ABSTRACT
Recently, we interrogated public microarray databases with regard to the expression patterns of metagenes corresponding to major immune cell subtypes present in malignant tumors. This analysis, which involved approximately 3,500 tumor samples, revealed organ-specific differences in the composition of the immune infiltrate as well as in the correlation among distinct cell-type specific metagenes, reflecting changes in the functional organization of the anticancer immune response.
KEYWORDS: Breast cancer, colorectal carcinoma, melanoma non-small cell lung cancer, meta-analysis of microarrays
There is no doubt that the density, composition and function of the immune infiltrate has a major impact on the prognosis of cancer patients, as well as on the therapeutic response to chemotherapy and radiotherapy.1,2 This general observation can be inscribed into the context of the immunosurveillance theory.3 This theory predicts that most (pre-) malignant lesions are eradicated by immune mechanisms at an early, subclinical stage (during the so-called ‘elimination’ phase). Smoldering lesions can stay for a variable period in a precarious balance between two antagonistic forces, namely, the tumor cells that attempt to overcome the immune response, or vice versa (during the so-called ‘equilibrium’ phase). Only at later stage, when tumor cells have undergone ‘immunoediting’ and have acquired the capacity to escape from the immune response or to actively suppress such a response, neoplasia can progress to become a lethal disease (during the so-called ‘escape’ phase). In this context, anticancer therapies only are successful if they reinstate immunosurveillance and hence reset the relationship between the tumor and the immune system from ‘escape’ to ‘equilibrium’, meaning that cancer at least transiently becomes a chronic disease. Ideally, however, anticancer therapy restore immunosurveillance to the phase of‘elimination’, hence leading to complete and permanent cure. This rule does not only apply to so-called immunotherapies but also to conventional chemotherapies and radiotherapies that rely on the contribution of the immune system to be able to mediate a success that last beyond the cessation of the treatment. Hence immune parameters do not only dictate the prognosis of cancers; they also determine the therapeutic response to antineoplastic therapies4-9
Although this general paradigm likely applies to many different histological and molecular types of malignancy, a systematic analysis of the immune infiltrate across distinct cancer types has been elusive. In an attempt to start such as systems biology approach, we studied publicly available microarray data on four distinct cancer types, namely, breast cancer, colorectal cancer, melanoma and non-small cell lung cancers.10 These cancers have in common that they are now known to be under strict immunosurveillance,1 yet differ in their intrinsic prognosis that is much more favorable for breast cancer (for which excellent treatment options exist) than for non-small lung cancer (for which chemotherapy has marginal if any effects on overall survival). Melanoma (that is now entering the era of broadly applied immunotherapy by checkpoint blockers) and colorectal cancers (which can respond to oxaliplatin-based chemotherapy) constitute intermediate cases.
We studied the expression of a series of metagenes originally described by Bindea et al.2 whose transcription indicates the infiltration of the tumor by defined subtypes of immune cells. The expression of each metagene was determined in 20 different cohorts (five for each cancer type included in the study) representing a total of close to 3,500 patient samples.10 The overall composition of the immune infiltrate turned out to be profoundly influenced by the cancer type (Fig. 1). For example, Th1 cells appear to be more abundant in mammary and colorectal carcinomas than in melanoma and non-small cell lung cancer. CD8+ T cells are less abundant in melanoma then in other cancer types. Conversely, the proportion of CD56bright, CD56dim and other NK cells appears rather different in breast cancer as compared to other neoplasias (Fig. 1).
Figure 1.
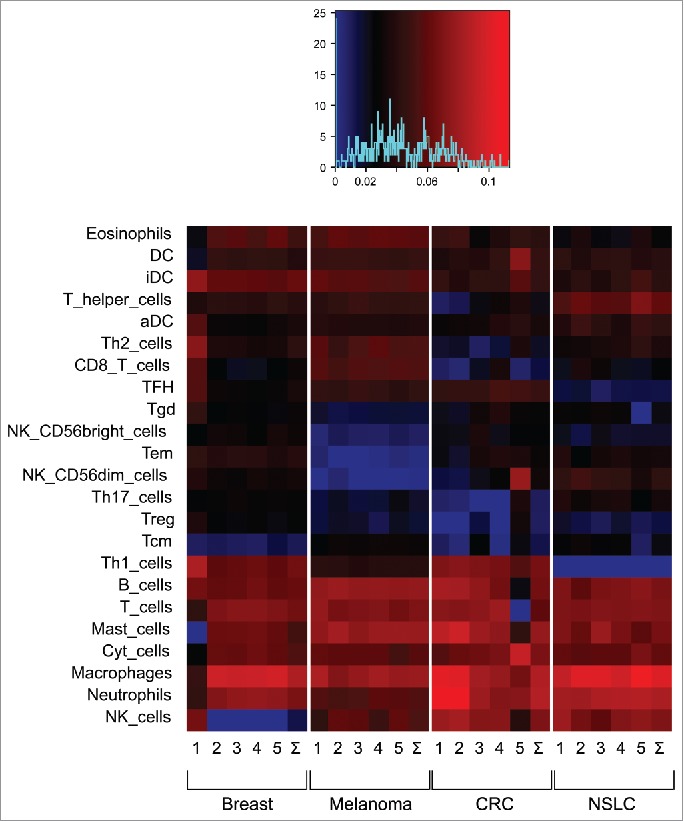
Composition of the immune infiltrate in four different major cancer types. Microarrays were analyzed for the abundance of metagenes reflecting the presence of distinct immune cell types within the malignant lesion. Each cancer type is represented by five different cohorts described in Stoll et al.10 The overall abundance of distinct immune-related metagenes has been normalized to a value of 100% for each sample contained in the different cohorts.
A detailed analysis of the correlation among different immune cell type-related metagenes revealed important differences between breast, colorectal, non-small cell lung cancer and melanoma. Only in breast cancer, both positive and negative correlations among distinct elements of the immune system were discernible across the five cohorts that were analyzed. In contrast, for all other tumor cells, most of the significant correlations were positive (not negative), indicating that not only the composition of the immune infiltrate but also its overall organization is quite strongly influenced by the cancer type.10
Although these data reveal fascinating differences among distinct human malignancies with regard to their immune infiltrate, the present study is afflicted by severe limitations. First of all, microarray data do not provide any insights on the histological architecture of the tissue, although the exact localization of distinct immune cell subtypes may have a major impact on their function in the immunosurveillance system.1 Also, public microarray data often are poorly annotated with respect to the evolution of the patient after therapy. Second, the methodology used to ‘deconvolute’ microarray data with respect to the immune infiltrate2 can be criticized because it conceives immune cells as static entities (with a constant cell-specific transcriptome) that can be classified according to a rigid scheme into a limited number of elements. This is certainly not a realistic scenario. Third, microarray data are being surpassed in quality by RNAseq, a method that provides a potentially much more accurate quantification of the abundance of different transcripts. Last but not least, our study was limited to only four cancer types. However, it can be expected that other malignancies may reveal a yet distinct pattern with regard to the state of local immunosurveillance.1
Future studies must overcome these limitations by integrating immunohistochemical and clinical data, by developing new analysis tools, by focusing on high-quality RNAseq data sets, as well as by including additional tumor types. It is only by a combination of such improvements that we will acquire the capacity to understand the prognostic and predictive impact of each immune subtype for each cancer subtype.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12:298-306; PMID:22419253; http://dx.doi.org/ 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 2.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A et al.. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39:782-95; PMID:24138885; http://dx.doi.org/ 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases–elimination, equilibrium and escape. Curr Opin Immunol 2014; 27:16-25; PMID:24531241; http://dx.doi.org/ 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M et al.. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012; 337:1678-84; PMID:23019653; http://dx.doi.org/ 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 5.Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity 2013; 39:74-88; PMID:23890065; http://dx.doi.org/ 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remédios C et al.. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 2014; 20:1301-9; PMID:25344738; http://dx.doi.org/ 10.1038/nm.3708 [DOI] [PubMed] [Google Scholar]
- 7.Michaud M, Xie X, Bravo-San Pedro JM, Zitvogel L, White E, Kroemer G. An autophagy-dependent anticancer immune response determines the efficacy of melanoma chemotherapy. Oncoimmunology 2014; 3:e944047; PMID:25610726; http://dx.doi.org/ 10.4161/21624011.2014.944047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoll G, Enot D, Mlecnik B, Galon J, Zitvogel L, Kroemer G. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014; 3:e27884; PMID:24790795; http://dx.doi.org/ 10.4161/onci.27884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vacchelli E, Aranda F, Eggermont A, Galon J, Sautes-Fridman C, Cremer I, Zitvogel L, Kroemer G, Galluzzi L. Trial Watch: Chemotherapy with immunogenic cell death inducers. Oncoimmunology 2014; 3:e27878; PMID:24800173; http://dx.doi.org/ 10.4161/onci.27878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoll G, Bindea G, Mlecnik B, Galon J, Zitvogel L, Kroemer G. Meta-analysis of organ-specific differences in the structure of the immune infiltrate in major malignancies. Oncotarget 2015; 6:11894-909; PMID:26059437; http://dx.doi.org/ 10.18632/oncotarget.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]


