Abstract
A microfluidic device was fabricated and characterized for studying cell lysis of Arcella vulgaris, a nonpathogenic amoeba, over time. The device contains a series of chambers which capture cells allowing them to be subsequently exposed to a constant flow of biocidal agent. With this microfluidic system, individual cells are observed as they undergo lysis. This allows high-throughput measurements of individual lysis events, which are not possible with conventional techniques. Differences in lysis and decay times for Arcella were seen at different flow rates and concentrations of benzalkonium chloride, a biocidal detergent. The efficacy of benzalkonium chloride, chlorhexidine digluconate, phenol, sodium dodecyl sulfate, and Triton X-100 were compared, revealing information on their mechanisms of action. The presented device allows cell capture, controlled exposure to chemical biocides, and observation of lysis with single-cell resolution. Observations at the single cell level give insight into the mechanistic details of the lysis of individual Arcella cells vs. the population; decay times for individual Arcella cells were much shorter when compared to a population of 15 cells.
INTRODUCTION
Microfluidic and lab-on-a-chip devices for analyzing a variety of single cells,1–6 embryos,7, 8 and whole animals9, 10 have been recently developed. Microfluidic systems are excellent tools for biological assays; they require small volumes of reagents and sample sizes, and allow for multiple steps of an analysis to be integrated into a single system. In addition, measurements on microfluidic chips can be multiplexed and/or automated allowing simultaneous analyses to be performed. Commonly, microfluidic devices are rapidly prototyped via soft lithography with poly(dimethylsiloxane) (PDMS),11 an inexpensive, biocompatible polymer. This soft lithographic fabrication technique allows for flexibility in device design which can be tailored to meet the needs of the assay, making these systems ideally suited for investigating biological phenomena.12 One such phenomenon is the reaction of biological cells following exposure to chemical biocides.
Biocidal agents (i.e., disinfectants and antiseptics) are an important class of chemical compounds that encompass a variety of molecular sizes, structures, elemental compositions, and modes of action.13–20 Biocides exhibit a large degree of diversity since their targets—fungi, protozoa, bacteria, viruses, and other parasites—all have unique cellular structure and composition. Aldehydes, for example, are biocidal as they induce cross-linking of proteins and DNA, whereas silver salts interact with thiol groups on membrane-bound enzymes, effectively killing the cell. Many detergents and related compounds (e.g., quaternary ammonium compounds and chlorhexidine digluconate) are employed as disinfectants and antiseptics whose mode of action is disruption and permeabilization of the cellular membrane.21–26 These common compounds are often found in commercially available products; hence, determining their efficacy and mode of action is important in evaluating their toxicity.
Current methods for evaluating biocidal efficacy and toxicity are carried out with either liposomes or cells. Liposomes are useful models as they are easy to prepare and the membrane composition can be controlled.27 Typical studies of membrane lysis in liposomes use spectrofluorimetry to monitor leakage of an encapsulated fluorophore.28–33 These assays are easy to perform and can provide valuable information; however, they involve a large population and do not allow individual liposomes to be observed microscopically. Liposomes used in these studies are often much smaller than cells (< 1 µm diameter), making direct microscopic imaging difficult or impossible. Furthermore, liposomes are only simple membrane models and do not contain all of the necessary components of biological membranes such as proteins. Toxicity studies can also be carried out by fluorescence or absorbance measurements in petri dishes or microtiter plates of cell populations incubated with detergents and disinfectants.34 However, these studies are limited as it is difficult to collect data if membrane disruption occurs in a short time frame. These assays do not allow cells to be imaged with single-cell resolution, preventing the observation of morphological changes. When a viability dye is released from lysed cells, there is no means of removing it from solution. Similarly, populations of luminescent bacteria or dinoflagellates have been employed as whole-cell biosensors to quantify toxicity in environmental water samples.35, 36 With these tests, cells are exposed to toxins, and decreases in luminescence are monitored over time with a photometer. Unfortunately, these assays suffer from many of the same challenges as assays performed using liposome and cell populations (vide supra). In order to observe responses of individual cells upon chemical exposure, compounds can be applied to cells with micropipettes and lysis observed via optical microscopy. However, the solution applied using micropipettes is rapidly diluted due to diffusion, thus forming irreproducible temporal and spatial concentration gradients37–39 leading to variation in the flux across cells. Additionally, micropipettes often expose a single cell or liposome at a time, limiting throughput. Lastly, each of the aforementioned techniques requires large volumes along with large cell populations that are not always available, which is another benefit of the microliter volumes utilized in microfluidics.
There are several techniques2, 40, 41 for lysing single cells in microfluidic devices, including the use of lasers and electrical pulses, application of heat, mechanical lysis with nanobarbs, and chemical lysis with detergents. However, these lysis applications do not focus on the use of detergents as biocidal agents with the goal of screening disinfectant efficacy on a microbial species. Here, a microfluidic device has been fabricated to quantify changes in membrane integrity after chemical biocide and detergent exposure on the amoeba Arcella vulgaris, a common, non-pathogenic unicellular protozoan species. Arcella serves as a model for several pathogenic amoebae responsible for diseases such as amoebiasis (dysentery),42 acanthamoeba keratitis,43, 44 granulomatous amoebic encephalitis,44, 45 and primary amoebic meningoencephalites.44, 46–48 The device presented here allows the real-time monitoring of multiple cells via fluorescence microscopy. In contrast to the application of biocides with micropipettes, the fluid flow is laminar, simultaneously exposing many cells to a uniform concentration of detergent over an extended period of time. The unique structure of the system contains chambers that capture cells and prevent them from moving while under continuous flow. The device was characterized by determining the effects of flow rate and concentration of biocides on trapped cells. Cell lysis rates among detergents and biocides were compared, yielding information on membrane integrity and efficacy of these compounds as biocidal agents on amoeboid species.
PROCEDURE
Chemicals
Poly(dimethylsiloxane) (PDMS/Sylgard 184) was obtained from Dow Corning Corp. (Midland, MI, USA). SU-8 photoresist and developer solution (1-methoxy-2-propyl acetate) were purchased from MicroChem Corp. (Newton, MA, USA), and 3-in silicon wafers were purchased from Silicon Quest International, Inc. (Santa Clara, CA, USA). Benzalkonium chloride, chlorhexidine digluconate (20% v/v in water), phenol, sodium dodecyl sulfate (SDS), and Triton X-100 were obtained from Sigma-Aldrich Co. (St. Louis, MO, USA). Chlorotrimethylsilane was obtained from Fluka/Sigma-Aldrich Co. (Seelze, Germany) and acetone was from EMD Chemicals, Inc. (Gibbstown, NJ, USA). Fluorescein diacetate and Alexa Fluor 647 were obtained from Invitrogen Corp. (Eugene, OR, USA). Water (18 MΩ) was purified through a Millipore Milli-Q system (Billerica, MA, USA).
Microfluidic device fabrication
Masks were designed in Illustrator CS3 (Adobe Systems, Inc., San Jose, CA, USA) and printed onto transparency films by a laser photoplotter (CAD/Art Services, Inc., Bandon, OR, USA). A layer of SU-8 25 photoresist (50 µm) was spun onto a silicon wafer, exposed through a mask with a mask aligner (Karl Suss MA6, Suss Microtec, Waterbury Center, VT, USA), and developed according to the photoresist manufacturer protocol. This master mold contains the capture channels and serves as the lower layer in the final device. A second mold was prepared with a 10-µm layer of SU-8 5 which does not contain any capture wells (upper layer channel). Both master molds were stored under vacuum in a desiccator with 1 mL of chlorotrimethylsilane for at least 1 h. A 10:1 (base:curing agent by weight) mixture of PDMS prepolymer was poured onto the master molds and cured in an oven at 70 °C for 2 h. PDMS was peeled away from the master molds and a 1-mm inlet hole was removed with a tissue punch (Harris Uni-Core) on the upper layer. The two PDMS layers were exposed to air plasma for 90 s at 18 W (PDC-32G plasma cleaner, Harrick Plasma, Ithaca, NY, USA). The lower layer channels were placed face up, and the upper layer channel was placed face down and conformally sealed (Figure 1a). The device was treated with air plasma prior to loading with fluids to increase hydophilicity and wetting of the channels.
Figure 1.
(a) Microfluidic device assembly in which two PDMS channels facing each other are sealed together. The bottom channels are 50 µm high and the top channel is 10 µm high. Drawing is not to scale, and only three of eight total chambers are shown. (b) Diagram of the chambers along with channel dimensions. (c) Image of Arcella incubated with fluorescein diacetate in capture chambers. Scale bar is 100 µm.
Fluorescence microscopy and flow experiments
A syringe pump (KD Scientific Inc., Holliston, MA, USA) was used to control the volumetric flow rates of solutions in the microfluidic device. The pump interfaced with the inlet holes of the PDMS microchannels via syringes connected to polyethylene tubing (0.58 mm i.d., 0.965 mm o.d.). Stock solutions of lysis agents (10 mM in water) were diluted in filtered Arcella media. Alexa Fluor 647 (1.33 mg mL−1 water) was diluted to 6.67 µg mL−1 in working lysis solutions to facilitate of the time at which the protozoa were exposed to detergents. Fluorescein diacetate (14.4 mM in acetone) was diluted to 72 µM in suspensions of Arcella vulgaris (Carolina Biological Supply Co., Burlington, NC, USA) for at least 3 min before introducing them into the device. Fluorescence images were acquired with a laser-scanning confocal microscope (TCS SP5, Leica Microsystems, Inc., Bannockburn, IL, USA). Fluorescein was excited with an argon-ion laser at 488 nm and fluorescence collected at 500–580 nm; Alexa Fluor 647 was excited with a HeNe laser at 633 nm and fluorescence collected at 640–700 nm. Fluorescence intensities were normalized according to the equation,
where Inorm is normalized intensity, I is raw intensity, Imax is maximum intensity before lysis, and Ib is background intensity. Lysis was quantified by measuring the decay time and lysis onset time. The decay time is defined as the time in which fluorescence intensity decreases from 95% to 5% (t0.95−t0.05). Lysis onset time represents the time at which there is a 50% decrease in fluorescence intensity due to lysis (t0.50) and is indicative of the time required for lysis to occur after initial exposure to detergent. All errors are expressed as standard error of the mean (SEM).
Computational fluid dynamics (CFD) simulations
CFD simulations were performed with Comsol Multiphysics 3.4 (Comsol, Inc., Burlington, MA, USA), a finite element method solver in order to determine the velocity flow profile in three dimensions. The density and dynamic viscosity of water were used (ρ = 1.0 g cm−3 and μ = 1.0 × 10−3 Pa s). Incompressible Navier-Stokes equations were solved, given by:
where u is velocity, t is time, ρ is density, p is pressure, and μ is dynamic (absolute) viscosity.
RESULTS AND DISCUSSION
Device design and characterization
A microfluidic system for quantifying the toxicity of several compounds was characterized by studying the effects of biocide flow rate and concentration on single cells. The microfluidic device for monitoring cell lysis is depicted in Figure 1. Figure 1a shows two layers of channels, each cast from an SU-8 master mold, which are sealed together resulting in a complete system. The bottom channel (50 µm high) has a 1-mm wide inlet which eventually diverges to a width of 2 mm (Figure 1b). At this point, the wide channel splits into eight separate capture chambers. The entrance to each chamber is 250 µm wide and narrows to 100 µm. On top of the capture chambers is a 10-µm high channel that extends beyond the end of the chambers. As fluid flows through the system, cells are captured in the chambers (Figure 1c) and fluid continues through the top channel so it can exit the device. The width and height of the capture chambers confines single cells or small groups of cells to the chambers during lysis. The number of cells trapped in each chamber was related to the cell density of the suspension and the amount of time spent flowing the cell suspension through the device. An average of 1.75 ± 0.13 cells (n = 23 runs) were captured in each chamber. The use of multiple chambers, each accommodating between one and eight cells, demonstrates the high-throughput screening capability with single-cell resolution. Although not quantified, cells occasionally escaped capture chambers during the loading process. No cells escaped during biocide perfusion in all experiments in which linear fluid velocities were 360, 1080, and 3600 µm s−1.
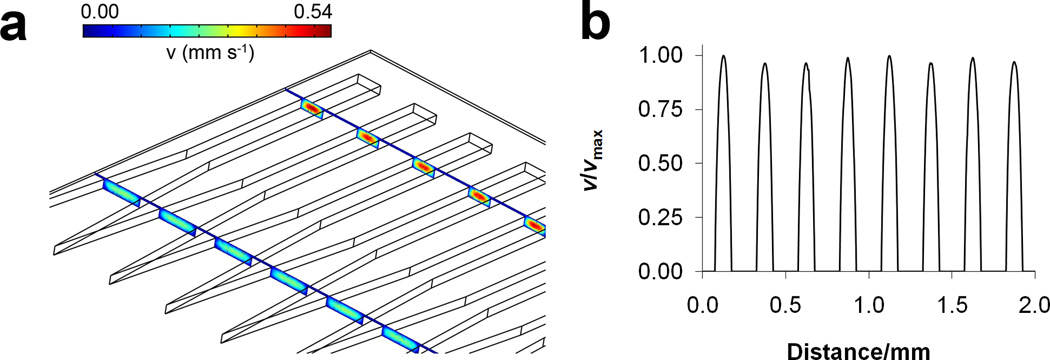
Hydrodynamic flow through microchannels results in a parabolic velocity profile, meaning the shear stress and the flux of molecules will vary across the width of a channel. The system presented here has a single, large channel that splits into eight separate ones, each 100 µm wide and able to accommodate protozoan cells. A computational fluid dynamics (CFD) simulation (Figure 2) shows the linear fluid velocity profile in the microfluidic system. Fluid velocity at the entrance of the channels is low and increases further downstream as the chamber width decreases, as illustrated in the cross-sectional velocity profiles (colored boxes) taken in Figure 2a. The velocity profile taken across the width of all eight channels possesses minimal channel-to-channel variation (Figure 2b, trace taken at a distance of 30 µm from the channel floor and channel ceiling). Areas of zero velocity in between parabolae correspond to the walls located between channels. The results of the simulation show that velocity profiles in all chambers are nearly identical, and therefore cells located in them would experience equal shear stress and fluid flux. The small deviations in maximum velocity (< 5%) for each channel are attributed to voxel size in the simulation which limits computational accuracy. Volumetric flow rates used in lysis studies were 5, 15, and 50 µL min−1, corresponding to average linear flow velocities of 360, 1080, and 3600 µm s−1, and Reynolds numbers (Re) of 0.036, 0.108, and 0.36, respectively. Therefore, the flow in this system is laminar (Re << 2300).
Figure 2.
(a) Three-dimensional CFD simulation displaying cross-sectional linear fluid velocity profiles in the microfluidic chambers corresponding to a volumetric flow rate of 5 µL min−1. Only five of eight total chambers are shown. (b) Normalized velocity line profile taken across the width of all eight chambers, 30 µm from the channel floor.
Single cell vs. average lysis
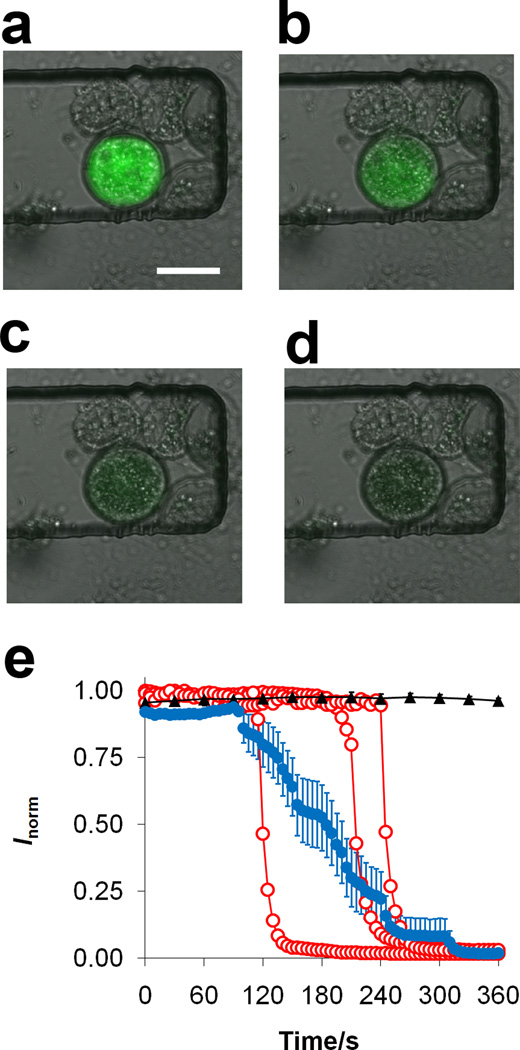
A motivating factor in using microfluidics for cellular assays is the ability to probe events at the single cell level. This is in contrast to data collected from a population of cells, which would yield an average value for the entire collection of cells. Here we explored the responses of single cells during cell lysis with benzalkonium chloride, a quaternary ammonium compound and cationic detergent. Arcella vulgaris cells were first incubated with fluorescein diacetate, a cell viability dye, which is nonfluorescent and can penetrate cell membranes. Once inside the cell, endogenous esterases cleave acetyl groups on the dye molecule, yielding fluorescein which has strong fluorescence emission at a wavelength of 520 nm. After incubation with the dye, the cells were loaded into the device and then exposed to benzalkonium chloride. Fluorescence images over time for a single Arcella exposed to 250-µM benzalkonium chloride at a flow rate of 5 µL min−1 are shown in Figure 3a–d. A marked decrease in fluorescence intensity over time, corresponding to the lysis event, is evident in these images. It is important to note that there is debris present around the cell pictured in Figure 3a–d, which was present in the suspension. Some of this debris consists of proteinous shells from dead Arcella in the original sample prior to loading into the device. Quantitative data regarding lysis is expressed as a plot of normalized fluorescence intensity over time in Figure 3e. In order to determine if shear stress caused cells to leak fluorescein, which would lead to decreased fluorescence over time, control flow experiments were carried out using only media (black trace in Figure 3e). The results confirm that the cells do not leak dye or lose integrity due to shear stress. Furthermore, the laser excitation did not cause photobleaching over the period of time corresponding to lysis.
Figure 3.
Fluorescence images of Arcella exposed to a 5 µL min−1 flow of 250 µM benzalkonium chloride after (a) 195 s, (b) 215 s, (c) 235 s, and (d) 255 s. Scale bar is 50 µm. (e) Normalized fluorescence of three representative individual Arcella cells (○) and average response (●, n = 15 cells) during exposure to 250 µM benzalkonium chloride at 5.0 µL min−1. Control cells (▲, n = 4) were exposed to media only. Error bars represent SEM.
Arcella cells in the channels were then exposed to benzalkonium chloride, and the fluorescence intensity monitored over time. The fluorescence intensities of three individual Arcella cells along with an average of 15 cells exposed to benzalkonium chloride are displayed in Figure 3e. The average trace was determined by averaging the fluorescence intensities of 15 individual cells at each time point. The average fluorescence reveals a broad decrease in intensity over time (decay time, t0.95−t0.05) and larger variation when compared to a single cell. The results for each individual cell lysis event illustrate that the decay time for single cells (41 ± 2 s, n = 19) is much faster than the decay time measured from the average trace of 15 cells (220 s). The decay time of the average trace appears to be a reflection of the variation in the lysis onset times (t0.50) for individual Arcella. There are several reasons why the single cell lysis onset times may vary. Arcella are a type of amoebae; their shape and membranes deform as pseudopods grow from the cell, meaning their sizes may vary. These differences in size can account for different lysis onset times; cells with larger intracellular volumes will take a longer time for the fluorophore, and thus the intracellular contents, to be released. Furthermore, Arcella are testate, meaning that they are protected by a porous, hemispherical shell. The orientation of the cell can account for the differences in lysis onset times since one side of the shell might be completely exposed while the other areas contain a porous coat. Although the cells have varying lysis onset times (176 ± 13 s, n = 19), the decay time for each cell is shorter and has less variation (41 ± 2 s, n = 19). This demonstrates that each lysis event, once begun, is relatively rapid.
Effect of flow velocity and concentration on lysis rates
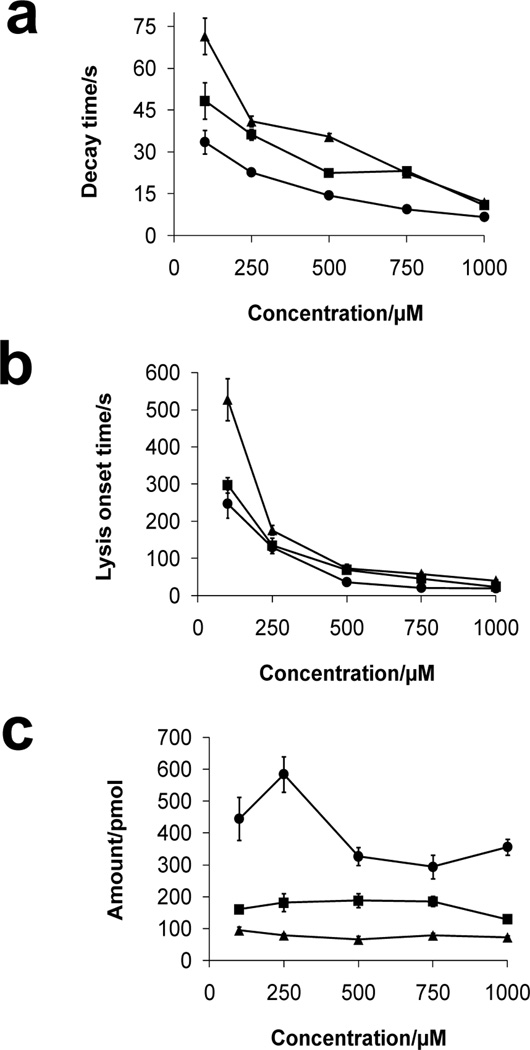
When employing conventional assays (vide supra) without hydrodynamic flow, lysis depends on concentration and diffusive flux. Convection, although present, is uncontrollable and thus highly variable in conventional assays. Conversely, in the microfluidic device, cells are exposed to a constant flow of detergent, meaning the mass transfer of detergent to the cell is faster than in a stagnant solution. The microfluidic system presented here allows accurate control of concentration and convection, thus lysis is dependent on the convective flux. Furthermore, flow enables detergent molecules that solubilize lipids in the membrane to be removed, exposing more membrane, which can be solubilized further. To investigate the influence of flow rate and concentration on cell lysis, solutions of benzalkonium chloride (100–1000 µM) were pumped through the device at different volumetric flow rates (5.0, 15.0, and 50.0 µL min−1) followed by monitoring the fluorescence intensity of individual cells over time. The results of these experiments are plotted in Figure 4a and b showing the effects of concentration and flow rate of benzalkonium chloride on lysis decay time and onset time. At low concentration, Arcella is exposed to less detergent, which yields significantly longer lysis onset and decay times (Figure 4a, two-way ANOVA, F = 9.8, p < 0.0001; Figure 4b, F = 8.4, p < 0.0001). Alternatively, higher detergent concentrations induce lysis in a shorter amount of time. The traces in Figure 4a and b decrease with increasing concentration and flow rate although at low concentrations the decrease in decay and lysis onset times is sharper than at higher concentrations where the decrease is less apparent. In addition to concentration, there are also significant differences in the rates of lysis versus flow rate. At higher flow rates, more detergent is delivered to cells in the device, whereas at lower flow rates, less detergent is delivered and there are longer lysis onset/decay times.
Figure 4.
Plots of (a) decay and (b) lysis onset times of Arcella cells exposed to benzalkonium chloride (100–1000 µM) at flow rates of 5.0 (▲), 15.0 (■), and 50.0 µL min−1 (●). (c) Amount of benzalkonium chloride delivered to cells calculated from lysis onset time at the same flow rates in parts (a) and (b). Values are expressed as mean ± SEM (the number of cells, n, ranges from 17 to 75 for each point).
The total amount of biocide required to cause cell lysis can be determined by calculating convective flux, J:
where N is the amount of biocide in moles, A is cross-sectional area of the microfluidic channel, t is the lysis onset time (defined as the time at which fluorescence intensity of intracellular viability dye reached 50%), c is bulk concentration, and v is average linear velocity determined by CFD simulation. This allows the amount of biocide delivered to a cell at each lysis onset time to be determined. The results in Figure 4c illustrate the differences in amount of benzalkonium chloride required to cause cell lysis at concentrations ranging from 100–1000 µM and flow rates 5.0, 15.0, and 50.0 µL min−1. In general, for each flow rate, the amount of benzalkonium chloride required to lyse the cell remained constant as the concentration is varied; straight lines fit to the data at each flow rate have slopes that are not significantly non-zero (p > 0.22 for each line). There was a difference in the amount of detergent required to induce lysis across the three flow rates, but within each flow rate the concentration did not matter. It is important to note that at 50.0 µL min−1, there was more variation in the amount of benzalkonium delivered to induce lysis (Figure 4c); however, the slope is not significantly non-zero. We hypothesize that at higher flow rates, there is less interaction time between the detergent and cell membrane; therefore, a higher amount of detergent was required for solubilization and lysis. At a flow rate of 50.0 µL min−1 and concentration of 250 µM, a much larger amount of detergent was required, compared to the other concentrations at the same flow rate.
Comparison of various equimolar detergent lysis profiles
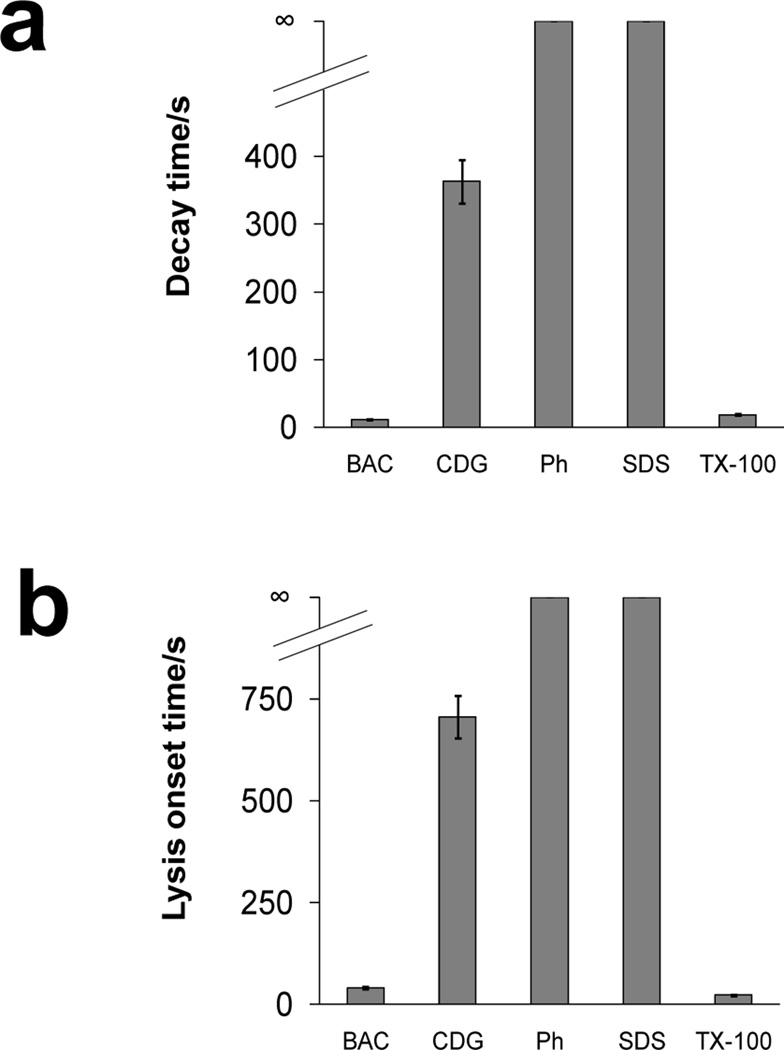
In addition to benzalkonium chloride, the lysis of Arcella during exposure to several biocides including chlorhexidine digluconate, phenol, sodium dodecyl sulfate, and Triton X-100 were also studied to compare their efficacy. With the microfluidic system, Arcella was exposed to equimolar concentrations (1 mM) of biocides and detergents at a constant flow rate of 5.0 µL min−1. The decay and lysis onset times of these biocides and detergents are shown in Figure 5. Neither SDS nor phenol induced lysis after 40 min of exposure. It has been reported that Amoeba proteus contains a mucopolysaccharide coating on the membrane which is negatively charged.49 Since Arcella is also an amoeboid species, it may similarly contain a negatively-charged membrane which hinders lysis by SDS. For the same reason, benzalkonium chloride is an effective lysis agent of Arcella as it is a cationic, quaternary ammonium detergent, enabling it to effectively penetrate and solubilize components of the membrane. This is characterized by the short decay and lysis onset times in Figure 5. Conversely, chlorhexidine digluconate had relatively long decay times. Chlorhexidine is known to cause congealment and coagulation in cell cytoplasm15 making it an effective biocide. Unlike benzalkonium chloride, which is characterized by short, sudden decay times, the loss of fluorescence began quickly after Arcella were exposed to chlorhexidine. Triton X-100, a nonionic detergent, was similarly effective in causing lysis, characterized by short decay and lysis onset times, when compared to benzalkonium chloride.
Figure 5.
(a) Decay and (b) lysis onset times of Arcella exposed to 1000 µM benzalkonium chloride (BAC, n = 26 cells), chlorhexidine digluconate (CDG, n = 16), phenol (Ph, n > 20), sodium dodecyl sulfate (SDS, n > 20), and Triton X-100 (TX-100, n = 15). Flow rate for all biocides was 5.0 µL min−1. ∞ indicates that lysis did not occur within 40 min. Values are expressed as mean ± SEM.
CONCLUSIONS
A microfluidic chip has been fabricated to carry out temporal analysis of individual cells exposed to various biocides at a constant, controlled concentration. These characteristics are difficult to achieve with conventional methods. Arcella vulgaris cells, a model for pathogenic amoebae, were lysed with different biocidal agents and the concentration and flow rate dependence was investigated. With this microfluidic system, lysis events from single cells were quantified, yielding information not easily obtained with population study using conventional techniques. Results show that lysis rates are dependent on both concentration and flow rate, and that there was a wide range of lysis rates among the biocides and detergents employed in these studies.
Acknowledgments
This work was funded by the National Institutes of Health (GM072432) and and the Swedish National Science Foundation (VR). A.G.E. is supported by a Marie Curie Chair from the European Union 6th Framework. This publication was also supported by the Pennsylvania State University Materials Research Institute Nano Fabrication Network and the National Science Foundation Cooperative Agreement No. 0335765, National Nanotechnology Infrastructure Network, with Cornell University.
References
- 1.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 2.Price AK, Culbertson CT. Anal. Chem. 2007;79:2614–2621. doi: 10.1021/ac071891x. [DOI] [PubMed] [Google Scholar]
- 3.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 4.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 5.Di Carlo D, Lee LP. Anal. Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 6.Zhu L, Zhang Q, Feng H, Ang S, Chau FS, Liu W-T. Lab Chip. 2004;4:337–341. doi: 10.1039/b401834f. [DOI] [PubMed] [Google Scholar]
- 7.Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucchetta EM, Munson MS, Ismagilov RF. Lab Chip. 2006;6:185–190. doi: 10.1039/b516119c. [DOI] [PubMed] [Google Scholar]
- 9.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Proc. Natl. Acad. Sci. USA. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chronis N, Zimmer M, Bargmann CI. Nat. Meth. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 11.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 12.McDonald JC, Whitesides GM. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 13.Maillard JY. J. Appl. Microbiol. 2002;92:16S–27S. [PubMed] [Google Scholar]
- 14.Block SS. Disinfection, Sterilization, and Preservation. Philadelphia: Lippincott Williams & Wilkins; 2000. [Google Scholar]
- 15.McDonnell G, Russell AD. Clin. Microbiol. Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denyer SP, Stewart GSAB. Intl. Biodeter. Biodegr. 1998;41:261–268. [Google Scholar]
- 17.Frayha GJ, Smyth JD, Gobert JG, Savel J. Gen. Pharmacol. Vasc. Sys. 1997;28:273–299. doi: 10.1016/s0306-3623(96)00149-8. [DOI] [PubMed] [Google Scholar]
- 18.Maris P. Revue Scientifique et Technique de L Office International des Epizooties. 1995;14:47–55. doi: 10.20506/rst.14.1.829. [DOI] [PubMed] [Google Scholar]
- 19.Khaw M, Panosian CB. Clin. Microbiol. Rev. 1995;8:427–439. doi: 10.1128/cmr.8.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffrey DJ. Revue Scientifique et Technique de L Office International des Epizooties. 1995;14:57–74. doi: 10.20506/rst.14.1.828. [DOI] [PubMed] [Google Scholar]
- 21.Seddon AM, Curnow P, Booth PJ. Biochim. Biophys. Acta - Biomembr. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 22.le Maire M, Champeil P, Moller JV. Biochim. Biophys. Acta - Biomembr. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 23.Neugebauer JM, Murray PD. Meth Enzymol. Academic Press; 1990. pp. 239–253. [Google Scholar]
- 24.Lichtenberg D, Robson RJ, Dennis EA. Biochim. Biophys. Acta - Rev. Biomembr. 1983;737:285–304. doi: 10.1016/0304-4157(83)90004-7. [DOI] [PubMed] [Google Scholar]
- 25.Helenius A, McCaslin DR, Fries E, Tanford C, Fleischer S, Lester P. Meth. Enzymol. Academic Press; 1979. pp. 734–749. [DOI] [PubMed] [Google Scholar]
- 26.Helenius A, Simons K. Biochim. Biophys. Acta - Rev. Biomembr. 1975;415:29–79. doi: 10.1016/0304-4157(75)90016-7. [DOI] [PubMed] [Google Scholar]
- 27.Jesorka A, Orwar O. Annu. Rev. Anal. Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- 28.Apel-Paz M, Doncel GF, Vanderlick TK. Langmuir. 2005;21:9843–9849. doi: 10.1021/la050568r. [DOI] [PubMed] [Google Scholar]
- 29.Castillo JA, Pinazo A, Carilla J, Infante MR, Alsina MA, Haro I, Clapes P. Langmuir. 2004;20:3379–3387. doi: 10.1021/la036452h. [DOI] [PubMed] [Google Scholar]
- 30.Carrillo C, Teruel JA, Aranda FJ, Ortiz A. Biochim. Biophys. Acta - Biomembr. 2003;1611:91–97. doi: 10.1016/s0005-2736(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 31.Apel-Paz M, Kyle Vanderlick T, Chandra N, Doncel GF. Biochem. Biophys. Res. Comm. 2003;309:724–732. doi: 10.1016/j.bbrc.2003.08.065. [DOI] [PubMed] [Google Scholar]
- 32.Apel-Paz M, Doncel GF, Vanderlick TK. Langmuir. 2003;19:591–597. [Google Scholar]
- 33.Sila M, Au S, Weiner N. Biochim. Biophys. Acta - Biomembr. 1986;859:165–170. [Google Scholar]
- 34.Catalone BJ, Ferguson ML, Miller SR, Malamud D, Kish-Catalone T, Thakkar NJ, Krebs FC, Howett MK, Wigdahl B. Biomed. Pharmacother. 2005;59:460–468. doi: 10.1016/j.biopha.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Rosen G, Osorio-Robayo A, Rivera-Duarte I, Lapota D. Arch. Environ. Contam. Toxicol. 2008;54:606–611. doi: 10.1007/s00244-007-9068-3. [DOI] [PubMed] [Google Scholar]
- 36.Lapota D, Osorio AR, Liao C, Bjorndal B. Marine Pollution Bulletin. 2007;54:1857–1867. doi: 10.1016/j.marpolbul.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Zheng JQ, Felder M, Connor JA, Poo M-m. Nature. 1994;368:140–144. doi: 10.1038/368140a0. [DOI] [PubMed] [Google Scholar]
- 38.Lohof AM, Quillan M, Dan Y, Poo MM. J. Neurosci. 1992;12:1253–1261. doi: 10.1523/JNEUROSCI.12-04-01253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown RB, Audet J. J. Royal Soc. Interface. 2008;5:S131–S138. doi: 10.1098/rsif.2008.0009.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi C, Li C-W, Ji S, Yang M. Anal. Chim. Acta. 2006;560:1–23. [Google Scholar]
- 42.Stanley SL. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 43.Marciano-Cabral F, Cabral G. Clin. Microbiol. Rev. 2003;16:273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma P, Visvesvara GS, Martinez AJ, Theodore FH, Daggett P-M, Sawyer TK. Rev. Infect. Dis. 1990;12:490–513. doi: 10.1093/clinids/12.3.490. [DOI] [PubMed] [Google Scholar]
- 45.Siddiqui R, Khan NA. Microbial Pathogen. 2008;44:89–97. doi: 10.1016/j.micpath.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Visvesvara GS, Moura H, Schuster FL. FEMS Immunol. Med. Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 47.Schuster FL, Visvesvara GS. Intl. J. Parasitol. 2004;34:1001–1027. doi: 10.1016/j.ijpara.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Ferrante A. Parasite. Immunol. 1991;13:31–47. doi: 10.1111/j.1365-3024.1991.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 49.Brewer JE, Bell LGE. Nature. 1969;222:891–892. doi: 10.1038/222891b0. [DOI] [PubMed] [Google Scholar]