Significance
The Min system of Escherichia coli uses the proteins MinD and MinE to form a standing wave oscillator on the membrane that prevents cell division at the cell poles. Using purified MinD and MinE, several dynamic patterns have been previously reconstituted on lipid bilayers. However, these dissimilar patterns occur under different reaction settings; therefore, the underlying mechanistic principles are unclear. By using a limiting supply of MinD, we reproduced standing wave oscillation on a flat bilayer. We find that periodic depletion of active MinD from solution is essential for the standing wave. Also, the MinD-to-MinE ratio on the bilayer acts as a toggle switch between membrane-binding and -release by MinD, which drives the oscillation.
Keywords: cell division, subcellular organization, pattern formation, self-organization, intracellular positioning
Abstract
The Escherichia coli Min system self-organizes into a cell-pole to cell-pole oscillator on the membrane to prevent divisions at the cell poles. Reconstituting the Min system on a lipid bilayer has contributed to elucidating the oscillatory mechanism. However, previous in vitro patterns were attained with protein densities on the bilayer far in excess of those in vivo and failed to recapitulate the standing wave oscillations observed in vivo. Here we studied Min protein patterning at limiting MinD concentrations reflecting the in vivo conditions. We identified “burst” patterns—radially expanding and imploding binding zones of MinD, accompanied by a peripheral ring of MinE. Bursts share several features with the in vivo dynamics of the Min system including standing wave oscillations. Our data support a patterning mechanism whereby the MinD-to-MinE ratio on the membrane acts as a toggle switch: recruiting and stabilizing MinD on the membrane when the ratio is high and releasing MinD from the membrane when the ratio is low. Coupling this toggle switch behavior with MinD depletion from the cytoplasm drives a self-organized standing wave oscillator.
The ParA/MinD family of adenosine triphosphatases (ATPases) forms dynamic patterns on biological surfaces, such as the nucleoid or the inner membrane, to spatially organize a variety of processes including the segregation of plasmids, chromosomes, and organelles as well as positioning the cell division machinery (1, 2). These ATPases are commonly associated with a partner protein that stimulates the local release of the ATPase from its binding surface, resulting in the formation of a dynamic ATPase pattern that imparts positional information to the cell (3). Despite the ubiquity of these minimal self-organizing positioning systems and their importance in a wide variety of essential processes throughout the microbial world, the patterning mechanism is not fully understood.
In Escherichia coli, the MinD ATPase forms a cell-pole to cell-pole oscillator on the membrane, as a standing wave with a node at the cell center, in response to its stimulator protein MinE (4–6). The final component of the Min system is the inhibitor of divisome assembly, MinC, which is a passenger protein on MinD and not required for oscillation (7, 8). Oscillations apparently result from the perpetual chase and release of MinD by MinE on the membrane, which produces a time-averaged concentration of MinC that is lowest at midcell (4, 8–11). The Min system therefore promotes symmetric cell division at midcell by inhibiting division near the poles (5).
The patterning reaction occurs from a series of ATP-driven protein–protein and protein–membrane interactions. ATP promotes MinD dimerization and membrane binding via its membrane-targeting sequence (MTS) (12–15). It is currently thought that membrane-bound MinD at a cell pole recruits MinE dimers to the membrane in the form of an E-ring residing on the periphery of the MinD-bound zone (5, 6, 16). MinE also has an MTS essential for the spatial regulation of cell division (17). Structural studies suggest that, in solution, the MTS and the adjacent MinD–interaction domain of MinE are sequestered in the hydrophobic core of the dimer (18, 19). Thus, under physiological conditions, MinE does not interact with MinD in the absence of membrane and shows only a weak affinity for E. coli membrane in the absence of MinD (17). This transient interaction with membrane apparently helps unveil the adjacent MinD interaction interface, which is bound and stabilized by membrane-bound MinD (18, 19). MinD interaction with MinE leads to ATP hydrolysis and membrane release (16). After MinD release, MinE dimers remain bound to the membrane (17, 20–22). This “lingering” MinE species in vivo could remain on the membrane at the cell pole from which MinD just disassembled, thereby providing positional memory to the system by preventing MinD from rebinding the same pole and directing de novo MinD binding to the opposing pole.
The role of lingering MinE in patterning via its membrane-binding activity has been debated. Previous in vitro reconstitutions suggest a role for MinE lingering on the membrane after releasing MinD (22, 23), but doubt remains as to whether this property is important for self-organization. For example, in vivo dynamics have been recapitulated by simulations based on different models either with (24) or without (25, 26) considering MinE membrane-binding activity. Here we address the molecular mechanism by directly studying the MinE membrane-binding requirement on patterning. We find that a MinE mutant lacking its MTS can still form patterns, but cannot form an E-ring or regulate the periodicity of the standing wave.
The apparent simplicity of this binary system makes it attractive for modeling, and many mathematical models recapitulate qualitative aspects of Min patterns (11, 23, 25, 27). Each model is predicated on significantly different molecular mechanisms, but all recapitulate the in vivo dynamics quite well, which makes it difficult to discriminate among the considered mechanisms. To date, the underlying mechanistic basis for patterning has not been uniquely constrained by in vivo or in vitro experiments. Specifically, to achieve self-organized spatiotemporal patterning, at least one nonlinear reaction term is required, e.g., a higher-order concentration-dependent rate, in addition to at least one energy-coupled reaction step that is irreversible. In previous modeling exercises, this critical nonlinearity has been included in a variety of manners as ad hoc cooperativity parameters without direct experimental validation. Therefore, it remains critically important to experimentally identify the molecular mechanistic source of this nonlinearity.
To experimentally dissect the mechanistic details of the system, we developed a cell-free imaging technique to visualize Min patterning on a supported lipid bilayer (SLB). Min patterning was first reconstituted in the form of spiraling wave trains of MinD chased by MinE on the bottom of an SLB-coated well (28). In our flowcell setup, several modes of Min patterning, including spirals, have been reconstituted (22, 29). However, in all of these previous reconstitutions, patterns are achieved with protein densities on the bilayer far in excess of those in vivo. Furthermore, the observed patterns lack standing wave dynamics with nodes where the time-averaged local MinD concentration is minimum, as observed at midcell in vivo. Recently, standing wave dynamics were reconstituted by isolating the reaction in small volumes (30, 31). However, the mechanistic basis for standing wave oscillation has not been experimentally addressed. We hypothesized that Min protein depletion from the cytoplasm as proposed earlier by Meinhardt and de Boer (11) is a critical, but experimentally unexplored, feature of the oscillation mechanism.
With our flowcell setup, we find that, under limiting MinD conditions, MinD and MinE form a previously unidentified pattern that we term “bursts.” Bursts share several features with in vivo Min patterns such as the ability to spontaneously form standing wave patterns even at a membrane-surface area-to-volume ratio much lower than a bacterial cell. Our observations indicate that pole-to-pole oscillation of Min proteins is correlated with periodic depletion of MinD dimers from the cytoplasm during the formation of MinD polar zones on the membrane. Together, the data reported here support a patterning mechanism that is largely governed by the local MinD:MinE stoichiometry on the membrane, which acts as a toggle switch promoting MinE-stimulated MinD recruitment to the membrane when MinD is in excess or MinE-stimulated MinD release from the membrane when MinE is in excess. We propose that this toggle switch mechanism, coupled with cytoplasmic MinD depletion, provides the crucial nonlinear coupling terms that support standing wave oscillations of the Min system.
Results
Local Surface Concentration of Min Proteins Dictates the Mode of Patterning.
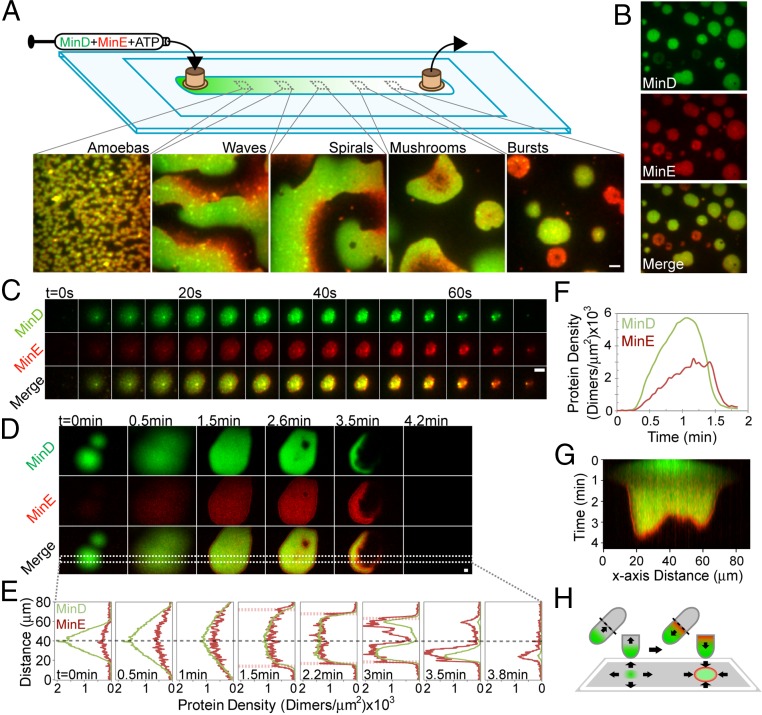
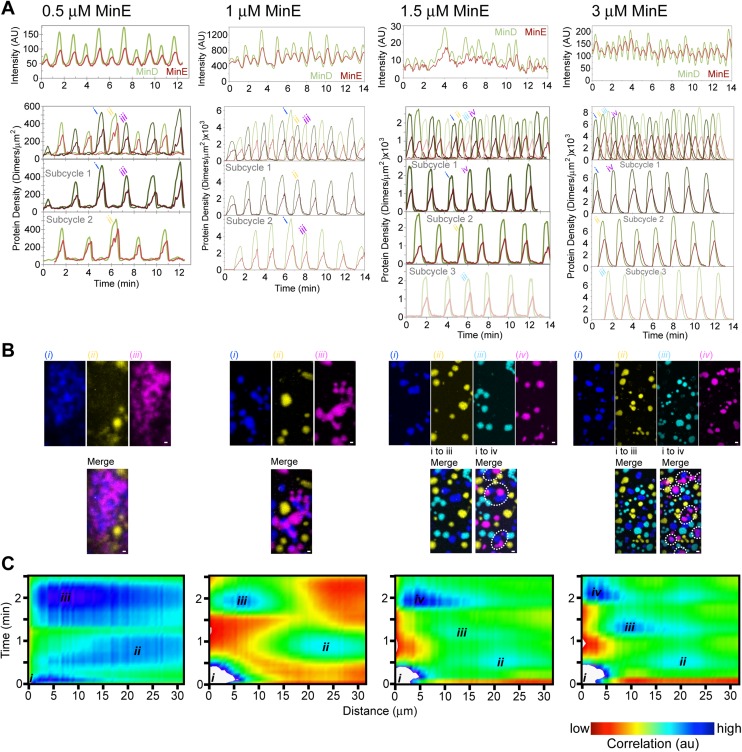
In our standard experimental protocol for this report, 1 μM GFP–MinD and 1.5 μM MinE mixed 1:19 with MinE-Alexa 647 (or an alternative MinE concentration as stated) were preincubated with ATP and infused into an ∼25-μm-thick flowcell coated with an SLB composed of E. coli lipid. ATP-bound MinD dimers that formed during the preincubation bound the SLB near the inlet of the flowcell. This preferential SLB binding of MinD (and MinE) near the inlet depleted the solution concentration of proteins, resulting in lower concentrations further downstream. Therefore, when the flow was stopped after infusion of ∼3 flowcell volumes, MinD and MinE formed a spectrum of patterns from inlet to outlet on the SLB due to the decreasing concentration gradient of MinD toward the flowcell outlet (Fig. 1A and Movie S1). Near the inlet, MinD and MinE formed densely packed “amoebas,” a previously observed pattern on the SLB (29) that consists of MinD-rich centers (D-cores) with perimeters that are surrounded by a well-defined MinE-rich ring (E-ring). Moving down the flowcell away from the inlet to lower MinD densities, amoebas were replaced by traveling waves that progressively became more regular spirals (Fig. 1A and Movie S1). Waves were composed of spatially periodic and staggered bands of MinD and MinE, as previously described (22–24, 28, 29). Closer to the outlet of the flowcell, where MinD concentration was limited, we observed the previously unidentified mode of patterning called “bursts,” described in detail below. Between spirals and bursts was an intermediate pattern of periodically growing and dissipating “mushrooms” (Movie S2 and Fig. S1 A–C). No patterns were observed immediately adjacent to the outlet.
Fig. 1.
MinD and MinE form bursts under limiting MinD conditions. (A) Reconstituting a spectrum of Min patterns. GFP-MinD (green) and MinE-Alexa647 (red) were preincubated with ATP and infused for 10 min into a flowcell coated with an SLB made with E. coli lipid. Still images of the spectrum of patterns on the SLB supported by the decreasing GFP-MinD density on the SLB from inlet to outlet are shown. (B) Still image of a population of bursts. (C) Time-lapse of a burst on E. coli lipid. (D) Time-lapse of a large burst on an mSLB. (E) Cross-section of MinD (green) and MinE (red) protein densities associated with the burst in D. The dashed gray and red lines indicate the burst center and regressing E-ring, respectively. (F) Time course of MinD and MinE densities within the burst in C. (G) Kymograph for MinD (green) and MinE (red) of the burst cross-section in D. (H) The burst dynamics resemble in vivo MinD polar zone dynamics. If an E. coli cell were cut in half and flattened onto a single plane, the formation and disassembly of a MinD polar zone by MinE in vivo equates to a burst cycle on the SLB. (Scale bars: 5 μm.) See also Movies S1–S4 and Fig. S1.
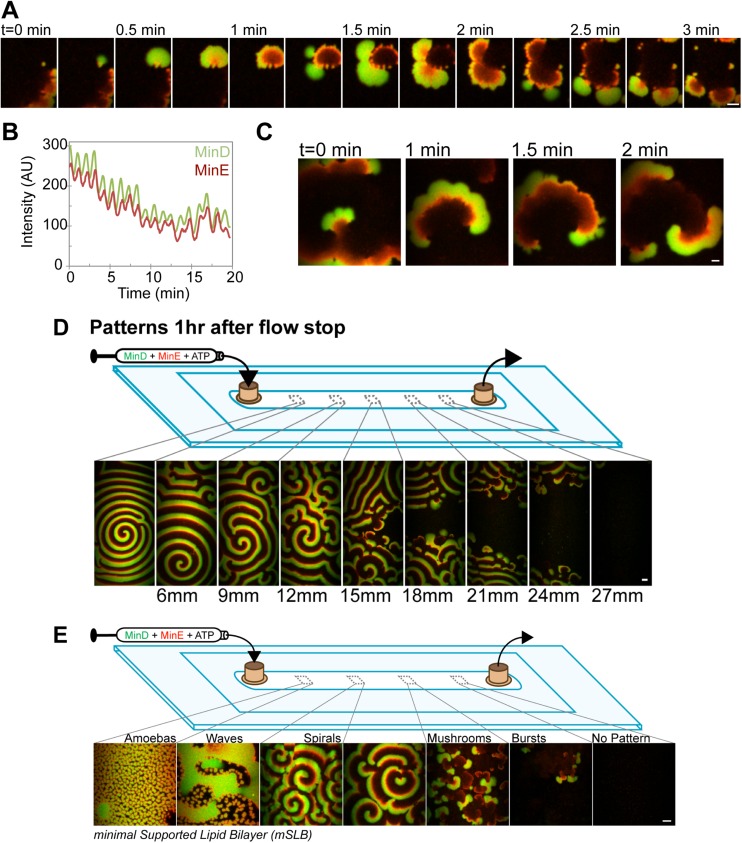
Fig. S1.
Bursts transition to spirals with increasing levels of SLB-bound Min proteins. (A) A time-lapse image series of “mushrooms”—an intermediate pattern between SLB regions with spirals and bursts. Mushrooms did not expand and dissipate symmetrically like bursts that were spatially disconnected from one another. Rather, mushrooms tended to bud from the disassembly phase of the previous set of mushrooms, resulting in asymmetric growth of the D-core and a spatially skewed disassembly by MinE. After the MinD binding front of a mushroom was stalled by the formation of an E-ring, the spatial asymmetry was propagated by subsequent mushrooms. GFP-MinD (green) and MinE-Alexa647 (red) channels are merged. (B) Like bursts, mushrooms temporally oscillated as radially expanding D-cores that were corralled and disassembled by MinE. GFP-MinD and MinE-Alexa647 intensities were plotted over time to emphasize the temporal periodicity of mushrooms. (C) A time-lapse image series of mushrooms merging to form Min spirals. With increasing locally averaged SLB-bound densities of Min proteins, mushrooms merged and developed into the continuous propagation of spirals. (Scale bars: 5 μm.) See also Movie S2. (D) Min spirals are stable at limiting MinD concentrations. Still images when scanning the SLB at 3-mm intervals 1 h after flow stoppage, under limiting MinD concentrations, show the narrow boundary between SLB regions with or without spirals. The abrupt spatial transition highlights several features of the patterning mechanism. First, spirals were the most stable pattern on the SLB. Regions with any other pattern eventually converted to segregated subareas—one with spirals and the other with no pattern. Therefore, even with limiting MinD, spirals are sustained by depleting the MinD supply from neighboring areas of the SLB where bursts initially oscillated. Our observations suggest that, once MinD and MinE are active, their propensity to associate where SLB-bound protein molecules are already situated, without requiring de novo nucleation of new binding zone initiation, locally sequesters the proteins and restricts diffusional loss to nonpatterned areas of the SLB. Patterned areas would also recruit active MinD dimers via bulk-phase diffusion from neighboring nonpatterned areas, thereby suppressing de novo pattern initiation. This dynamic phase-separation under MinD-limiting conditions evolves over long spatial (hundreds of micrometers to millimeters) and temporal (hours to days) scales. In contrast, the burst pattern that we describe in the main text shows spatiotemporal scales that compare well with the in vivo Min pattern dynamics. (Scale bar: 40 μm.) See also Movie S3. (E) Reconstituting a spectrum of patterns on a mSLB. GFP-MinD (1 μM; green) and MinE-Alexa647 (1.5 μM; red) were preincubated with ATP and infused for 10 min into the mSLB-coated flowcell. Flow was stopped before acquisition. Freeze-frame images of the spectrum of Min patterns on the mSLB are shown from inlet to outlet. (Scale bar: 40 μm.) The mSLB was composed of 67% phosphotidylcholine and 33% phosphatidylglycerol and supported the same spectrum of patterns as that found on E. coli lipid (Fig. 1A).
MinD and MinE Form Radial Burst Patterns Under Limiting MinD Conditions.
The membrane surface area in vivo (∼10 μm2) can accommodate a far greater number of MinD dimers than exists in the cell (∼2,000 molecules) (32). Therefore, cytoplasmic depletion of active MinD dimers likely plays a critical role in the oscillatory dynamics on the membrane. Accordingly, we focused on SLB regions with lower MinD concentration where the newly discovered burst patterns emerged immediately after the flow was stopped (Fig. 1B). Our method provided a reliable way to study bursts for roughly 1 h before the SLB segregated into an area with spirals and an area with no pattern (Movie S3 and Fig. S1D).
Min bursts started with GFP-MinD, in the presence of MinE, cooperatively binding the SLB as radially expanding circular initiation centers (Fig. 1C). MinE binding immediately followed, but at a slower rate. Burst expansion then halted, presumably due to slowed MinD binding as active dimers were depleted from solution. Finally, as the D-core started to shrink, an E-ring formed around the perimeter where the MinE concentration exceeded that of MinD. This was followed by the rapid dissipation of the D-core (Fig. 1C).
A “minimal” supported lipid bilayer (mSLB), composed of 67% phosphotidylcholine and 33% phosphatidylglycerol (22), supported patterns essentially identical to those found on E. coli lipid (Fig. S1E) except that the improved uniformity of the mSLB facilitated observation of burst pattern details (Fig. 1D and Movie S4). As on E. coli lipid, D-core expansion stalled and regressed as MinE accumulated to form an E-ring (Fig. 1 D and E). The E-ring followed the receding perimeter of the D-core with a MinE-to-MinD ratio that significantly exceeded one (Fig. 1E). Within the D-core, the initial rapid binding of MinD to the SLB was accompanied by a slower accumulation of MinE up to a threshold density that led to the rapid release of both proteins, with MinE release lagging that of MinD (Fig. 1 F and G).
The geometry of individual small bursts closely mirrored Min oscillation dynamics in vivo. If an E. coli cell was cut in half and the resulting cylinder was flattened onto a plane, the initiation and expansion of a MinD polar zone, followed by the chase and release accompanied by an E-ring, would correspond to a burst cycle (Fig. 1H).
Clusters of Min Bursts Form Standing Waves.
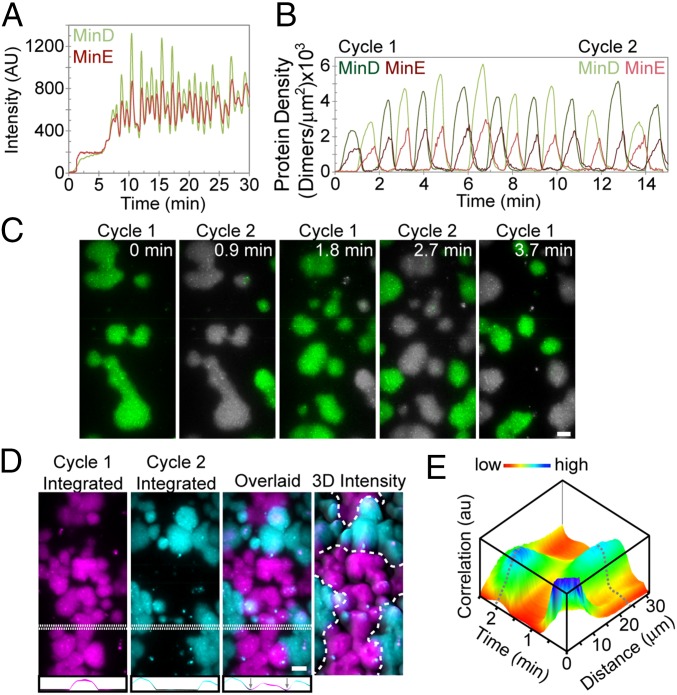
Aside from the geometric similarities with in vivo Min patterns, the most striking feature of bursts was that they did not occur randomly on the SLB (Fig. 2A and Movie S5). Rather, spatially separated clusters of bursts would expand and dissipate in unison across a region of the SLB with the dissipation phase of one set temporally overlapping with the initiation phase of the next (Fig. 2B). Burst cycles were therefore composed of temporally staggered subsets, each with a highly regular period of 2.1 ± 0.2 min. While one set of bursts dissipated, the next set developed on regions of the SLB devoid of lingering MinE. Therefore, successive phase-shifted subcycles never spatially overlapped (Fig. 2C). The MinD pattern, when integrated over several burst cycles, clearly shows a standing wave with a boundary node spatially separating the subcycles (Fig. 2D). By performing a 2D cross-correlation image analysis of the burst cycles over time, we found that successive burst phases were spatially separated by a characteristic distance of 24 μm (Fig. 2E and Materials and Methods). We conclude that, under limiting MinD conditions, the Min system self-organizes into a standing wave oscillator even at the significantly lower surface-to-volume ratio of our flowcell compared with that of a bacterial cell.
Fig. 2.
Bursts spatiotemporally oscillate. (A) GFP-MinD and MinE-Alexa647 intensities over time for the entire imaged region (150 × 75 μm) to emphasize the temporal periodicity of bursts. (B) MinD and MinE densities associated with individual bursts over time. The two phase-shifted subcycles are distinguished by dark- and light-colored lines. (C) Still images of successive burst clusters at their peak MinD protein density. The image of the previous burst was overlaid in gray to emphasize that consecutive subcycles never spatially overlap. (D) Bursts form a standing wave oscillator. Still images of burst clusters at their peak MinD protein densities, as shown in C, were integrated for subcycle 1 (magenta) and subcycle 2 (cyan) and then overlaid. Cross-sections of the intensities were graphed below to emphasize the MinD nodes (arrows). The dashed line in the 3D intensity plot outlines the MinD minima node line between spatially segregated subcycle zones. (Scale bars: 10 μm.) (E) Two-dimensional image cross-correlation analysis of movie frames from the MinD channel quantifying the spatial and temporal periodicity and phase relation of burst cycles (Materials and Methods). Dashed gray lines highlight the peak positions in time and space. See also Movie S5.
Solution Concentration of Min Proteins Oscillates in the Burst Zone as Predicted by the Active MinD Depletion Hypothesis.
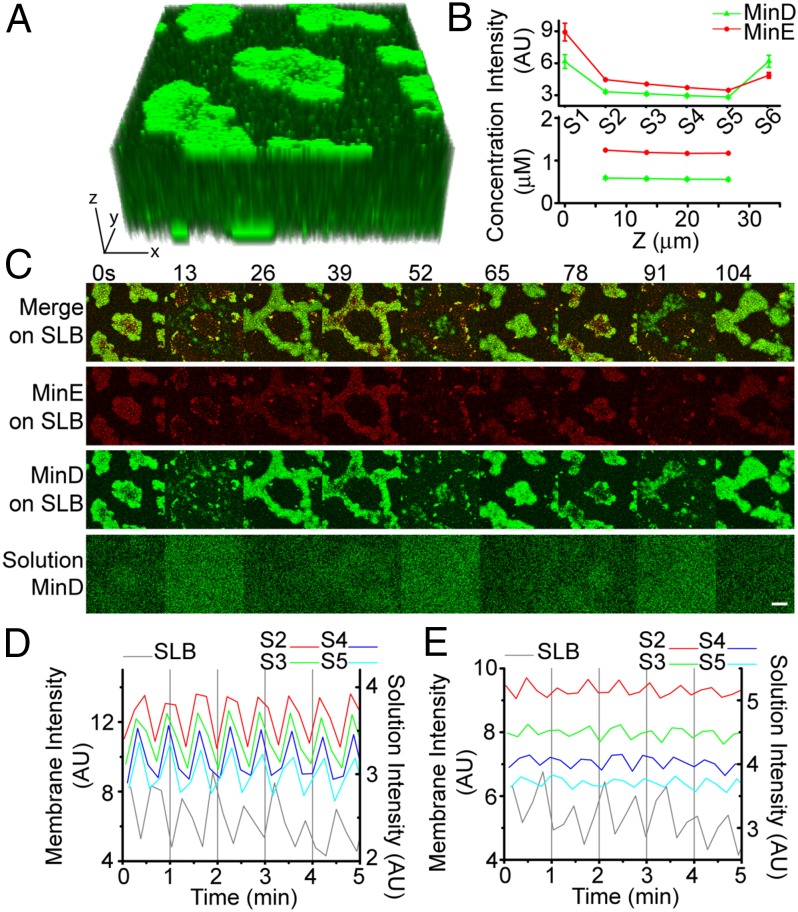
The distribution of Min proteins in solution remains relatively homogeneous for Min patterns that form when the protein supply is not limiting, such as amoebas or spirals (Fig. 1A) (22, 28, 29). However, in vivo, the total number of MinD and MinE molecules are essentially fixed at subsaturating concentrations. We suspected, therefore, that the cytosolic protein concentration is inversely related to the membrane-bound protein concentration. We propose that this is also the case for the burst conditions studied in our 25-μm-thick flowcell. To verify this supposition, we used confocal microscopy to measure the distribution of Min proteins in the solution phase over bursts on the SLB (Fig. 3A). Throughout the entire solution depth of the flowcell, the time-averaged concentrations of MinD and MinE monomers were ∼0.55 and ∼1.25 μM, respectively—less than the input concentrations of 1 μM MinD and 1.5 μM MinE (Fig. 3B). As expected from the principle of mass conservation, the solution concentrations of Min proteins oscillated 180° out of phase with burst cycles on the SLB (Fig. 3C). When a burst cluster reached its peak protein density on the SLB, the solution concentration of Min proteins decreased to a minimum throughout the entire depth above the burst cluster, and vice versa (Fig. 3 D and E).
Fig. 3.
Min proteins oscillate in solution under limiting MinD conditions. (A) A Z-stack image (100 × 100 μm) of a flowcell with bursts. (B) Min protein intensities in arbitrary units from each slice in A averaged over 12 frames (two cycles) and plotted as a function of slice number from the bottom (S1) to top (S6) surface (Top). Solution intensities were converted to monomer concentrations and plotted as a function of z-position in the flow cell (Bottom) (SI Materials and Methods). (C) Time-lapse images of MinD (green) and MinE (red) bursts on the SLB (S6) or in solution (S5). Note that during MinD release (0, 39, and 78 s) the diffuse patterns of MinD in solution (S5) match those on the SLB (S6). (D) MinD intensity temporally oscillates in-phase throughout the solution (S2–S5), but 180° out-of-phase with MinD on the SLB (S6). (E) MinE intensity in solution oscillates in-phase (S2–S5) but 180° out-of-phase with MinE on the SLB (S6). (Scale bars: 10 μm.)
The solution oscillation amplitude was small (∼20% fluctuation around the average concentration), but was consistent with the number of MinD dimers bound in bursts on the SLB. This percentage likely reflects the total amount of active MinD dimers that accumulate in, or deplete from, solution when bursts dissipate or expand, respectively. We conclude that depletion of active MinD from the entire cytosol, which accompanies MinD polar zone formation, is a critical determinant of Min oscillation dynamics in vivo.
Accumulation and Dissipation Cycle of MinE on the Membrane Sets the Oscillation Period.
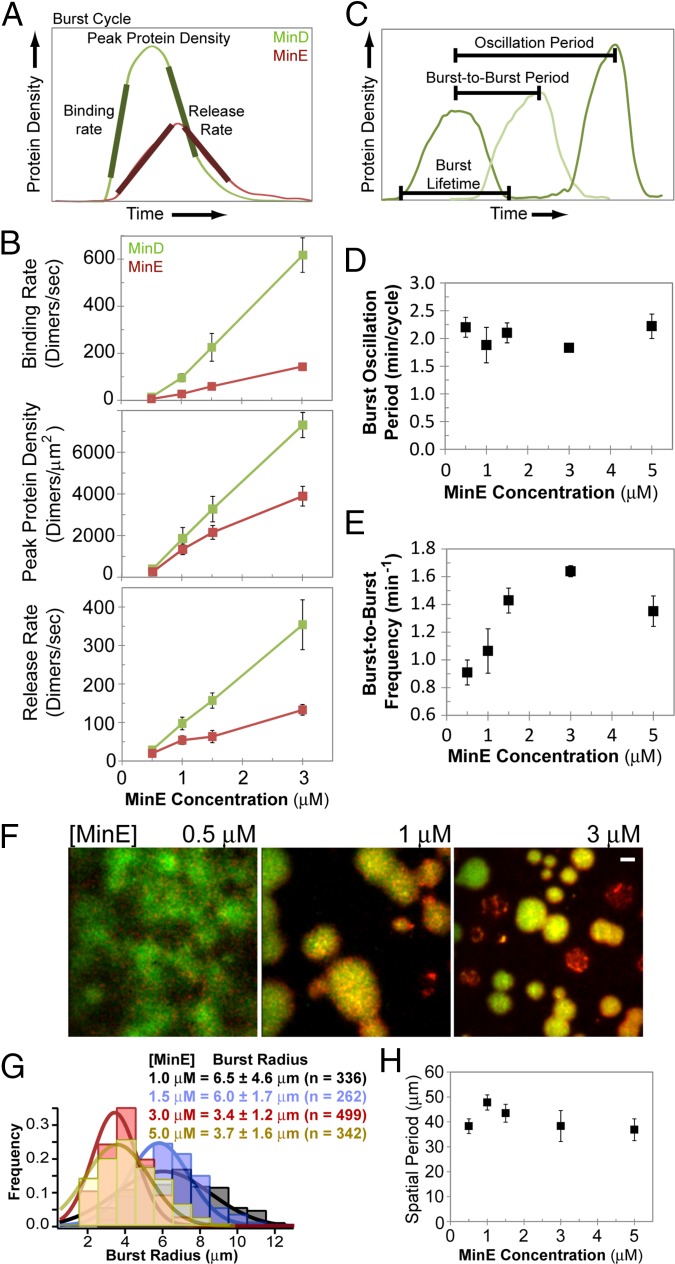
The MinD-to-MinE ratio has been shown to influence Min patterning both in vivo (6) and in vitro (22, 28). How Min protein stoichiometry controls the patterning mechanism is unclear. We studied the effects of varying the protein ratio on burst cycle dynamics (Fig. 4A) by changing the initial input MinE concentration while holding MinD concentration constant. Unexpectedly, at higher MinE concentrations, the binding rate of not only MinE, but also MinD, increased during burst expansion (Fig. 4 A and B). However, the time to reach the peak protein density remained roughly the same, which resulted in higher protein densities within bursts at higher MinE concentrations. The release rate of both proteins also increased with increasing MinE. The burst oscillation period remained constant at ∼2 min with the initiation of a new burst cycle occurring after the dissipation of MinE at the same SLB zone (Fig. 4 C and D). Current models posit MinE as a stimulator of MinD release from the membrane. However, our MinE titration data suggest that the role of MinE goes much further in the patterning mechanism. Our results indicate MinE as both a membrane association and dissociation catalyst for MinD.
Fig. 4.
MinE concentration affects burst dynamics. (A) Three aspects of a burst cycle were studied: the average binding rate, peak protein density, and release rate of both MinD (green) and MinE (red). (B) MinE is a catalyst for MinD membrane binding and release. Rates were obtained from averaging the slope of the linear portions of the expansion and dissipation phases of bursts over at least two experiments per MinE concentration. (C) Three aspects of burst cycles were studied over time. “Burst lifetime” is the duration between burst initiation and dissipation. “Burst-to-burst period” is the temporal separation between successive spatially nonoverlapping bursts over a wide region of the SLB. “Oscillation period” is the duration between successive burst clusters occupying the same region of the SLB. (D) The oscillation period remained constant with varying MinE. (E) The burst-to-burst frequency (inverse of burst-to-burst period) increases with MinE concentration. (F) Burst size decreased with higher MinE. Still images of bursts are shown at the indicated MinE concentrations. (Scale bar: 5 μm.) (G) Histograms of individual burst radii at the indicated MinE concentrations. The mean burst radius and SD are based on Gaussian fits. The SE (mean uncertainty) is ±0.3 μm. (H) The average spatial period between successive burst zones remained constant with varying MinE. The spatial period is twice the average distance between successive burst zones. Also see Movie S6 and Fig. S2.
Although the oscillation period remained constant, the frequency of out-of-phase burst clusters increased with higher MinE concentrations (Fig. 4 C and E; Fig. S2A; and Movie S6). Because MinD binding was faster within bursts with higher MinE, this led to a more rapid depletion of MinD from the local solution and therefore smaller individual bursts, as well as smaller burst zones (Fig. 4F and Movie S6). Above 1.5 μM MinE, the average burst radius was ∼3.5 μm (Fig. 4G). At 1 μM MinE, the average radius increased to 6.5 μm with a broad distribution. At 0.5 μM MinE, bursts expanded to the point of overlap, making radius quantification difficult (Fig. 4F).
Fig. S2.
MinE concentration effects on the spatiotemporal periodicity of bursts. (A) GFP-MinD (1 μM input concentration) and MinE-Alexa647 surface intensities associated with burst cycles over time at the MinE concentration indicated. Intensities were converted to MinD and MinE protein densities associated with individual bursts over the time course above. Overlapping subcycles were separated on independent graphs for clarity. MinE at 0.5 or 1 μM supported two overlapping subcycles, whereas 1.5 and 3 μM MinE supported three. The labels i, ii, iii… track the still frames used for the analyses in B and C. (B) Still images of successive bursts at their peak MinD densities. Images were merged to highlight which of the successive burst clusters spatially overlap. With 0.5 or 1 μM MinE, burst clusters from i overlap with iii. With 1.5 or 3 μM MinE, burst clusters from i overlap with iv. (Scale bar: 5 μm.) (C) Cross-correlation analysis showing the temporal and spatial periodicity of overlapping burst subcycles. The spatial period is twice the average spatial distance between the auto-correlation peak (0) and the following cross-correlation peak (ii). Irrespective of the MinE concentration in solution, the temporal and spatial period was ∼2 min and ∼40 μm (2 × correlation peak ii ∼ 20 μm), respectively. This figure is related to Fig. 4.
Despite the smaller burst zones with higher MinE, the spatial separation between these zones remained relatively constant (Fig. 4H) as deduced from 2D image cross-correlation analysis (Fig. S2C). This disparity opened up space on the SLB for the formation of additional phase-shifted burst clusters (Fig. S2 B and C). For example, at 0.5 or 1 μM MinE, there were only two alternating phases of burst cycles occupying two spatially segregated burst zones (Fig. S2). At or above 1.5 μM MinE, three phase-shifted burst cycles occurred, each occupying one of three spatially segregated burst zones (Fig. S2). Therefore, although the overall burst frequency increased with increasing MinE (Fig. 4E), both the spatial and temporal oscillation periods of bursts at a given area of the SLB were insensitive to the MinE concentration, irrespective of the number of spatially segregated burst zones (Fig. 4 D and H). We conclude that MinE lingering after a burst disassembles inhibits MinD in solution from forming a burst on the same area of the SLB, which causes the temporally staggered burst cycles to spatially segregate.
Membrane Binding by MinE Is a Key Regulator for Min System Oscillation.
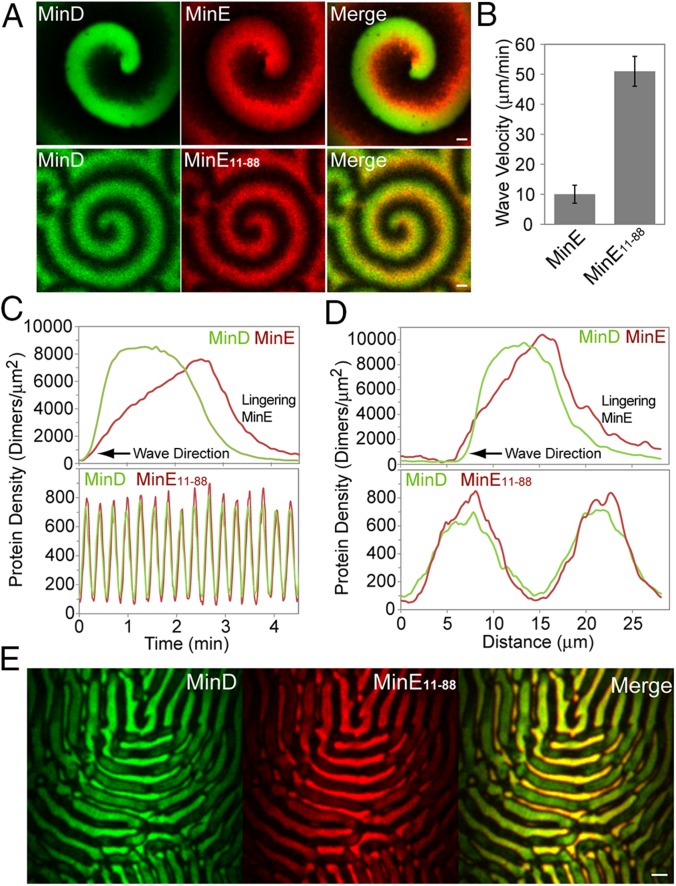
The role of MinE membrane binding in determining the system dynamics has become a major focus of in vivo and in vitro study (17, 19, 20, 22, 33, 34) and computational modeling (24), as well as a matter of considerable debate (35, 36). To further examine the role of lingering MinE in patterning, we studied a MinE mutant lacking its MTS, MinE11–88. At locations in the flowcell where the MinD supply was not limiting, MinE11–88 supported spirals resembling those formed with full-length MinE, except that the wave trains had closer spacing (Fig. 5A) and the wave front velocity was fivefold faster (Fig. 5B), resulting in a 15-fold shorter wave periodicity and significantly lower Min protein densities (Fig. 5C). Another striking difference was that MinE11–88 dissociated in phase with the release of MinD (Fig. 5D), indicating that, as predicted, it cannot linger on the SLB after MinD is released. The data show that MinE11–88 can associate with MinD on the membrane, stimulate MinD recruitment, and accumulate to a density that triggers MinD dissociation. These activities are sufficient to support Min spirals. However, without direct membrane binding, the MinE mutant cannot linger on the SLB after MinD release, which explains the diminished lag period between successive waves and the increased velocity of wave front propagation.
Fig. 5.
A membrane-binding mutant of MinE forms spirals, but not bursts. (A) Still images of the SLB where the MinD and MinE protein densities are high enough to support spirals. (Top) Spirals supported by full-length MinE. (Bottom) Spirals supported by the membrane-binding mutant MinE11–88. (Scale bar: 5 μm.) (B) Velocities of the spiraling wave trains supported by MinE or MinE11–88. (C) Time-course of MinD and MinE protein densities at a fixed location within the wave trains in A. Time 0 was chosen arbitrarily. (D) Cross-section of the MinD and MinE protein densities within the wave trains in A. (E) At the low protein densities that support bursts on the SLB when using full-length MinE, MinE11–88 formed a dynamic zebra pattern. (Scale bar: 40 μm.) Also see Movie S7.
We hypothesized that the membrane-binding activity of MinE that allows it to linger at the rear of a wave is also essential for E-ring formation around bursts. Consistently, MinE11–88 could not form bursts under MinD-limiting conditions on the SLB (Fig. 5E and Movie S7). Instead, we observed a dynamic “zebra” pattern composed of oscillating miniwaves of MinD that were rapidly disassembled by MinE11–88. MinD immediately bound all available regions of the SLB, which is in stark contrast to the well-separated spirals or bursts supported by full-length MinE that can linger and inhibit MinD binding. Together, the data show that, without the ability to linger on the membrane after MinD dissociation, MinE11–88 cannot form a defined E-ring or control the spatiotemporal parameters needed for burst oscillation, which also explains why membrane-binding mutants of MinE cannot support proper oscillation in vivo (17).
MinE Stabilizes MinD on the Membrane Before Stimulating Its Release.
Previous models for Min patterning are predicated on a dimer of MinE interacting with a dimer of MinD (D2E2) to stimulate MinD ATPase activity and release from the membrane (19, 24, 25, 37). However, our finding that MinE accelerates MinD recruitment to the SLB before stimulating MinD release is inconsistent with this model. Furthermore, the steady increase of MinD and MinE on the membrane during the early phase of patterning suggests that D2E2 is actually a stable complex on the SLB. The D2E2 structure indicates that the monomers of a MinE dimer cannot simultaneously interact with both sides of a MinD dimer (19). Rather, a MinE dimer asymmetrically binds one side of a membrane-bound MinD dimer, with one MTS in position to bind the membrane and further stabilize the D2E2 complex. We therefore hypothesized that two dimers of MinE symmetrically bound to both sides of a MinD dimer, E2D2E2, is the MinD dissociation complex that stimulates ATP hydrolysis and membrane release. According to this hypothesis, the local stoichiometry of MinD and MinE on the membrane changes the state of the toggle switch from MinE-stimulated MinD recruitment to MinD release from the membrane as detailed in Discussion.
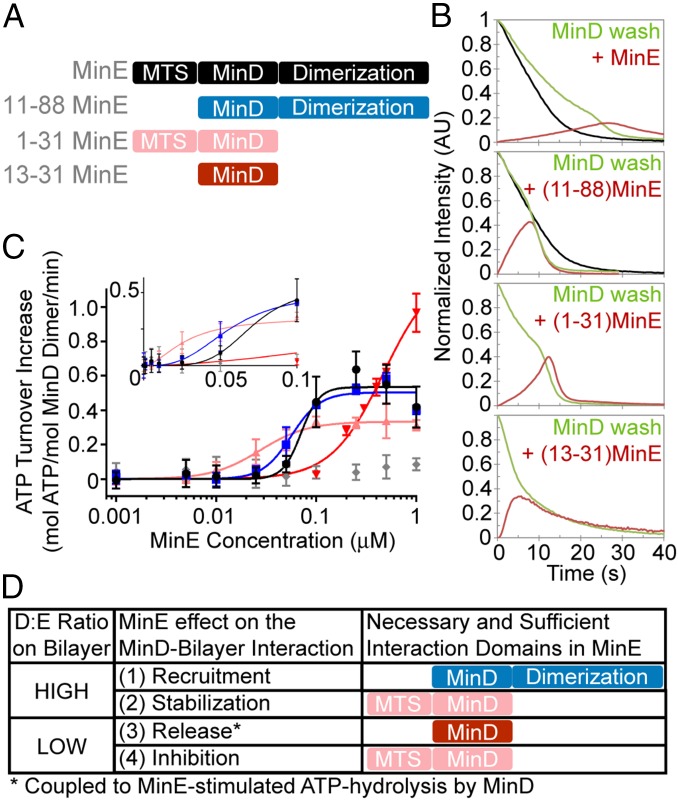
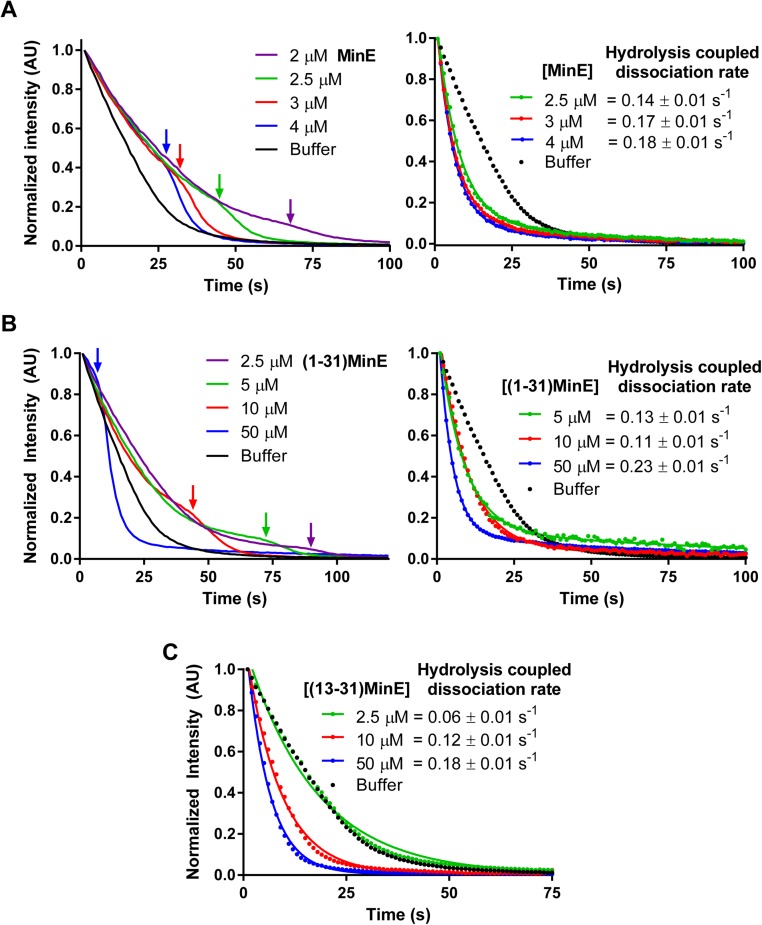
We first tested this model by determining if MinE can stabilize MinD membrane binding at low relative concentrations before stimulating MinD release at higher relative concentrations. We studied the dissociation kinetics of membrane-bound MinD washed with buffer containing full-length MinE or truncated MinE variants deficient for membrane binding, dimerization, or both (Fig. 6A). Consistent with our model, MinD release slowed as MinE accumulated on the mSLB up to a density close to that of MinD, at which point MinD release accelerated with MinE release lagging behind (Fig. 6B, Top, and Fig. S3A). MinD stabilization was less pronounced with the MinE variant lacking its MTS, MinE11–88. However, MinE11–88 still stimulated MinD dissociation when its density approached that of MinD, at which point both proteins released in phase. The dissociation kinetics are consistent with the dynamic patterning observed with this membrane-binding mutant of MinE (Fig. 5). Monomeric MinE1–31, which interacts with membrane and MinD, stabilized MinD on the membrane before stimulating its release, after which it lingered on the membrane (Fig. 6B and Fig. S3B). The MinE variant containing the MinD interaction interface but lacking both the dimerization and MTS domains, MinE13–31, stimulated the fastest MinD release rate and neither stabilized MinD nor lingered after MinD release (Fig. 6B and Fig. S3C). Neither dimerization-deficient MinE variants supported patterning. These findings support our proposal that MinE interacting with membrane-bound MinD stabilizes the D2E2 complex (or D2E, in the case of monomeric MinE mutants) with the help of the MTS of MinE.
Fig. 6.
Evidence for a higher-order MinE concentration dependence on stimulating MinD ATPase activity and membrane release. (A) Schematic of the interaction domains of full-length MinE and truncation variants: MTS, membrane targeting sequence; MinD, MinD interaction interface; dimerization, dimerization interface. (B) MinE stabilizes MinD on the SLB before stimulating its release. MinD was prebound to the SLB in the presence of ATP and then washed with buffer alone (black line, Top two panels) or with buffer containing 2.5 μM of the MinE variant specified. When a MinE variant was in the wash buffer, MinD (green) and MinE (red) intensities on the SLB were monitored and normalized to the MinD density on the SLB at the start of the wash (t = 0 s). (C) MinE stimulation of MinD ATPase activity shows higher-order concentration dependency. ATPase activity of 1 μM MinD with full-length or truncated MinE was measured in the presence of 0.5 mg/mL E. coli lipid as small unilamellar vesicles and 1 mM [γ-32P] ATP. Basal ATPase activity of MinD (0.1 mol/mol MinD dimer/min) was subtracted and the data were fit to a Hill equation. Line colors correspond to the full-length and truncated forms of MinE as illustrated in A. The gray data indicate full-length MinE stimulation of 5 μM MinD in the absence of lipid. (Inset) A linear scaling of the x-axis. (D) Summary of MinE effects on the MinD–bilayer interaction at a high or low MinD-to-MinE (D:E) ratio on the SLB. The necessary and sufficient interaction domains of MinE supporting the specified activity are illustrated. Also see Fig. S3.
Fig. S3.
MinE concentration effect on the MinD dissociation kinetics from membrane. (A) MinD (1 μM) was prebound to the mSLB in the presence of ATP to the steady-state density and then washed with buffer alone (black line) or with buffer containing full-length MinE (unlabeled) at the indicated concentration. (Left) The transition between the initial slow phase of MinD release, where MinE stabilizes MinD binding to the SLB, and the fast phase, where MinE stimulates the rapid release of MinD. Arrows highlight the transition point between the slow and fast phase. (Right) The fast phases of MinD release at the indicated MinE concentrations in the wash buffer. The data were normalized to a time point after the arrows on the Left graph. Curves were fit to a double-exponential decay function, and the MinD dissociation rate constants that contributed 90 ± 5% of the curves are presented. We propose that the MinD dissociation rate during this fast phase is coupled to the MinE-stimulated ATP-hydrolysis activity of MinD. (B) As in A, except MinE1–31 was in the wash buffer. (C) As in A, except MinE13–31 was in the wash buffer. MinE13–31 did not support the clearly separated slow MinD stabilization phase. This figure is related to Fig. 6.
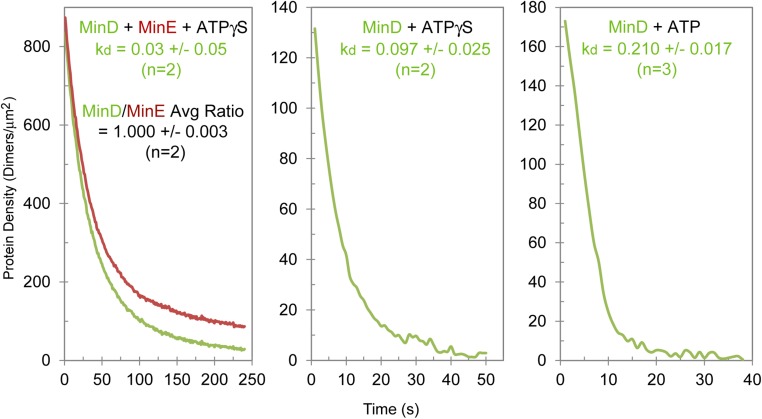
We sought to directly detect the MinDE complex stably bound on the membrane in the absence of ATP hydrolysis. MinD was mixed with an excess of MinE in a buffer containing ATPγS and flowed onto the mSLB. Membrane dissociation of the complex was then monitored during a buffer wash (Fig. S4). The MinDE complex that released from the SLB exhibited a 1–1 ratio with an off-rate constant of ∼0.03 s−1. The release rate of MinD-ATPγS in the absence of MinE was 3.3-fold faster (∼0.1 s−1) (Fig. S4). The finding supports the idea that D2E2 can accumulate on the membrane and dissociates slowly in the absence of ATP hydrolysis.
Fig. S4.
Membrane dissociation and stoichiometry of the MinD–MinE complex with ATPγS. GFP-MinD (0.5 μM) and 1 mM ATPγS were preincubated with or without 2.5 μM MinE-Alexa647 (∼4% labeled) and infused into an mSLB-coated two-inlet flowcell at a flow rate of 5 μL/min from one inlet. In parallel, the other inlet had wash buffer (no protein) flowing at a rate of 0.5 μL/min. When the surface MinD density reached ∼500–1,000 dimers/μm2, the flow rates from the two inlets were switched to shift the laminar boundary between the binding solution and the wash buffer to commence MinD–MinE complex dissociation from the mSLB (Left). The dissociation curves were fit to a double-exponential function to extract the MinD off-rate from the major component (∼80%). The amplitude of the slowly dissociating minority fraction varied depending on SLB quality, protein prep, and other incidental variables and was not further analyzed. The dissociation rates of the two proteins matched well, supporting the notion that they are in complex, and the stoichiometry of the two proteins in this complex was calculated using a conversion parameter estimated by the procedure described below. Dissociation curves in the absence of MinE with either ATPγS (Center) or ATP (Right) were obtained in essentially the same way, except the starting surface MinD density was lower and the dissociation rate constants were estimated based on single exponential fit. To obtain the conversion parameter to estimate the number of MinD and MinE molecules from the arbitrary camera unit of fluorescence intensity for this experiment, 2.5 μM of GFP-MinD and MinE-Alexa647 dimers (∼4% labeled) were mixed in a reaction buffer without ATP and flowed into a flowcell coated with 1,2-dioleoyl-sn-glycero-3-phosphocholine at 5 μL/min. Fluorescence intensity data were collected before the protein arrival (background), with the protein sample in the flowcell, and after washing the flowcell with buffer (background). The fluorescence signal of the protein sample in the solution volume within the evanescence illumination depth (47 AU for GFP-MinD and 56.5 AU for 4% MinE-Alexa647 under the instrument parameter settings used) was obtained by subtracting the background signal with buffer only (mostly camera dark noise). From the wavelength, the refractive indices of fused silica and the reaction buffer and the illumination angle, the evanescence penetration depths were calculated to be 131 and 170 nm for 488- and 633-nm excitation beams, respectively. Thus, the fluorescence intensity observed above corresponded to 197 MinD dimers/μm2 and 255 MinE dimers/μm2 on the surface.
Two MinE Dimers Release a MinD Dimer from the Membrane.
If one MinE dimer stabilizes a MinD dimer on the membrane, then perhaps it takes two MinE dimers interacting on both sides of a MinD dimer to stimulate ATP hydrolysis and release from the membrane. Consistently, stimulation of MinD ATPase activity showed higher-order MinE concentration dependency (Fig. 6C), suggesting that more than one MinE dimer is necessary for full ATPase stimulation of a MinD dimer. Similar results were found with MinE11–88 that cannot bind membrane and even with MinE1–31 that can interact with membrane and MinD but cannot dimerize. The MinE variant with only the MinD interaction interface, MinE13–31, also demonstrated higher-order concentration dependency, albeit with reduced affinity that required a higher concentration for full stimulation. The data show that the super-linear MinE stimulation of MinD ATPase activity does not require membrane binding or dimerization by MinE. In all cases, MinE-stimulated MinD ATPase activity required lipid (Fig. 6C), indicating that membrane binding by the MinD dimer is a prerequisite for ATPase stimulation by MinE. The data are consistent with full stimulation of MinD ATPase activity driven by MinE dimers on both sides of the membrane-bound MinD dimer. To summarize our interpretation of these findings, at a high MinD-to-MinE ratio on the SLB, MinE stabilizes MinD on the membrane in a D2E2 complex, which can further recruit MinD from the cytoplasm (Fig. 6D). As MinE accumulates and the MinD-to-MinE ratio drops, an additional MinE dimer can join D2E2 to form the E2D2E2 complex, which hydrolyzes ATP and releases the MinD dimer. The remaining MinE dimers linger on the bilayer and locally inhibit MinD binding. Taken together, our data support the proposal that MinE successively recruits, stabilizes, releases, and inhibits MinD interaction with membrane to drive Min oscillation.
Discussion
Spiraling wave trains of MinD and MinE on an SLB were the first patterns to be reconstituted in a cell-free reaction (28), but many of their features are quite different from the in vivo dynamics. For example, the protein densities (∼10,000 MinD dimers/μm2, Fig. 5C) far exceed those of the in vivo patterns [∼200 MinD dimers/μm2 (32)], and the wavelength and wave velocities are higher by roughly one order of magnitude. A previously described amoeba pattern (29) geometrically resembled in vivo polar zones of MinD corralled by E-rings, but lacked oscillatory properties. Thus, it has been difficult to decipher the mechanistic principles shared by these dissimilar patterns occurring under significantly different reaction settings in vivo and in vitro.
Periodic Solution Depletion of Active MinD Is Essential for Standing Wave Oscillations.
Our data suggest that Min oscillation in vivo requires cytosolic depletion of active MinD dimers as a MinD-binding zone develops at a cell pole. Programmed shuttling of the active MinD dimer pool from cytosol to membrane temporarily depletes the cytosolic pool, which prevents the continued expansion of the MinD polar zone. As the polar zone disassembles, the cytosolic supply recovers, allowing for de novo initiation of MinD binding at the opposing cell pole. In previous reconstitutions, the active MinD supply in solution was never significantly depleted, which explains why the expanding circular binding zones continued expanding to form a variety of surface-saturating patterns (22, 28, 29) instead of stalling and turning into a standing wave as seen in vivo.
By limiting the locally available amount of Min proteins in the flowcell, we reproduced many in vivo features of the Min system including standing wave oscillations. De novo initiation of a radially expanding MinD-binding zone was followed by the stalling of expansion, development of an E-ring, and dissipation of the binding zone (Fig. 1). These Min bursts showed highly regular spatial and temporal periodicities, expanding and dissipating in unison on regions of the SLB that were then restricted from MinD binding during the next set of bursts (Fig. 2). These phase-shifted binding cycles on spatially segregated zones of the SLB were separated by a node line of time-averaged MinD concentration minima, consistent with standing wave dynamics. We believe these oscillating bursts on the flat surface of an SLB are the cell-free equivalent of in vivo Min oscillation (Fig. 1H), albeit at a lower surface-to-volume ratio.
Once burst expansion initiates, the local solution concentration of active MinD rapidly depletes (Fig. 3). Depletion progresses until bursts approach their peak protein density ∼20 s later. Assuming that the suppression of a de novo binding zone begins ∼5 s after the onset of depletion, MinD dimers in solution with an estimated diffusion coefficient of 60 μm2/s (20) will diffuse an average distance of ∼35 μm in 2D (or ∼24 μm in 1D). This estimate of the diffusion distance of active MinD in solution is consistent with the spatial periodicity obtained from our cross-correlation analysis of burst cycles (Fig. 4H). Thus, in our ∼25-μm-thick flowcell, the diffusion distance of active MinD dimers in solution controls the spatial parameter of the standing wave. In a 1- × 3-μm E. coli cell, however, Min protein diffusion in the cytosol would not be a major limiting parameter of the system dynamics, considering that the estimated pole-to-pole diffusion time is less than a second (38).
Lingering MinE Suppresses MinD Membrane Binding and Establishes the Min Oscillation Period.
We propose that the accumulation–dissipation cycle of MinE on the membrane, coupled with the MinD reactivation rate in solution, is the principal biochemical timing mechanism that establishes the oscillation period. This MinE cycle on the membrane encompasses MinE dimers binding to MinD dimers on the membrane, accumulating up to the local surface density of MinD, stimulating MinD release, lingering after MinD is gone, and, finally, dissociating from the membrane upon reverting to the inactive form.
MinE lingering from a dissipated burst cycle suppresses MinD binding to the SLB. Therefore, as one set of bursts dissipate, another set initiates on areas where the lingering MinE density has decreased to the threshold value of ∼100 dimers/μm2 (Fig. 2B). The data provide further evidence that MinE lingering on the membrane after MinD release is a key regulator of the oscillatory mechanism, preventing MinD from rebinding the cell pole from which it dissociated and driving de novo binding to the opposing pole (19, 20, 22). We find that MinD-binding suppression by lingering MinE is critically dependent upon the membrane-binding activity of MinE via its MTS (Fig. 5).
Min Patterning by a Stoichiometry-Mediated Toggle Switch.
Previous models postulate that the primary role of MinE is to stimulate MinD ATPase activity and membrane release. This class of models predicts that both the protein density and oscillation period of a burst would decrease with higher MinE concentration. Instead, we found that the peak MinD density actually increased at higher MinE concentrations (Fig. 4B), whereas the burst oscillation period remained essentially constant (Fig. 4D). This result unveils the ability of MinE to accelerate both MinD membrane binding during burst expansion as well as MinD release during burst dissipation. Our findings suggest that MinE first catalyzes MinD recruitment to form the MinD polar zone in vivo before stimulating its ATPase activity and membrane release.
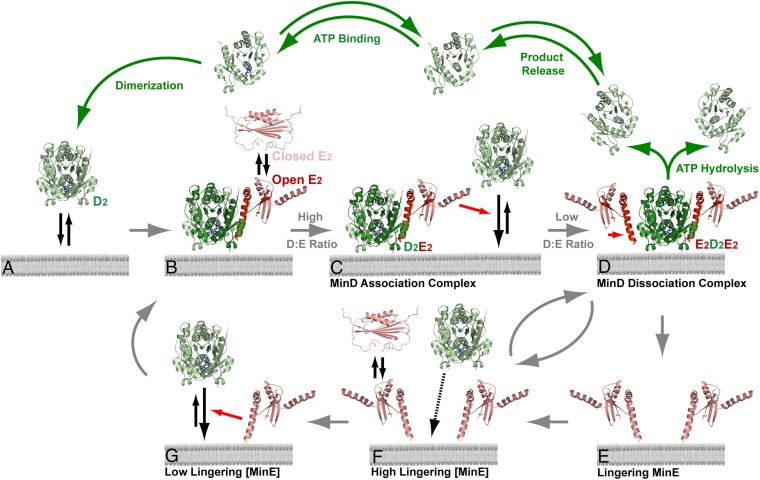
We propose that the local stoichiometry of MinE and MinD on the membrane acts as a toggle switch between MinE-stimulated MinD recruitment and MinD release from the membrane (Fig. 7). ATP-bound MinD forms a sandwich dimer (D2) that slowly and uniformly binds the membrane with no sign of cooperativity (Fig. 7A) (29). MinE binds, stabilizes MinD on the membrane, and accumulates during burst expansion (Fig. 7B). When the MinD-to-MinE ratio on the membrane is high, essentially all MinE dimers on the membrane would exist in a 1–1 complex with MinD dimers (D2E2). Our data suggest that ATP hydrolysis is not stimulated in this D2E2 complex. Rather, D2E2 remains stably associated with the membrane. We propose that D2E2 acts as a catalyst for further D2 recruitment (Fig. 7C). The D2E2 cocrystal structure inspired this aspect of our model (19) whereby D2E2 would be stably anchored to the SLB via three MTSs: two from D2 and one from E2. The D2E2 structure also suggests that the other potential MinD interface of E2 is available for recruiting D2 from solution (19). However, the arriving D2 would not be able to bind the membrane without deforming the D2E2 complex. Thus, we believe that D2E2D2 does not exist as a stable complex. Instead, we propose that by transiently interacting with D2 from solution, D2E2 plays a catalytic role in recruiting MinD to the membrane without stimulating its ATPase activity. This would explain the observed MinE-stimulated radial expansion of MinD binding on the membrane from a nucleation point. The proposal is also consistent with our finding that MinE1–31 can efficiently stabilize as well as dissociate MinD from membrane (Fig. 6B and Fig. S3B), but, as a monomer, it cannot catalyze MinD recruitment to the membrane and therefore does not support patterning.
Fig. 7.
MinD/MinE stoichiometry on the membrane drives Min patterning. (A) MinD (green) binds ATP, dimerizes, and binds membrane as D2. (B) A “closed” or inactive dimer of MinE (red), E2, “opens” to asymmetrically bind D2 on membrane. (C) At a high MinD-to-MinE ratio on the SLB, the available MinD interaction interface of E2 acts as a catalyst for further MinD recruitment. Therefore, D2E2 is the “MinD association complex.” (D) As MinE accumulates and the MinD-to-MinE ratio decreases, two MinE dimers symmetrically bind a MinD dimer to form E2D2E2, the “MinD dissociation complex,” which stimulates MinD ATPase activity and membrane release. (E) MinE can linger on the bilayer after MinD release, thus acting as a local self-amplifying catalyst. (F) Lingering MinE prevents MinD binding to the membrane (D). Lingering MinE eventually releases from the membrane as it reverts back to its closed form. (G) As lingering MinE diminishes, MinD can bind the SLB. MinE can once again stimulate MinD binding to the membrane. The monomer units undergoing a MinD–MinE interaction are shaded dark. Green arrows indicate the ATP cycle of inactive MinD in the cytoplasm. Structures for open MinE, MinD, and the MinD–MinE cocomplex were adapted from Protein Data Bank (PDB) ID 3R9J (19) for conceptual illustration purpose only. The closed MinE structure was adapted from PDB ID 2KXO (18). The MTS domains of MinD and MinE were not included in the crystal structures and are not shown in the model.
As the rate of MinD binding slows and the density of MinE increases to approach that of MinD on the membrane, the D2E2 complex will get the chance to capture another MinE dimer on the membrane to form E2D2E2, which triggers ATP hydrolysis. Because the ATP molecules from each MinD monomer “glue” the sandwich dimer together, ATP hydrolysis results in MinD monomerization and membrane release (Fig. 7D). MinD is then inactive for a significant period as it releases ADP, rebinds ATP, dimerizes, and possibly undergoes further ATP-dependent transitions before becoming competent for membrane binding (5). The two MinE dimers, on the other hand, remain on the membrane (Fig. 7E) and can locally remove the remaining D2E2. Consequently, the local relative concentration of MinE over MinD further increases. In other words, lingering MinE acts as a local self-amplifying catalyst. When a high level of lingering MinE remains, MinD dimers arriving at the membrane can form D2E2 and are transiently stabilized, but then quickly encounter another lingering MinE dimer to form E2D2E2, which hydrolyzes ATP and releases MinD (Fig. 7F). This scenario explains the sudden switch-like transition from the slow accumulation of MinE to the quick dissociation phase.
At high density, lingering MinE provides a positional memory that spatially organizes the system by quickly dissociating MinD. Lingering MinE that does not encounter D2 arriving from solution, or D2E2 already on the membrane, eventually dissociates and reverts back to its inactive form after several seconds (Fig. 7F). As the local lingering MinE density diminishes toward a certain level, and the concentration of active MinD in solution recovers, another round of MinD binding becomes possible (Fig. 7G). At this low lingering MinE density, the probability of MinD being recruited from solution by D2E2 can locally exceed the probability of forming E2D2E2. In other words, at a critical lingering MinE density, for a given solution concentration of active MinD, the kinetic competition shifts, resulting in the nucleation and accelerating expansion of a circular binding zone as the local MinD-to-MinE ratio increases. This stoichiometry-mediated toggle switch uniquely confers the critical nonlinearity to the system necessary for dynamic pattern self-organization.
The mechanism that we propose showing E2D2E2 as the MinD dissociation complex is supported by the biphasic kinetics of MinE-stimulated MinD disassembly from the SLB (Fig. 6B). In addition, MinD ATPase stimulation exhibited a higher-order MinE concentration dependency, which has also been reported for the Neisseria gonorrheae and E. coli Min systems (18, 39). If our proposed mechanism is correct, why did we not detect a significant accumulation of E2D2E2 stably bound on the membrane in the presence of ATPγS (Fig. S4)? We propose that binding of the second MinE dimer to form E2D2E2 is much slower compared with the binding of the first MinE dimer to form D2E2. Formation of E2D2E2 perhaps is accompanied by an energetically unfavorable transition in the MinD dimer that is necessary for ATP hydrolysis. This hypothesis would explain the clear kinetic separation of the reaction steps of MinD stabilization by MinE, in the form of D2E2, and ATP-hydrolysis–coupled MinD dissociation by MinE, in the form of E2D2E2.
Our mechanism is at odds with the “Tarzan of the Jungle” model proposed by the Lutkenhaus group (19). A series of elegant experiments involving Min protein heterodimers compromised for MinD–MinE interaction, ATPase stimulation, and/or ATP hydrolysis supported the conclusion that a one-sided interaction with MinE, D2E2, is sufficient for MinD ATPase stimulation and release from the membrane (37). However, this model fails to explain the MinE-stimulated recruitment and stabilization of MinD on the membrane, the biphasic kinetics of MinE-stimulated MinD release from the SLB, and the higher-order MinE concentration-dependent stimulation of MinD ATPase activity reported here. These discrepancies need to be clarified, but at present we suspect that the interpretation of the heterodimer results are complicated by the likely cooperativity between the two ATPase active sites within a MinD dimer and by the difficulty in obtaining mutant proteins that are completely inactive for one aspect of function and uncompromised in others.
Comparing Min Oscillation Dynamics in Vivo and in Vitro.
The most obvious difference between our flowcell and an E. coli cell is reaction vessel geometry. With the high membrane surface-to-volume ratio inside a cell and the limiting number of MinD molecules, the cytosolic pool of active MinD dimers would deplete when a MinD polar zone develops. The total MinD concentration in vivo (∼3 μM) is higher than that in our flowcell (∼0.6 µM). The lower concentration was needed to compensate for the lower surface-to-volume ratio of our flowcell. In vivo, active MinD depletion would take place more quickly because of the higher surface-to-volume ratio. The MinD reactivation rate for membrane binding is also likely faster at higher concentrations. Combined with cell geometry, the cytosolic depletion of MinD and the suppression of MinD membrane binding by lingering MinE can readily explain the in vivo dynamics that exhibit a shorter oscillation period (0.5–1 min) compared with the 2-min periodicity observed here (Fig. 3C).
Recently, the Schwille group has shown that Min spirals on the bottom of an SLB-coated well are converted to an in vivo-like oscillating pattern (30, 31). This was achieved by aspirating the sample solution so that the remaining volume was confined to 10-μm-deep × 10-μm-wide troughs with varying lengths (30, 31) . This observation is fully consistent with, and lends support to, the mechanism that we have identified, which predicts that a high membrane surface-to-volume ratio promotes standing wave oscillations.
At present, many of the critical biochemical parameters necessary for realistic numerical modeling of Min patterning remain unknown. We are currently studying the kinetic parameters necessary to formulate a numerical model that globally explains the fascinating variety of patterns supported by this “simple” system.
Materials and Methods
Proteins.
Protein expression, purification, and labeling were performed as previously described (22).
Flowcell Assembly.
Flowcell assembly and bilayer coating with E. coli polar lipid extract or monounsaturated (18:1) synthetic lipids were previously described (22). The mSLB was composed of 67% 1,2-dioleoyl-sn-glycero-3-phosphocholine (catalog no. 850375), and 33% 1,2-dioleoyl-sn-glycero-3-[phosphor-rac-(1-glycerol)] (catalog no. 840475). All lipids were purchased from Avanti and dissolved in chloroform at 25 mg/mL.
Sample Handling and Preparation.
Experiments were performed in Min buffer: 25 mM Tris⋅HCl, pH 7.4, 150 mM KCl, 5 mM MgCl2, 2 mM DTT, and 0.5 μg/mL ascorbate. Five millimolar phosphoenolpyruvate (Sigma) and 10 μg/mL pyruvate kinase (Sigma) provided ATP regeneration.
When Min proteins were preincubated at the concentrations indicated, His6-eGFP-MinD was mixed with MinE-His6 (mixed 1:19 with MinE-Alexa 647) in Min buffer for 15 min at 23 °C before addition of 2.5 mM ATP in a final reaction volume of 500 μL. The sample was passed through a 0.2-μm Amicon filter and loaded into a 1-mL syringe. TFZL tubing (1/16 × 0.02 inch; UpChurch) connected the syringe to the flowcell inlet nanoport (UpChurch). Samples were infused into the 3-μL flowcell (∼25 μm × 4 mm × 30 mm) with a neMESYS pump (Cetoni) at 1 μL/min (cross-sectional velocity of ∼0.17 mm/s) for 10 min. Flow was stopped before movie acquisition.
Total Internal Reflection Fluorescence Microscopy.
Total Internal Reflection Fluorescence (TIRF) illumination and microscopy as well as camera settings were as previously described (22). Prism-type TIRF microscopy was used with an Eclipse TE2000E microscope (Nikon) with a PlanApo 10× (N.A. = 0.45, air) or 40× (N.A. = 1.0, oil-immersed) objective lens. The TIRF illumination had a Gaussian shape in the field of view; therefore, intensity data for Min protein density estimations were measured at or near the middle of the illumination profile.
Movies were acquired using Metamorph 7 (Molecular Devices) and transferred to ImageJ (National Institutes of Health) for analysis and conversion to AVI file format. Brightness and contrast were set for each image or movie acquisition individually to best represent the features discussed. However, paneled acquisitions in the same movie share the same settings. All data were acquired at 5 s/frame when using full-length MinE. When using MinE11–88, the frame rate was 1 s/frame. Accelerations are indicated in the movie legends.
TIRF Microscopy Protein Density Estimation.
The average fluorescence intensity of single GFP-MinD or MinE-Alexa647 molecules was measured as previously described (22) to calculate the Min protein density on the SLB expressed as dimers/μm2.
Burst Cross-Correlation Analysis.
Image cross-correlation analysis was performed with a custom written LabVIEW program (program available upon request). A normalized 2D cross-correlation between a reference MinD image at a peak of a burst and a source MinD image was generated using the LabView function “IMAQ Correlate VI.” The reference image was initially auto-correlated and then cross-correlated with subsequent sequential images from the movie. The 2D correlation maps were converted into one-dimensional radial profile curves by calculating the average correlation over successive, non-overlapping 2-pixel-wide annuli centered on the (0,0) point in the correlation. To prevent cropping, the maximum radial profile was set by the shortest dimension of the images.
ATPase Assays.
ATPase activity of 1 μM MinD, with the indicated concentration of full-length or truncated MinE, was measured in 25 mM Tris⋅HCl (pH 7.4), 150 mM KCl, 5 mM MgCl2, 0.5 mg/mL E. coli lipid as small unilamellar vesicles, and 1 mM [γ-32P]ATP purified as previously described (40). Samples were incubated for 3 h at 30 °C and analyzed by TLC as previously described (41). Error bars represent the SD for at least three independent experiments.
Other Procedures.
Additional procedures are described in SI Materials and Methods.
SI Materials and Methods
Confocal Microscopy.
The Min reaction containing 1 μM GFP-MinD, 1.5 μM MinE-Alexa647 (∼50% labeled), and 2.5 mM ATP in 500 μL Min buffer was loaded into a 1-mL syringe and infused into a flowcell coated with E. coli lipids at 0.8 μL/min for 10 min. Flow was stopped and patterns were imaged using a Zeiss LSM 780 microscope with a Zeiss EC Plan-Neofluor 40× Objective lens/1.30 Oil DIC M27, N.A. 1.4.
For image quantification, image quality was optimized by setting the digital gain at 1.0 and adjusting the background level using digital offset. The dynamic range of the images was optimized using laser settings at 4% for both 488 and 633 nm, and the photomultiplier tube master gain at 700 and 800 for the green and red channels, respectively. The pinhole size was set at 1 Airy unit, 33 μm. Dual-color Z-stack images were acquired by scanning six slices from the bottom to top surfaces of the flowcell at equal intervals. Depending on the thickness of the flowcell (25–35 μm), the interval between slices was between 5 and 7 μm. The 512 × 512 pixel (0.69 μm/pixel), 16-bit images were acquired with each slice taking 2 s to scan. The Z-stacks were scanned continuously. Image analysis was carried out using Zen 2010 software (Zeiss), ImageJ (NIH), and data were analyzed using OriginPro 9.1 (OriginLab).
Photometric concentration calibrations of GFP-MinD and MinE-Alexa647 in solution phase were performed in a flowcell coated with E. coli lipids. GFP-MinD at 0, 0.1, 0.2, 0.4, 0.6, 0.8, and 1 μM were infused without ATP into the flow cell, and a Z-stack of six slices was measured from the bottom to top surface of the flowcell. The mean intensity for each slice (512 × 512 pixels) was averaged over three measurements. The averaged intensities of each of the six slices were plotted against GFP-MinD concentration with a linear fit to obtain calibration curves at each depth. This was repeated for MinE-Alexa647 (50% labeled) from 0 to 1.5 μM. The slopes of the calibration curves decreased from the bottom to top slices. This decrease in image intensity is presumably due to spherical aberration from the refractive index mismatch that decreases photon yield at the pinhole with increasing imaging depth. Using the depth-specific calibration curves, Min protein concentrations in solution averaged over the full camera chip and two oscillation cycles (12 frames) obtained in arbitrary units were converted to molar concentrations (Fig. 3B). To quantify the amount of crosstalk between slices, 20 μL of 1 μM GFP-MinD or 1.5 μM MinE-Alexa647 (50% labeled) was allowed to dry on the surface of a clean coverslip and solvated again in buffer after several washes. Z-stack images at 1-μm-interval slices were acquired from the surface of the coverslip. The intensity of each slice was plotted against z-position. Intensities dropped to background levels at 7–8 μm from the surface. The percentage of crosstalk from the surface of the coverslip to the first solution slice (5–7 μm) was 1.4–3% for GFP-MinD and 2–2.3% for MinE-Alexa647, showing that crosstalk from adjacent slices is insignificant.
MinD Dissociation from SLB.
GFP-MinD (1 μM) in Min buffer (without ascorbate) containing 1 mM ATP was infused into the mSLB-coated two-inlet flowcell at a flow rate of 5 μL/min from one inlet. In parallel, the other inlet had the same buffer in the presence (or absence) of the specified MinE variant flowing at a rate of 0.5 μL/min. GFP-MinD was bound to the mSLB up to an apparent steady-state surface density of 11,000 ± 3,000 GFP-MinD dimers/μm2. The flow rates from the two inlets were then switched to shift the laminar boundary between the binding solution and the washing solution to commence the wash of GFP-MinD from the mSLB (t = 0 in Fig. 6B). GFP-MinD dissociation kinetics were then recorded. Fluorescence intensity of the Alexa 647-labeled MinE variants were also tracked during the wash. Alexa 647 labeling efficiencies for each MinE variant were different, but accounted for when normalizing the MinE-Alexa647 fluorescence intensity in Fig. 6B to compare the molecular surface density of MinD and MinE in the graph.
Supplementary Material
Acknowledgments
This work was supported by the intramural research fund for National Institute of Diabetes and Digestive and Kidney Diseases (K.M.); the National Heart, Lung, and Blood Institute (K.C.N.); National Institutes of Health; Department of Health and Human Services; and the Nancy Nossal Fellowship (A.G.V. and L.C.H.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 2803.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600644113/-/DCSupplemental.
References
- 1.Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012;20(9):411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vecchiarelli AG, Mizuuchi K, Funnell BE. Surfing biological surfaces: Exploiting the nucleoid for partition and transport in bacteria. Mol Microbiol. 2012;86(3):513–523. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiekebusch D, Thanbichler M. Spatiotemporal organization of microbial cells by protein concentration gradients. Trends Microbiol. 2014;22(2):65–73. doi: 10.1016/j.tim.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34(1):82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 5.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76(1):539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 6.Raskin DM, de Boer PAJ. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999a;96(9):4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Lutkenhaus J. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol. 2000;182(14):3965–3971. doi: 10.1128/jb.182.14.3965-3971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raskin DM, de Boer PAJ. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999b;181(20):6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X, Shih Y-L, Zhang Y, Rothfield LI. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci USA. 2001;98(3):980–985. doi: 10.1073/pnas.031549298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale CA, Meinhardt H, de Boer PAJ. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001;20(7):1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meinhardt H, de Boer PAJ. Pattern formation in Escherichia coli: A model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci USA. 2001;98(25):14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Lutkenhaus J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol. 2003;47(2):345–355. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- 13.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA. 2002;99(24):15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W, Park K-T, Holyoak T, Lutkenhaus J. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Mol Microbiol. 2011;79(6):1515–1528. doi: 10.1111/j.1365-2958.2010.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Lutkenhaus J. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol. 2003;185(15):4326–4335. doi: 10.1128/JB.185.15.4326-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA. 2002;99(10):6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh C-W, et al. Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli. Mol Microbiol. 2010;75(2):499–512. doi: 10.1111/j.1365-2958.2009.07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghasriani H, et al. Appropriation of the MinD protein-interaction motif by the dimeric interface of the bacterial cell division regulator MinE. Proc Natl Acad Sci USA. 2010;107(43):18416–18421. doi: 10.1073/pnas.1007141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park K-T, et al. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146(3):396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loose M, Fischer-Friedrich E, Herold C, Kruse K, Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat Struct Mol Biol. 2011;18(5):577–583. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- 21.Renner LD, Weibel DB. MinD and MinE interact with anionic phospholipids and regulate division plane formation in Escherichia coli. J Biol Chem. 2012;287(46):38835–38844. doi: 10.1074/jbc.M112.407817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vecchiarelli AG, Li M, Mizuuchi M, Mizuuchi K. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol Microbiol. 2014;93(3):453–463. doi: 10.1111/mmi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweizer J, et al. Geometry sensing by self-organized protein patterns. Proc Natl Acad Sci USA. 2012;109(38):15283–15288. doi: 10.1073/pnas.1206953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonny M, Fischer-Friedrich E, Loose M, Schwille P, Kruse K. Membrane binding of MinE allows for a comprehensive description of Min-protein pattern formation. PLOS Comput Biol. 2013;9(12):e1003347. doi: 10.1371/journal.pcbi.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halatek J, Frey E. Highly canalized MinD transfer and MinE sequestration explain the origin of robust MinCDE-protein dynamics. Cell Reports. 2012;1(6):741–752. doi: 10.1016/j.celrep.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Huang KC, Meir Y, Wingreen NS. Dynamic structures in Escherichia coli: Spontaneous formation of MinE rings and MinD polar zones. Proc Natl Acad Sci USA. 2003;100(22):12724–12728. doi: 10.1073/pnas.2135445100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard M, Kruse K. Cellular organization by self-organization: Mechanisms and models for Min protein dynamics. J Cell Biol. 2005;168(4):533–536. doi: 10.1083/jcb.200411122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320(5877):789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci USA. 2010;107(18):8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zieske K, Schwille P. Reconstitution of pole-to-pole oscillations of min proteins in microengineered polydimethylsiloxane compartments. Angew Chem Int Ed Engl. 2013;52(1):459–462. doi: 10.1002/anie.201207078. [DOI] [PubMed] [Google Scholar]
- 31.Zieske K, Schwille P. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife. 2014;3:e03949. doi: 10.7554/eLife.03949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih YL, Fu X, King GF, Le T, Rothfield L. Division site placement in E.coli: Mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 2002;21(13):3347–3357. doi: 10.1093/emboj/cdf323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih Y-L, et al. The N-terminal amphipathic helix of the topological specificity factor MinE is associated with shaping membrane curvature. PLoS One. 2011;6(6):e21425. doi: 10.1371/journal.pone.0021425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng M, et al. Self-assembly of MinE on the membrane underlies formation of the MinE ring to sustain function of the Escherichia coli Min system. J Biol Chem. 2014;289(31):21252–21266. doi: 10.1074/jbc.M114.571976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonny M, et al. 2014. Response to Halatek and Frey: Effective two-dimensional model does account for geometry sensing by self-organized proteins patterns. arXiv: 1406.1347.
- 36.Halatek J, Frey E. Effective 2D model does not account for geometry sensing by self-organized proteins patterns. Proc Natl Acad Sci USA. 2014;111(18):E1817. doi: 10.1073/pnas.1220971111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park K-T, Wu W, Lovell S, Lutkenhaus J. Mechanism of the asymmetric activation of the MinD ATPase by MinE. Mol Microbiol. 2012;85(2):271–281. doi: 10.1111/j.1365-2958.2012.08110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr Opin Biotechnol. 2011;22(1):117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli: Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell. 2001;7(6):1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 40.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA–DNA interactions: A key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78(1):78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fung E, Bouet J-Y, Funnell BE. Probing the ATP-binding site of P1 ParA: Partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 2001;20(17):4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.