Significance
DNA is a canonical target for chemotherapeutic intervention, and several DNA-reactive natural products are in clinical use. An understanding of the mode of DNA binding of these agents is an essential component of translational development. Here we show that (–)-lomaiviticin A (1), a naturally occurring DNA cleavage agent undergoing preclinical evaluation, binds DNA by an unusual mode of association involving insertion of two complex polycyclic arene fragments into the duplex, with concomitant disruption of base pairing. Additionally, our studies suggest that DNA binding activates the DNA cleavage activity of 1. This study provides a structural basis for the activity of 1 and for the development of synthetic DNA-damaging agents capable of recapitulating this mechanism of association and activation.
Keywords: DNA, natural product, double-strand break, NMR
Abstract
(–)-Lomaiviticin A (1) is a complex antiproliferative metabolite that inhibits the growth of many cultured cancer cell lines at low nanomolar–picomolar concentrations. (–)-Lomaiviticin A (1) possesses a C2-symmetric structure that contains two unusual diazotetrahydrobenzo[b]fluorene (diazofluorene) functional groups. Nucleophilic activation of each diazofluorene within 1 produces vinyl radical intermediates that affect hydrogen atom abstraction from DNA, leading to the formation of DNA double-strand breaks (DSBs). Certain DNA DSB repair-deficient cell lines are sensitized toward 1, and 1 is under evaluation in preclinical models of these tumor types. However, the mode of binding of 1 to DNA had not been determined. Here we elucidate the structure of a 1:1 complex between 1 and the duplex d(GCTATAGC)2 by NMR spectroscopy and computational modeling. Unexpectedly, we show that both diazofluorene residues of 1 penetrate the duplex. This binding disrupts base pairing leading to ejection of the central AT bases, while placing the proreactive centers of 1 in close proximity to each strand. DNA binding may also enhance the reactivity of 1 toward nucleophilic activation through steric compression and conformational restriction (an example of shape-dependent catalysis). This study provides a structural basis for the DNA cleavage activity of 1, will guide the design of synthetic DNA-activated DNA cleavage agents, and underscores the utility of natural products to reveal novel modes of small molecule–DNA association.
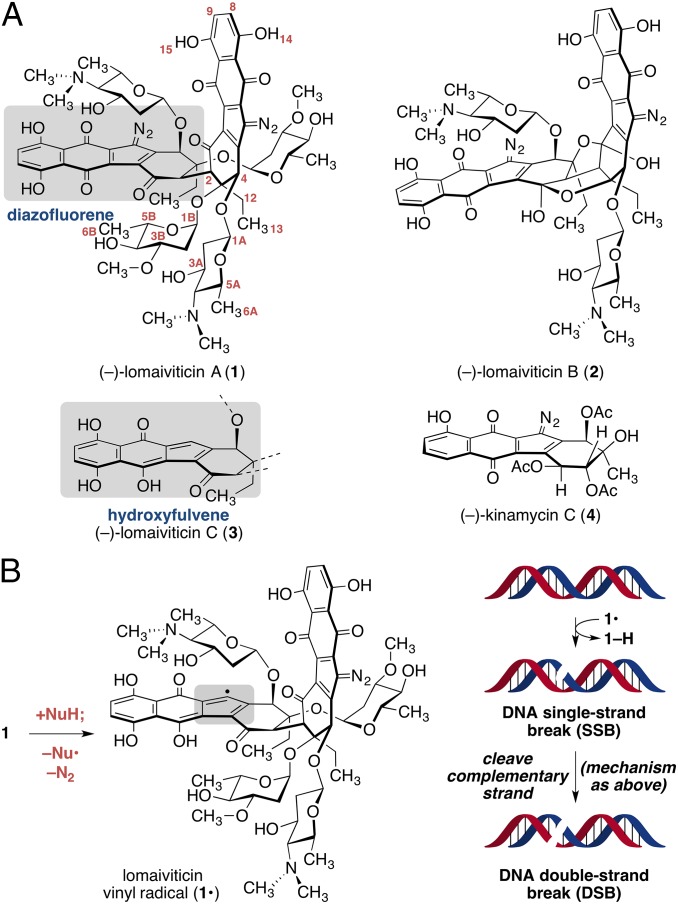
The complex bacterial metabolites (–)-lomaiviticins A–C (1–3, Fig. 1A) (1–3) and (–)-kinamycin C (4) (4–11) are produced by certain strains of Salinispora and Streptomyces. The metabolites 1–4 contain a diazotetrahydrobenzo[b]fluorene (diazofluorene, gray box in 1), which comprises a diazocyclopentadiene fused to naphthoquinone and oxidized cyclohexenone rings (12–17). Lomaiviticins A (1) and B (2) possess two diazofluorenes and C2-symmetric structures containing 2–4 deoxyglycoside residues. (–)-Lomaiviticin C (3) contains a hydroxyfulvene (below 1, Fig. 1A) in place of one diazofluorene, but is otherwise identical to 1.
Fig. 1.
(A) Structures of (–)-lomaiviticins A–C (1–3), (–)-kinamycin C (4), and positional numbering of 1. (B) Reductive activation of lomaiviticin A (1) generates the vinyl radical 1•. Hydrogen atom abstraction is believed to initiate formation of an SSB. Reductive activation of the remaining diazofluorene, followed by hydrogen atom abstraction from the cDNA strand, induces DSB formation.
(–)-Lomaiviticin A (1) possesses half-maximal inhibitory potencies (IC50s) in the low nanomolar–picomolar range against several cultured cancer cell lines (1, 2). (–)-Lomaiviticin C (3) and (–)-kinamycin C (4) are several orders of magnitude less potent, whereas (–)-lomaiviticin B (2) is ∼10–100-fold less potent. The cytotoxicity of 1 derives from the induction of double-strand breaks (DSBs) in DNA (18, 19). K562 cells exposed to 5 nM of 1 for 30 min accumulated DSB levels that were comparable to 40 Gy of ionizing radiation. This DNA cleavage activity is not recapitulated by 3 or 4, suggesting both diazofluorenes of 1 are essential for cleavage activity. DNA DSBs are exceedingly cytotoxic (20), and these data provide an explanation for the remarkable potency of 1.
The molecular mechanism of DNA DSB induction by (–)-lomaiviticin A (1) has been studied (18). Cell-free deuterium-labeling experiments are consistent with a pathway comprising reductive activation of one diazofluorene (Fig. 1B) to generate the vinyl radical intermediate 1•, followed by hydrogen atom abstraction from the deoxyribose chain, a process known to lead to single-strand breaks (SSBs) (21). Reductive activation of the remaining diazofluorene, followed by hydrogen atom abstraction, is believed to cleave the complementary DNA (cDNA) strand, leading to the observed DSB.
However, the mode of interaction of (–)-lomaiviticin A (1) with DNA has not been elucidated. As C(sp2) radicals are high-energy species, the mechanistic model outlined above presupposes that (i) both diazofluorenes are simultaneously oriented in close proximity to each nucleotide chain or that (ii) 1 undergoes dynamic reorganization to place each diazofluorene in proximity to each strand before vinyl radical generation. Given the size of 1 and the fidelity of its DSB induction activity [the ratio of SSBs:DSBs formed by 1 = (5.3 ± 0.6):1] (18), substantial reorganization (ii) seems less plausible. Certain synthetic diazofluorenes and (–)-kinamycin C (4) intercalate DNA, but with low affinity (DC50 values, the concentration of ligand required to displace 50% of a bound intercalator, are >8 μM) (22). Herein, we report studies aimed at determining the DNA binding affinity and sequence preferences of 1. Using these data, we have generated a structural model for the interaction of 1 with the palindromic duplex d(5′-GCTATAGC-3′)2. This structure reveals that both diazofluorene residues of 1 penetrate the duplex and disrupt base pairing about the binding region. This mode of interaction orients each diazo carbon in close proximity to opposing DNA strands and provides a structural basis for the DNA cleavage activity of 1.
Results
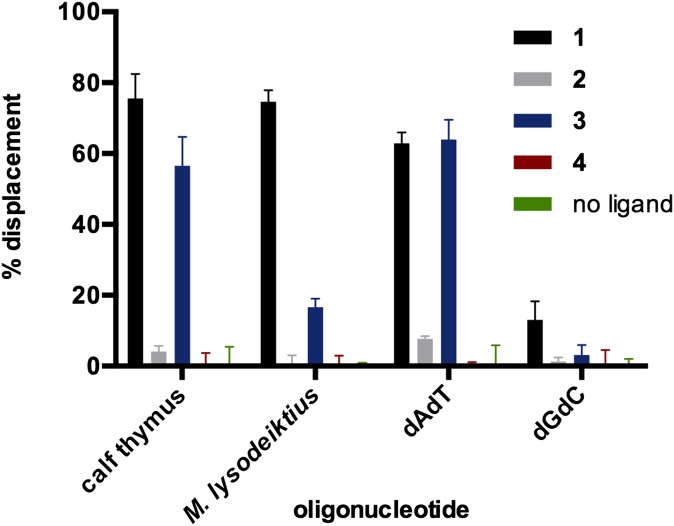
We first performed fluorescent intercalator displacement (FID) assays (23–25) to probe the interaction and binding preferences of (–)-lomaiviticins A–C (1–3) and (–)-kinamycin C (4) to duplex DNA (Fig. 2). We evaluated the affinity of each compound at an equimolar concentration (880 nM) for calf thymus (58% AT content) and Micrococcus lysodeiktius (25% AT content) DNA, as well as poly-dAdT and poly-dGdC, using thiazole orange (26) as an intercalator probe. (–)-Lomaiviticin A (1) associated with AT-containing polynucleotides with five- to sixfold higher affinity than poly-dGdC [63−76% vs. 13% ± 5% decrease in fluorescence (DIF), respectively]. Although 3 displayed high affinity for calf thymus DNA (57% ± 8% DIF) and poly-dAdT (64% ± 6% DIF), the affinity for M. lysodeiktius DNA was ∼fourfold lower than that of 1. The basis for this discrepancy is not known but may relate to the lower AT content of M. lysodeiktius DNA, which may disfavor binding of the more electron-rich species 3. Under the conditions examined, 2 and 4 did not possess significant displacement activity (<12% DIF).
Fig. 2.
Equimolar FID assays using 1–4 and thiazole orange as the intercalator probe. Conditions: [1–4] = 0.88 μM, [thiazole orange] = 1.25 μM, [base pairs] = 0.88 μM, 1 h, 24 °C. Mean percent displacement from three experiments and error bars representing 1 SD from the mean are shown.
The DC50 values of (–)-lomaiviticin A (1) toward several DNA structures were determined by titrating a standard solution of 1 into thiazole orange-saturated duplexes. DC50 values in the 235–770-nM range were obtained using dAdT, dA•dT, and dGdC polynucleotides with ratios of base pairs-to-ligand (rbl) of 1.25–3.0 (Table 1, entries 1–3). To further examine sequence selectivity, 1 was titrated against thiazole orange-saturated 12-bp DNA duplexes containing a variable central six base pair region (entries 4–7). In accord with the FID displacement studies above, 1 displayed higher affinity for duplexes containing AT base pairs. For example, 1 bound alternating AT sequences (entry 5, DC50 = 1.87 ± 0.14 μM) with ninefold higher affinity than alternating GC sequences (entry 4, DC50 = 18.3 ± 1.4 μM). The affinity of 1 for AT tracts decreased with decreasing alternation of the base pairs (entries 5–7). A ratio of duplex to ligand (rdl) of 1.0 was obtained for 1 and the decameric duplex d(CGCATATGCG)2, (entry 8) thereby unequivocally establishing a binding site size of three alternating AT base pairs (compare entries 5 and 8). The affinity of 1 for this duplex was higher than any of the other oligonucleotides examined (DC50 = 0.85 ± 0.05 μM). Based on these data, we selected the DNA duplex d(GCTATAGC)2 for our structural studies. An earlier NMR spectroscopic characterization of this duplex aided resonance assignments in our work (27).
Table 1.
DC50 and rbl or rdl values of (−)-lomaiviticin A (1) and duplexed DNA
| Entry | Polynucleotide DNA | DC50 (per base pair) | rbl |
| 1 | dAdT | 235 ± 10 nM | 3.0 |
| 2 | dGdC | 770 ± 44 nM | 1.25 |
| 3 | dA•dT | 245 ± 15 nM | 2.1 |
| Oligonucleotide duplexes | DC50 (per duplex) | rdl | |
| 4 | GCGC: d(CGCGCGCGCGCG)2 | 18.3 ± 1.4 μM | 0.09 |
| 5 | ATAT: d(CGCATATATGCG)2 | 1.87 ± 0.14 μM | 0.5 |
| 6 | Dickerson: d(CGCAAATTTGCG)2 | 2.14 ± 0.18 μM | 0.5 |
| 7 | A6T6: d(CGCAAAAAAGCG) d(GCGTTTTTTCGC) | 2.70 ± 0.38 μM | 0.2 |
| 8 | 10mer: d(CGCATATGCG)2 | 0.85 ± 0.05 μM | 1.0 |
rbl = ratio of base pairs to ligand. rdl = ratio of duplex to ligand. (–)-Lomaiviticin A (1) was incrementally added to a solution of DNA (2.50 μM/base pair for polynucleotides, 1.25 μM/duplex for oligonucleotides) saturated with thiazole orange (1.25 μM/base pair for polynucleotides, 2.50 μM/duplex for oligonucleotides) at 24 °C. The fluorescence was measured after incubation for 5 min, and the addition of 1 was continued until no further changes in fluorescence were observed.
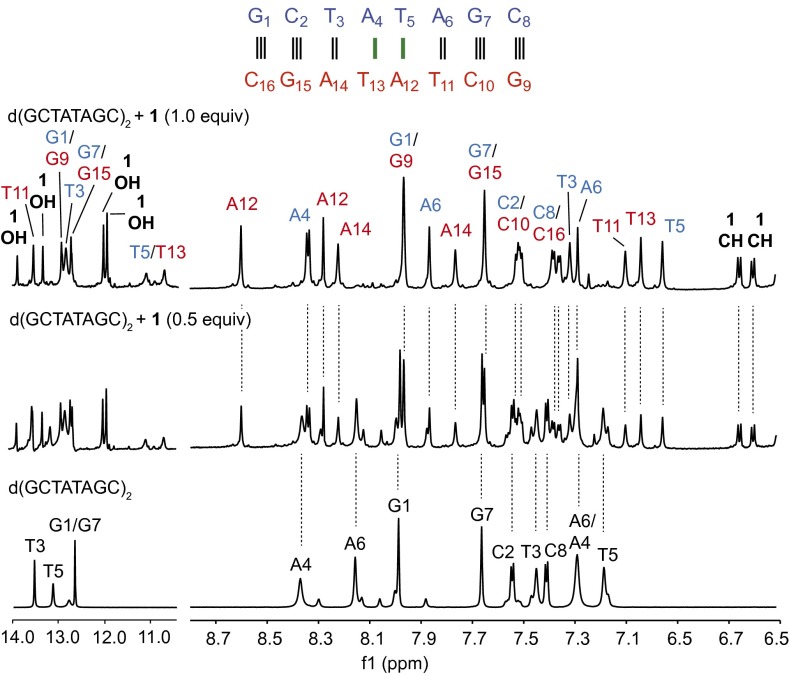
To initiate complex formation, a solution of (−)-lomaiviticin A (1; 10 mM phosphate buffer, pH 7.5, 25 mM sodium chloride) was added to a solution of d(GCTATAGC)2 (10 mM phosphate buffer, pH 7.5, 25 mM sodium chloride) at 24 °C and monitored by 1H NMR spectroscopy. As shown in Fig. 3, a new complex was observed upon the addition of 1 (for the annotated H1′ region of the spectrum, see SI Appendix, Fig. S1). To elucidate the structure of this complex, we determined the structure of the unbound and bound oligomer. The unbound oligonucleotide exhibited a B conformation, as expected (resonance assignments are presented in SI Appendix, Table S1). The complex with 1 was characterized in phosphate-buffered D2O by 1H, NOESY, HSQCAD, and DQF-COSY NMR spectroscopy. An additional sample was prepared in 10% D2O–90% H2O and analyzed by Watergate NOESY and DQF-COSY. Resonance assignments were made using established NMR solution structure methods and are presented in full in SI Appendix, Table S2 (28–30).
Fig. 3.
Selected regions of the 1H NMR spectrum of d(GCTATAGC)2 and (−)-lomaiviticin A (1; 0, 0.5, or 1 equiv, bottom to top). Green bars denote the site of binding. The 14.0–10.5-ppm spectra were obtained in 10% D2O–90% H2O using the Watergate scheme. The 8.7–6.5-ppm spectra were obtained in phosphate-buffered D2O. Experimental conditions: 10 mM phosphate buffer (pH 7.5), 25 mM sodium chloride, 800 MHz, 24 °C. For additional spectroscopic data, see SI Appendix, Fig. S1.
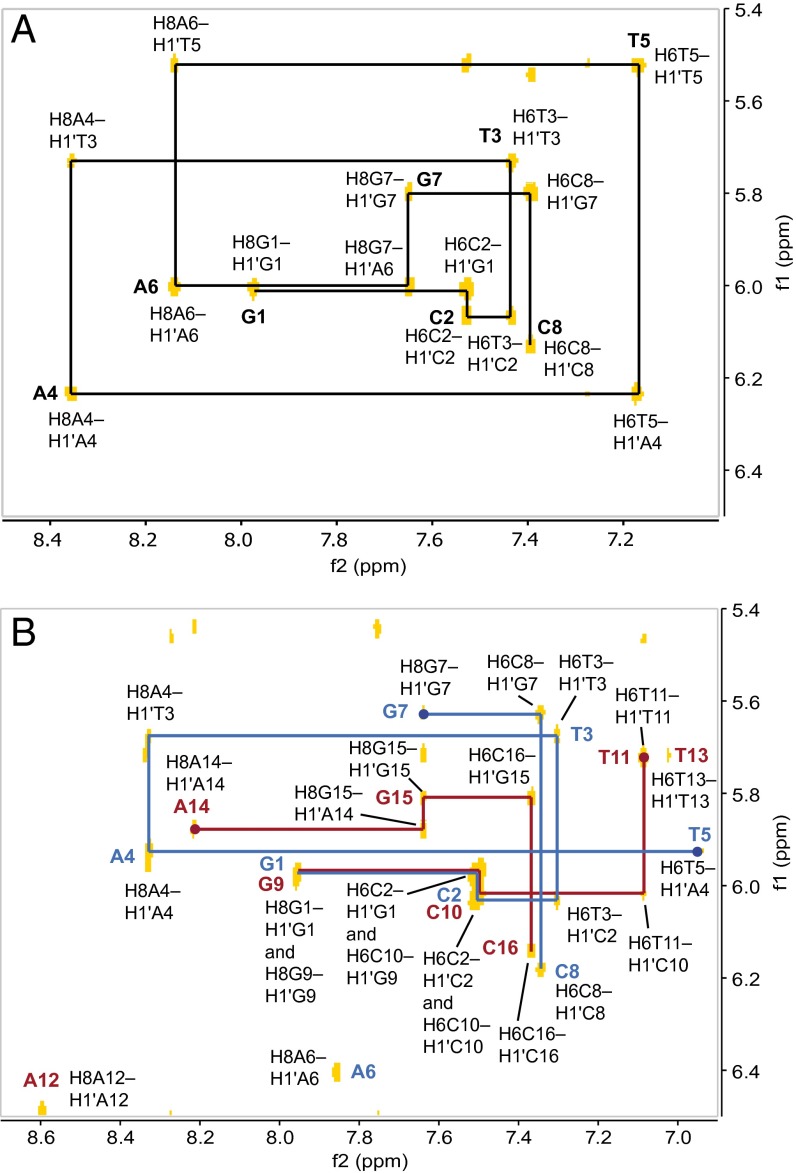
An NOE walk along the A/G H8→H1′(n–1) or T/C H6→H1′(n–1) region of the oligonucleotide can be followed through the entire sequence of the uncomplexed duplex, as expected (Fig. 4A). After addition of (–)-lomaiviticin A (1), significant changes in the central AT region of the oligomer were observed, resulting in disruption of the NOE walk (highlighted by circular nodes, Fig. 4B). This conformational change was corroborated by changes in the A/G H8→H2′(n–1) or T/C H6→H2′(n–1) walk, as well as changes in the internucleotide contacts between A/G H8 or T/C H6 and H2′′(n–1) (SI Appendix, Figs. S2 and S3). Strong intranucleotide contacts were observed between T5 H6 and T13 H6 and both H5′ and H5′′ of their respective carbohydrate residues, suggesting that 1 substantially disrupts the orientation of the central bases. Notably, these NOE contacts were not observed in earlier studies that used an identical duplex and acridinium- or fluorenone-based intercalative ligands (27, 31). These latter ligands were shown to bind by a typical intercalative mode and the differences between these complexes and that formed from 1 suggest a novel mode of association. Disruption of the A/G H8→H1′(n–1) or T H6→H1′(n–1) NOESY walk between residues T5–G7 and T11–A14, and disruption of the A H8 or T H6→H2′(n–1) and A H8 or T H6→H2′′(n–1) contacts between residues A4–A6 and A12–A14 identified these residues as the site of binding. Furthermore, the T5 and T13 exchangeable protons lost hydrogen-bonding character and were observed as broad peaks in a range (11.2–11.7 ppm) typical of free thymine, indicative of complete disruption of base pairing between A4/T13 and T5/A12. Base pairing at the ends of the complexed duplex (G1/C16, C2/G15, G7/C10, C8/G9) was intact as evidenced by minimal perturbation of the NOE contacts and nominal (<0.25 ppm) differences in the chemical shifts of the imino protons of these bases between the free and complexed duplex.
Fig. 4.
NOE walk of the A/G H8→H1′(n-1) or T/C H6→H1′(n-1) region for (A) d(GCTATAGC)2 only and (B) the complex between (−)-lomaiviticin A (1) and d(GCTATAGC)2 (1:1). Breaks in the walk are indicated by circular nodes.
Fifty-five NOE contacts between (–)-lomaiviticin A (1) and d(GCTATAGC)2 were detected (Table 2; see Fig. 1 for positional assignments within 1). NOE contacts between the naphthoquinone (H8 or H9) (designation of positions 8 and 9 is arbitrary and could not be established based on the NMR data) protons of 1 and T11 and A12 of one strand support this region as the intercalation site. In addition, the amino sugar residues of 1 [positions 1A–6A and N(CH3)2] displayed extensive contacts to A4, T5, and A6. Other NOE contacts of note include an interaction between H4 of 1 and A14 H8 and H2, interactions between H13 of 1 and T13 H3′, and interactions between the phenolic protons of 1 (positions 14 and 15) and A4, T5, T13, and A14. Notably, the oleandrose residues of 1 displayed only a limited number of contacts to the nucleotides in proximity to the binding site (T13 H1′, H2′′, H6; T5 H6; A14 H1′), suggesting these residues are largely exposed to solvent.
Table 2.
Intra- and intermolecular NOE contacts in the complex between (−)-lomaiviticin A (1) and d(GCTATAGC)2
| Position | Selected NOEs* | δH, side 1 (base 1–8) | δH, side 2 (base 9–16) |
| 2 | (13, 2B, 4B, N(CH3)2) | 3.85 | 3.81 |
| 4 | A4 H2, A14 H8, A14 H2 (13, 2A,0 3A, 5A, 1A, 3A, 12, 2A, 2, 5A, 1A) | 5.72 | 5.45 |
| 8 | A12 H2, T11 H6, T11 H2′, T11 H2′′, T11 CH3 (9, 8) | 6.65 | 6.60 |
| 9 | (8) | 5.97 | 5.47 |
| 12 | (13) | 2.21/2.04 | 2.11/1.97 |
| 13 | T13 H3′ | 1.21 | 1.15 |
| 14† | T5 H6, T5 CH3, A4 H1′, T13 H6, T13 CH3 (8) | 13.44 | 13.90 |
| 15† | — | 12.36 | 12.30 |
| 1A | A4 H2, A4 H5′, A14 H2 (2A, 5A, 3A) | 5.04 | 4.99 |
| 2A | A4 H2, T3 CH3, A14 H8, A14 H2 | 2.52/2.13 | 2.23/2.51 |
| 3A | A12 H3′ | 4.25 | 4.17 |
| 4A | T13 H6 (1A) | 3.13 | 2.98 |
| N(CH3)2 | A4 H8, A6 H2, T5 H6, A4 H1′, A4 H3′, A4 H4′, T13 H6, T13 H1′ (2B) | 3.13 | 2.98/3.10 |
| 5A | A4 H3′ | 4.13 | 4.04 |
| 6A | A6 H8, A4 H8, A4 H3′, A12 H8, A12 H2, A14 H2, A12 H1′, A12 H3′ | 2.10 | 2.09 |
| 1B | T13 H2′′ (13, 6B, 2B, 4B, 3A, 3B-OCH3) | 6.02 | 6.02 |
| 2B | T5 H6, T13 H6, A14 H1′, T13 H1′ (13, 6B) | 2.04/1.71 | 2.04/1.71 |
| 3B | (13, 2B, 12, 6A, 3A, 2A, 2B, N(CH3)2) | 4.04 | 4.04 |
| 3B OCH3 | (13, 6B, 2B, N(CH3)2, 4B) | 3.73 | 3.61 |
| 4B | (6B, 2B) | 3.28 | 3.28 |
| 5B | — | 4.12 | 4.12 |
| 6B | — | 1.31 | 1.31 |
Selected NOE contacts and chemical shifts of 1 in the complex between (–)-lomaiviticin A (1) and d(GCTATAGC)2. Conditions: 10 mM phosphate buffer, pH 7.5, 25 mM sodium chloride, 800 MHz, 24 °C. The number of contacts between 1 and DNA, within 1 alone, within DNA alone, and the total number of contacts = 55, 92, 278, and 425, respectively.
Contacts between 1 and DNA are listed in bold, intramolecular contacts within 1 only are listed in parentheses.
Assigned by Watergate NOESY, 800 MHz, 24 °C.
Complexation of (–)-lomaiviticin A (1) to DNA abolishes the degeneracy of the metabolite’s native C2-symmetric structure, rendering identical positions on each monomeric unit of 1 chemically inequivalent and thereby providing additional opportunities to infer structural information from the NMR data. First, despite this desymmetrization, the 1H NMR resonances attributed to 1 for matching (but now nondegenerate) positions on either monomeric unit were surprisingly similar; many (although not all, see below) of these pairs of positions displayed resonances within 0.20 ppm of each other. This pseudo-C2 symmetry suggested both halves of 1 interact with DNA in a similar fashion. In addition, each naphthoquinone C–H signal of 1 (positions 8, 9, 8′, and 9′) was resolved and shifted upfield relative to the unbound metabolite (7.23 and 7.12 ppm for free 1; 6.65, 6.60, 5.97, and 5.47 ppm in the complex). The vicinal relationships between these signals were readily established by DQF-COSY analysis, and NOE interactions were observed between the naphthoquinone residues (SI Appendix, Fig. S4). Collectively these data are consistent with the formation of a specific pseudo-C2-symmetric complex between the oligomer and 1 that involves penetration of both diazofluorene residues into the DNA duplex.
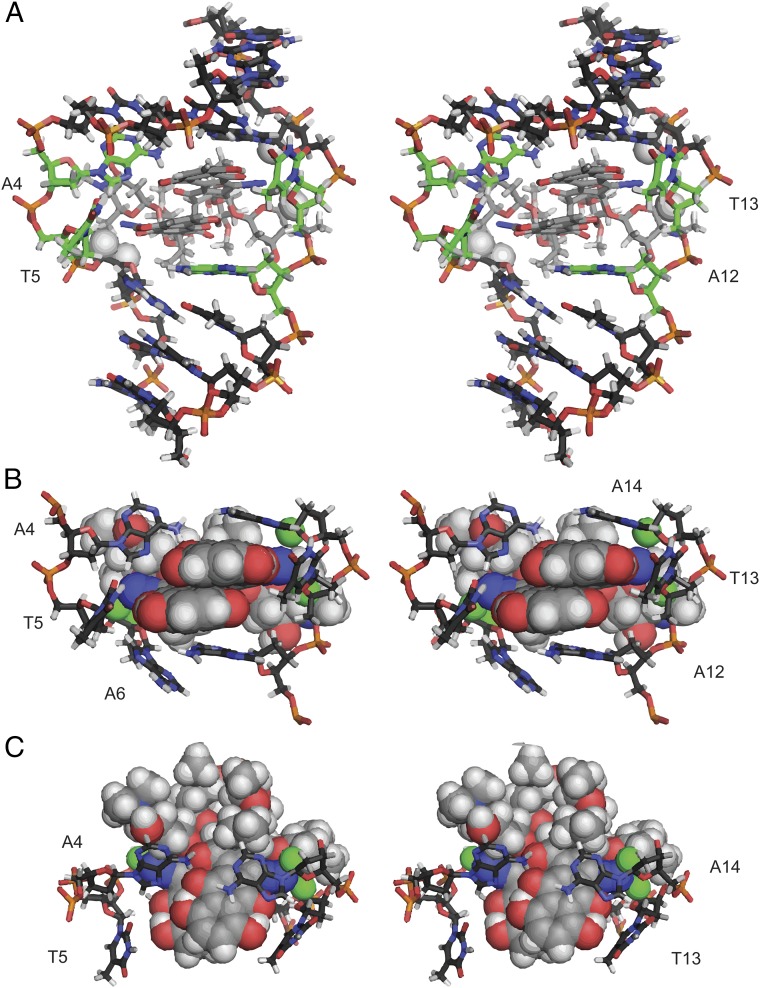
A computational model of the complex was then generated. A lowest-energy conformation of 1 was obtained by molecular mechanics optimization (100,000 starting conformers) using Spartan (32). This conformer was then subjected to density functional theory (DFT) optimization in Gaussian 09 [B3LYP 6–31G(d,p)+] (33). The solution structure was calculated using AMBER 14 (34, 35) by sampling 10 starting conformations of the Gaussian-minimized structure of 1 placed 15–40 Å from the DNA duplex in an explicit hydration environment. Following two steps of relaxation, each structure was annealed for 30 or 100 ps (20 structures total) using the NMR restraints, and the best structures were annealed for an additional 1 ns. To constrain the annealing, 425 NMR-derived distance restraints were used. The complex that best fit the NMR restraints was subjected to 1 ns of molecular dynamics, during which the root-mean-square deviation (RMSD) converged (SI Appendix, Fig. S6). Six structures were selected from the last 100 ps of modeling as representative of the final NMR solution structure (SI Appendix, Fig. S5). The overall RMSD was 0.74 ± 0.14 Å, and the RMSD at the intercalation site was 0.72 ± 0.18 Å (SI Appendix, Fig. S6). Three perspectives of the final structure are shown in Fig. 5 A–C. The RMSD between the annealed complex and the molecular dynamics-minimized structure, with the distance constraints removed, is 3.1 Å and contains changes to areas with large degrees of freedom, such as the oleandrose residues and the nucleotides that have been twisted out of the duplex. A single-point calculation reveals the bound conformation of 1 as 0.40 kcal/mol higher in energy than that of the minimized structure in solution (18).
Fig. 5.
Solution structure of (–)-lomaiviticin A (1) complexed to 5′-G1C2T3A4T5A6G7C8-3′. (A) Full stereoview. Bases flipped out of the duplex are shown in green. (B) Intercalation site, front view. (C) Intercalation site, top view. A6 H5′, A6 H4′, A14 H1′, and T13 H4′ are shown as white balls in A and as green balls in B and C and are located 4.2, 4.3, 4.2, and 4.9 Å, respectively, from the nearest diazo carbon atom.
Discussion
Prior cell free and tissue culture studies of (–)-lomaiviticin A (1) have established that its potent cytotoxic effects derive from the induction of DSBs in DNA (18, 19). Deuterium-labeling experiments are consistent with a mechanism involving the generation of vinyl radical intermediates at each diazo carbon of 1, followed by hydrogen atom abstraction from DNA (18). Although many natural products induce DNA DSBs via indirect mechanisms (for example, by inhibition or poisoning of topoisomerase complexes) (36), comparatively fewer natural products directly cleave DNA, the enediynes (37) and bleomycins (38) being notable exceptions. However, an understanding of the sequence preferences and mode of binding of 1 to DNA had remained conspicuously absent.
The data shown in Fig. 2 and Table 1 demonstrate that (–)-lomaiviticin A (1) binds AT tracks with higher affinity than GC tracks. A 63−76% decrease in fluorescence was observed for AT-containing duplexes whereas only an ∼13% decrease in fluorescence was observed for poly-dGdC. In addition, the DC50 of 1 toward poly-dAdT was 3.3-fold lower than that of poly-dGdC. This selectivity is consistent with a mode of binding involving disruption of base pairing. A single binding site for 1 was observed using the 10-mer duplex d(CGCATATGCG)2 (Table 1, entry 8).
Our data suggest a mode of interaction wherein (–)-lomaiviticin A (1) is inserted from the minor groove and the diazofluorene residues are positioned adjacent to each other, in a π-stacked orientation. We had anticipated intercalation of a single diazofluorene, and the discovery that both of these residues penetrate the duplex was unexpected. Unequivocal evidence for this derives from the observation that all of the arene C–H bond resonances in 1 (positions 8, 9, 8′, and 9′) are shifted upfield by 0.47–1.36 ppm in the presence of the duplex (Table 2). In addition, DNA binding abolishes the C2 symmetry of 1 and this leads to fortuitous resolution of the 8, 9, 8′, and 9′ resonances in the NMR spectrum of the complex, thereby permitting elucidation of vicinal relationships within these signals by DQF-COSY analysis. In turn, we detected an interdiazofluorene NOE contact (SI Appendix, Fig. S4), which provides additional and compelling evidence that these residues are in close proximity. Although somewhat more circumstantial, the observation that many of the resonances for each (now chemically inequivalent) pair of positions in 1 are similar (Δδ < 0.20 ppm) is consistent with a pseudosymmetrical mode of binding. This mode of binding represents a rare instance in which two aromatic units have been demonstrated to penetrate a duplex adjacent to each other without covalent binding. In the minimized structure the bases A4, T5, and T13 are twisted out of the duplex and base pairing has been disrupted for A12. Such base flipping has been reported for noncovalent (e.g., actinomycin) (39) and covalent (e.g., trioxicarcin A) (40) small molecule DNA binding agents. This base flipping may underlie the sequence selectivity of 1, as the stability gained from A–T pairs (hydrogen bonding and base stacking) is less than that gained from G–C pairs (41). An electrostatic interaction between the aminosugar residues of 1 and the phosphate backbone may help to compensate for the energetic penalty associated with base flipping; in the optimized structure these residues are within 4.3 Å of the nearest phosphate. Consistent with these data, (−)-lomaiviticin B (2), which cannot adopt the same mode of binding owing to the presence of a rigid bis(tetrahydrofuranol) structure, did not display high affinity for any of the oligonucleotides or duplexes examined (Fig. 2).
This mode of binding places each diazo carbon of 1 in close proximity to each DNA strand. A6 H5′ and A6 H4′ are located 4.2 and 4.3 Å from the nearest diazo carbon, respectively, whereas A14 H1′ and T13 H4′ are located 4.2 and 4.9 Å from the alternate diazo carbon atom, respectively (Fig. 5). These distances are within the range required for hydrogen-atom abstraction and initiation of strand cleavage with only a minor reorganization of 1, although the proximity of the second strand is likely to be altered following SSB formation. The ratio of DNA SSBs to DSBs produced by 1 in a plasmid cleavage assay was determined to be (5.3 ± 0.6):1 (18). The antitumor agent bleomycin mediates DSB formation via the stepwise generation of complementary SSBs (42). The ratio of SSBs:DSBs produced by bleomycin is in the range of 3.3–6.0:1, depending upon the sequence and assay conditions (43, 44). Thus, our data are consistent with stepwise cleavage of DNA by 1 and suggest that dissociation of 1 from the duplex is competitive with nucleophilic activation of the remaining diazofluorene following induction of the first SSB.
These studies also provide essential insights into structure–function relationships within (–)-lomaiviticin A (1). The amine substituents of 1 enhance DNA binding through electrostatic stabilization, but the nature and location of these substituents may be subject to refinement. On the other hand, the oleandrose residues of 1 may be amenable to simplification (or elimination), as these are not involved in extensive interactions with the duplex. Finally, these data provide further evidence that both diazofluorenes of 1 are essential for DNA cleavage activity. Incorporation of these insights into the design of wholly synthetic analogs of 1 capable of potently cleaving DNA is an area of current investigation in our laboratory.
It has been suggested that vinyl radicals are formed from (–)-lomaiviticin A (1) via nucleophilic addition to the diazofluorene, to generate a diazo intermediate, followed by loss of dinitrogen (18). This process was shown to be efficiently mediated by thiols, but intriguingly was observed to occur on warming aqueous solutions of 1 and DNA alone, suggesting that DNA itself may serve as a nucleophile toward 1. It was also noted that the first hydrodediazotization of 1 proceeded faster than reaction of the remaining diazofluorene or isolated monomeric diazofluorenes. DFT calculations suggested the high reactivity of 1 might be attributed to a short (3.8 Å) interdiazofluorene distance, which allows for delocalization of the developing negative charge across both diazofluorene substituents (18). In the complex characterized herein, this interaction may be enhanced by reducing the degrees of freedom within 1 and by compressing the interdiazofluorene distance (to 3.5 Å). Although detailed kinetic studies are needed to fully establish this point, this result suggests that 1 may become activated toward nucleophilic addition and DNA cleavage on binding to DNA. Such “shape-dependent catalysis” is reminiscent of the DNA-mediated activation of CC-1065 and duocarmycin elegantly elucidated by Boger and coworkers (45–47).
In conclusion, we have determined the sequence selectivity and mode of binding of (–)-lomaiviticin A (1) to DNA. Our data support a model involving penetration of both diazofluorene residues of 1 into the DNA duplex. This binding places each diazo carbon in close proximity to the DNA strands, and provides a structural foundation to study DNA DSB production by 1. These data also establish essential structure–function relationships that will guide the design of synthetic derivatives of 1. These results underscore the opportunities to elucidate novel modes of small molecule–DNA recognition through the study of DNA-reactive natural products (48).
Materials and Methods
Supplementary figures and tables are presented in the SI Appendix. In addition, the SI Appendix contains detailed experimental procedures for fluorescence intercalator displacement, NMR spectroscopic, and computational methods used in this study.
Supplementary Material
Acknowledgments
We thank Mr. Herman Nikolayevskiy for conducting a single-point energy calculation of 1. Financial support from the National Institute of General Medical Sciences (R01GM090000) is gratefully acknowledged. (−)-Kinamycin C (4) was a gift from the Developmental Therapeutics Program of the National Cancer Institute (NSC 138425).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID code 2n96).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519846113/-/DCSupplemental.
References
- 1.He H, et al. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J Am Chem Soc. 2001;123(22):5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 2.Woo CM, Beizer NE, Janso JE, Herzon SB. Isolation of lomaiviticins C-E, transformation of lomaiviticin C to lomaiviticin A, complete structure elucidation of lomaiviticin A, and structure-activity analyses. J Am Chem Soc. 2012;134(37):15285–15288. doi: 10.1021/ja3074984. [DOI] [PubMed] [Google Scholar]
- 3.Kersten RD, et al. Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica. ChemBioChem. 2013;14(8):955–962. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itŏ S, Matsuya T, Ōmura S, Otani M, Nakagawa A. A new antibiotic, kinamycin. J Antibiot (Tokyo) 1970;23(6):315–317. doi: 10.7164/antibiotics.23.315. [DOI] [PubMed] [Google Scholar]
- 5.Hata T, Ōmura S, Iwai Y, Nakagawa A, Otani M. A new antibiotic, kinamycin: Fermentation, isolation, purification and properties. J Antibiot (Tokyo) 1971;24(6):353–359. doi: 10.7164/antibiotics.24.353. [DOI] [PubMed] [Google Scholar]
- 6.Ōmura S, et al. Structure of kinamycin C, and the structural relation among kinamycin A, B, C, and D. Chem Pharm Bull (Tokyo) 1971;19(11):2428–2430. [Google Scholar]
- 7.Furusaki A, et al. Crystal and molecular structure of kinamycin C p-bromobenzoate. Isr J Chem. 1972;10(2):173–187. [Google Scholar]
- 8.Ōmura S, Nakagawa A, Yamada H, Hata T, Furusaki A. Structures and biological properties of kinamycin A, B, C, and D. Chem Pharm Bull (Tokyo) 1973;21(5):931–940. doi: 10.1248/cpb.21.931. [DOI] [PubMed] [Google Scholar]
- 9.Cone MC, Seaton PJ, Halley KA, Gould SJ. New products related to kinamycin from Streptomyces murayamaensis. I. Taxonomy, production, isolation and biological properties. J Antibiot (Tokyo) 1989;42(2):179–188. doi: 10.7164/antibiotics.42.179. [DOI] [PubMed] [Google Scholar]
- 10.Seaton PJ, Gould SJ. New products related to kinamycin from Streptomyces murayamaensis. II. Structures of pre-kinamycin, keto-anhydrokinamycin, and kinamycins E and F. J Antibiot (Tokyo) 1989;42(2):189–197. doi: 10.7164/antibiotics.42.189. [DOI] [PubMed] [Google Scholar]
- 11.Gould SJ, Hong ST, Carney JR. Cloning and heterologous expression of genes from the kinamycin biosynthetic pathway of Streptomyces murayamaensis. J Antibiot (Tokyo) 1998;51(1):50–57. doi: 10.7164/antibiotics.51.50. [DOI] [PubMed] [Google Scholar]
- 12.Gould SJ. Biosynthesis of the Kinamycins. Chem Rev. 1997;97(7):2499–2510. doi: 10.1021/cr9600215. [DOI] [PubMed] [Google Scholar]
- 13.Marco-Contelles J, Molina MT. Naturally occurring diazo compounds: The kinamycins. Curr Org Chem. 2003;7(14):1433–1442. [Google Scholar]
- 14.Arya DP. Diazo and diazonium DNA cleavage agents: Studies on model systems and natural product mechanisms of action. In: Lee M, editor. Heterocyclic Antitumor Antiobiotics, Topics in Heterocyclic Chemistry. Vol 2. Springer; Berlin, Heidelberg: 2006. pp. 129–152. [Google Scholar]
- 15.Nawrat CC, Moody CJ. Natural products containing a diazo group. Nat Prod Rep. 2011;28(8):1426–1444. doi: 10.1039/c1np00031d. [DOI] [PubMed] [Google Scholar]
- 16.Herzon SB. The kinamycins. In: Li JJ, Corey EJ, editors. Total Synthesis of Natural Products. At the Frontiers of Organic Chemistry. Springer; Berlin: 2012. [Google Scholar]
- 17.Herzon SB, Woo CM. The diazofluorene antitumor antibiotics: Structural elucidation, biosynthetic, synthetic, and chemical biological studies. Nat Prod Rep. 2012;29(1):87–118. doi: 10.1039/c1np00052g. [DOI] [PubMed] [Google Scholar]
- 18.Colis LC, et al. The cytotoxicity of (–)-lomaiviticin A arises from induction of double-strand breaks in DNA. Nat Chem. 2014;6(6):504–510. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colis LC, Hegan DC, Kaneko M, Glazer PM, Herzon SB. Mechanism of action studies of lomaiviticin A and the monomeric lomaiviticin aglycon. Selective and potent activity toward DNA double-strand break repair-deficient cell lines. J Am Chem Soc. 2015;137(17):5741–5747. doi: 10.1021/ja513117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hühn D, Bolck HA, Sartori AA. Targeting DNA double-strand break signalling and repair: Recent advances in cancer therapy. Swiss Med Wkly. 2013;143:w13837. doi: 10.4414/smw.2013.13837. [DOI] [PubMed] [Google Scholar]
- 21.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98(3):1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 22.Woo CM, Ranjan N, Arya DP, Herzon SB. Analysis of diazofluorene DNA binding and damaging activity: DNA cleavage by a synthetic monomeric diazofluorene. Angew Chem Int Ed Engl. 2014;53(35):9325–9328. doi: 10.1002/anie.201404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boger DL, Fink BE, Hedrick MP. Total synthesis of distamycin A and 2640 analogues: A solution-phase combinatorial approach to the discovery of new, bioactive DNA binding agents and development of a rapid, high-throughput screen for determining relative DNA binding affinity or DNA binding sequence selectivity. J Am Chem Soc. 2000;122(27):6382–6394. [Google Scholar]
- 24.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J Am Chem Soc. 2001;123(25):5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 25.Tse WC, Boger DL. A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Acc Chem Res. 2004;37(1):61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 26.Lee LG, Chen C-H, Chiu LA. Thiazole orange: A new dye for reticulocyte analysis. Cytometry. 1986;7(6):508–517. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura T, et al. DNA binding of tilorone: 1H NMR and calorimetric studies of the intercalation. Biochemistry. 2007;46(27):8156–8163. doi: 10.1021/bi602402m. [DOI] [PubMed] [Google Scholar]
- 28.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley and Sons; New York: 1986. [Google Scholar]
- 29.Kim S-G, Lin L-J, Reid BR. Determination of nucleic acid backbone conformation by 1H NMR. Biochemistry. 1992;31(14):3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- 30.Cordier C, Pierre VC, Barton JK. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: An NMR solution study. J Am Chem Soc. 2007;129(40):12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhury JR, Bierbach U. Characterization of the bisintercalative DNA binding mode of a bifunctional platinum-acridine agent. Nucleic Acids Res. 2005;33(17):5622–5632. doi: 10.1093/nar/gki869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spartan 10 for PC (2010) (Wavefunction, Inc., Irvine, CA)
- 33.Frisch MJ, et al. 2009. Gaussian 09, Revision A.1 (Gaussian, Inc., Wallingford, CT)
- 34.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheatham TE, 3rd, Case DA. Twenty-five years of nucleic acid simulations. Biopolymers. 2013;99(12):969–977. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borders DB, Doyle TW. Enediyne Antibiotics as Antitumor Agents. Marcel Dekker; New York: 1995. [Google Scholar]
- 38.Sidney MH. Bleomycin Group Antitumor Agents. Anticancer Agents from Natural Products. 2nd Ed. CRC; Boca Raton, FL: 2011. pp. 451–478. [Google Scholar]
- 39.Chou S-H, Chin K-H, Chen F-M. Looped out and perpendicular: Deformation of Watson-Crick base pair associated with actinomycin D binding. Proc Natl Acad Sci USA. 2002;99(10):6625–6630. doi: 10.1073/pnas.102580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfoh R, Laatsch H, Sheldrick GM. Crystal structure of trioxacarcin A covalently bound to DNA. Nucleic Acids Res. 2008;36(10):3508–3514. doi: 10.1093/nar/gkn245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34(2):564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stubbe J, Kozarich JW, Wu W, Vanderwall DE. Bleomycins: A structural model for specificity, binding, and double strand cleavage. Acc Chem Res. 1996;29(7):322–330. [Google Scholar]
- 43.Povirk LF, Wübter W, Köhnlein W, Hutchinson F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 1977;4(10):3573–3580. doi: 10.1093/nar/4.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Absalon MJ, Kozarich JW, Stubbe J. Sequence-specific double-strand cleavage of DNA by Fe-bleomycin. 1. The detection of sequence-specific double-strand breaks using hairpin oligonucleotides. Biochemistry. 1995;34(6):2065–2075. doi: 10.1021/bi00006a029. [DOI] [PubMed] [Google Scholar]
- 45.Boger DL, Garbaccio RM. Catalysis of the CC-1065 and duocarmycin DNA alkylation reaction: DNA binding induced conformational change in the agent results in activation. Bioorg Med Chem. 1997;5(2):263–276. doi: 10.1016/s0968-0896(96)00238-6. [DOI] [PubMed] [Google Scholar]
- 46.Boger DL, Garbaccio RM. Shape-dependent catalysis: Insights into the source of catalysis for the CC-1065 and duocarmycin DNA alkylation reaction. Acc Chem Res. 1999;32(12):1043–1052. [Google Scholar]
- 47.Wolkenberg SE, Boger DL. Mechanisms of in situ activation for DNA-targeting antitumor agents. Chem Rev. 2002;102(7):2477–2495. doi: 10.1021/cr010046q. [DOI] [PubMed] [Google Scholar]
- 48.Tse WC, Boger DL. Sequence-selective DNA recognition: Natural products and nature’s lessons. Chem Biol. 2004;11(12):1607–1617. doi: 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.