive mechanism that contributes to its improved drought tolerance.
Fig. S3.
Comparison of anthocyanin contents in arr1,10,12 and WT plants. (A) The arr1,10,12 and WT plants were grown and exposed to drought stress as described in Fig. 1A. At the indicated time points, plants were harvested for measurement of leaf RWC. Error bars represent SEs (n = 5). (B) Anthocyanin content of the arr1,10,12 and WT plants exposed to drought as described in A and in those grown under well-watered conditions. Error bars represent SEs (n = 4). Asterisks indicate significant differences between the genotypes under well-watered control or drought conditions as determined by a Student’s t test analysis (*P < 0.05; **P < 0.01; ***P < 0.001). DW, dry weight.
ARR1, ARR10, and ARR12 Are Redundant Regulators of Drought Response, with ARR1 Being the Most Critical Gene.
We next examined the drought tolerance of lower-order mutant combinations to rank ARR1, ARR10, and ARR12 in terms of their importance in controlling drought response. We first subjected all three arr1,10, arr1,12, and arr10,12 double-mutant plants to a drought-tolerance assay for comparison with WT. As shown in Fig. S4, all three double mutants showed improved tolerance to drought at different levels compared with WT, demonstrating the redundant and negative regulatory functions of the ARR1, ARR10, and ARR12 in drought responses. Furthermore, among the three double mutants, arr1,12 exhibited the highest drought tolerance, followed by arr1,10 and arr10,12. The data also suggested that ARR1 plays the most prominent role in regulation of plant responses to drought, followed by ARR12 and ARR10. Additionally, although the single mutant arr1 exhibited higher drought tolerance relative to WT, both the arr1,10 and arr1,12 double mutants still displayed higher drought tolerance than the arr1 single mutant (Fig. S4), further strengthening that ARR1, ARR10, and ARR12 function in combination to effectively regulate drought responses in plants.
Fig. S4.
Comparison of drought tolerance of arr1,10, arr1,12, and arr10,12 double, arr1 single mutant, and WT plants. (A) Two-week-old arr1,10, arr1,12, arr10,12, arr1, and WT plants were transferred from GM plates to soil and grown for an additional week. (B) Three-week-old plants were exposed to a drought treatment by water withholding and then rewatered for 3 d. Photographs were taken subsequent to rewatering and after the removal of inflorescences. (C) For control purposes, 2-wk-old arr and WT plants were transferred from GM plates to trays and grown under well-watered control conditions in parallel with the drought tolerance test as shown in A. (D) Survival rates and SEs (error bars) were calculated from the results of three independent experiments (n = 30 plants per genotype). Asterisks indicate significant differences as determined by a Student’s t test analysis (*P < 0.05; **P < 0.01; ***P < 0.001). Red numbers above the asterisks represent the fold-change over the WT or arr1 or arr1,10.
Down-Regulation of ARR1, ARR10, and ARR12 by Dehydration and ABA Treatment.
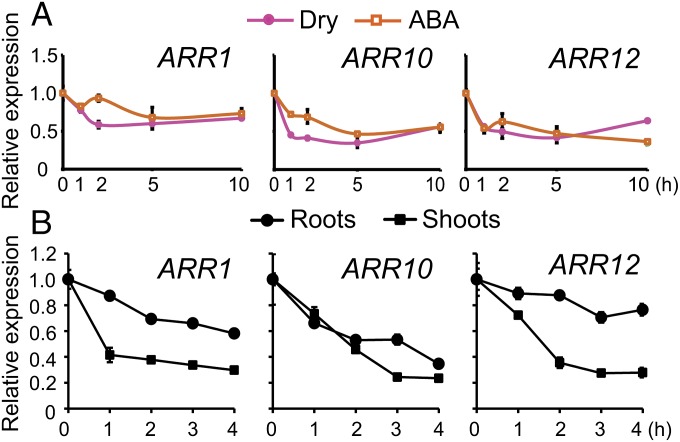
Based on their role in mediating drought responses, we investigated the expression patterns of the three ARR genes in stress-exposed plants. We treated Arabidopsis WT plants with dehydration or ABA over a time-course and analyzed the expression patterns of the three ARR genes. Quantitative RT-PCR (RT-qPCR) data indicated that expression of the three ARR genes was affected by both dehydration and ABA treatments. At the whole-plant level, all three ARR genes were repressed by dehydration and ABA treatments (Fig. 3A). Furthermore, in a tissue-specific expression assay, all three ARR genes were substantially down-regulated in shoots (>twofold), whereas only ARR10 showed a comparable decrease in transcript level (>twofold) in roots (Fig. 3B). ARR1 was determined as the most rapidly down-regulated in shoots, followed by ARR12 and ARR10 (Fig. 3B). The differential expression of ARR1, ARR10, and ARR12 under dehydration and ABA treatments corroborates their involvement in regulating plant adaptation to drought, and indicates that these genes may also act in an ABA-dependent manner.
Fig. 3.
Expression of ARR1, ARR10, and ARR12 genes under stress conditions. (A) Expression of ARR1, ARR10, and ARR12 genes in 14-d-old WT plants exposed to dehydration and ABA treatments for indicated time points. (B) Expression of ARR1, ARR10, and ARR12 genes in roots and shoots of 17-d-old WT plants exposed to a dehydration treatment for indicated time points. Relative expression levels were normalized to a value of 1 in the respective control plants. Data represent the means and SEs of three independent biological replicates (n = 3). UBQ10 was used as reference gene.
Differential Gene Expression in Shoots of arr1,10,12 and WT Plants Under Normal and Dehydration Conditions.
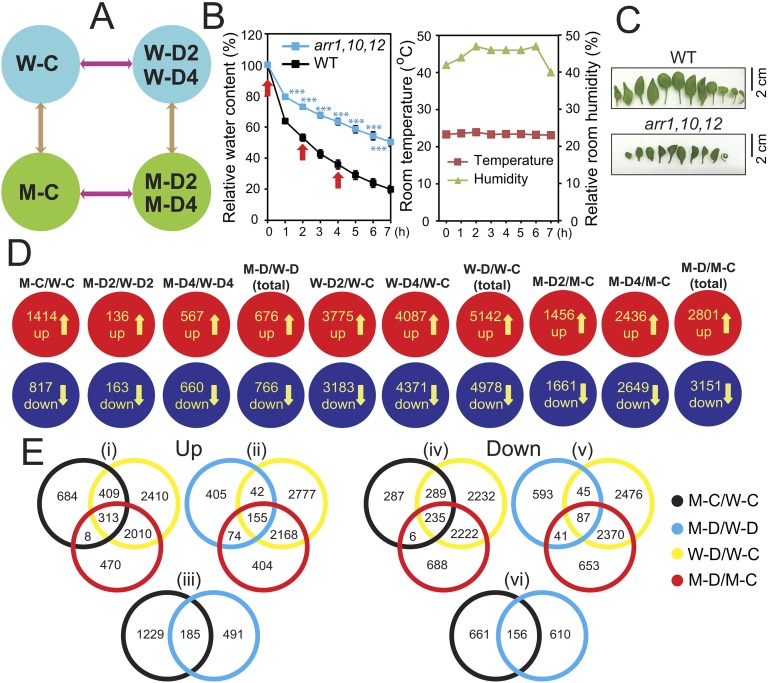
A comparative transcriptome analysis of leaves of arr1,10,12 and WT plants was performed under both normal and dehydration conditions to identify effector genes regulated by the three ARR genes. Our experimental design is illustrated in Fig. S5 A–C, and results of the microarray analysis are summarized in Dataset S1. A search for differentially expressed genes (DEGs) identified 1,414 up-regulated and 817 down-regulated genes in arr1,10,12 vs. WT under unstressed conditions (M-C/W-C) (Fig. S5D and Dataset S2 A and E). With regard to the DEGs derived from comparative analysis of arr1,10,12 and WT under dehydration (M-D/W-D), a total of 676 induced genes were identified, of which a higher number of genes were found to be up-regulated by the long-term 4-h (M-D4/W-D4) dehydration than the short-term 2-h (M-D2/W-D2) dehydration treatment (Fig. S5D and Dataset S2 B–D). As for the down-regulated DEGs obtained from arr1,10,12 vs. WT under dehydration, a total of 766 down-regulated genes were noted in M-D/W-D comparison. In a similar manner, more down-regulated genes were identified in the prolonged dehydration rather than in the short-term dehydration (Fig. S5D and Dataset S2 F–H).
Fig. S5.
Illustration of experimental design and summary of the results of the microarray analysis. (A) Diagrams showing experimental design and all relevant comparisons (arrows). (B) RWC of aerial portions of the 24-d-old Arabidopsis response regulator arr1,10,12 and WT plants exposed to a dehydration treatment. Data represent the means and SEs (n = 5). Asterisks indicate significant differences as determined by a Student's t test (***P < 0.001). Rosette leaf samples collected at 0, 2, and 4 h (arrows) were used for transcriptome analysis. Room temperature and relative room humidity were recorded during the dehydration treatment. (C) Detached representative leaves from well-watered WT and arr1,10,12 plants. (D) Diagrams showing up- and down-regulated genes by dehydration treatment identified in each genotype. (E) Venn diagrams showing the overlapping and nonoverlapping up- and down-regulated genes among the major DEG sets. Data were obtained from the results of microarray analyses of three biological repeats. W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h; M-C, arr1,10,12 well-watered control; M-D2, arr1,10,12 dehydrated 2 h; M -D4, arr1,10,12 dehydrated 4 h. M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; W-D/W-C represents W-D2/W-C and/or W-D4/W-C; M-D/M-C represents M-D2/M-C and/or M-D4/M-C.
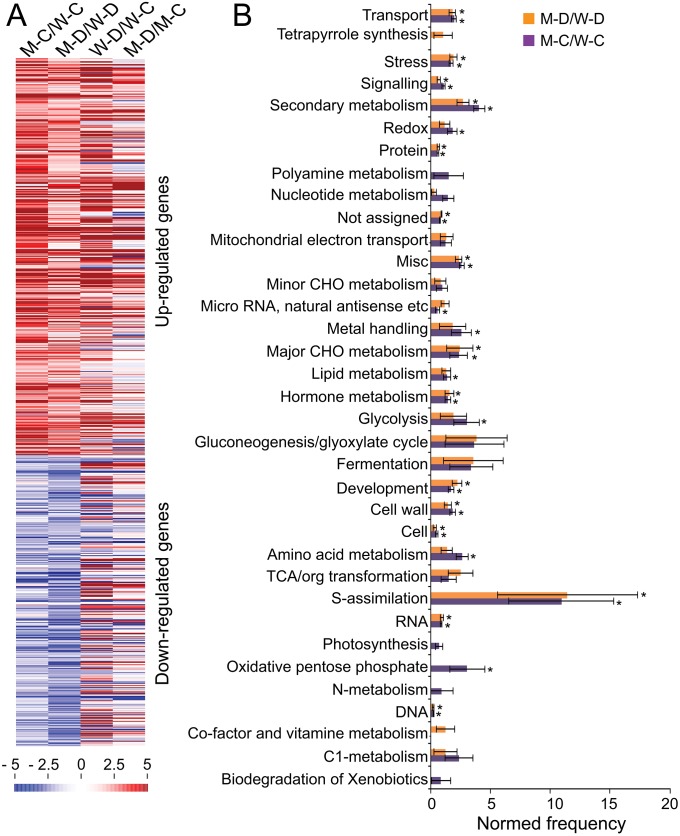
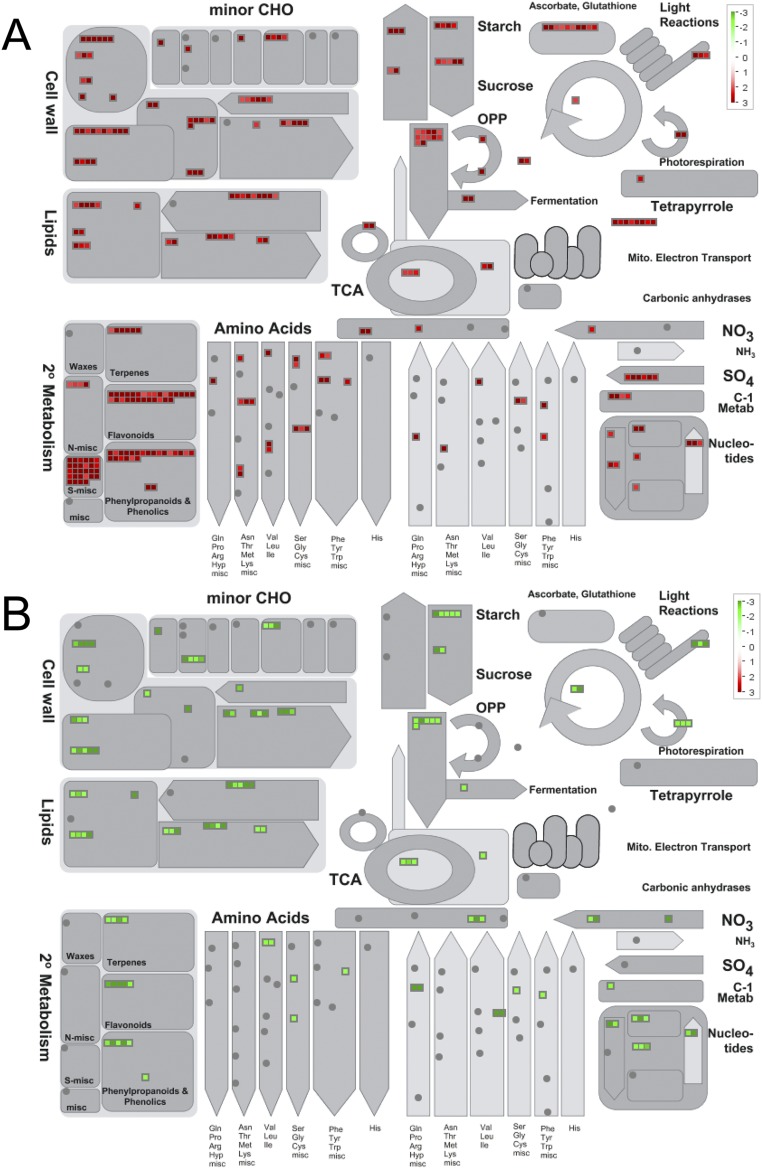
Venn diagram and MapMan analyses indicated that a significant number of the up-regulated genes identified in M-C/W-C and M-D/W-D comparisons are dehydration-inducible (Figs. S5 E, i and ii, and S6A, and Datasets S3–S5), whereas a substantial portion of down-regulated genes noted in the same comparisons are dehydration-repressible (Figs. S5 E, iv and v, and S6A, and Datasets S3, S4, and S6). A number of overlapped genes were also identified between the up-regulated DEGs (Fig. S5 E, iii and Dataset S5H), as well as between the down-regulated DEGs (Fig. S5 E, vi and Dataset S6H) obtained from comparisons M-C/W-C and M-D/W-D. These transcriptomic changes might have an impact on the improvement of drought tolerance of arr1,10,12 plants. MapMan was then used to classify the up-regulated genes obtained from M-C/W-C and M-D/W-D comparisons into functional categories, which indicated that “S-assimilation” was the most highly enriched category (Fig. S6B), suggesting the importance of sulfur assimilation in enhancement of drought tolerance of arr1,10,12. Increasing evidence has indicated that sulfur has a high demand during drought because it plays important role in drought responses by acting in ABA signaling or serving as a precursor for synthesis of several osmoprotectants and antioxidants, such as choline-O-sulfate, polyamines, and glutathione (16). Several DEGs were then selected for RT-qPCR to verify the microarray data (Fig. S7).
Fig. S6.
MapMan-based analysis of DEGs identified in Arabidopsis response regulator arr1,10,12 versus WT under nonstressed (M-C/W-C comparison) and dehydration (M-D/W-D comparison) conditions. (A) Heatmap representation of DEGs obtained from M-C/W-C, M-D/W-D, W-D/W-C, and M-D/M-C comparisons using MapMan. Changes in transcription abundance are indicated by color scales with saturation at fivefold. Blue and red colors indicate down- and up-regulation, respectively. (B) MapMan-based functional classification of the up-regulated genes identified in M-C/W-C and M-D/W-D using the web-tool Classification Superviewer (bar.utoronto.ca) with normalized class score option. *P < 0.05. CHO, carbohydrate; TCA, tricarboxylic acid; M-C/W-C, arr1,10,12 well-watered control versus WT well-watered control; M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; W-D/W-C represents W-D2/W-C and/or W-D4/W-C; M-D/M-C represents M-D2/M-C and/or M-D4/M-C; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h; M-C, arr1,10,12 well-watered control; M-D2, arr1,10,12 dehydrated 2 h; M-D4, arr1,10,12 dehydrated 4 h.
Fig. S7.
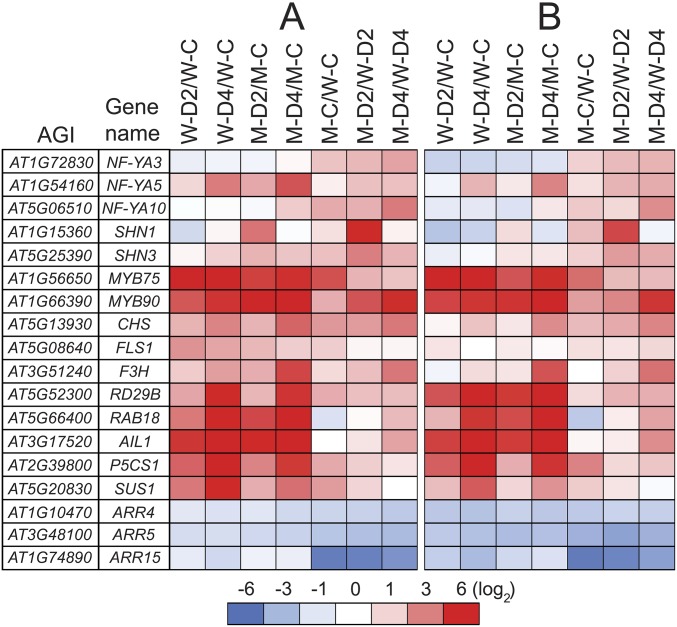
Validation of microarray data by RT-qPCR. Expression of selected genes was assessed by RT-qPCR in leaf samples obtained from nonstressed and dehydrated Arabidopsis response regulator arr1,10,12 and WT plants to validate the microarray data. (A) Heatmap presentation of the fold-changes obtained from microarray analysis. (B) Heatmap presentation of the fold-changes obtained by RT-qPCR of three independent biological replicates. UBQ10 was used as reference gene. Changes in transcription abundance are indicated by intensities of colors expressed in log2 scales with saturation at 6. Blue and red colors indicate down- and up-regulation, respectively. W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h; M-C, arr1,10,12 well-watered control; M-D2, arr1,10,12 dehydrated 2 h; M-D4, arr1,10,12 dehydrated 4 h.
A closer look of the up-regulated genes obtained from M-C/W-C and M-D/W-D comparisons identified many genes that may contribute to enhanced drought tolerance of arr1,10,12 (Dataset S2 A and D). Notably, almost all of the major nuclear factor Y subunit A (NF-YA) genes, members of the NF-Y-type TF family, which are well-known to confer drought tolerance when being overexpressed in Arabidopsis transgenic plants (17, 18), were up-regulated in arr1,10,12 under well-watered and/or dehydration conditions (Fig. S7 and Dataset S7A). Up-regulation of SHINE1 (SHN1) and SHN3, two members of the AP2/EREBP-type TF family that activate epidermal wax biosynthesis (19), was also observed in stressed arr1,10,12 plants (Fig. S7 and Dataset S7A). An enhanced wax synthesis might contribute to protect the plants from cellular dehydration during drought (20). Additionally, a subset of the up-regulated genes identified in the arr1,10,12 mutant with respect to the WT under unstressed and/or stressed conditions were found to be involved in anthocyanin/flavonoid biosynthesis, including CHS, F3H, FLS1-5, PAP1/MYB75, PAP2/MYB90, and MYB112 genes (Figs. S7–S9 and Dataset S7B) (15, 21, 22), supporting the results of the anthocyanin assay (Fig. S3). Furthermore, an increase in transcript levels of late embryogenesis abundant (LEA) genes, such as responsive to dehydration 29B (RD29B), responsive to ABA 18 (RAB18), and ABA-inducible LEA class gene 1 (AIL1) (23, 24), and osmolyte synthesis-related δ1-pyrroline-5-carboxylate synthase 1 (P5CS1) and sucrose synthase 1 (SUS1) genes in the arr1,10,12 mutant relative to WT might also contribute to the better survival of arr1,10,12 plants under drought through protection of membrane structure and osmotic adjustment (Fig. S7 and Dataset S7C) (13, 14). Most of these genes are induced by dehydration, some of which are also induced by ABA (Dataset S7), providing evidence that ARR1, ARR10, and ARR12 regulate drought responses through both ABA-dependent and ABA-independent stress-responsive pathways. Regarding the down-regulated DEGs identified in M-C/W-C and M-D/W-D comparisons, it is worth mentioning that 8 of 10 type A ARR genes (except ARR3 and ARR8) are down-regulated in M-C/W-C and/or M-D/W-D comparisons (Fig. S7 and Dataset S7D), providing a hint for their potential function in drought responses.
Fig. S9.
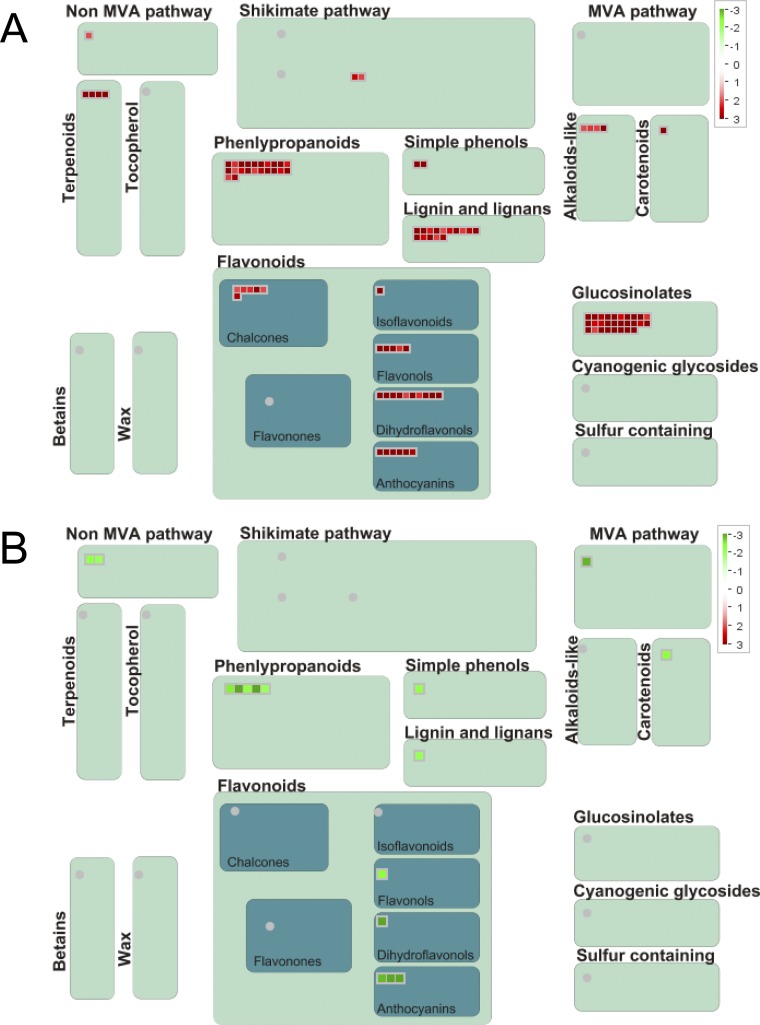
MapMan-based illustration of DEGs in M-C/W-C and/or M-D/W-D comparisons associated with secondary metabolism. (A) Up-regulated genes in M-C/W-C and/or M-D/W-D. (B) Down-regulated genes in M-C/W-C and/or M-D/W-D. Green and red colors indicate down- and up-regulation, respectively. Colored bars in each panel indicate fold-changes in gene expression. M-C/W-C, arr1,10,12 well-watered control versus WT well-watered control; M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; M-C, arr1,10,12 well-watered control; M-D2, arr1,10,12 dehydrated 2 h; M-D4, arr1,10,12 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h. For repetitive genes, their highest fold-change was used in the analysis.
Fig. S8.
MapMan-based illustration of DEGs in M-C/W-C and/or M-D/W-D comparisons involved in different metabolic processes. (A) Up-regulated genes in M-C/W-C and/or M-D/W-D. (B) Down-regulated genes in M-C/W-C and/or M-D/W-D. Green and red colors indicate down- and up-regulation, respectively. Colored bars in each panel indicate fold-changes in gene expression. M-C/W-C, arr1,10,12 well-watered control versus WT well-watered control; M-D/W-D represents M-D2/W-D2 and/or M-D4/W-D4; M-C, arr1,10,12 well-watered control; M-D2, arr1,10,12 dehydrated 2 h; M-D4, arr1,10,12 dehydrated 4 h; W-C, WT well-watered control; W-D2, WT dehydrated 2 h; W-D4, WT dehydrated 4 h. For repetitive genes, their highest fold-change was used in the analysis.
Discussion
In this study, we explored the function of the type B ARR1, ARR10, and ARR12 in drought responses by investigating the consequences of their loss-of-function on the adaptation of Arabidopsis plants to drought. Our data demonstrate that ARR1, ARR10, and ARR12 redundantly act as negative regulators of plant responses to drought, with ARR1 appearing to be the most critical protein (Fig. 1 and Figs. S2 and S4). This study has therefore taken one more step into fulfilling the challenging task of dissecting the roles of different components of the CK signaling pathway in drought responses. Previously, upstream components AHK2 and AHK3, as well as AHP2, AHP3, and AHP5 were identified as negative regulators in drought tolerance (9–11), which together with the results of this study demonstrate that repression of the CK response, and thus CK signaling, is one of the strategies plants use to cope with water deficit.
In accordance with its enhanced drought tolerance (Fig. 1 A–C and Fig. S2), the arr1,10,12 mutant maintained higher RWC than WT (Fig. 1D), which can be attributed to its enhanced ability to protect the membrane structure (Fig. 1 F and G), reduced stomatal opening (Fig. 2B) and enhanced sensitivity to ABA (Fig. 2E), rather than an alteration in stomatal density (Fig. 2D). In agreement with this finding, the repression of CK response through down-regulation of CK signaling-related genes or reduction of endogenous CK also improved drought tolerance of Arabidopsis mutant plants by protecting membrane structures and enhancing ABA sensitivity, independently from the status of stomatal density (4, 11). An enhanced ABA response may lead to up-regulation of downstream ABA-responsive genes, such as LEA and osmolyte biosynthesis-related genes, to activate protective mechanisms, including damage repair, protection of membrane structure, and maintenance of osmotic homeostasis in plants during water deficit, which in turn improves the overall performance of plants (14, 23, 24). For example, constitutive overexpression of the active form of AREB1 gene (AREB1ΔQT) and the AHK1 gene improved drought tolerance in Arabidopsis, which might be ascribed to the enhanced cellular dehydration tolerance through up-regulation of ABA-responsive LEA class genes and osmolyte synthesis-related P5CS1 and SUS1 genes, respectively (13, 23). Concurrently, a higher expression of LEA class genes, including RD29B, RAB18, and AIL1, as well as P5CS1 and SUS1, was found in arr1,10,12 relative to WT under normal or dehydration conditions (Fig. S7 and Dataset S7C), which might partially explain the enhanced drought tolerance of the arr1,10,12 mutant. Furthermore, up-regulation of SHN1 and SHN3 genes detected in the arr1,10,12 mutant (Fig. S7) might result in a reduction in electrolyte leakage noted in the arr1,10,12 mutant (Fig. 1 F and G), through enhancement of the membrane permeability barrier by activating cuticular wax biosynthesis, thereby enhancing drought tolerance (19, 20). Additionally, transcriptome analysis also identified a number of anthocyanin biosynthesis-related genes up-regulated in the arr1,10,12 mutant under both unstressed and dehydration conditions (Fig. S7 and Dataset S7B), which is in agreement with anthocyanin accumulation in the arr1,10,12 mutant under the same conditions (Fig. S3). A previous study also detected many anthocyanin/flavonoid biosynthesis-related genes induced in the drought-tolerant ahp2,3,5 mutant in comparison with WT under well-watered and/or drought conditions (Dataset S7B) (11). Taken together, these results strengthen that repression of CK signaling leads to increased anthocyanin biosynthesis, contributing to plant adaptation to drought, as anthocyanins can protect cells from various stresses by acting as reactive oxygen species-scavenging antioxidants and absorbing UV and high light irradiation (15, 22).
The impairment of stomatal opening observed in the arr1,10,12 mutant might help the plant avoid dehydration (Fig. 2 A and B) (17); however, it may cause aberrant photosynthesis as a trade-off, resulting in its dwarf phenotype (Fig. S1) (12). This finding also implies that CK signaling positively regulates light-stimulated stomatal opening. In support of our finding, a recent study reported that the ahk2,3 double mutant also showed a significantly narrower stomatal aperture than WT (25), which correlates with its enhanced drought tolerance and dwarf-shoot phenotype (Fig. S1) (9, 10). Taken together, these data provide genetic evidence that CK and CK signaling regulate plant drought adaptation through controlling stomatal closure. In line with this finding, the up-regulation of 7 of 10 NF-YA genes (except NF-YA1, -4, and -9) in arr1,10,12 under normal and/or dehydration conditions deserves mention. Several reports studying NF-YA2, -3, -5, -7, and -10 showed that overexpression of these NF-YA genes remarkably enhanced drought tolerance in Arabidopsis transgenic plants (17, 18). Moreover, detailed analysis of NF-YA5 overexpressor plants indicated a link between enhanced drought tolerance and impairment in stomatal opening (17), as observed with the arr1,10,12 mutant (Fig. 2 A and B), further supporting that CK signaling is involved in regulation of NF-YA genes and stomatal closure in plant acclimation to drought. Some of the overexpressor plants, such as those overexpressing NF-YA2, -7, and -10 (18), also exhibited growth retardation, which together with the down-regulation of the CYCD3;1 gene, encoding a D-type cyclin implicated in regulating CK effects on cell division (12), may explain the reduced plant size of the arr1,10,12 mutant as well. It was also interesting to discover that the majority of type A ARR genes were down-regulated in the arr1,10,12 mutant under both normal and dehydration conditions (Fig. S7 and Dataset S7D). Several lines of evidence indicate, although indirectly, that drought tolerance correlates with down-regulation of type A ARR genes. For example, it was reported that arr4 and arr7 mutants exhibited smaller stomatal aperture than WT (26), which also suggests that impairment in stomatal opening of arr1,10,12 may be attributed to the down-regulation of type A ARR genes. More evidence is that CK-deficient mutant and type C ARR22 overexpressor plants exhibited drought tolerance, which coincided with down-regulation of type A ARR5 and ARR7, and ARR4, ARR5, ARR6, ARR7, ARR9, and ARR16 genes, respectively (4, 27). The biological functions of the type A ARRs under drought remain to be determined to directly provide further mechanistic explanation.
In agreement with the negative regulatory functions of ARR1, ARR10, and ARR12 in drought responses, the expression of all three genes was found to be repressed by dehydration and ABA treatments (Fig. 3). This finding suggests that during drought exposure, plants may down-regulate ARR1, ARR10, and ARR12 genes, and thereby the CK response to enhance survival. The differential down-regulation of the three ARR genes in shoots and roots points to a particular role for drought in reducing the CK sensitivity in shoots (Fig. 3B), which is consistent with the results of our physiological analyses of shoot-related phenotypes. Plants with reduced endogenous CK levels (e.g., CKX overexpressor plants) or CK sensitivity (e.g., ahk2,3 mutant) display retarded shoot growth but enhanced root growth, which correlates with their enhanced drought tolerance (4, 6, 9, 10). Restraint of shoot growth is known as an adaptive strategy of plant responses to water deficit, through which plants can reallocate limited energy resources from high energy-demanding processes, such as photosynthesis, for stimulating defense pathways to deal with adverse environmental conditions (4, 6, 7). Moreover, enhanced root growth may help plants respond to drought by obtaining water from deeper soil layers or foraging subsoil surface moisture (28). Mutations in various type B ARR genes—including ARR1, ARR10, and ARR12—result in shoot retardation or root enhancement (12, 29), strengthening the importance of morphological adjustment through CK signaling in drought responses. The finding that CK levels are reduced by drought at some periods during long-term stress (2, 4) implies that plants may trigger mechanisms to suppress the negative effect of CK signaling to survive drought by either reducing endogenous CK content or directly downregulating CK-signaling genes, including type B ARR genes. This stress-induced suppression of CK signaling appears to be a yin-yang strategy facilitating environmental acclimation/adaptation of plants. Detailed functional analysis of all of the components of CK signaling in plant responses to drought, as well as other stresses, remains an important future task to elucidate this complex network underlying plant adaptation to stress.
Materials and Methods
Plant Materials, Dehydration, and Treatments.
The arr1 single, arr1,10, arr1,12, arr10,12 double, and arr1,10,12 triple mutants used in this study are different combinations of arr1-3, arr10-5, and arr12-1 mutants. All of the arr mutants are in Columbia genetic background and were obtained from Argyros et al. (12). For dehydration and ABA treatments, WT plants were grown on germination medium (GM) agar plates for 14 or 17 d (22 °C, 16-h light/8-h dark cycle, 60 µmol⋅m–2⋅s–1 photon flux density) and treated with dehydration, water (hydroponic control), or 100 µM ABA for the indicated time periods. Samples were collected in whole plants or separately in shoot and root fractions in at least three biological replicates, and immediately frozen in liquid nitrogen for further analyses.
Assessment of Drought Tolerance.
The same-tray method that ensures a valid comparison of genotypes with different growth rates was used to evaluate drought tolerance, as previously described (4). Rewatering was performed when the greatest difference in appearance was observed between the mutant and control plants. Three days after rewatering, trays were photographed after removal of inflorescences from the survived plants.
Measurements of RWC and Electrolyte Leakage from Drought-Stressed Plants.
RWC and electrolyte leakage of the detached aerial parts of plants during exposure to soil drying treatment were measured according to the previously described methods (4).
Stomatal Movement Assays.
Assay for ABA-mediated stomatal closure and measurement of stomatal density were conducted as previously described (4).
Assay for Sensitivity to ABA.
Germination and growth inhibition assays were performed on GM medium containing 1% sucrose and various concentrations of ABA, as described in Nishiyama et al. (4).
Determination of Anthocyanin Content.
Plants were subjected to a drought tolerance treatment on the same tray as described above. At indicated time points, aerial parts (without inflorescence) of well-watered control and stressed plants were separately collected in two sets for measurement of fresh weight, dry weight, and RWC (4), and anthocyanin content (30), respectively. For comparison, anthocyanin contents per plant dry weight were estimated using the ratios of fresh and dry weights of collected plant parts.
Expression Analyses.
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen). Methods described in ref. 31 were used for cDNA synthesis and RT-qPCR. UBQ10 was used as a reference gene in analysis of RT-qPCR data. Gene-specific primers used for RT-qPCR are listed in Dataset S8.
Dehydration Treatment and Transcriptome Analysis.
Fourteen-day-old arr1,10,12 and WT plants germinated on GM agar plates were transferred to soil (30 plants/each in the same tray) and grown under well-watered conditions for an additional 10 d. Aerial portions of 24-d-old plants were then detached and subjected to dehydration by placing them on paper towels on a laboratory bench. During dehydration, RWC of treated samples was determined at the indicated time points (n = 5) (23). Rosette leaves collected in three biological repeats from arr1,10,12 and WT plants treated by dehydration for 0, 2, and 4 h were used for microarray and expression analyses. Transcriptome analysis using the Arabidopsis Oligo 44K DNA microarray (v4.0, Agilent Technology) was performed as described in ref. 32. The criteria of fold-change ≥ 2 and a false-discovery rate-corrected P value (q-value) < 0.05 were used in microarray analysis to search for DEGs. The detailed protocol and raw microarray data were deposited in the Gene Expression Omnibus database (GSE74880). MapMan (mapman.gabipd.org/web/guest;jsessionid=B426E14286E9DFBEF97E1474C00C2D95.ajp13_mapman_gabipd_org), Genevestigator (https://www.genevestigator.com), and Arabidopsis eFP browser (bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) were used to analyze the data.
Supplementary Material
Acknowledgments
We thank M. Toyoshima (Japan International Research Center for Agricultural Sciences) for excellent technical support. This project was supported in part by the Core Research for Evolutional Science and Technology, Japan Science and Technology (M.S.); and National Science Foundation Grant IOS-1022053 (to G.E.S.). The PhD study of K.H.N. and C.V.H. was supported by the International Program Associate of RIKEN.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE74880).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600399113/-/DCSupplemental.
References
- 1.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14(3):290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Zwack PJ, Rashotte AM. Interactions between cytokinin signalling and abiotic stress responses. J Exp Bot. 2015;66(16):4863–4871. doi: 10.1093/jxb/erv172. [DOI] [PubMed] [Google Scholar]
- 3.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124(4):509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama R, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23(6):2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 6.Werner T, et al. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell. 2010;22(12):3905–3920. doi: 10.1105/tpc.109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17(3):172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book. 2014;12:e0168. doi: 10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran LS, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(51):20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang NY, Cho C, Kim NY, Kim J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J Plant Physiol. 2012;169(14):1382–1391. doi: 10.1016/j.jplph.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama R, et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc Natl Acad Sci USA. 2013;110(12):4840–4845. doi: 10.1073/pnas.1302265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argyros RD, et al. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20(8):2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wohlbach DJ, Quirino BF, Sussman MR. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20(4):1101–1117. doi: 10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45(4):523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 15.Nakabayashi R, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77(3):367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ. Balancing metabolites in drought: The sulfur assimilation conundrum. Trends Plant Sci. 2013;18(1):18–29. doi: 10.1016/j.tplants.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Li WX, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One. 2012;7(10):e48138. doi: 10.1371/journal.pone.0048138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aharoni A, et al. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, et al. OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS One. 2015;10(1):e116676. doi: 10.1371/journal.pone.0116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito K, et al. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Lotkowska ME, et al. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015;169(3):1862–1880. doi: 10.1104/pp.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita Y, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17(12):3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin X, et al. Transcription factor OsAP21 gene increases salt/drought tolerance in transgenic Arabidopsis thaliana. Mol Biol Rep. 2013;40(2):1743–1752. doi: 10.1007/s11033-012-2228-1. [DOI] [PubMed] [Google Scholar]
- 25.Marchadier E, Hetherington AM. Involvement of two-component signalling systems in the regulation of stomatal aperture by light in Arabidopsis thaliana. New Phytol. 2014;203(2):462–468. doi: 10.1111/nph.12813. [DOI] [PubMed] [Google Scholar]
- 26.Mira-Rodado V, et al. Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Signal Behav. 2012;7(11):1467–1476. doi: 10.4161/psb.21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang NY, Cho C, Kim J. Inducible expression of Arabidopsis response regulator 22 (ARR22), a type-C ARR, in transgenic Arabidopsis enhances drought and freezing tolerance. PLoS One. 2013;8(11):e79248. doi: 10.1371/journal.pone.0079248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50(7):1260–1276. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 29.Mason MG, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17(11):3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S, et al. Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS One. 2015;10(3):e0119724. doi: 10.1371/journal.pone.0119724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le DT, et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18(1):17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama R, et al. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.