Abstract
Goal
We demonstrate a novel and robust approach for visualization of upper airway dynamics and detection of obstructive events from dynamic 3D magnetic resonance imaging (MRI) scans of the pharyngeal airway.
Methods
This approach uses 3D region growing, where the operator selects a region of interest that includes the pharyngeal airway, places two seeds in the patent airway, and determines a threshold for the first frame.
Results
This approach required 5 sec/frame of CPU time compared to 10 min/frame of operator time for manual segmentation. It compared well with manual segmentation, resulting in Dice Coefficients of 0.84 to 0.94, whereas the Dice Coefficients for two manual segmentations by the same observer were 0.89 to 0.97. It was also able to automatically detect 83% of collapse events.
Conclusion
Use of this simple semi-automated segmentation approach improves the workflow of novel dynamic MRI studies of the pharyngeal airway and enables visualization and detection of obstructive events.
Significance
Obstructive sleep apnea is a significant public health issue affecting 4-9% of adults and 2% of children. Recently, 3D dynamic MRI of the upper airway has been demonstrated during natural sleep, with sufficient spatio-temporal resolution to non-invasively study patterns of airway obstruction in young adults with OSA. This work makes it practical to analyze these long scans and visualize important factors in an MRI sleep study, such as the time, site, and extent of airway collapse.
Index Terms: Biomedical image processing, image segmentation, magnetic resonance imaging, medical diagnostic imaging, sleep apnea
I. Introduction
Obstructive sleep apnea (OSA) syndrome is a common breathing disorder in which the airflow pauses during sleep due to physical collapse of pharyngeal airway [1]. It affects approximately 4-9% of adults [2] and 2% of children [3] in the United States; in particular, OSA has been reported in 13-66% of obese children [4]. OSA is linked to decreased productivity, accidents, and increased risk of cardiovascular disease [5]. It is widely recognized that determining the accurate location of obstruction sites may benefit treatment planning and patient outcome [6].
The current gold standard for diagnosing OSA is overnight polysomnography (PSG). PSG involves monitoring and continuously recording several physiological signals that reflect sleep and breathing, for roughly 7 to 8 hours. The single most important output of overnight PSG is the apnea-hypopnea index (AHI)—the average number of complete or partial obstructive events that occur per hour during sleep. PSG also helps to elucidate the impact of these events on sleep pattern and gas exchange. An overnight PSG requires interpretation by a skilled sleep specialist for visual scoring; in contrast, computer scoring has not been proven accurate.
Objective monitoring is required because patient history and physical findings suggestive of OSA are not considered sufficiently sensitive or specific to make a diagnosis. Often, uncomplicated cases of OSA in children can be diagnosed and treated with removal of the tonsils and adenoids or by the provision of positive airway pressure therapy. While PSG provides a detailed physiologic diagnosis, no information regarding the site, or sites, of obstruction can be inferred from the recording. This is especially important for complicated cases of OSA, including obesity and craniofacial abnormalities. In these cases surgical intervention directed at relieving sites of obstruction may be needed.
Endoscopy is one imaging approach that provides site information. It involves the use of cameras inserted through the nose, and anesthesia to mimic sleep, hence the alternative name drug induced sleep endoscopy (DISE). Endoscopy provides information about one site of obstruction and nothing distal to it. Furthermore, it cannot replicate natural sleep or the various sleep cycles or sleep stages.
Magnetic resonance imaging (MRI) is an alternative imaging approach that has shown significant potential for airway assessment and collapse site identification in these patients. It is non-invasive, involves no ionizing radiation, and can resolve all relevant soft tissues in three dimensions. It has been shown to yield valuable insight into the dynamics and shape of airways for patients with OSA [7]–[9], and the primary limitation until now has been imaging speed.
Previous MRI studies have focused on static 3D or dynamic 2D imaging that can easily be performed on current clinically available MRI scanners [10]. Static 3D MRI provides a complete picture of the upper airway (UA) anatomy in patients with sleep disorders [7], but does not depict changes with tidal breathing or during collapse events. Wagshul et al. recently showed that tidal breathing could be resolved using retrospective gating [11], however this approach would not apply to collapse events. Single and multi-slice dynamic imaging captures the dynamics of tidal breathing [12] and collapse events [13] [14]. These methods suffer from gaps in coverage and are susceptible to imperfect placement of the imaging slices.
Recently, real-time 3D UA MRI during natural sleep has been demonstrated with 1.6-2.0 mm isotropic spatial resolution, 2 temporal frames per second, and synchronized recording of physiological signals [15], [16]. Each scan provides between 240 and 2400 temporal frames, corresponding to 2 to 20 minutes of continuous recording. In these studies, the shorter 2 minute scans were used for tests that involve a stimulus to encourage collapse [13] whereas longer 20 minute scans were used to capture natural collapse events. We anticipate a potential need for even longer scans, on the order of hours, for the study of sleep state changes. With such large 3D dynamic data sets, analysis becomes a significant bottleneck. Using software assistance, manual upper airway segmentation requires roughly 10 minutes per 3D volume. Visual inspection of all data is not practical, and therefore automated segmentation and collapse detection tools could provide significant value. Such tools would enable identification of time periods of airway obstruction, visualization of collapse sites, and volumetric analysis of airway changes.
Previously, automatic segmentation of the upper airway has been applied to 3D computed tomography (CT) and MR images. 3D static CT images of the upper airway were segmented for volumetric analysis using level-set based deformable models [17] and region growing algorithm [18][19]. In [3][20] static 3D T2-weighted MR images, were segmented using a fuzzy connectedness-based algorithm that required 4 min/study. This framework required significant operator and processing time, even for short scans. More recently, Wagshul et al. used threshold-based segmentation to segment and depict 3D upper airway images of tidal breathing [11]. Their static 3D and retrospectively gated upper airway images had significantly higher contrast to noise ratio and reduced artifacts compared to what is seen in real-time 3D UA MRI. Additionally, these previous methods were not designed for or applied to collapsed airways.
In this work, we demonstrate visualization of upper airway dynamics and detection of natural obstructive apneas during sleep. The key development is a method for semi-automated analysis of real-time 3D UA MRI using multi-seeded 3D region growing [21]. Region growing was chosen for its simplicity to demonstrate a new application where segmentation facilitates the workflow of a novel study. Multiple-seeds were found to enable more accurate segmentation and visualization during collapse events when the airway is divided into two (or more) patent sections. We narrow the analysis volume to just the pharyngeal airway, and propagate seeds from one time frame to the next using the observation that the nasal cavity and base of the airway remain patent at all times. We demonstrate that this approach performs as well as manual segmentation. Errors were comparable to intra-operator variability of manual segmentation. We also show that it enables analysis of long scans and visualization of important factors in an MRI sleep study, such as the time, site, and extent of airway collapse.
II. Methods
A. Segmentation and Analysis
Fig. 1 illustrates the proposed segmentation approach. A reference frame is selected to be the first frame with a visibly patent airway in each data set. A region of interest (ROI) is selected from the hard palate to the epiglottis in the mid-sagittal slice of the reference frame as shown in Fig. 1a. Two seeds are placed within the mid-sagittal slice, one in the nasopharyngeal airway and one in the oropharyngeal airway as shown in Fig. 1a. The first frame is segmented using multi-seeded 3D region growing [21]. The threshold is manually set to separate air and tissue signals in the ROI, which was between 7th-15th percentiles of pixel intensities for all cases studied. Seeds for subsequent frames are generated automatically by dividing segmented airway into five sections and placing two new seeds for the next time frame at the centroid of the sub sections with the largest volume, as illustrated in Fig. 1a. The two initial seeds plus the two new seeds are used for segmentation. This process repeats until all frames are processed. Segmentation is performed using an open-source implementation of multi-seeded 3D region growing [22] in MATLAB (MathWorks Inc., Natick, MA).
Fig. 1.
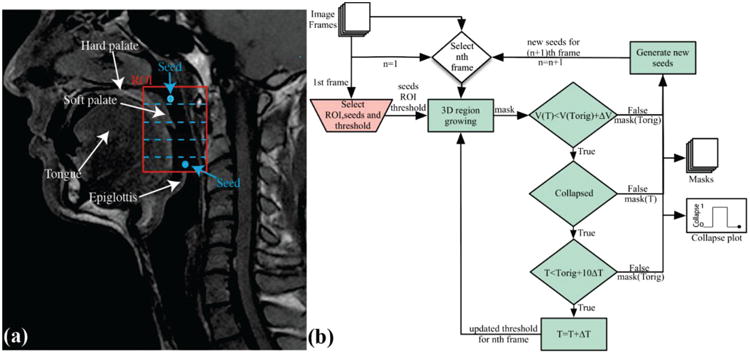
Segmentation Procedure. a) Graphical representation of region of interest (ROI) placement, seed placement, and division of airway into five sections for generation of new seeds. b) Flowchart illustrating the entire procedure. An ROI is manually selected (red), two seeds are placed within the ROI and a threshold (Torig) is determined based on the first frame. The first frame is segmented using automatic region growing (green). The threshold is increased by ΔT and segmentation is repeated if volume of airway is less than the segmented volume using original threshold plus ΔV, collapse is detected and threshold is less than original threshold plus 10ΔT. Two new seeds for subsequent frames are generated using segmented masks.
The images have non-zero intensity in the airway, due to noise, which results in under-estimation of the patent airway. This could also result in false positive collapse detection if the threshold is set too low. Therefore the initial threshold is increased by an operator-determined ΔT and segmentation is repeated with an updated threshold if two conditions are met: 1) if a collapse is detected, and 2) if the threshold is less than the original threshold plus 10ΔT. This process is repeated until one of the conditions becomes false or the volume of the mask with updated threshold is greater than the original volume plus 1 cm3. We found that this significantly improved collapse detection. If the collapse is detected for all thresholds the mask from the original threshold is used. However, if the collapse is resolved by updating the threshold the mask from the updated threshold value is saved and is used to generate seeds for the subsequent frame.
When the airway is patent, region growing results in a single contiguous region. Airway collapse is therefore detected based on the existence of two (or more) disconnected regions that were larger than 0.2 cm3. This threshold excludes small-disconnected regions that are not part of the airway. The binary airway collapse waveform can be displayed alongside other physiological recordings. 3D renderings of airway were generated for frames before, during, and after collapse events, using MATLAB.
B. Evaluations
Dynamic 3D upper airway data was analyzed from 3D RT-MRI sleep studies in ten adolescents with sleep-disordered breathing, described in Table I. Our Institutional Review Board approved the imaging protocol, and written informed consent was provided. Images were acquired using a 3DFT gradient echo sequence with golden angle radial view order, and constrained reconstruction, as described by Kim et al. [15], [16].
Table I.
Patient information.
| Patient Info | Polysomnography | |||
|---|---|---|---|---|
| # | Age/Sex | BMI | oAHI* | Phenotype |
| 1 | 14/F | 45.0 | 0.3 | Primary snorer |
| 2 | 16/M | 30.0 | 0.2 | High Arousal |
| 3 | 14/M | 23.7 | 4.0 | Sleep related hypoxemia / hypoventilation |
| 4 | 17/F | 26.9 | 1.7 | Primary snorer |
| 5 | 14/F | 30.2 | 2.5 | Sleep related hypoxemia / hypoventilation |
| 6 | 14/F | 32.8 | 1.1 | Primary snorer |
| 7 | 17/F | 32.0 | 2.0 | Sleep related hypoxemia / hypoventilation |
| 8 | 20/F | 24.9 | 4.0 | Sleep related hypoxemia / hypoventilation |
| 9 | 13/F | 32.0 | 70 | Obstructive sleep apnea |
| 10 | 15/M | 29.2 | 0.7 | Primary snorer |
OAHI= Obstructive apnea hypopnea index (i.e. # of events per hour) measured from an overnight polysomnography. The use case columns show that subject 1 and 2 were used for dice coefficient analysis and subject 3 was used for testing the accuracy of collapse detection.
Relevant imaging parameters for short scans are: scan time 2 min, field of view (FOV) 16.0×12.8×6.4 cm3; spatial resolution 1.6×1.6×1.6 mm3, echo time (TE) 1.74 ms, and repetition time (TR) 3.88 ms. Relevant imaging parameters for long scans are: scan time 14-18 min, field of view (FOV) 20.0×16.0×8.0 cm3; spatial resolution 2.0×2.0×2.0 mm3, echo time (TE) 1.72 ms and repetition time (TR) 6.02 ms. The body coil was used for radio frequency (RF) transmission, and a six-channel carotid coil was used for signal reception.
Respiratory effort, heart rate, oxygen saturation, and mask pressure were recorded during the MRI scan as described by Kim et al. [15]. Two respiratory transducers placed over the abdomen were used to measure respiratory effort. An optical fingertip plethysmograph (Biopac Inc., Goleta, CA) was used to measure oxygen saturation and heart rate. A facial mask (Hans Rudolph Inc., Kansas City, MO) along with a Validyne pressure transducer (Validyne Engineering Inc., Northridge, CA) was used to measure airway pressure at the mouth and for airway occlusion test.
Segmentation accuracy was evaluated using manual segmentation as the gold standard. Ten datasets were chosen from the work of Kim et al. [15], [16].These provided a range of expected data quality for this nascent imaging approach. In five patients (Patients # 1, 2, 8, 9, and 10), inspiration was blocked for three breaths as part of an airway compliance test [13]. In all other patients, the scan was performed during free breathing. Each dynamic 3D data set was segmented using semi-automated 3D pharyngeal airway segmentation. Twelve frames were selected and manually segmented twice by one MR physicist with knowledge of the upper airway anatomy.
Dice coefficient analysis [23] was used to compare the semi-automatic segmentations with manual segmentations, and to evaluate the intra-operator variability of manual segmentation. Manual segmentations were performed using a commercial package (Slice-o-Matic, TomoVision Inc. Magog, Canada). For two segmented volumes A and B, the DC is given by (2|A + B|)/(|A| + |B|) and quantifies the amount of overlap between the two sets. The DC ranges from 0 to 1, where 1 signifies exact overlap between A and B, and 0 signifies no overlap. A DC of 0.85 or greater is considered to be “very good”.
DC analysis was also used to evaluate differences in estimated airway clearance between manual and semi-automatic segmentations at four different axial cross-sections of the UA. The locations were chosen to be at the level of the uvula, because that is where the airway is typically the narrowest and is where we have observed the largest number of collapse events in our patient cohort.
Sensitivity of the method to seed selection was also evaluated; using 10 different pairs of seed points to segment each data set with semi automated segmentation. DC analysis was used to compare the different semi-automated segmentations.
Five long datasets during natural sleep, each 18-22 min and 1500-1800 frames, were selected to demonstrate visualization of airway dynamics and detection of collapse events. These particular datasets were more likely to contain central and obstructive apnea events. A clinical sleep expert identified these events using physiological signals acquired during the MRI scan. Obstructive apneas were identified based on the presence of respiratory effort detected by respiratory bellows with no change in mask pressure measured at the mouth. Central apneas were identified by the absence of respiratory effort coincident with no change in mask pressure. The timing of obstructive events was compared to the events detected by automatic collapse detection.
III. Results
Semi-automated segmentation of 2 minutes of data (240 frames) required 2 minutes of operator time, and 20 minutes of CPU time (Intel Xeon 2.93Ghz, 192 GB RAM). Manual segmentation of 6 seconds of data (12 frames) required roughly 2 hours (10min/frame). Table II contains Dice Coefficients and Table I contains baseline subject information, sleep-disordered breathing phenotype, and MRI analysis of airway collapse. DC for manual vs. manual was comparable to DC for manual vs. automatic for both the ROI and airway clearance at 4 slices around the uvula. The accuracy of semi-automated segmentation was comparable to the intra-operator variability of manual segmentation. The method was not sensitive to initial seed placement resulting in DC of 1 for all the semi-automated segmentations performed using different initial seeds.
Table II.
Dice coefficient comparison of semi-automatic and manual segmentation. A DC of 0.85 or greater indicates good agreement. First column contains Patient numbers from Table I.
| # | Manual vs. Manual | Manual vs. Automated |
|---|---|---|
| 1 | 0.95 | 0.87, 0.87 |
| 2 | 0.94 | 0.84, 0.85 |
| 3 | 0.97 | 0.93,0.94 |
| 4 | 0.97 | 0.87,0.86 |
| 5 | 0.89 | 0.86,0.91 |
| 6 | 0.91 | 0.92,0.94 |
| 7 | 0.94 | 0.91,0.91 |
| 8 | 0.94 | 0.92,0.89 |
| 9 | 0.95 | 0.93,0.94 |
| 10 | 0.94 | 0.91,0.93 |
The DC values for manual vs. automatic segmentation were low (<0.85) for 22% (9/40) of 2D slices. These slices had narrow airway clearance. Because the segmented airway consisted of less than 175 pixels in these slices, very small segmentation errors result in lower DC values.
Natural obstructive events were observed in two Patient's scan data (patient 3 and 10 from Table I). Patient 10 only showed 1 naturally occurring obstructive event. Patient 3 was used to test automatic collapse detection because he showed the most number of naturally occurring collapses. Of the 18 obstructive events, 15 were detected automatically. Of the three cases where obstructive events were not detected, two suffered from poor image quality and one showed a narrowed but patent airway. In addition, there were 9 false positives. A series of consecutive frames showing collapse was regarded as an event. False positive events had at-most 4 frames of consecutive collapse compared to 15 or more frames in actual events with the exception of two true events that had 4 and 8 consecutive frames showing collapse due to poor image quality.
Fig. 2 contains surface renderings illustrating upper airway dynamics during one collapse from validation Subject 1. Such anatomical information along with other physiological signals can be used for treatment planning and to monitor treatment outcome. The information from the plots of airway collapse also makes it easier to navigate to frames that show airway collapse and allows for more efficient data processing. In Fig. 2 collapses were observed in three frames for manual segmentation 1 and in two frames for manual segmentation 2. This was due to the difficulty in segmenting airway with high signal intensity that made it difficult for the operator to decide if a particular region is part of the airway or tissue.
Fig. 2.
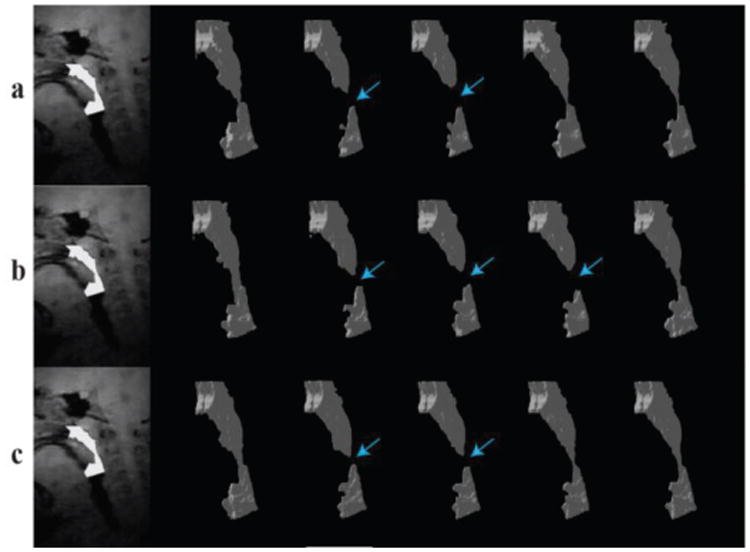
Visualization of a retropalatal airway collapse event during natural sleep. a) Automatic segmentation b) manual segmentation #1 and c) manual segmentation #2.The manual segmentations differ in the number of collapse frames detected due to low contrast to noise ratio (CNR) in the image data from patient 1. The number of detected collapse frames in automatic segmentation are comparable to manual segmentations.
Fig. 3 and Fig. 4 show data from patients experiencing a natural central apneic event, and a natural mixed apneic event, respectively. Mixed apneic event consists of a central apnea followed by an obstructive apnea in this case. The volume plot and binary collapse plot are displayed along with physiological signals to simplify the process of identifying and classifying events. Obstructive events are identified based on the presence of respiratory effort detected by respiratory bellows with no change in mask pressure measured at the mouth. Central events were identified by the absence of respiratory effort coincident with no change in mask pressure. In Fig. 3, the decrease in airway volume, flat respiratory effort (black arrows), zero mask pressure and subsequent decrease in oxygen saturation indicate a central apneic event that resulted in airway narrowing, shown by the volume plot and visualizations. In Fig. 4, flat respiratory effort and zero mask pressure indicate a central apnea followed by fluctuations in respiratory effort in the abdomen, zero mask pressure and the binary collapse plot (black arrows) indicating an obstructive apneic event. The binary collapse plot makes it easier to navigate to collapse events and the visualizations allow us to observe the precise site and extent of collapse.
Fig. 3.
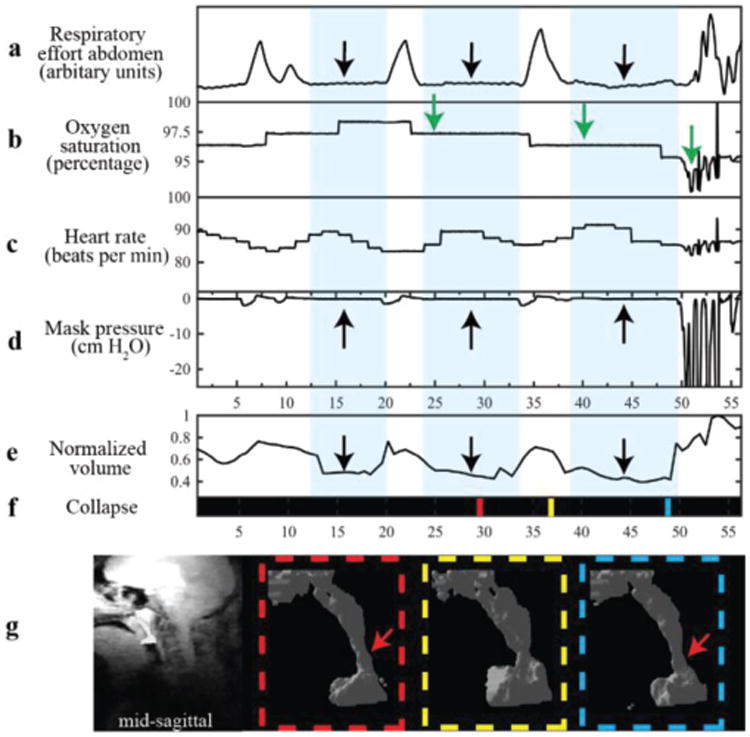
Example use from a 14/M with central apnea event. Waveforms shown are (a) abdominal respiratory effort, (b) oxygen saturation, (c) heart rate, (d) mask pressure, (e) normalized volume (f) collapse and (g) 3D visualizations. The first four a-d were simultaneously measured during an MRI scan, and e-g were displayed based on calculations from segmented masks using the proposed approach. The light blue background in (a)-(e) highlights the durations of the central apneas. The black arrows indicate flat bellow signal, zero mask pressure and the drop in volume. The apneas are followed by drop in oxygen saturation indicated by the green arrows. The colored lines in f) represent times when the visualizations of UA were generated. The red arrow in visualizations points to site of airway narrowing during the apneic event. In f) black indicates a patent airway.
Fig. 4.
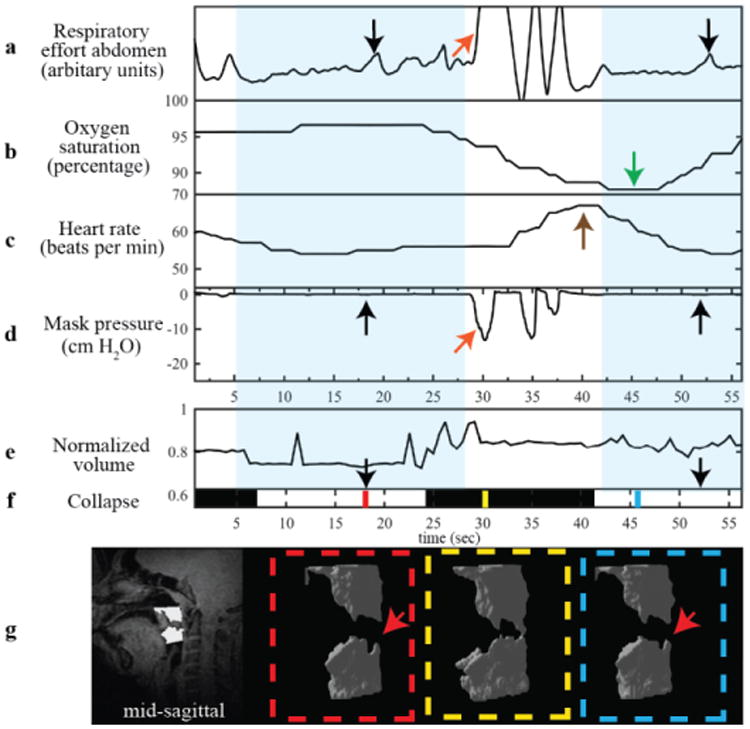
Example use from a 14/M with obstructive apnea event. Waveforms shown are (a) abdominal respiratory effort, (b) oxygen saturation, (c) heart rate, (d) mask pressure, (e) normalized volume (f) collapse and (g) 3D visualizations. The first four a-d were simultaneously measured during an MRI scan and e-g are calculated from segmented masks using the proposed approach with 3D RT-MRI. The blue background highlights the length of the mixed (central and obstructive) apneas. The black arrows indicate change in respiratory effort, flat mask pressure and the white collapse plot. The mixed apnea is followed by a drop in oxygen saturation indicated by the green arrow and an increase in heart rate indicated by the brown arrow. The orange arrows indicate breathing evident from changes in respiratory effort and mask pressure. The colored lines on f) represent times when the visualizations of UA were generated. The red arrows in visualization point to site of airway collapse during the apneic event. In f) black indicates a patent airway and white indicates a collapsed airway.
IV. Discussion
The proposed semi-automated segmentation drastically reduces the processing time of 3D real-time upper airway MRI. It takes a trained operator approximately 10 min/frame to perform manual segmentation, not including training time. Comparatively, semi-automated segmentation requires just 2 min of operator time, and 5 seconds of CPU time per frame. For a typical dataset containing 1800 frames, manual segmentation would require 100+ hours, while semi-automated segmentation requires 2 minutes of manual adjustments in the initial frame and 3 hours of CPU time in subsequent frames.
The specific data acquisition and reconstruction methods used in this work are nascent and evolving. Optimal k-space sampling patterns and reconstruction constraints have not yet been determined, and may improve the underlying image quality. The current images do have residual signal in the airway, suffer from coil roll off in the head-foot direction, suffer from motion artifacts in frames acquired during arousal following an apneic event and appear spatially blurred in some frames. These quality issues make automated segmentation with high accuracy a challenging problem. As the acquisition and reconstruction methods are improved so will the accuracy of segmentation due to the improvement in image quality.
Rapid involuntary motion such as that following an arousal after and apneic event or from swallowing will result in motion artifacts. We did not address these artifacts in the present study, however they may be resolved by identification and rejection of some temporal frames, or by incorporating non-rigid motion correction into the MRI acquisition and reconstruction methods.
Another limitation of the RT-MRI approach is that the scanning itself is loud and subjects have reported difficulty falling asleep. Currently, the sleep state is determined by manual inspection of heart rate, mask pressure, oxygen saturation, and respiratory signals by experienced pediatric pulmonologists. A sustained regular heart rate and breathing
pattern, free of artifact, serve as a surrogate for identifying sleep. Arousal from sleep and wakefulness were assumed with any variability in the heart rate, irregular respiratory breath intervals and/or amplitude, and/or movement artifact contaminating the signals. Manual inspection is subjective and not very accurate where as EEG is the best way to objectively assess sleep state. Due to difficulty using (EEG) in an MRI scanner it was not used in the current protocol. Since sleep state was determined retrospectively, the scan was completed as is even if the patient did not fall asleep or woke up in the middle of the scan. Efforts are being made to use electroencephalogram (EEG) in an MRI scanner. Knowledge of sleep state will enable us to determine the duration of our scan to allow subjects to attain a sufficiently deep level of sleep in the MR scanner for future studies. Efforts are also being made to reduce the acoustic noise from the scanner to help subjects fall asleep more easily because in many OSA patients, obstructive events tend to occur more frequently during rapid eye movement sleep.
This work has demonstrated the feasibility of semi-automated airway segmentation in dynamic 3D airway MRI and there is much room for improvement. The present segmentation method requires manual selection of ROI, seeds and the intensity threshold. Manual selection is subjective and prone to intra- and inter- observer variability. Therefore, further investigation is needed to make this algorithm fully automated and to explore best methods for segmentation.
Recent advances in automated landmark detection algorithms may help reduce variability by automating ROI and seed selection. Recently Wang et al. presented an evaluation and comparison of four robust anatomical landmark detection methods for cephalometry 2D X-ray images [24]. These methods rely on sophisticated image analysis and pattern-recognition principles. They all require annotated training datasets to train the landmark detectors before use in non-labeled datasets [25]–[28]. Automatic land-mark detection has also been used for automated seed placement for colon segmentation using an automated heuristic approach that doesn't rely on training datasets but used knowledge of anatomy for seed placement [29]. This approach was specifically designed for colon segmentation and may not be easily translated to 3D upper airway data. In [30] an automated segmentation plus landmark detection approach is presented for 2D upper airway MR images. Most of these methods were developed for high-resolution 2D images with much higher CNR than in our datasets. However, they may still be applicable to dynamic 3D upper airway images presented in this work and allow for further automation of the segmentation algorithm presented here.
Furthermore, more sophisticated methods may improve the false positive rate. For example, in some frames from the long scan of Subject 1 the airway boundaries were not well preserved and resulted in over-segmentation. The segmented region grew to include the tissue that had lower signal intensity. Such errors might be resolved using region-growing algorithms that detect breach of periphery as it grows, and ensures that segmented region stays within the airway. A technique that addressed this issue has been previously demonstrated in brain lesion segmentation [31].
The 3D renderings generated from our segmentation method can be used to study the dynamics of airflow in the upper airway and its interaction with airway walls during events. Current studies have used static MRI to characterize UA using computational modeling and fluid structure interactions [32]. However, static MRI lacks information about dynamics of UA during collapse. This information can be provided by 3D RT-MRI and may help with improved characterization of UA using computational modeling and fluid structure interactions
V. Conclusion
We have demonstrated a novel and simple semi-automated segmentation approach for dynamic 3D MRI of the pharyngeal airway. The proposed method has shown very good agreement with manual segmentation. This approach, for the first time, enables time-efficient identification of collapse events, and enables visualization of the pharyngeal airway dynamics throughout all events. The segmented dynamic MR data can provide previously unavailable anatomical landmarks for clinical interventions such as surgery or oral appliances in the treatment of OSA, or for patient-specific modeling of the airway collapse pattern
Table III.
Dice coefficient comparison of airway clearance between semi-automated and manual segmentation using four axial slices. DC values for all four slices are shown. In column 3 the average of DC for semi-automated and the two manual segmentation is shown. A DC of 0.85 or greater indicates good agreement. First column contains Patient numbers from Table I.
| # | Manual vs. Manual | Manual vs. Automated |
|---|---|---|
| 1 | 0.88,0.93,0.95,0.96 | 0.72,0.86,0.89,0.88 |
| 2 | 0.91,0.84,0.96,0.96 | 0.74,0.63,0.89,0.91 |
| 3 | 0.98,0.97, 0.98,0.95 | 0.95,0.93,0.94,0.90 |
| 4 | 0.95,0.96,0.97,0.96 | 0.79,0.86,0.89,0.83 |
| 5 | 0.87,0.86,0.91,0.91 | 0.85,0.71,0.81,0.92 |
| 6 | 0.88,0.90,0.95,0.93 | 0.92,0.93,0.96,0.94 |
| 7 | 0.93,0.93,0.92,0.97 | 0.87,0.85,0.87,0.94 |
| 8 | 0.93,0.91,0.93,0.96 | 0.88,0.81,0.83,0.93 |
| 9 | 0.97,0.94,0.91,0.91 | 0.95,0.92,0.89,0.90 |
| 10 | 0.96,0.96,0.95,0.93 | 0.94,0.94,0.94,0.91 |
Acknowledgments
We gratefully acknowledge Ziyue Wu and Ximing Wang for help with data collection, Leonardo Nava, Aaron Bernardo, and Winston Tran for help with the occlusion devices, and Drs. Roberta Kato, Biswas Joshi, Shirleen Loloyan, and Tom Keens for their help with patient recruitment, consent, identifying events and feedback on image quality.
We acknowledge grant support from the National Institutes of Health (R01-HL105210). YCK received support from an American Heart Association postdoctoral fellowship (13POST17000066).
Contributor Information
Ahsan Javed, Email: ahsanjav@usc.edu, Ming Hseih Department of Electrical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA, USA.
Yoon-Chul Kim, Email: yoonckim1@gmail.com, Ming Hseih Department of Electrical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA 90007 USA. He is now with Samsung Medical Center, Seoul, South Korea.
Michael C.K. Khoo, Email: khoo@bmsr.usc.edu, Department of Biomedical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA 90007, USA.
Sally L. Davidson Ward, Email: sward@chla.usc.edu, Department of Pediatrics, Keck School of Medicine, University of Southern California, Children's Hospital Los Angeles, Los Angeles, California, USA.
Krishna S. Nayak, Email: knayak@usc.edu, Department of Electrical Engineering and the Department of Biomedical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA, USA.
References
- 1.Malhotra A, White DP. Obstructive sleep apnoea. Lancet. 2002 Jul;360(9328):237–45. doi: 10.1016/S0140-6736(02)09464-3. [DOI] [PubMed] [Google Scholar]
- 2.Arens R, Marcus CL. Pathophysiology of upper airway obstruction: a developmental perspective. Sleep. 2004 Aug;27(5):997–1019. doi: 10.1093/sleep/27.5.997. [DOI] [PubMed] [Google Scholar]
- 3.Arens R, et al. Upper airway size analysis by magnetic resonance imaging of children with obstructive sleep apnea syndrome. Am J Respir Crit Care Med. 2003 Jan;167(1):65–70. doi: 10.1164/rccm.200206-613OC. [DOI] [PubMed] [Google Scholar]
- 4.Verhulst SL, et al. Sleep-disordered breathing in overweight and obese children and adolescents: prevalence, characteristics and the role of fat distribution. Arch Dis Child. 2007 Mar;92(3):205–8. doi: 10.1136/adc.2006.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009 Jan;373(9657):82–93. doi: 10.1016/S0140-6736(08)61622-0. [DOI] [PubMed] [Google Scholar]
- 6.Rama AN, et al. Sites of obstruction in obstructive sleep apnea. Chest. 2002 Oct;122(4):1139–47. doi: 10.1378/chest.122.4.1139. [DOI] [PubMed] [Google Scholar]
- 7.Schwab RJ, et al. Upper airway and soft tissue anatomy in normal subjects and patients with sleep-disordered breathing. Significance of the lateral pharyngeal walls. Am J Respir Crit Care Med. 1995 Nov;152(5 Pt 1):1673–89. doi: 10.1164/ajrccm.152.5.7582313. [DOI] [PubMed] [Google Scholar]
- 8.Trudo FJ, et al. State-related changes in upper airway caliber and surrounding soft-tissue structures in normal subjects. Am J Respir Crit Care Med. 1998 Oct;158(4):1259–70. doi: 10.1164/ajrccm.158.4.9712063. [DOI] [PubMed] [Google Scholar]
- 9.Schwab RJ, et al. Identification of upper airway anatomic risk factors for obstructive sleep apnea with volumetric magnetic resonance imaging. Am J Respir Crit Care Med. 2003 Sep;168(5):522–30. doi: 10.1164/rccm.200208-866OC. [DOI] [PubMed] [Google Scholar]
- 10.Nayak K, Fleck R. Seeing Sleep: Dynamic imaging of upper airway collapse and collapsibility in children. Pulse, IEEE. 2014;5(5):40–44. doi: 10.1109/MPUL.2014.2339398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagshul ME, et al. Novel retrospective, respiratory-gating method enables 3D, high resolution, dynamicimaging of the upper airway during tidal breathing. Magn Reson Med. 2013 Dec;70(6):1580–90. doi: 10.1002/mrm.24608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arens R, et al. Changes in upper airway size during tidal breathing in children with obstructive sleep apnea syndrome. Am J Respir Crit Care Med. 2005 Jun;171(11):1298–304. doi: 10.1164/rccm.200411-1597OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YC, et al. Measurement of upper airway compliance using dynamic MRI. ISMRM 20th Scientific Sessions, Melbourne. 20:3688. [Google Scholar]
- 14.Kim YC, et al. Real-time MRI can differentiate sleep-related breathing disorders in children Introduction. Proc ISMRM 21st Scientific Sessions, Salt Lake City. 21(c):251. [Google Scholar]
- 15.Kim YC, et al. Real-time 3D magnetic resonance imaging of the pharyngeal airway in sleep apnea. Magn Reson Med. 2014 Apr;71(4):1501–10. doi: 10.1002/mrm.24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YC, et al. Investigations of upper airway obstruction pattern in sleep apnea benefit from real-time 3D MRI. ISMRM 22nd Scientific Sessions, Milan. 2013(5):4364. [Google Scholar]
- 17.Yeoa SY, et al. Segmentation of human upper airway using a level set 496 based deformable model. Proc 13th Med Image Underst Anal. 2009:174–178. [Google Scholar]
- 18.Salerno S, et al. Semi-Automatic Volumetric Segmentation of the Upper Airways in Patients with Pierre Robin Sequence. 2014:487–494. doi: 10.15274/NRJ-2014-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Water VR, et al. Measuring upper airway volume: Accuracy and reliability of dolphin 3d software compared to manual segmentation in craniosynostosis patients. J Oral Maxillofac Surg. 2014;72(1):139–144. doi: 10.1016/j.joms.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, et al. System for upper airway segmentation and measurement with MR imaging and fuzzy connectedness. Acad Radiol. 2003;101:13–24. doi: 10.1016/s1076-6332(03)80783-3. [DOI] [PubMed] [Google Scholar]
- 21.Adam R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell. 1994 Jun;16(6):641–647. [Google Scholar]
- 22.Daniel Region Growing (2D/3D grayscale) [Accessed: 20-Sep-2013];2011 [Online]. Available: http://www.mathworks.com/matlabcentral/fileexchange/32532-region-growing--2d-3d-grayscale-
- 23.Dice L. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 24.Wang CW, et al. Evaluation and Comparison of Anatomical Landmark Detection Methods for Cephalometric X-Ray Images: A Grand Challenge. IEEE Trans Med Imaging. 2015:1–1. doi: 10.1109/TMI.2015.2412951. [DOI] [PubMed] [Google Scholar]
- 25.Geurts P. Automatic Cephalometric X-Ray Landmark Detection Challenge 2014 : A machine learning tree-based approach. 2014 [Google Scholar]
- 26.Chu C, et al. Fully Automatic Cephalometric X-Ray Landmark Detection Using Random Forest Regression and Sparse shape composition. Proc ISBI International Symposium on Biomedical Imaging 2014: Automatic Cephalometric X-Ray Landmark Detection Challenge. 2014 [Google Scholar]
- 27.Mirzaalian H, Hamarneh G. Automatic Globally-Optimal Pictorial Structures with Random Decision Forest Based Likelihoods For Cephalometric X-Ray Landmark Detection. Proc ISBI International Symposium on Biomedical Imaging 2014: Automatic Cephalometric X-Ray Landmark Detection Challenge. 2014 [Google Scholar]
- 28.Ibragimov B, et al. Automatic cephalometric X-ray landmark detection by applying game theory and random forests. Proc ISBI International Symposium on Biomedical Imaging 2014: Automatic Cephalometric X-Ray Landmark Detection Challenge. 2014 [Google Scholar]
- 29.Iordanescu G, Pickhardt PJ, Choi JR, Summers RM. Automated seed placement for colon segmentation in computed tomography colonography. Acad Radiol. 2005;12(2):182–190. doi: 10.1016/j.acra.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Raeesy Z, Rueda S, Udupa JK, Coleman J. Automatic Segmentation of Vocal Tract Mr Images. 2013:1316–1319. [Google Scholar]
- 31.Hojjatoleslami SA, Kruggel F. Segmentation of Large Brain Lesions. 2001;20(7):666–669. doi: 10.1109/42.932750. [DOI] [PubMed] [Google Scholar]
- 32.Ward SD, Amin R, Arens R. Pediatric Sleep-Related Breathing Disorders: Advances in imaging and computational modeling. Pulse, IEEE. 2014;5(5):33–39. doi: 10.1109/MPUL.2014.2339293. [DOI] [PMC free article] [PubMed] [Google Scholar]


