Abstract
Changes in the phosphorylation status of the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAPII) correlate with the process of eukaryotic transcription. The yeast protein regulator of transcription 1 (Rtr1) and the human homolog RNAPII-associated protein 2 (RPAP2) may function as CTD phosphatases; however, crystal structures of Kluyveromyces lactis Rtr1 lack a consensus active site. We identified a phosphoryl transfer domain in Saccharomyces cerevisiae Rtr1 by obtaining and characterizing a 2.6 Å resolution crystal structure. We identified a putative substrate-binding pocket in a deep groove between the zinc finger domain and a pair of helices that contained a trapped sulfate ion. Because sulfate mimics the chemistry of a phosphate group, this structural data suggested that this groove represents the phosphoryl transfer active site. Mutagenesis of the residues lining this groove disrupted catalytic activity of the enzyme assayed in vitro with a fluorescent chemical substrate, and expression of the mutated Rtr1 failed to rescue growth of yeast lacking Rtr1. Characterization of the phosphatase activity of RPAP2 and a mutant of the conserved putative catalytic site in the same chemical assay indicated a conserved reaction mechanism. Our data indicated that the structure of the phosphoryl transfer domain and reaction mechanism for the phosphoryl transfer activity of Rtr1 is distinct from those of other phosphatase families.
INTRODUCTION
Posttranslational modifications (PTMs) of the C-terminal domain (CTD) of RNA polymerase II (RNAPII) correlate with different stages of eukaryotic transcription—promoter binding, initiation, pausing, elongation, mRNA processing, termination, and RNAPII recycling (1, 2). CTD kinases and phosphatases, methyltransferases and demethylases, acetyltransferases and deacetylases, and prolyl isomerases mediate the modifications that occur on the CTD during transcription and release of RNAPII, although the physiological roles of methylation and acetylation remain not well understood (1, 2). Intriguingly, the consensus sequence of CTD is composed of multiple heptad repeats of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7.
Global chromatin immunoprecipitation (ChIP) analysis of RNAPII in yeast and human cells using PTM-specific antibodies [antibodies recognizing phosphorylation of Tyr1, Ser2, Thr4, Ser5, or Ser7 residues in the heptad repeat (2), methylation of Arg residues (3), and acetylation of Lys residues (4)] provided information about the PTM patterns of the CTD and enabled predictions regarding the proteins that interact with and modify RNAPII, thereby producing a model of PTM dynamics of RNAPII as transcription progresses (5). In this model, transcription starts with unphosphorylated RNAPII forming a preinitiation complex (PIC) enabled by RNAPII-associated transcription factors. Phosphorylation of Ser5 residues in the heptad repeat is an indication that the PIC has dissociated and RNAPII has been removed from the promoter, which is referred to as “promoter clearance” (6). During the elongation phase of transcription, as RNAPII moves toward the 3′ end of a gene, phosphatase activity reduces phosphorylation of Ser5 residues in the heptad repeat, whereas phosphorylation of Ser2 residues in the heptad repeat increases and becomes the dominant modification of the CTD. Complete dephosphorylation of the CTD is necessary for RNAPII to initiate a new round of transcription (6, 7).
Although two phosphatases, the Scps (small CTD phosphatases) family and Ssu72, target the CTD, neither is responsible for the dephosphorylation of Ser5 residues in the heptad repeat during the transition from transcription initiation to elongation. Scps are transcriptional corepressors in human cells that silence the expression of certain neuronal genes and are, therefore, not involved in active transcription, but rather prevent transcription from even initiating (8). Ssu72 is a protein conserved from yeast to human that mediates mRNA coprocessing and transcription termination, events that occur at the end of transcription (2). Ssu72 is also localized near the 3′ end of the coding portions of genes, which is consistent with the reduction of phosphorylated Ser5 residues in the heptad repeat observed at the end of the transcription cycle (2). However, the identity of the phosphatase that accounts for this reduction in Ser5 phosphorylation in the heptad repeat during the transition from initiation to elongation is controversial.
Three criteria must be met for a phosphatase to be matched with a physiological substrate: (i) the protein must have an in vitro phosphatase activity; (ii) the elimination of the protein must result in the in vivo accumulation of the phosphorylated substrate; and (iii) the substrate and putative phosphatase must be colocalized in the cell at some point during the cell cycle. Rtr1, an RNAPII-associated protein in yeast, meets these three requirements for the phosphatase responsible for dephosphorylating Ser5 in the heptad repeat of the CTD: (i) Rtr1 shows invitro phosphatase activity against the CTD peptide repeats phosphorylated by transcription kinase TFIIH invitro (9) and dephosphorylates a general phosphatase substrate, 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), in vitro (10); (ii) in yeast, the abundance of CTD phosphorylated at Ser5 residues in the heptad repeat increased upon Rtr1 deletion (9, 11); and (iii) ChIP analysis indicated that RNAPII and Rtr1 were associated during the transition from transcription initiation to elongation (9). Western blotting analysis, using phosphorylation site-specific antibodies recognizing the CTD, showed that Rtr1-mediated dephosphorylation of glutathione S-transferase (GST)–tagged CTD [phosphorylated by either Saccharomyces cerevisiae CTD kinase 1 (CTDK1) or mitogen-activated protein kinase (MAPK)] occurred preferentially at phosphorylated Ser5 compared with its activity at phosphorylated Ser2 or Ser7 residues in the heptad repeat (9). Consistent with conserved activity, the human homolog of Rtr1, RNAPII-associated protein 2 (RPAP2), dephosphorylates GST-CTD that had been phosphorylated by the cyclin-dependent kinase P-TEFb (12) in vitro. Knockdown of RPAP2 increases the abundance of RNAPII phosphorylated at Ser5 residues in the heptad repeat of CTD (12), and RPAP2 physically and functionally associates with RNAPII (13).
Although invivo and invitro studies of Rtr1 and RPAP2 provide evidence for phosphatase activity (10, 12), the primary sequences of Rtr1 and RPAP2 lacks any recognizable phosphatase motif. Although a structure for S. cerevisiae Rtr1 was obtained, the resolution was low (4 Å) (10, 14). A crystal structure of Rtr1 from the yeast Kluyveromyces lactis did not have any structural resemblance to structures of known phosphatase families, and no putative active site pocket was identified (14). Furthermore, K. lactis Rtr1 lacked activity for phosphorylated CTD in vitro under the conditions used to detect this activity for S. cerevisiae Rtr1 (14). Thus, the structural and biochemical analysis of K. lactis Rtr1 casts doubt on the identification of Rtr1 as an RNAPII CTD phosphatase.
To resolve the controversy about the phosphoryl transfer activity of Rtr1, we identified the phosphoryl transfer domain of S. cerevisiae Rtr1 and crystallized it at a resolution of 2.6 Å. A deep groove formed between the zinc finger core and an insertion α-helical pair trapped a phosphate-mimicking sulfate group, indicating the location of the active site. Residues aligning this deep groove play an essential role for phosphoryl transfer reaction, and Rtr1 with mutations in these residues slowed cell growth in yeast. The availability of the full-length phosphoryl transfer domain structure allowed us to compare it to all known phosphatase families. Rtr1 presents a distinct fold compared to that of other phosphatase families and also seems to utilize a unique catalytic mechanism different from other phosphatases, which suggests the existence of a novel phosphatase family.
RESULTS
Biochemical characterization of the S. cerevisiae Rtr1 phosphoryl transfer domain
We focused on the characterization of S. cerevisiae Rtr1 phosphatase activity. The substrate para-nitrophenyl phosphate (pNPP) is commonly used to assay typical phosphatases. This compound produces a yellow color when cleaved by a phosphatase. However, the colorimetric pNPP assay does not work for RPAP2 (15). Consistent with the study of RPAP2, we observed no activity with S. cerevisiae Rtr1 in the pNPP assay, unless the protein and substrate were incubated overnight (fig. S1). The necessity for such a prolonged incubation indicated that Rtr1 can dephosphorylate pNPP but with very low activity. Therefore, we switched to a more sensitive assay that uses the fluorescent compound DiFMUP, which is 500 times more sensitive than the pNPP assay (16) and has been used to assay the activity of K. lactis Rtr1 (10).
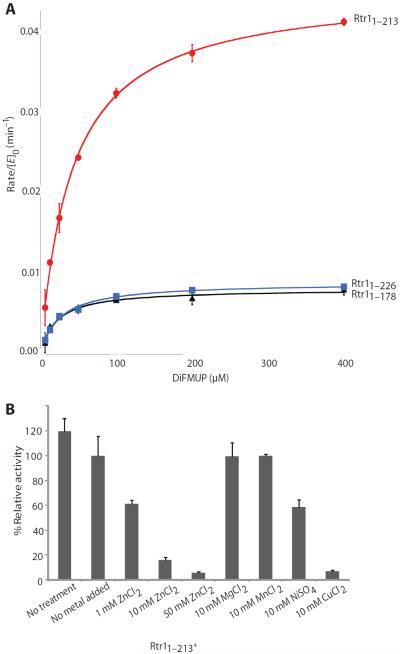
Because we wanted to crystallize the catalytically active region of Rtr1 and because S. cerevisiae Rtr1 had been difficult to crystallize previously (10, 14), we sought to identify the boundaries of the catalytic domain of S. cerevisiae Rtr1 by analyzing the activity of truncated forms of the enzyme in vitro with DiFMUP. Secondary structure prediction guided the truncations (17). Along with polyhistidine (His)–tagged full-length Rtr (amino acids 1 to 226), we cloned and attempted to express five truncated forms of His-tagged Rtr1 in Escherichia coli: Rtr11–129, Rtr11–137, Rtr11–167, Rtr11–178, and Rtr11–213 (table S1). We could not express the three shortest forms in E. coli, suggesting issues in protein folding. Full-length Rtr1, Rtr11–178, and Rtr11–213 exhibited robust expression, and we purified these to homogeneity and removed the His tag by protease digestion (figs. S2 and S3). Rtr11–213 was the most active, with a Michaelis constant (Km) almost twice that of full-length Rtr1 and kcat ~5 times that of full-length Rtr1 or Rtr11–178 (Fig. 1A and Table 1). Because Rtr11–178 had activity similar to that of full-length Rtr1 (Fig. 1A), we reasoned that this region contained the catalytic activity and was sufficient to mediate phosphoryl transfer.
Fig. 1. Phosphatase activity of purified recombinant Rtr1 in the DiFMUP assay.
(A) Kinetic parameters of phosphatase activity, Km and kcat (Table 1), of Rtr1 were determined by fitting the data to the Michaelis-Menten equation for the two truncation mutants and full-length Rtr1 (Rtr11–226). The error bars represent SD from triplicate samples, and data are representative of five independent experiments. (B) Influence of metals on the phosphatase activity of Rtr11–213. Except in the first assay, proteins in all other assays were treated with 10 mM EDTA and then dialyzed in a buffer with no EDTA overnight. The activity of Rtr1 after EDTA without metal added is set to 100% (no metal added). Data shown are representative of three independent experiments. Values indicate the average from three replicates and errors bars indicate SD.
Table 1. Steady-state kinetic parameters of the full-length and truncated forms of S. cerevisiae Rtr1 and human RPAP2 assayed in vitro with DiFMUP as the substrate.
The K. lactis Rtr values (10) are included for comparison.
| Protein | Km (μM) | 10−3 kcat (min−1) | kcat/Km |
|---|---|---|---|
| K. lactis Rtr1 | 587 ± 70 | 60 ± 1.8 | 0.10 |
| Rtr11–178 | 20.44 ± 2.70 | 8.06 ± 0.31 | 0.40 |
| Rtr11–213 | 41.15 ± 1.28 | 45.25 ± 0.51 | 1.10 |
| Rtr11–226 | 24.71 ± 1.60 | 8.85 ± 0.18 | 0.36 |
| RPAP232–274 | 30.95 ± 1.75 | 1.45 ± 0.06 | 0.05 |
| RPAP232–345 | 34.20 ± 0.84 | 0.55 ± 0.01 | 0.02 |
Effect of metal ions on the phosphoryl transfer activity of S. cerevisiae Rtr1
The core structure of K. lactis Rtr1 is a conserved zinc finger motif. Previous reports showed that K. lactis Rtr1 obtained by nickel affinity purification displayed no detectable phosphatase activity until the nickel was removed by incubation of the recombinant protein with the divalent cation chelating agent EDTA and subsequent addition of zinc ion (kcat/Km = 0.10mM−1 min−1) (9, 10). Although our purification methods were similar to those used for isolating K. lactis Rtr1 and relied on nickel affinity purification, we found that without EDTA washing, S. cerevisiae Rtr1 exhibited a 10-fold higher phosphatase activity than that reported for K. lactis (compare kcat/Km in Table 1). Indeed, instead of increasing the activity of S. cerevisiae Rtr11–213, EDTA exposure reduced Rtr1 activity (Fig. 1B). After removal of divalent metal ions with EDTA, we supplemented the reaction with zinc ions, which produced a dose-dependent decrease in Rtr11–213 activity (Fig. 1B), similar to the effect of Zn2+ ions on RPAP2 (15). Supplementation with Mg2+ or Mn2+ resulted in a similar amount of Rtr11–213 activity as buffer alone after EDTA exposure (Fig. 1B). Ni2+ reduced Rtr1 activity ~50% compared to the Rtr11–213 preparation that was not exposed to EDTA, and Cu2+ almost abolished the activity (Fig. 1B). Because EDTA treatment provided no increase in activity, we did not expose the affinity-purified S. cerevisiae Rtr1 proteins to EDTA or supplemented the reaction mixture with metal ions.
Effect of phosphatase inhibitors on S. cerevisiae Rtr1
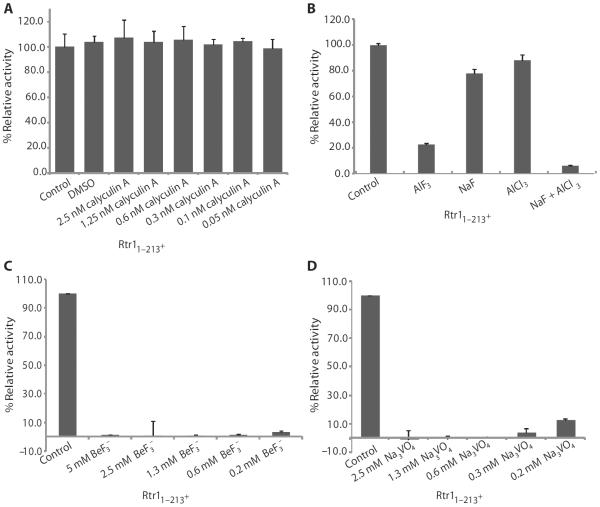
To confirm that the activity of Rtr1 measured in the DiMFUP assay resulted from the phosphatase activity, we tested the effect of various phosphatase inhibitors on Rtr1 activity in this assay. Two categories of phosphatase inhibitors exist, which target different phosphatases on the basis of the phosphatase catalytic mechanism. Metal-dependent phosphatases, such as protein phosphatase 1 (PP1) and PP2A, activate a water molecule that bridges between two metal ions to attack the phosphorus atom in the substrate (18) and are usually inhibited by natural product toxins. Phosphatases that mediate the phosphoryl transfer reaction through a phosphoryl-enzyme intermediate, such as cysteine-based phosphatases (tyrosine phosphatases and dual-specificity phosphatases) or aspartate-based phosphatases [the haloalkanoic acid dehalogenase (HAD) superfamily], are inhibited by compounds mimicking inorganic phosphate (19). Because these pharmacological differences arise from the phosphatase catalytic mechanism, inhibitors of these two different categories can provide insight regarding catalytic mechanism. Therefore, we tested the effects of inhibitors from each category on Rtr11–213 in the DiFMUP assay (Fig. 2). The PP1 and PP2 inhibitor calyculin A did not inhibit Rtr1 when tested through a range of concentrations that inhibit PP1 and PP2 (20) (Fig. 2A).
Fig. 2. The effects of phosphatase inhibitors on the phosphoryl transfer reaction of Rtr1 in the DiFMUP assay.
(A) Effect of an inhibitor of metal-dependent phosphatases. Activity of Rtr11–213 in the presence of the indicated concentrations of the PP1 and PP2 inhibitor calyculin A. DMSO, dimethyl sulfoxide. (B) Effect of aluminum fluoride, an inhibitor of cysteine- and aspartate-based phosphatases. Activity of Rtr11–213 in the presence of aluminum fluoride (AlF3) provided directly or by including both sodium fluoride (NaF) and aluminum chloride (AlCl3) in the reaction. Control experiments included adding sodium fluoride or aluminum chloride individually or reactions performed in the absence of added compounds (Control). (C) Effect of beryllium fluoride, an inhibitor of cysteine- and aspartate-based phosphatases. Activity of Rtr11–213 in the presence of the indicated concentrations of beryllium fluoride ion in solution (BeF3−). (D) Effect of sodium vanadate, an inhibitor of cysteine- and aspartate-based phosphatases. Activity of Rtr11–213 in the presence of the indicated concentrations of sodium vanadate (NaVO4). Activity is shown relative to the control sample, which was set to 100%. The error bars represent SD from triplicate samples, and data are representative of three independent experiments.
In contrast, aluminum fluoride (AlF3), which resembles the chemical structure of inorganic phosphate, reduced the activity of Rtr11–213 (Fig. 2B). The inhibitory effect is due to the formation of AlF4−, because reactions containing either NaF or AlCl3 showed no effect on Rtr1 activity (Fig. 2B); however, the presence of both NaF and AlCl3, which generates AlF4−, effectively inhibited Rtr1. We also tested other phosphatase inhibitors, beryllium fluoride (BeF3) and sodium orthovanadate (Na3VO4), that mimic different stages of the phosphoryl transfer reaction, and these abolished Rtr1 activity with only low amounts of Rtr1 activity detected at the lowest concentrations of the inhibitors tested (Fig. 2, C and D). Therefore, inhibitors mimicking the chemical structure of inorganic phosphate were effective at inhibiting Rtr1 activity, suggesting that the phosphoryl transfer mechanism of this phosphatase proceeds through a phosphoryl-enzyme intermediate.
Identification of important residues for phosphoryl transfer in S. cerevisiae Rtr1
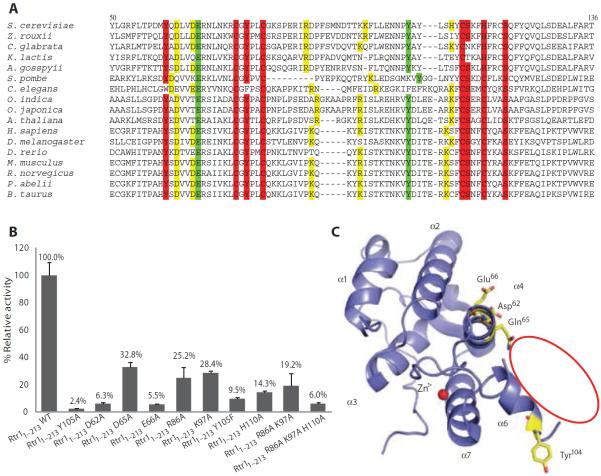
Our kinetic and inhibitor studies indicated that Rtr1 has phosphoryl transfer activity, yet the sequence of Rtr1 lacks a consensus phosphoryl transfer motif. To gain insight into the reaction mechanism of Rtr1, we aligned sequences of Rtr1 and its homologs in species ranging from plants to animals to insects and other yeast to identify conserved residues (Fig. 3A). We introduced point mutations in each of the 15 conserved sites into Rtr11–213. We selected those that expressed well in E. coli and exhibited an appropriate biochemical profile consistent with proper folding by size exclusion chromatography for in vitro phosphatase activity in the DiFMUP assay (fig. S4, A and B, and table S1).
Fig. 3. Identification of conserved residues important for Rtr1 phosphoryl transfer activity.
(A) Multiple sequence alignment of 17 Rtr1 homologs in the zinc-binding region, representing the phosphoryl transfer region and showing residue conservation. Red color highlights residues that cause the loss of protein expression upon single mutation to Ala. The mutation variants of green residues have undetectable phosphatase activity in the DiFMUP assay. The mutation variants of yellow residues have less than 30% phosphoryl transfer activity than nonmutated Rtr11–213. Mutants were introduced in Rtr11–213. (B) Effect of mutation of the indicated residues in the phosphoryl transfer activity of Rtr11–213. The percentage of activity relative to nonmutated Rtr11–213 [wild type (WT)] is shown as bar graph, with error bar showing the SD of three independent biological experiments. (C) Cartoon representation of the crystal structure of K. lactis Rtr1 [Protein Data Bank (PDB) identifier (ID): 4FC8]. The four amino acid residues that are conserved among the 17 different species shown in (A) and identified as important for phosphoryl transfer activity shown in (B) are shown as sticks. The red oval line indicates the disordered region.
Mutating Tyr60, Tyr75, Ser113, or Ser120, as well as any of the four residues that coordinate a zinc ion (Cys73, Cys78, Cys112, or His116), to alanine resulted in an inability to express Rtr1 in E. coli, suggesting that these residues may be necessary for the structural integrity of the protein. The remaining mutated proteins were expressed and could be purified and assayed for phosphatase activity in the DiFMUP assay (Fig. 3B). The Rtr11–213 with Y105A, D62A, E66A, or Y105F exhibited less than 10% of the activity of the wild-type Rtr11–213. We analyzed these proteins by size exclusion gel filtration and SDS—polyacrylamide gel electrophoresis (SDS-PAGE) to confirm that the proteins were not degraded (fig. S4 and table S1). The D65A, R86A, and K97A mutant Rtr11–213 exhibited ~20 to 30% of the activity of the wild-type Rtr11–213, using bovine serum albumin (BSA) as negative control (Fig. 3B), indicating that these proteins were likely stable and properly folded. These results identified residues important for the phosphoryl transfer reaction mediated by Rtr1, especially Tyr105, Asp62, and Glu66.
To understand the possible role of these putative catalytic residues, we mapped these amino acids to the structure of K. lactis Rtr1 (14). All catalytically important residues locate adjacent to a disordered region (residues 88 to 100 in K. lactis Rtr1) in the crystal structure (Fig. 3C); thus, no information could be gained. However, we postulated that the catalytic site may be within the disordered region.
Crystal structure of S. cerevisiae Rtr1
Because full-length S. cerevisiae Rtr1 does not diffract to high resolution even after extensive surface entropy optimization and seeding trials, consistent with independent efforts (10), we used the shortest active S. cerevisiae Rtr1 truncation mutant Rtr11–178, which had activity similar to wild-type Rtr1 in the DiFMUP assay (Fig. 1A), to generate crystals. We determined a 2.6 Å resolution structure of Rtr11–178 by molecular replacement using K. lactis Rtr1 as the initial search model (PDB ID: 4FC8) (14) and refined the structure (21) to produce a final model (Table 2).
Table 2. Summary of crystallographic statistics for S. cerevisiae Rtr1.
Statistics for the highest-resolution shell are shown in parentheses. RMS, root mean square.
| S. cerevisiae Rtr11–178 | |
|---|---|
| Data collection statistics | |
| Synchrotron source | Advanced Photon Source (APS) beamline GM-CAT |
| Wavelength (Å) | 1.28849 |
| Resolution range (Å) | 50–2.6 (2.64–2.60) |
| Space group | P 43 21 2 |
| Unit cell (a, b, c) (Å) | 118.965, 118.965, 69.414 |
| Unique reflections | 14,886 (545) |
| Multiplicity | 5.6 (2.3) |
| Completeness (%) | 93.6(71.2) |
| Mean I/sigma(I) | 9.24(1.32) |
| R sym | 0.169 (0.675) |
| Refinement statics | |
| Resolution limits (Å) | 45.18–2.60 |
| Number of reflections per test | 14,868 (1486) |
| R work | 0.207(0.315) |
| R free | 0.262 (0.354) |
| Number of nonhydrogen atoms | 2,884 |
| Macromolecules | 2,846 |
| Ligands | 21 |
| Water | 18 |
| Protein residues | 346 |
| RMS bonds (Ǻ) | 0.009 |
| RMS angles (°) | 1.21 |
| Ramachandran favored (%) | 98.25 |
| Ramachandran outliers (%) | 0 |
| Clashscore calculated from MolProbity | 6.32 |
| Average B-factor | 45.8 |
| Macromolecules | 37.4 |
| Ligands | 56.5 |
| Solvent | 34.4 |
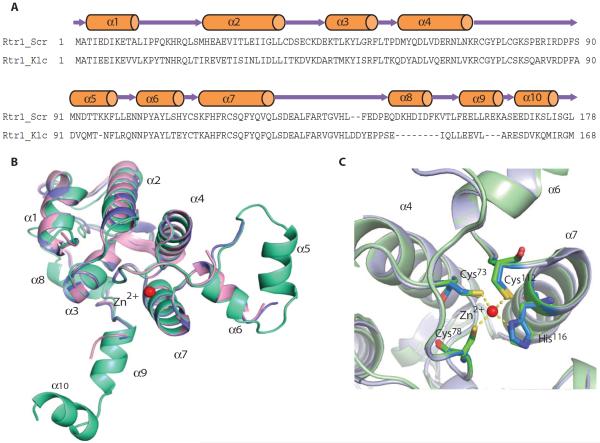
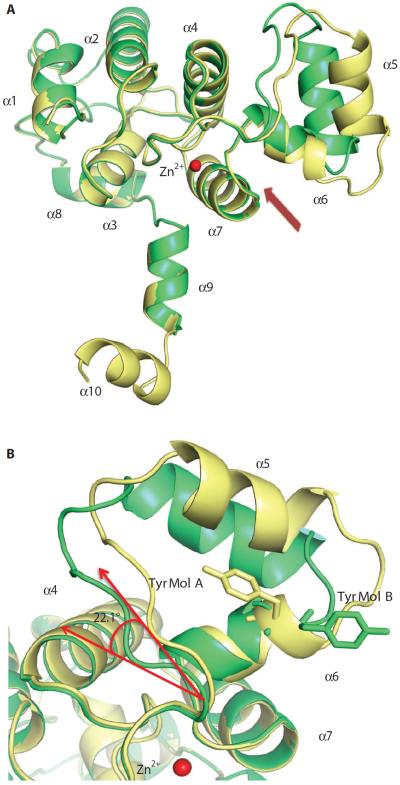
Compared to the K. lactis homolog, the region adjacent to the residues that we identified as important for phosphoryl transfer activity was ordered in the S. cerevisiae Rtr11–178 structure (Fig. 4, A and B). Both K. lactis and S. cerevisiae Rtr1 are α-helical proteins, but in K. lactis Rtr1, 65% of the amino acids adopt a random coil configuration, whereas only 35% of the amino acids are present in flexible coils in S. cerevisiae Rtr1. The electron density of S. cerevisiae Rtr1 is continuous from residues 1 to 176, with only the last two amino acids disordered. The N-terminal domain of Rtr1 (residues 1 to 153 in S. cerevisiae) is formed by a four-helical bundle coordinating a zinc ion with a pair of short helices (α5 and α6) inserted between the third (α4) and fourth helix (α7) coordinating zinc (Fig. 4B). This nonconsensus zinc finger motif (C-X4-C-X27-C-X3-H/C) is conserved throughout the Rtr1 homologs (Fig. 3A). In the S. cerevisiae Rtr11–178 structure, a zinc atom displaying a tetrahedral geometry is well coordinated by three conserved cysteine residues (Cys73, Cys78, and Cys112) and one histidine residue (His116) (Fig. 4C). We confirmed the identity of the zinc ion through fluorescence scans of Rtr1 crystals at APS, in which the signal represented the anomalous signal absorption edges of zinc (fig. S5). Single mutations at any of the zinc-coordinating cysteines compromised protein expression in E. coli, suggesting that zinc coordination is essential for protein folding or stability (10).
Fig. 4. Crystal structure of S. cerevisiae Rtr11–178.
(A) Schematic diagram of the secondary structure of the phosphoryl transfer domain of S. cerevisiae Rtr1 shown above the primary sequence of S. cerevisiae Rtr1 (Rtr1_Scr) and K. lactis Rtr1 (Rtr1_Klc). (B) Superimposition of the K. lactis (PDB ID: 4FC8 in violet and PDB ID: 4M3O in magenta) and the S. cerevisiae (green) Rtr1 structures. (C) Superimposition of the conserved zinc finger motif in S. cerevisiae (green) and K. lactis (light violet) Rtr1. Residues coordinating the zinc are shown as sticks and numbered using the sequence of S. cerevisiae Rtr1.
The structures of the regions after the zinc-binding helices show great differences between the two yeast species (Fig. 4B). Little secondary structure was observed in K. lactis Rtr1 with residues 126 to 152 modeled as an extended loop. S. cerevisiae Rtr1 has three short helices in this region (α8 to α10) (Fig. 4B). These secondary elements extend in a different orientation from the core zinc finger motif, suggesting a separate domain at the C terminus (fig. S6). This separate domain may be the autoinhibitory CTD reported for these phosphatases (10), which is not present in the truncated Rtr1 used for crystallization. Consistent with this interpretation, the Rtr11–213 exhibited greater activity than full-length Rtr1 in the DiFMUP assay (Fig. 1A), suggesting that loss of the autoinhibitory domain without loss of other sections of the protein (such as those not present in the Rtr11–178 protein) may enhance phosphatase activity.
Identification of the substrate-binding pocket
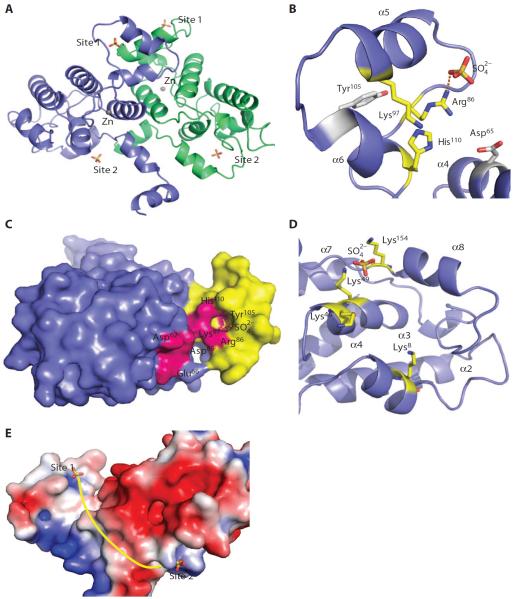
The Rtr11–178 structure contains two tetrahedral-shaped electron density elements with intensities that are too strong to be modeled as water molecules (Fig. 5A). These could represent putative phosphate-binding sites. Because we used a high concentration of lithium sulfate as the precipitant in the crystallization condition, we built a sulfate ion into each electron density, which fit well (Fig. 5A). Binding of the sulfate group suggested that the local environment of the protein has a high propensity to bind a negatively charged chemical group with tetrahedral geometry, consistent with a phosphate group in the physiological environment. These results are also consistent with the mass spectrometry data in which as many as two phosphate molecules can be associated with Rtr1 (fig. S2).
Fig. 5. Identification of putative phosphate-binding sites in the structure of S. cerevisiae Rtr11–178.
(A) The asymmetric unit contains two Rtr1 molecules. Sulfate-binding sites are indicated as sticks with the sulfate atom colored yellow. Site 1 and site 2 are labeled in each molecule per asymmetric unit. (B) The site 1 sulfate-binding site represents the first putative phosphate-binding site. The positively charged triad (Arg86, Lys97, and His110) is shown as sticks with the carbon atoms colored yellow. The structure contains a sulfate group close to the triad, ~3 Å from the positively charged side chains (dotted line represents a potential hydrogen bond). Two residues important for catalytic activity, Asp65 and Tyr105, are shown as sticks with the carbon atoms colored gray. (C) Surface representation of S. cerevisiae Rtr11–178. The zinc finger motif domain is shown in blue, and the inserted helical pair is colored yellow. The residues important for the phosphatase activity are colored hot pink. A sulfate ion from the crystallization buffer is found close to Tyr105. (D) The site 2 sulfate-binding site is modeled with a sulfate ion into an electron dense region, which is present in a shallow pocket surrounded by positively charged residues (Lys8, Lys46, Lys49, and Lys154) on several different helices. (E) An electrostatic surface representation of S. cerevisiae Rtr11–178 showing the position of the site 1 and site 2 putative phosphate-binding sites, based on the positions of the bound sulfate ions. The sulfate groups are shown as sticks with a yellow dotted line indicating the grove along the protein that could potentially accommodate the CTD of the RNAPII.
The asymmetric unit contains two Rtr1 molecules, each with two sulfate groups bound. These sulfate molecules cluster around two surface regions of the enzyme (Fig. 5A). The first region (site 1) locates close to the groove formed between the core of the zinc-binding helices and the insertion helical pair (α5 and α6), near the residues important for catalytic activity during the DiFMUP assay. At the elbow of the helix-turn-helix of the inserted helical pair, a sulfate group is adjacent to a small deep pocket next to the groove between the insertional helical pair and the zinc finger motif (Fig. 5B and fig. S7). All of the residues near this putative phosphate-binding pocket are conserved. Three residues (Arg86, Lys97, and His110) form a positive triad at the entrance of the pocket and associate with the negatively charged sulfate (Fig. 5B). The positive properties of these residues are well conserved among species (Fig. 3A), and mutation of any of these residues to Ala compromised phosphoryl transfer activity of Rtr11–213 in the DiFMUP assay (Fig. 3B). For example, 15 of the 17 homologs examined have Arg or Lys corresponding to the position at Arg86. For Lys97, 14 of the 17 homologs have positively charged residues in the same approximate position. Although S. cerevisiae Rtr1 was the only one with His at position 110, in all but four homologs examined, the same residue was replaced by Lys. Single mutation to alanine at any of these residues compromised activity (loss of 75% activity with Arg86Ala, 72% with Lys97Ala, and 86% with His110Ala) (Fig. 3B), and combination of the three mutations of this triad reduced activity to less than 10% that of Rtr11–213 (Fig. 3B). These results suggested that the positive charge of this pocket is important for Rtr1 function. At the bottom of this deep pocket is the conserved Tyr105, which was conserved in all species except Caenorhabditis elegans (Fig. 3A). Tyr105 mutation to Ala reduced the phosphoryl transfer activity to less than 5%, and the conservative mutation to Phe also resulted in low activity (Fig. 3B). Both mutated proteins were produced in E. coli (fig. S4) and were stable and folded as evaluated by gel filtration chromatography (table S1). The loss of catalytic activity by the mutants indicated that Tyr105, in particular its hydroxyl group, is important for the phosphatase activity of Rtr1. Facing the exit of this putative phosphate-binding pocket is Glu66, which is conserved in all homologs that we examined (Fig. 3A) and is essential for phosphoryl transfer in the DiFMUP assay (Fig. 3B). Mutational analysis also showed that Asp62 and Asp65, additional conserved residues near this putative phosphate-binding pocket, were important for activity (Fig. 3B). Overall, this region is conserved and contains residues that can disrupt phosphoryl transfer upon mutation (Fig. 5C), suggesting that this sulfate-bound pocket is the catalytic active site for the phosphoryl transfer reaction of Rtr1.
At site 2, the sulfate group is present in a shallow pocket with several positively charged residues (Lys8, Lys46, Lys49, and Lys154) (Fig. 5D). However, the residues of this pocket are not well conserved. The affinity of this pocket for negatively charged groups may enhance the interaction with the CTD by functioning as a secondary binding site for additional phosphate groups in the CTD. Indeed, Rtr1 has weak activity against phosphorylated Ser5 in the heptad repeat of the CTD peptide in vitro but seems to function more effectively when phosphorylation occurs at multiple sites on the CTD substrate (9, 10). Additional phosphorylation at the Ser7 or Tyr1 residue in the heptad repeat promotes the dephosphorylation of Ser5 in the heptad repeat by Rtr1 (9, 10). In our structure, an elongated groove extends along the protein surface connecting the two sulfate-bound regions (~40 Å long). This groove could potentially accommodate one and a half CTD heptad repeats with multiple phosphorylated residues (Fig. 5E), a possibility that awaits the complex structure of Rtr1 with phosphorylated CTD peptides.
The coexistence of two conformations of the insertion domain
Intriguingly, the orientations of the inserted helical pair (α5 and α6) relative to the core domain are different for the two molecules in the same asymmetric unit of S. cerevisiae Rtr1 (Fig. 6A). A hinge-like motion between the helical pair and zinc finger core could result in a rotation of as much as 22.1° (Fig. 6B), allowing the groove between the two domains to undergo movement between an opened or closed conformation. Because this groove is the putative substrate-binding site, this movement would alter the accessibility of the active site to substrate. Noticeably, molecule B in S. cerevisiae Rtr1 is less well ordered compared to molecule A with a partial loss of secondary structure, resulting in a shorter helix 6. The side chain of Tyr105, which is now part of a longer loop connecting helices 5 and 6, adapts a different orientation from molecule A and sways outward away from the putative substrate-binding groove (Fig. 5B). The flexibility of this helical pair is also consistent with the structure of K. lactis Rtr1, which is mostly disordered in this region (PDB ID: 4FC8 and 4M3O). These data suggested that the active site of Rtr1 is less well formed than that of other phosphatases in the absence of substrate, other binding proteins, or both.
Fig. 6. The flexibility of the inserted helical pair as seen in two monomers of Rtr11–178.
(A) Superimposition of the two molecules in each asymmetric unit of the S. cerevisiae Rtr1 structure (yellow and green). The arrow points to the putative binding groove for the substrate between the zinc finger motif and the α-helical pairs. (B) Tyr105 residues in the two monomers of asymmetric unit have different conformations (yellow, molecule A; green, molecule B). The angle of movement of the α4 helix from the zinc finger core is indicated.
Phosphoryl transfer activity of Rtr1 in S. cerevisiae growth
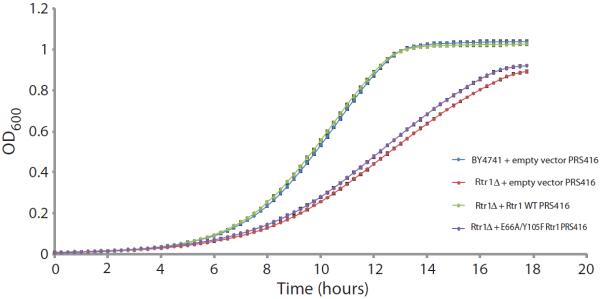
To explore if the phosphoryl transfer activity involving the putative phosphate-binding site that we identified for Rtr1 is required for the biological function of Rtr1, we assessed the ability of wild-type full-length Rtr1 or the double mutant E66A/Y105F to restore growth of yeast deficient for Rtr1. Whereas the Rtr1-deficient strain transformed with an empty vector grew at a lower rate than wild-type yeast, consistent with previous reports (22), transformation with wild-type Rtr1 rescued the knockout yeast strain and provided for normal growth (Fig. 7). Transformation with Rtr1 E66A/Y105F failed to rescue growth of yeast lacking Rtr1 (Fig. 7), indicating that the phosphoryl transfer activity dependent on site 1 is biologically important.
Fig. 7. The putative active site is important for the function of Rtr1 in vivo.
Growth curves of yeast strains BY4741 + empty PRS416 vector, rtr1Δ + empty pRS416 vector, rtr1Δ + Rtr1 WT in pRS416, and rtr1Δ+Rtr1 E66A/Y105F in pRS416. All strains were grown in uracil-deficient media. The y axis indicates the optical density at 600 nm (OD600 nm) of the 200-μl volume culture in a 96-well plate. SE is shown from three independent biological experiments. Data are representative of one of the duplicate experiments.
Phosphoryl transfer activity of RPAP2
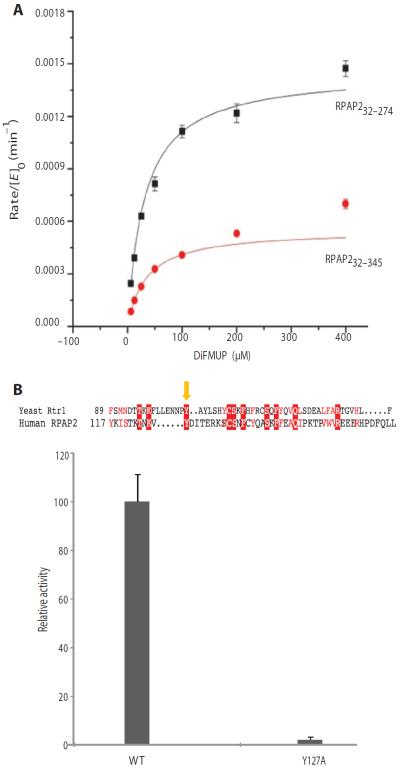
Rtr1 is conserved from yeast to human. Human RPAP2 is 612 amino acids. To test if a similar reaction mechanism is responsible for the activity of human Rtr1 homolog RPAP2, we created RPAP232–274 and RPAP232–345 based on secondary structure prediction, expressed the proteins in E. coli, and purified them (table S2). We removed the first 32 amino acids to increase the amount of soluble protein in the bacterial expression system. Kinetic characterization of the two variants in the DiFMUP assay showed that both have similar Km for this substrate, but that the turnover of RPAP232–274 was 2.5 times higher than that of RPAP232–345 (Fig. 8A). This result is consistent with our studies of S. cerevisiae and K. lactis Rtr1 that the C-terminal region of the Rtr1 homologs negatively regulates phosphatase activity (10).
Fig. 8. Phosphoryl transfer activity of human RPAP2 in the DiFMUP assay.
(A) Kinetic parameters of phosphatase activity of RPAP232–274 and RPAP232–345. (B) Tyr105 function is conserved for phosphoryl transfer reaction of Rtr1 and RPAP2. The insert shows the sequence alignment of yeast (S. cerevisiae) Rtr1 and RPAP2, with the yellow arrow indicating the conserved tyrosine at the 105 position of Rrt1 and 127 in RPAP2. Effect of Y127A mutation on the phosphatase activity of RPAP2 in DiFMUP assay is shown at the bottom. The error bars represent SD from triplicate samples, and data are representative of three independent experiments.
To identify if the putative phosphate-binding site that involves Tyr105 in S. cerevisiae Rtr1 is conserved in RPAP2, we mutated the corresponding residue Tyr127 in RPAP232–274 and assessed its activity in the DiFMUP assay (Fig. 8B). Although the protein was stable and had a similar profile to the nonmutated protein by gel filtration (table S2), activity of RPAP232–274 Y127A was undetectable (Fig. 8B), indicating that a similar reaction mechanism is utilized from yeast to human in the Rtr1 family.
DISCUSSION
Our in vitro kinetic study of S. cerevisiae Rtr1 in the DiFMUP assay established that the first 178 residues of the protein are sufficient to perform the phosphoryl transfer reaction. We compared the crystal structure of Rtr11–178 to structures of representatives of the three known protein phosphatase families: PTP1b (protein tyrosine phosphatase 1b) [PDB ID: 2CM7 (23)] of the cysteine-based phosphatase family, Scp1 [PDB ID: 3L0C (24)] of the HAD phosphatase family, and PP2A [PDB ID: 2NYM (25)] of the metal-dependent phosphatase family (fig. S8). Rtr1 shows no structural resemblance to any of the three phosphatase families. Protein structure data-base search using the Dali server (26) with the phosphoryl transfer domain of Rtr1 identified no other proteins with structural similarity.
We also considered the possibility that the active-site geometry of this phosphoryl transfer domain may resemble other phosphatases even though the overall fold is different. Rtr1 does not have the dimetal active site like PP1 and PP2 families. Cysteine residues, used as the nucleophile in most cysteine-based phosphatases, are coordinating zinc in Rtr1 and, therefore, cannot mediate a nucleophilic phosphoryl transfer reaction. The catalytic importance of Asp62, Asp65, and Glu66 in Rtr1 motivated us to evaluate if this DXXDE motif resembled the DXDE motif found in HAD phosphatase family (27). However, the spatial arrangement of the residues in Rtr1 does not seem to support the phosphoryl transfer mechanism used by HAD phosphatases (fig. S9). Furthermore, we determined that magnesium ion, which is essential for HAD phosphatase catalytic activity, had no effect on Rtr1 function and was not associated with the protein. With the exception of His116 that coordinates a zinc ion, histidine residues, which can function as nucleophiles, are not well conserved in Rtr1.
The in vitro analysis of the mutated Rtr1 proteins showed that the conserved residue Tyr105 was essential for the catalytic function for Rtr1, as well as the activity of the human homolog RPAP2. The double mutant Rtr1 E66A/Y105F also compromised growth in yeast, consistent with the importance of this residue in vivo. Enzymes such as type I DNA topoisomerase relieve the supercoil of DNA utilizing the formation of a phosphoryltyrosine intermediate (28). The conserved tyrosine residue at the active site replaces one of the phosphodiester bonds in the DNA backbone and forms a bond between the enzyme and the cut DNA, thereby enabling the rotation of DNA strand and subsequent reannealing of the DNA with no energy cost (28). In the Rtr1 structure, Tyr105 is present at the bottom of the putative phosphate-binding pocket, suggesting a catalytic mechanism in which tyrosine functions as a nucleophile to target the phosphorus to mediate the phosphoryl transfer. However, we did not detect the existence of this phosphoryl-tyrosine intermediate species by crystallography or mass spectrometry. The presence of a metal ion in Rtr1 may indicate a metal-dependent mechanism for phosphoryl transfer. The reaction mechanism of this atypical phosphoryl transfer domain warrants more study.
The intrinsic phosphatase activity of recombinant Rtr1 is weak when tested in vitro, showing a low activity against general phosphatase substrates, as well as physiological substrate CTD of RNAPII (9). The Rtr11–178 structure provides an explanation for this. The inserted helical pair (α5 and α6) that forms the wall of the active site is flexible and adopts two different conformations. The flexibility is even higher in the structure of K. lactis Rtr1, in which this region is disordered (10, 14). Therefore, recombinant purified Rtr1 has a poorly formed substrate-binding groove. In contrast, invivo Rtr1 is part of the RNAPII complex and interacts with binding partners, which may facilitate the folding and formation of the active site. Indeed, two human CTD-interacting proteins, RPRD1A and RPRD1B, form a heterodimer and “clamp” onto RPAP2 (15). Both of these CTD-interacting proteins exhibit strong binding with CTD, and their interaction with RPAP2 targets the phosphatase to CTD. Such a CTD-interacting protein(s) has yet to be characterized in yeast, but it is possible that a similar mechanism occurs across species. Furthermore, other CTD phosphatases, such as Ssu72, function in multiple phases of transcription (29). Overall, our S. cerevisiae Rtr1 structure provides a putative active site with a substrate-binding pocket for this family of enzymes and an explanation for the poor catalytic activity of the purified protein in vitro.
MATERIALS AND METHODS
Materials
DiFMU (6,8-difluoro-7-hydroxy-4-methylcoumarin), which is used as the reference standard, and DiFMUP were purchased from Life Technologies. RPAP2 open reading frame was purchased from Open Biosystems (GE Life Sciences). All other chemicals and reagents were purchased from Sigma-Aldrich unless specified.
Cloning, protein expression, and purification
Yeast genomic DNAwas used as template to amplify full-length Rtr11–226 and was cloned into a modified pET28 vector with a C protease cleavage site between restriction sites NdeI and XhoI, resulting in a His-tagged construct containing the non-native residues MGSSHHHHHHHHLEVLFQGPH as an affinity tag. This full-length construct was used as template to generate other Rtr1 truncated constructs. Cloned constructs were verified using DNA sequencing.
Plasmids were transformed into E. coli BL21 (DE3) cells and selected by kanamycin (50 mg/ml) resistance. Colonies were tested for protein expression in small scale in 1-ml cultures, and high-expressing colonies were inoculated into 30 ml of Luria-Bertani (LB) medium and grown at 37°C with antibiotics overnight. Overnight culture was inoculated into 1 liter of LB medium at 37°C until OD600nm reached 0.6 and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 22°C overnight. Cells were harvested and lysed by sonication in lysis buffer [20 mM tris (pH 8), 500 mM NaCl, 25 mM imidazole]. Lysate was clarified by centrifugation (22,000 rpm for 1 hour in Sorvall RC-5B centrifuge). The filtered lysate was loaded onto lysis buffer–equilibrated Ni-NTA beads, washed extensively, and eluted in lysis buffer containing 500 mM imidazole. Protein molecular weight and purity were confirmed using SDS-PAGE. Protein was pooled and incubated with 3C protease (1 U/mg of protein) (GE Healthcare) in dialysis buffer (50 mM tris, 150 mM NaCl, and 2 mM β-mercaptoethanol) for 16 hours at 4°C to ensure removal of the His affinity tag. After confirming cleavage by SDS-PAGE, excess His tag and un-cleaved protein were removed by incubating the sample with Ni-NTA beads and eluting (QIAGEN). The pooled protein was dialyzed against 20 mM tris (pH 8.0) overnight and subjected to ion-exchange chromatography using the Mono Q column (Bio-Rad). Protein was eluted from ion exchange using a gradient of NaCl concentrations ranging from 0 to 1 M. Fractions containing Rtr1 were verified using SDS-PAGE, pooled, and concentrated. The protein was then loaded onto a Superdex 75 size exclusion column (GE Healthcare Life Sciences) preequilibrated with 20 mM tris and 150 mM NaCl (pH 8.0). Peak fractions were analyzed on SDS-PAGE and concentrated to ~10 mg/ml.
Various His-tagged RPAP2 constructs were generated on the basis of secondary structure analysis and tested for expression and solubility. Constructs yielding soluble protein were used in kinetic analysis. The amount of protein produced in E. coli was similar to that of Rtr1, and the recombinant proteins were purified using the identical protocol used to purify Rtr1.
Rtr1 and RPAP2 mutants were made by site-directed mutagenesis and confirmed by sequencing. These constructs were purified using an identical protocol to wild-type Rtr1 protein.
DiFMUP phosphatase assay
The phosphatase activities of Rtr1 and RPAP2, as well as their mutation variants, were characterized by detecting the hydrolysis of DiFMUP. DiFMUP generates a fluorescent signal once the associated phosphate is removed. Florescent DiFMU, the hydrolysis product of DiFMUP, was monitored at excitation/emission wavelengths of 358/455 nm using a TECAN plate reader (Infinite 200). A standard curve was generated using various concentrations of DiFMU. Both DiFMU and DiFMUP were dissolved in 100% DMSO to make 10 mM stock. For each experiment, fresh substrate was prepared by diluting compounds in buffer containing 20 mM tris-acetate (pH 6.0), 150 mM NaCl, and BSA (0.1 mg/ml) to desired concentrations. Background activity in the presence of BSA alone was subtracted from all values. Assays were carried out in 384-well amber plates in a 20-μl reaction volume. Michaelis-Menten kinetic parameters were determined by measuring initial reaction rates at various DiFMUP concentrations in assay buffer with 7 μM Rtr1 protein. Initial rates were calculated using the Magellan 6 software (TECAN). The datawere fit to the Michaelis-Menten equation using nonlinear curve fit in KaleidaGraph (Synergy), and kcat and Km were determined.
Phosphatase inhibition assay
To study the effect of inhibitors, 7 μM S. cerevisiae Rtr1 protein was incubated with buffer or inhibitors in various concentrations before adding 200 μM substrate for 10 min. Five different nonselective phosphatase inhibitors were tested: Na3VO4, AlF3, BeF3−, NaF, and AlCl3. Activity in the presence of the inhibitors with respect to the control is represented as relative activity in bar graph. BeF3− ion is prepared by mixing BeCl2 and NaF at a ratio of 1:3. All experiments were carried out in triplicate in a single experiment and repeated thrice.
Protein crystallization, data collection, and structure determination
Rtr1 variants (1 to 178, 1 to 213, and 1 to 226 amino acids) at ~10 mg/ml were screened using the Phoenix robotic protein crystallization system (Art Robbins Instruments). Conditions were tested at both 4°C and room temperature and at various protein–to–reservoir solution ratios (1:1, 1:2, and 2:1). After optimization, needle-shaped crystals grew at room temperature for the Rtr1(1 to 178) construct in a condition containing 0.1 M Hepes sodium (pH 7.5) and 1.5 M lithium sulfate monohydrate in sitting drop vapor diffusion. Individual crystals were flash-frozen directly in liquid nitrogen after brief incubation with a cryoprotectant-containing reservoir solution supplemented with 25% (v/v) glycerol. Diffraction data were collected at GM-CAT BM-23 microfocus beamline in APS (Chicago, IL), which was optimal for small crystals. Diffraction was processed to 2.6 Å resolution using HKL2000 (30). The data collection statistics are provided in Table 2.
Phases were obtained by molecular replacement using the CCP4 (Collaborative Computational Project No. 4) program Phaser (31) with K. lactis Rtr1 (PDB ID: 4FC8) (14) as the initial search model. Extensive manual building in Coot (32) coupled with iterate cycle of refinement using autoBUSTER (33) finally completed the model. The final rounds of the refinement applied automatic LSSR NCS (local structure similarity restraints noncrystallographic symmetry) restraints and translation, liberation, and screw motion in autoBUSTER. The final refined model comprises residues 1 to 176 for monomer A and 1 to 166 for monomer B and resulted in a final crystallographic Rwork of 0.187 and Rfree of 0.245 (Table 2). In the Ramachandran plot, 98.25% of residues fell in the most favored regions with no outliers. The final crystallographic refinement statistics are shown in Table 2.
Crystallization effort to obtain Rtr1 in complex with phosphoryl-mimicking inhibitors was carried out by cocrystallization as well as soaking. Soaking experiments caused Rtr1 crystals to crack and lose diffraction. Cocrystallization with 10-fold excess inhibitors (aluminum fluoride, sodium vanadate, or sodium tungstate) failed to produce crystals.
Metal and EDTA assay
To test the effect of EDTA on protein activity, an additional EDTA incubation step was conducted. An identical procedure was utilized for K. lactis Rtr1 (10). Briefly, we added an additional dialysis step against 20 mM tris-HCl (pH 8), 150 mM NaCl, and 10 mM EDTA after protein eluted from nickel affinity column. Protein was purified further using gel filtration chromatography. To test the effect of other metal ions on Rtr1 activity, this EDTA-treated protein was supplemented with various concentrations of divalent metals during DiFMUP phosphatase assay. The effect of various metals on Rtr1 activity was tested under the same conditions as those used for the inhibitor experiments.
Alternatively, the incorporation of metal ions was also accomplished through supplementation of the growth media. Divalent metal ions were added to LB at concentrations of 1 mM at induction to promote their incorporation. The purified protein was then tested using DiFMUP phosphatase assay. Both methods produced similar results; all data shown used the method in which protein was treated with EDTA and then reconstituted with various metal ions.
Yeast growth assay
The S. cerevisiae Rtr1 gene (−266 to +681) was amplified out from BY4741 yeast genomic DNA and cloned into a pRS416 vector between Sac I and Bam HI restriction sites. The E66A/Y105F double mutant was prepared by site-directed mutagenesis of this cloned plasmid. The yeast strains BY4741 and Rtr1 knockout (rtr1Δ) were transformed with empty vector pRS416, vector containing Rtr1 insert, and vector with E66A/Y105F double mutant Rtr1 insert. Transformants were selected on plates lacking uracil. Yeast growth was analyzed in 200-μl cultures at 37°C in 96-well plates with shaking. Cultures were inoculated to an OD600nm of 0.05 using a saturated overnight culture. Measurements were made every 15 min over 18 hours using a TECAN plate reader (Infinite 200).
Supplementary Material
Acknowledgments
Crystallographic data collections were conducted at APS (BL23-ID-B), U.S. Department of Energy national user facility.
Funding: This work is supported in part by grants from NIH (R01 GM104896 to Y.Z.) and The Welch Foundation (F-1778 to Y.Z.).
Footnotes
SUPPLEMENTARY MATERIALS www.sciencesignaling.org/cgi/content/full/9/417/ra24/dc1
Author contributions: S.I. and S.D.Y. determined the crystal structure. S.I. and S.D.Y. conducted the kinetic analysis. S.I. conducted the yeast experiments. M.Z. designed the kinetic assay. Y.Z. constructed the truncation library of Rtr1. W.L.M. and G.N. made wild-type and mutation variants of Rtr1. N.A.P. conducted the pNPP assay. Y.J.Z. designed the experiments with the intellectual input from S.I., M.Z., and J.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The structures have been deposited in PDB with accession code 5C2Y.
REFERENCES AND NOTES
- 1.Corden JL. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem. Rev. 2013;113:8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 3.Sims RJ, III, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, III, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, Schnölzer M, Cole PA, Geyer M, Bruneau BG, Adelman K, Ott M. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol. Cell. 2013;52:314–324. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausmann S, Shuman S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 2002;277:21213–21220. doi: 10.1074/jbc.M202056200. [DOI] [PubMed] [Google Scholar]
- 8.Yeo M, Lee S-K, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 9.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell. 2009;34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu PL, Yang F, Smith-Kinnaman W, Yang W, Song J-E, Mosley AL, Varani G. Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J. Mol. Biol. 2014;426:2970–2981. doi: 10.1016/j.jmb.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M, Suh H, Cho E-J, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell. 2012;45:111–122. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang K, Manley JL, Tong L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat. Commun. 2012;3:946. doi: 10.1038/ncomms1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo W-HW, Li J, Young P, Olsen JB, Wan C, Loppnau P, Bakkouri M. El, Senisterra GA, He H, Huang H, Sidhu SS, Emili A, Murphy S, Mosley AL, Arrowsmith CH, Min J, Greenblatt JF. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat. Struct. Mol. Biol. 2014;21:686–695. doi: 10.1038/nsmb.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sotoud H, Gribbon P, Ellinger B, Reinshagen J, Boknik P, Kattner L, El-Armouche A, Eschenhagen T. Development of a colorimetric and a fluorescence phosphatase-inhibitor assay suitable for drug discovery approaches. J. Biomol. Screen. 2013;18:899–909. doi: 10.1177/1087057113486000. [DOI] [PubMed] [Google Scholar]
- 17.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Yogesha SD, Mayfield JE, Gill GN, Zhang Y. Viewing serine/threonine protein phosphatases through the eyes of drug designers. FEBS J. 2013;280:4739–4760. doi: 10.1111/febs.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fagerholm AE, Habrant D, Koskinen AM. Calyculins and related marine natural products as serine-threonine protein phosphatase PP1 and PP2A inhibitors and total syntheses of calyculin A, B, and C. Mar. Drugs. 2010;8:122–172. doi: 10.3390/md80100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibney PA, Fries T, Bailer SM, Morano KA. Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot. Cell. 2008;7:938–948. doi: 10.1128/EC.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ala PJ, Gonneville L, Hillman MC, Becker-Pasha M, Wei M, Reid BG, Klabe R, Yue EW, Wayland B, Douty B, Polam P, Wasserman Z, Bower M, Combs AP, Burn TC, Hollis GF, Wynn R. Structural basis for inhibition of protein-tyrosine phosphatase 1B by isothiazolidinone heterocyclic phosphonate mimetics. J. Biol. Chem. 2006;281:32784–32795. doi: 10.1074/jbc.M606873200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Liu J, Kim Y, Dixon JE, Pfaff SL, Gill GN, Noel JP, Zhang Y. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 2010;19:974–986. doi: 10.1002/pro.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen KN, Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr. Opin. Struct. Biol. 2009;19:658–665. doi: 10.1016/j.sbi.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 29.Rosado-Lugo JD, Hampsey M. The Ssu72 phosphatase mediates the RNA polymerase II initiation-elongation transition. J. Biol. Chem. 2014;289:33916–33926. doi: 10.1074/jbc.M114.608695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.