Abstract
Arylamines (AA) and heterocyclic aromatic amines (HAA) are structurally related carcinogens formed during combustion of tobacco or cooking of meat. They undergo cytochrome P450 mediated N-hydroxylation to form metabolites which bind to DNA and lead to mutations. The N-hydroxylated metabolites of many AA also can undergo a co-oxidation reaction with oxy-hemolgobin (HbO2) to form methemoglobin (met-Hb) and the arylnitroso intermediates, which react with the β-Cys93 chain of Hb to form Hb-arylsulfinamide adducts. The biochemistry of arylamine metabolism has been exploited to biomonitor certain AAs through their Hb arylsulfinamide adducts in humans. We examined the reactivity of HbO2 with the N-hydroxylated metabolites of 4-aminobiphenyl (ABP, HONH-ABP), aniline (ANL, HONH-ANL), and the HAA 2-amino-9H-pyrido[2,3-b]indole (AαC, HONH-AαC), 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP, HONH-PhIP) and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx, HONH-MeIQx). HONH-ABP, HO-ANL, and HONH-AαC induced methemoglobinemia and formed Hb sulfinamide adducts. However, HONH-MeIQx and HONH-PhIP did not react with the oxy-heme complex, and met-Hb formation and chemical modification of β-Cys93 residue was negligible. Molecular modeling studies showed that the distances between the H-ON-AA or H-ON-HAA substrates and the oxy-heme complex of HbO2 were too far away to induce methemoglobinemia. Different conformational changes in flexible helical and loop regions around the heme pocket induced by the H-ON-AA or H-ON-HAAs may explain the different proclivities of these chemicals to induce methemoglobinemia. Hb-Cys93β sulfinamide and sulfonamide adducts of ABP, ANL, and AαC were identified, by Orbitrap MS, following proteolysis of Hb with trypsin, Glu-C, or Lys-C. Hb sulfinamide and sulfonamide adducts of ABP were identified in blood of mice exposed to ABP, by Orbitrap MS. This is the first report of the identification of intact Hb sulfinamide adducts of carcinogenic AAs in vivo. The high reactivity of HONH-AαC with HbO2 suggests that the Hb sulfinamide adduct of AαC may be a promising biomarker of exposure to this HAA in humans.
Keywords: Tobacco smoke, cooked meat, carcinogens, aromatic amines, heterocyclic aromatic amines, methemoglobin, hemoglobin adducts
Graphical Abstract
INTRODUCTION
Aromatic amines (AA) and heterocyclic aromatic amines (HAA) are structurally related classes of environmental carcinogens.1,2 Occupational exposures to AAs can occur in the rubber, textile, leather, dye, and cosmetic industries.3–5 4-Aminobiphenyl (ABP), 2-naphthylamine (2-NA), and o-toluidine are also present in tobacco smoke and contribute to the pathogenesis of bladder cancer in smokers.6 Several HAAs, 2-amino-9H-pyrido[2,3-b]indole (AαC) in particular, occur as atmospheric pollutants7 and also arise in tobacco smoke.8,9 Other prominent HAAs, including 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) are formed in during the cooking of meat.1,10
AA and HAA undergo metabolic activation by cytochrome P450 (P450) oxidation to form genotoxic N-hydroxylated intermediates.11,12 These metabolites can react with DNA or undergo further biotransformation by conjugating enzymes to form unstable esters.13,14 The esters undergo heterolytic cleavage to form the presumed nitrenium ions,15 which readily form DNA adducts that can lead to mutations.16
DNA adducts of carcinogens have been employed to assess chemical exposures. However, the measurement of DNA adducts in human specimens remains an analytical challenge because many DNA lesions are rapidly repaired, and the trace levels of DNA adducts present in tissues can be difficult to measure, even with current MS instruments.2,17 Reactive metabolites of AAs, HAAs, and other procarcinogens also bind to nucleophilic sites in proteins to form covalent adducts.18,19 Stable carcinogen protein adducts do not undergo repair and are expected to follow the kinetics of the lifetime of the protein. Thus, adduct formation will accumulate during chronic exposure.20,21 Hemoglobin (Hb) and serum albumin are the two major proteins in blood: 1 mL of blood contains about 150 mg Hb, and 30 mg albumin as compared to 0.003–0.008 mg of DNA.21 Therefore, carcinogen adducts of Hb or albumin may be more abundant and facile to detect than DNA adducts in blood samples. Both Hb and albumin have served as sources of protein to biomonitor the exposure to a number of genotoxicants and reactive electrophiles.19,22–25
Hemoglobin is an α2β2 tetramer protein.26 The N-terminal valine of the β-chain of Hb has served as a nucleophile to trap alkylating agents.21 Hb contains six reduced Cys residues. The two β-Cys93 residues are surface-exposed to solvent and capable of adducting to electrophiles,26,27 whereas the β-Cys112 and α-Cys104 reside in the interior portion of Hb and react more slowly with electrophiles.26,28,29 The Cys34 residue of human albumin is also accessible to the solvent and a strong nucleophile, which reacts with many electrophiles of diverse structure.25 Adducts formed at these sites of Hb and albumin have been employed for biomonitoring exposures to hazardous chemicals.19,22,23,25
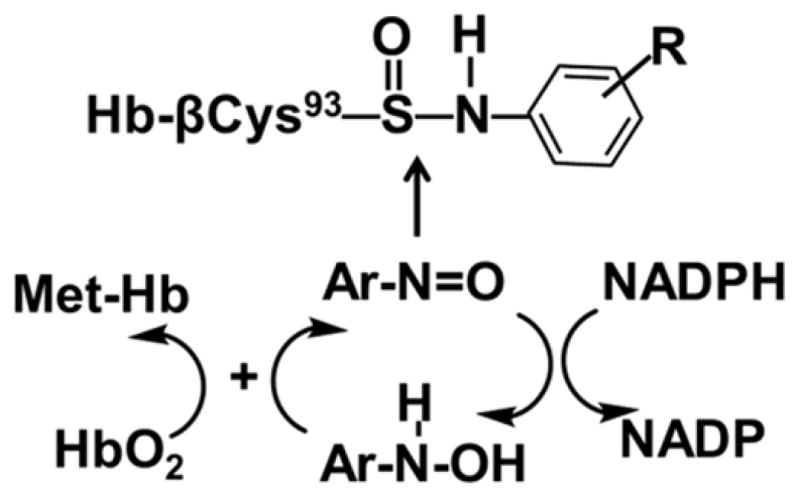
The biochemistry of arylamine-induced toxicity and methemoglobinemia is well documented.30,31 The arylhydroxylamine metabolites, largely produced by cytochrome P450s,2,32,33 can penetrate the erythrocyte and undergo a co-oxidation reaction with HbO2 to form met-Hb and the arylnitroso intermediates.31,34 The arylnitroso compounds undergo enzymatic redox cycling by NADPH reductase within the erythrocyte to reform the aryhydroxylamine and commence another round of co-oxidation with HbO2, leading to methemoglobinemia (Scheme 1). Arylnitroso intermediates are electrophiles, some of which react selectively with the β-Cys93 residue of Hb chain to form sulfinamide adducts.31,35,36 Hb-arylamine sulfinamide adducts are fairly stable in vivo,20,35 but upon acid or base treatment in vitro, the adducts undergo hydrolysis to yield the parent amine and Hb-Cys93β sulfinic acid.34,37 The released aromatic amine (AA) can be readily measured by mass spectrometry (MS) methods. The Hb sulfinamide adduct of 4-aminobiphenyl (ABP) has been employed extensively as a biomarker to monitor occupational and environmental exposures to ABP and to assess the risk of urinary bladder cancer of smokers.38–41 The elevated levels of Hb adducts of ABP, and other primary arylamines, such as 3-aminobiphenyl, 2-NA, o- and p-toluidine, 2,4-dimethylaninline, and 2-ethylaniline are associated with cigarette smoking.39
Scheme 1.
Co-oxidation of HbO2 by N-hydroxylated aromatic amines and heterocyclic aromatic amines in RBC
PhIP and MeIQx also undergo extensive N-oxidation in humans;2,42-44 however, the reactivity of HONH-PhIP and HONH-MeIQx with Hb is very low, and Hb adducts of these procarcinogens are not promising biomarkers for exposure assessment.45–49 To our knowledge, the reactivity of HONH-AαC with Hb has not been investigated. With our interest to develop biomarkers of HAAs with macromolecules for human risk assessment, we sought to understand the reactivity of N-hydroxylated intermediates of ABP, aniline (ANL), AαC, MeIQx, and PhIP (Figure 1) with HbO2 in red blood cells (RBC), and compare the capacity of these N-hydroxylated species to induce methemoglobinemia and form adducts with Hb. AαC was found to be a more potent inducer of met-Hb formation in RBC than any of the other AAs or HAAs tested. The structures of the Hb adducts were elucidated following proteolysis, by ultraperformance liquid chromatography/tandem mass spectrometry (UPLC/MS2). Sulfinamide and sulfonamide adducts of ABP, ANL, and AαC were identified at the β-Cys93 residue, whereas HONH-MeIQx and HONH-PhIP failed to react with HbO2 and did not form adducts.
Figure 1.
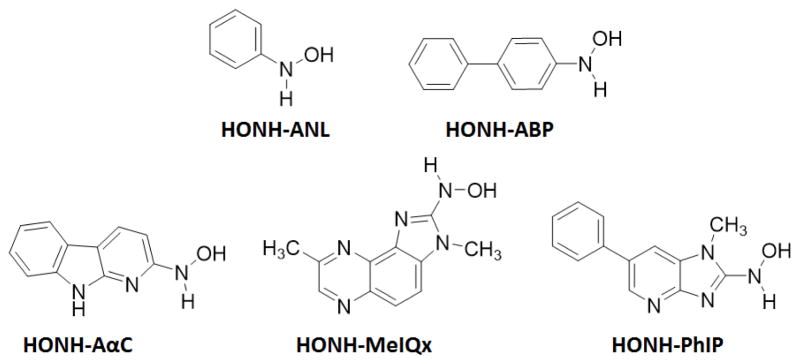
Chemical structures of N-hydroxylated metabolites of ABP, ANL, MeIQx, PhIP, and AαC
MATERIALS AND METHODS
Caution
Many AAs, HAAs and, their derivatives are carcinogens and, must be handled in well ventilated hood with appropriate clothing.
Chemicals and Materials
Phenylhydroxylamine (HONH-ANL), trypsin, iodoacetamide (IAM) and, Pd/C were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). AαC, MeIQx and, PhIP were procured from the Toronto Research Chemicals (Toronto, ON, Canda). [4b,5,6,7,8,8a-13C6]-AαC was a gift of Dr. Daniel Doerge, National Center for Toxicological Research (Jefferson, AR). 4-Nitrobiphenyl was obtained from Apolo Scientific, Stockport, UK. MS grade Pierce™Lys-C endoproteinase was purchased from Thermo Scientific (Grand Island, NY). Glu-C (endoproteinase) was purchased from Worthington Biochemical Corporation (Lackwood, NJ). Amicon Ultra Centrifugal Filters (10 kDa cut off) were purchased from Milllipore (Billerica, MA). LC-MS grade solvents were obtained from Fisher Scientific (Pittsburg, PA). 4-Chloromercuribenzoic acid (PMB) and tetrahdyrofuran (THF) were purchased from Alfa Aeser (Ward Hill, MA). Human whole blood was purchased from Bioreclamation LLC (Hicksville, NY). Isolute C18 solid phase extraction (SPE) columns (25 mg) were obtained from Biotage (Charlotte, NC). All other chemicals and solvents were ACS grade and procured from Sigma-Aldrich.
Synthesis of N-Oxidized Metabolites of AA and HAA
2-Nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO2-PhIP) and 2-nitro-3,8-dimethylimidazo[4,5-f]quinoxaline (NO2-MeIQx) were prepared by diazotization of PhIP and MeIQx.50 2-Nitro-9H-pyrido[2,3-b] indole (NO2-AαC) and [13C6]-NO2-AαC were synthesized by the oxidation of AαC with dimethyldioxirane.51 The synthesis of 2-hydroxyamino-9H-pyrido[2,3-b]indole (HONH-AαC), [13C6]-HONH-AαC, 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP) and 4-hydroxyaminobiphenyl (HONH-ABP) was carried out by reduction of their respective nitro derivatives in THF, using hydrazine with Pd/C as a catalyst.50,51 2-Hydroxyamino-3,8-dimethylimidazo[4,5-f]quinoxaline (HONH-MeIQx) was prepared by reduction of nitro derivative of MeIQx with ascorbic acid.50 The hydroxyamino derivatives of PhIP and MeIQx were enriched by C-18 SPE, whereas the hydroxyamino derivatives of ABP and AαC were concentrated by vacuum centrifugation. All compounds were stored in ethanol except HONH-MeIQx and HONH-ABP, which were stored in DMSO/ethanol (3/1) in liquid nitrogen.
RBC and Hb Preparation
The RBC were prepared from whole blood as described previously.35 Briefly, whole blood from healthy donors was centrifuged (800 × g for 10) at 4 °C to obtain packed RBC. The RBC were washed 3 times with cold Ringer’s buffer. To determine Hb concentration, RBC suspension was lysed by adding distilled water followed by sonication with a Branson 3800 ultra sonicator for 5 min. There after cell lysate was centrifuged at 25,000 × g at 4 °C. The HbO2 in solution was measured at 542 nm using ε (M−1 cm−1) = 57.1.52
Met-Hb Formation and Hb Adduct Formation with N-Hydroxy Derivatives of AAs and HAAs
HONH-AαC, HONH-ABP, HONH-PhIP, HONH-ANL or HONH-MeIQx (0.09 μM) was reacted with RBC containing 10 molar excess HbO2 tetramer (0.9 μM) diluted in 0.5 mL of Ringer’s buffer (pH 7.4). The reaction mixture was incubated at 37 °C for 4 h. At various time points, an aliquot of RBC was lysed and met-Hb formation was estimated at λ631nm using ε (M−1 cm−1) = 15.2.52 Met-Hb formation also was determined by incubating N-hydroxy derivatives of AAs and HAAs (0.09μM) with a 10 molar excess of purified HbO2 (0.9 μM).
More concentrated solutions were employed to examine Hb adduct formation with AAs and HAAs. The RBC suspension contained tetrameric HbO2 (1 nmol) in 0.25 mL of Ringer’s buffer (pH 7.4) and a 3-fold molar excess of HONH-ABP (3 nmol), HONH-ANL (3 nmol), or HONH-AαC (3 nmol). Incubations were conducted at 37 °C for a time period ranging from 15 to 120 min. The RBC were lysed and unreacted metabolites were removed by centrifugation with Amicon Ultra 10k cut-off filters. The Hb was alkylated with IAM (24 nmol) at 37 °C for 1 h. The excess of IAM was removed by Amicon Ultra centrifugal filters. The Hb was resuspended in 0.5 or 1 mL of either 100 mM potassium phosphate buffer (pH 7.4) or 50 mM ammonium bicarbonate containing 1 mM CaCl2 (pH 8.5). In some studies, the modification of Hb was performed at 1/10th and 1/100th fold lower concentrations of HONH-AαC or HONH-ABP in Ringer’s buffer (1 mL).
For mass tag-data dependent mass spectrometry analysis of adducts of AαC, the RBC (1 nmol) was reacted with a 3-fold molar excess of a 1:1 ratio of HONH-AαC (1.5 nmol): HNOH-[13C6] AαC (1.5 nmol) in 100 mM potassium phosphate buffer (pH 7.4, 0.25 mL) at 37 °C for 30 min.
Titration of Free Hb-β-Cys93
Unmodified Hb and Hb modified with N-hydroxylated AAs and HAAs were titrated with p-chloromercuribenzoic acid (PMB) according to Boyer,27 by measuring the increase in absorbance at 250 nm attributed to the mercaptide adduct formed at Cys34.
Optimization of Proteolytic Digestion
The unmodified and modified Hb (5μg) were digested using trypsin or Lys-C at protease:protein ratio of 1:50 (w/w) or 1:25 (w/w) in 50 mM ammonium bicarbonate buffer (pH 8.5) containing CaCl2 (1 mM) in a volume of 1 mL. For Glu-C digestion, Hb (5 μg) was digested with 1:50 (w/w) or 1:25 (w/w) Glu-C in 100 mM potassium phosphate buffer (pH 7.4, 1 mL). All the digestions were carried out at 37 °C for 18 h. After digestion, peptide adducts were desalted and enriched by C-18 SPE as described previously.53 The eluted peptide adducts were concentrated to 100 μL under gentle stream of nitrogen in presence of 50 μL of DMSO.
Animal Study
The guidelines established by the National Institutes of Health Office of Laboratory Animal Welfare were adhered to for the use of animals with housing conditions. All protocols were reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee. Eleven weeks old adult male B6C3F1/J mice (Jackson Laboratory, Bar Harbor, ME) were used in this study. The animals (N = 5) were given intraperitoneal injection of ABP at a dose of 40 mg/kg body weight (100 μL of 80% DMSO : 20% H2O). The blood was collected from the abdominal aorta into vacutainers containing potassium EDTA (Becton Dickinson), 24 h following dosing. The Hb was processed as described.35
UPLC-Mass Spectrometry
A Thermo Dionex Ultimate 3000 Nano/Cap UPLC system (Thermo Scientific, San Jose, CA) with an Atlantis C18 nanoACQUITY column (0.3 × 150 mm, 3μm particle size, 100 Å, Waters Corp., Milford, MA) was used for separation of proteolytic digests of Hb. Solvent A contained 5% CH3CN, 94.99% H2O, and 0.01% HCO2H, and solvent B was comprised of 95% CH3CN, 4.99% H2O, and 0.01% HCO2H. For data dependent acquisition (DDA), a 50 min linear gradient was employed and commenced at 99% solvent A, and increased to 60% solvent B at 40 min, and then increased to 90 % solvent B at 50 min. For targeted measurements, a 20 min linear gradient was used and commenced at 99% solvent A and reached 60% solvent B for 15 min, and then arrived at 90 % solvent B at 20 min. The flow rate was set at 5 μL/min. Mass spectral data were acquired on an Orbitrap Elite™ hydrid ion trap-Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) with a Michrom Advance CaptiveSpray™ source (Auburn, CA). Typical tuning parameters were as follows: capillary tube temperature, 270 °C, spray voltage, 2.0 kV for positive ion mode; S-lens RF level, 67.5%; in-source fragmentation, 5V; normalized collision energy, 35 eV; activation Q, 0.3; for studies with HCD, the collision energy used was 20 eV. The full scan MS spectral data acquisition was carried out at a resolving power of 60,000 (at 400 m/z) over a mass range of 100–1500 m/z whereas, MS/MS data were acquired either with the iontrap or Orbitrap (resolving power 30,000) with an isolation width m/z = 2. All manipulations were conducted with Xcalibur version 3.0.63 software.
Data Analysis of Hb Adducts
The identification of Hb adducts was conducted with the BumberDash platform (Version 1.4.115) with Myrimatch search algorithm (Version 2.1.138) containing 32 protein subset of RefSeq human protein database, version 37.3 with forward and reverse protein sequences.54,55 The dynamic modifications used for AαC-Hb adducts (AαC, +181.0640 on Cys, Ser, Thr, Val, His, Glu; AαC sulfinamide (AαCSO, +197.0589), AαC sulfonamide (AαCSO2, +213.0538) and, Hb oxidation (sulfinic acid, +31.9898 and sulfonic acid, +47.9982) on Cys were previously described.53 Dynamic modifications for ANL-Hb adducts; Aniline (ANL, +91.0422) on Cys, Ser, Thr, Val, His, Glu; Aniline sulfinamide (SOANL, +107.0371) and Aniline sulfonamide (SO2ANL, +123.0320) on Cys whereas, for ABP-Hb adducts; ABP (+167.0735) on Cys, Ser, Thr, Val, His, Glu; ABP sulfinamide (SOABP, 183.0684) and ABP sulfonamide (SO2ABP, 199.2059) on Cys were used in Myrimatch configuration file. For Trypsin, Lys-C digests, two missed cleavages were allowed with semi specificity and, mass tolerances 1.25 m/z (precursor ions) and 0.5 m/z (product ions). In Myrimatch algorithm, Glu-C enzyme was not present in the library; therefore, enzyme specificity was changed to nonspecific. The spectral identity was filtered through IDPicker algorithm with 1% false discovery rate.
Molecular Modeling
The molecular modeling work presented in this paper was carried out using MOE 2013.08 (Chemical Computing Group, Inc., Montreal, Canada.) Protein superposition was done using the “Protein Superpose” application,56 generating pairwise root-mean-square deviation (RMSD) values.57 The molecular docking simulation was conducted using the induced fit protocol within the MOE Docking application.58,59 This protocol allows the active site residue side chains to be included in the final minimization stage of docking. PyMOL (Schrödinger, LLC) was used to color code the secondary structure of human hemoglobin (2DN1.pdb) according to a crystallographic temperature factor (B-factor) spectrum.
RESULTS
N-Oxidized metabolites of AAs undergo co-oxidation with HbO2 to form met-Hb and the arylnitroso intermediate, which can react with the β-Cys93 chain of Hb to form Hb-arylsulfinamide adducts (Scheme 1). The reactivities of HONH-ABP and HONH-ANL with HbO2 and formation of the Hb-arylsulfinamide adducts are well documented.31,37 We compared the reactivity of HbO2 with these arylhydroxylamines and the N-hydroxylated intermediates of PhIP, MeIQx, and AαC, which are three prominent HAAs formed in cooked meat or tobacco smoke.1 Met-Hb formation and titration of available β-Cys93 were examined as a preliminary measure of reactivity of N-hydroxy-HAAs with Hb and adduct formation.
Kinetics of Met-Hb Formation in RBC Treated with N-Hydroxy-AAs or N-Hydroxy-HAAs
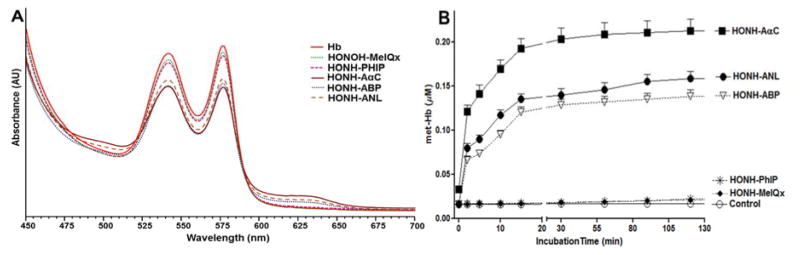
The incubation of RBC with 0.1 mol of HONH-ABP, HONH-ANL or HONH-AαC per mol HbO2 tetramer resulted in a rapid oxidation of HbO2, based on the increase in absorption at λ631nm attributed to met-Hb formation (Figure 2A). In contrast, the reaction of HONH-MeIQx or HONH-PhIP with RBC did not show appreciable change in visible spectrum of HbO2 (Figure 2A). The amount of met-Hb formation induced by HONH-AαC was ~1.4 – 1.6-fold higher than that level induced by HONH-ABP and HONH-ANL, whereas the formation of met-Hb was negligible for HONH-PhIP and HONH-MeIQx (Figure 2B).
Figure 2.
Met-Hb formation in RBC (0.9 μM HbO2 tetramer in Ringer’s phosphate buffer, pH 7.4) incubated with N-hydroxy metabolites of AαC, ABP, ANL, MeIQx, PhIP (0.09 μM), or the control (C2H5OH as solvent vehicle). (A) Visible spectra of HbO2 and met-Hb after 120 min. B) Kinetics of met-Hb formation in RBC over 120 min.
Free Thiols in Hb in RBC Treated with N-Hydroxy-AAs or N-Hydroxy-HAAs
The Hb in RBC was treated with a 30-fold higher concentration of N-hydroxy-AA or N-hydroxy-HAA (3 mol N-hydroxy substrate per mol HbO2 tetramer). Following lysis of the RBC, the titration of untreated Hb with PMB revealed the presence of ~2 thiols per Hb tetramer, a value consistent to those previously reported in the literature (Table l).31,60 These two thiols are attributed to the β-Cys93 chain residues, which are surface-exposed and rapidly react with PMB, whereas the β-Cys112 and α-Cys104 residues are partially shielded and react far more slowly with PMB.28–30,35 The treatment of RBC with HONH-ABP or HONH-AαC resulted in conversion of ~39 – 46% of HbO2 to met-Hb, and the titration of the Hb with PMB revealed that the available β-Cys93 residues were reduced to less than 0.1 mol “SH” per mol Hb tetramer (Table 1). These findings reveal that HONH-AαC, like primary arylhydroxylamines, undergoes co-oxidation with HbO2 in the erythrocyte to form 2-nitroso-9H-pyrido[2,3-b]indole (NO-AαC). The NO-AαC undergoes redox cycling with NADPH reductase in the erythrocyte to reform HONH-AαC, resulting in a new round of co-oxidation with HbO2, leading to methemoglobinemia.30 The oxidation of HbO2 and the diminution of the available β-Cys93 residue of Hb treated with HNOH-MeIQx and HNOH-PhIP were negligible (Table 1).
Table 1.
Determination of Free Cys and met-Hb Formation by N-Hydroxy Metabolites of AαC, ABP, PhIP, and MeIQx
| Substrate | Reduced β-Cys93 | met-Hb (%) | mol met-Hb formed per mol of HONH-Substrate or H2Oa |
|---|---|---|---|
| Hb + H2O | 2.00 ± 0.10 | 2.5 ± 0.14 | 0.03 |
| Hb +HONH-MeIQx | 1.96 ± 0.10 | 3.7 ± 0.18 | 0.05 |
| Hb +HONH-PhIP | 1.81 ± 0.10 | 4.7 ± 0.20 | 0.06 |
| Hb +HONH-ABP | <0.10 ± 0.01 | 41.4 ± 0.33 | 0.55 |
| Hb+HONH-AαC | <0.10 ± 0.02 | 48.8 ± 0.31 | 0.64 |
HbO2 (tetramer) content of RBC was adjusted to 2 nmol and RBC were incubated with 6 nmol of HONH-AA or HONH-HAA for 30 min, followed by titration with PMB.
met-Hb formation in the presence or absence of HONH-AA or HONH-HAA (mol/mol); mol of met-Hb formation in the H2O control was normalized to the ratio of the HONH-substrate treated cells by dividing amount of met-Hb formed by 6. Each value represents the mean and standard deviation of three experiments.
The poor reactivity of HONH-MeIQx and HONH-PhIP with HbO2 could be due to the inability of the metabolites to penetrate the RBC membrane and react with the oxy-heme complex. Alternatively, the HONH-HAAs may be unstable in the RBC environment and react with small molecular weight nucleophiles, such as thiols, or the HONH-HAAs may undergo rapid enzymatic reduction to the parent amines.61 Therefore, we examined the reactivity of N-hydroxy metabolites of PhIP, MeIQx, as well as AαC, ANL, and ABP with purified HbO2. We observed the same pattern of reactivity: HONH-AαC, HONH-ANL, and HONH-ABP oxidized HbO2 to the met-Hb form, and the free thiol content was decreased to less than 0.1 mol per mol of HbO2 tetramer. In contrast, HONH-PhIP and HONH-MeIQx did not induce oxidation of HbO2, and the number of titratable thiols remained unchanged from the control (Supporting Information, Table S-1). Thus, HONH-PhIP and HONH-MeIQx do not efficiently react with the oxy-heme complex of HbO2 and this explains their poor binding to Hb.
Mass Spectrometric Characterization of Hb Adducts of AαC
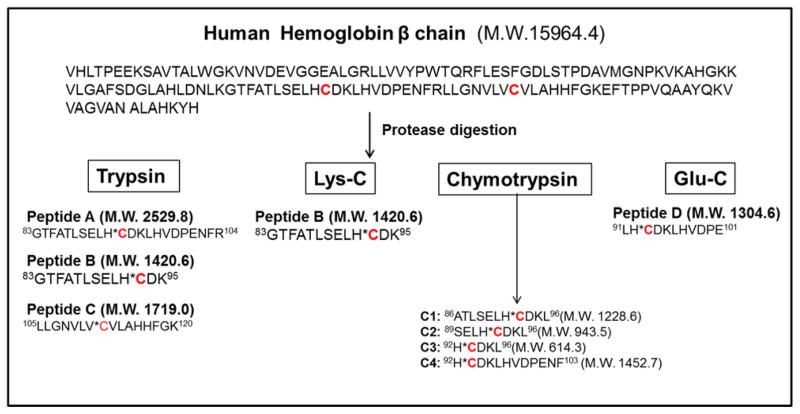
We sought to determine if the reduction of titratable β-Cys93 residues in RBC treated with HNOH-AαC was attributed to Hb adduct formation or oxidation of the thiol group to the Cys-sulfinic acid (CysSO2H) or Cys-sulfonic acid (CysSO3H), by the H2O2 generated during the co-oxidation reaction of HONH-AαC with HbO2.30 UPLC/MS2 was employed to characterize the Hb β-Cys93 chain containing the presumed adducts of ABP, ANL, and AαC, following digestion with different proteases. Trypsin, Lys-C, chymotrypsin, and Glu-C were examined to determine optimal digestion conditions for recovery of the β-Cys93 arylsulfinamide linkages. Trypsin cleaves peptide chain at C-terminal of Lys and or Arg, whereas Lys-C specifically cleaves at C-terminal of Lys.62 Glu-C is an endoprotease that cleaves peptide chain at the C-terminal acidic amino acids Glu and Asp, with higher specificity at Glu.63 The peptide sequences recovered from the β-chain of Hb, by the different proteases, are depicted in Scheme 2.
Scheme 2.
Schematic of Hb-β chain digestion with trypsin, Lys-C, chymotrypsin and Glu-C and peptide sequence with β-Cys93.
Trypsin and Lys-C digestion
The MS-tag DDA was performed Hb modified with 3-fold molar excess of 1:1 HONH-AαC and HONH-[13C6]AαC, following digestion of Hb at a ratio of trypsin to Hb of 1:10. The peptide adducts identified by the Myrimatch algorithm are summarized in Table 2. Two tryptic peptides encompassing β-Cys93, the single missed-cleavage peptide A) 84GTFATLH*CDKLHVDPENFR105 (M.W: 2529.8) and fully digested peptide B) 84GTFATLH*CDK96, (M.W: 1420.6) containing AαC sulfinamide (SOAαC) and AαC sulfonamide (SO2AαC) adducts were identified (Scheme 2). These were the sole adducts of AαC detected by Myrimatch. This search engine also identified sulfinic (-SO2H) and sulfonic acid (-SO3H) at β-Cys93, β-Cys112 and α-Cys104 residues of Hb; however, these thiol oxidation products were identified both in untreated and AαC-treated Hb (Table 2)
Table 2.
Peptide adducts and Oxidized Peptides Identified by Mass-tag Data Dependent Acquisition
| Peptide | Observed precursor ions (m/z) | Z | M.W. | tR (min) | AαC-peptide adduct | Site of modification | Enzyme |
|---|---|---|---|---|---|---|---|
| α-Chain | |||||||
| α | 754.7 | 4 | 3014.6 | 19.8 | 100LLSHC[SO3H]LLVTAAHLPAFTPAVHASLDK126 | α-Cys104 | Trypsin |
| 750.7 | 4 | 2998.6 | 20.5 | 100LLSHC[SO2H]LLVTAAHLPAFTPAVHASLDK126 | |||
| β-Chain | |||||||
| A | 686.3 | 4 | 2741.2 | 19.12 | 83GTFATLSELHC[SO2AαC]DKLHVDPENFR104a | β-Cys93 | Trypsin |
| 682.3 | 4 | 2725.2 | 19.54 | 83GTFATLSELHC[SOAαC]DKLHVDPENFR104a | |||
| 641.1 | 4 | 2560.2 | 14.95 | 83GTFATLSELHC[SO2H]DKLHVDPENFR104a | |||
| 645.1 | 4 | 2577.2 | 15.59 | 83GTFATLSELHC[SO3H]DKLHVDPENFR104a | |||
| B | 540.2 | 3 | 1617.7 | 16.91 | 83GTFATLSELHC[SOAαC]DK95a | β-Cys93 | Trypsin or Lys-C |
| 545.6 | 3 | 1633.7 | 16.55 | 83GTFATLSELHC[SO2AαC]DK95a | |||
| 485.2 | 3 | 1452.7 | 11.40 | 83GTFATLSELHC[SO2H]DK95a | |||
| 490.6 | 3 | 1468.7 | 11.41 | 83GTFATLSELHC[SO3H]DK956a | |||
| D | 501.6 | 3 | 1501.7 | 13.26 | 91LHC[SOAαC]DKLHCDPE101a | β-Cys93 | Glu-C |
| 506.9 | 3 | 1517.7 | 14.10 | 91LHC[SO2AαC]DKLHCDPE101a | |||
| 496.9 | 3 | 1487.7 | 14.94 | 91LHC[SOABP]DKLHCDPE101b | |||
| 502.2 | 3 | 1503.7 | 15.14 | 91LHC[SO2ABP]DKLHCDPE101b | |||
| 471.6 | 3 | 1411.7 | 12.26 | 91LHC[SOANL]DKLHCDPE101c | |||
| 476.9 | 3 | 1427.7 | 12.59 | 91LHC[SO2ANL]DKLHCDPE101c | |||
| 446.5 | 3 | 1337.6 | 08.39 | 912LHC[SO2H]DKLHCDPE101a.b.c | |||
| 451.9 | 3 | 1352.6 | 08.43 | 91LHC[SO3H]DKLHCDPE1012a.b.c | |||
| E | 584.7 | 3 | 1767.0 | 14.90 | 105LLGNVLVC[SO3H]VLAHHFGK120a | β-Cys112 | Trypsin |
| 590.3 | 3 | 1751.0 | 15.41 | 105LLGNVLVC[SO2H]VLAHHFGK120a | |||
Z – Charge state; tR- Retention time; M.W- Molecular weight;
HONH-AαC-Hb modified sample;
HONH-ABP-Hb modified sample;
HONH-ANL-Hb modified sample.
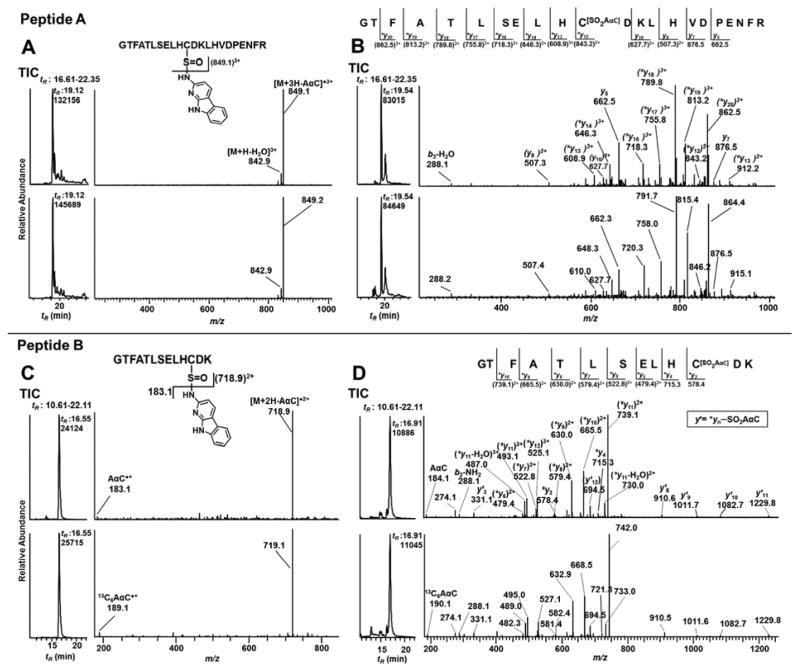
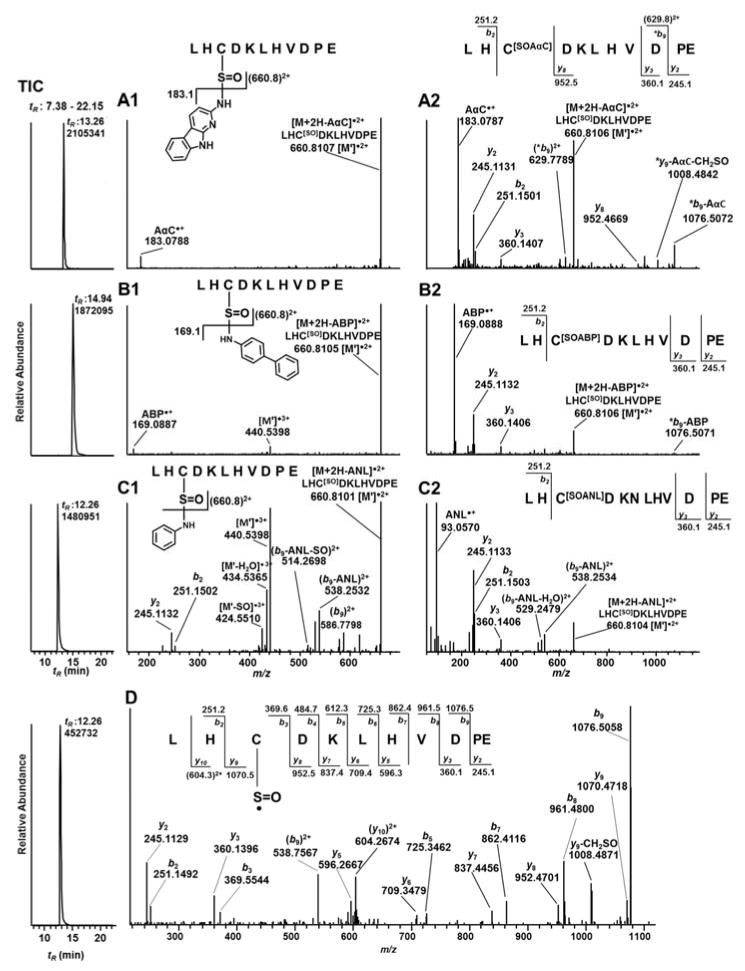
The product ion spectra of the β-Cys93SOAαC ([M+4H]4+ at m/z 682.8, upper panel) and β-Cys93SO[13C6]AαC (M+4H]4+ at m/z 684.3, lower panel) of peptide A, 84GTFATLH*CDKLHVDPENFR105, contain one major common product ion as the base peak at m/z 849.1 (Figure 3A). This fragment ion is the triply charged radical cation species GTFATLHC[SO]DKLHVDPENFR, which is formed by the cleavage of the sulfinamide linkage ([M+3H-AαC•]3+ and [M+3H-[13C6]AαC•]3+). The AαC•+ isotopomers are not observed in the product ion spectra because of the low mass cutoff or the so-called “1/3 Rule”.64 The product ion spectra of β-Cys93SOAαC ([M+3H]3+ at m/z 540.2, upper panel) and β-Cys93SO[13C6]AαC ([M+3H]3+ at m/z 542.3, lower panel) of the fully digested tryptic peptide B, 84GTFATLH*CDK96, are shown in Figure 3C. In this case, product ions of AαC•+ and [13C6]AαC•+ are observed, respectively, at m/z 183.1 and 189.1, along with the common ion at m/z 718.9, which is the doubly charged radical cation species GTFATLHC[SO]DK.
Figure 3.
TIC and CID-MS2 spectra of AαC-peptide A (GTFATLSELH*CDKLHVDPENFR) adducts (A) [M+4H]4+ of β93Cys-SO-AαC at m/z 682.3 (upper panel) and 93Cys-SO-[13C6]AαC at m/z 683.8 (lower panel) at tR 19.2 min; (B) [M+4H]4+ of β93Cys-SO2-AαC at m/z 686.3 (upper panel) and 93Cys-SO2-[13C6]AαC at m/z 687.8 (lower panel) at tR 19.5 min. TIC and CID-MS2 spectra of AαC-peptide B (GTFATLSELH*CDK) adducts (C) [M+3H]3+ of β93Cys-SO-AαC (upper panel) at m/z 540.2 and 93Cys-SO-[13C6]AαC (lower panel) at m/z 542.2 at tR 16.55 min; (D) [M+3H]3+ of β93Cys-SO2-AαC (upper panel) at m/z 545.6 and 93Cys-SO2-[13C6]AαC (lower panel) at m/z 547.6 at tR 16.91 min of trypsin digest of Hb modified with 3 molar excess of HONH-AαC and HONH-[13C6]AαC. Mass spectral data are analyzed using front end ion trap MS.
The Hb sulfonamide linkage of AαC is considerably more stable towards CID than the Hb sulfinamide linkage. The product ion spectra of the β-Cys93SO2AαC and β-Cys93SO2[13C6]AαC adducts ([M+4H]4+ at m/z 686.3 and 687.8 of peptide A) in Figure 3B, and the product ion spectra Cys93SO2AαC and β-Cys93SO2[13C6]AαC ([M+3H]+3 at m/z 545.6 and 547.6 of peptide B) in Figure 3D display a series of -yn ions of the respective GTFATLHC[SO2AαC]DKLHVDPENFR and GTFATLHC[SO2AαC]DK structures. Characteristic -y′n product ion series corresponding to the loss of SO2AαC or SO2[13C6]AαC ion from -yn ion series are also observed in the product ion spectra of the AαC sulfonamide adducts of B peptide (Figure 3D).
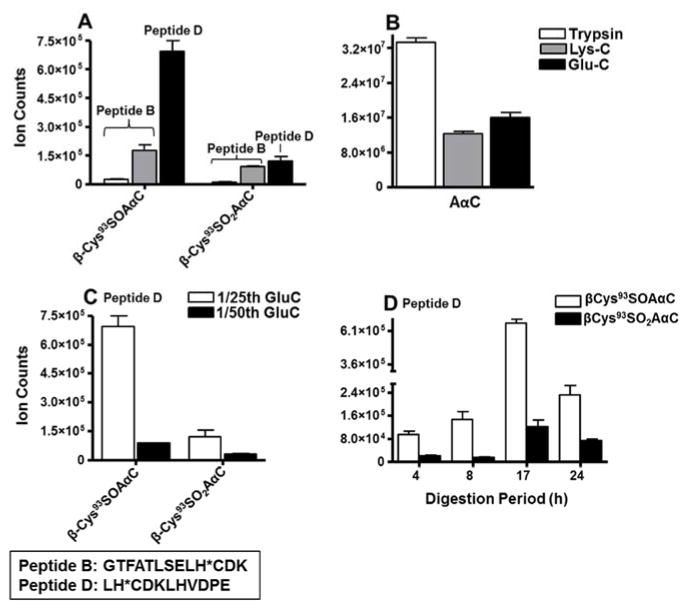
The AαC adducts of tryptic peptide A, based on ion counts, was 5 to 8 -fold higher than the fully digested AαC adducts of tryptic peptide B (Figure 1S1 in Supporting Information). However, peptide B with an optimal length (13 amino acid residues) for chromatography may be the preferred biomarker candidate peptide over larger sized single missed-cleavage peptide A with 21amino acid residues. The digestion of Hb-AαC adducts with a trypsin to Hb ratio of 1:10 yielded highest amount of adducted peptides A and B, but the large amount of protease also resulted in a large recovery of AαC, presumably as a result of hydrolysis of the sulfinamide adducts (Figure S1A–S1B in Supporting Information). Thus, we investigated the efficacy of other proteases to recover β-Cys93 containing peptides of AαC in high yield, while minimizing the hydrolysis of sulfinamide linkage.
Lys-C, an endoproteinase, which cuts primarily at C-terminal Lys residues,62 produced B peptides containing SOAαC, SO2AαC adducts, or -SO2H and -SO3H modifications at β-Cys93 (Table 2, Scheme 2). Employing Lys-C for proteolysis at protease to Hb ratio of 1:50, increased the recovery of β-Cys93SOAαC and β-Cys93SO2AαC adducts of peptide B by 7 and 8.4-fold in comparison to the digestion done with trypsin (Figure 4A). In contrast, the amount of AαC recovered from Hb digested with Lys-C was also 3-fold lower than that amount recovered from digestion of Hb with trypsin (Figure 4B).
Figure 4.
Ion counts of (A) β93Cys-SO-AαC and β93Cys-SO2-AαC adducts of peptide B (GTFATLSELH*CDK) and peptide D (LH*CDKLHVDPE), (B) free AαC recovered from 1/50 Trypsin, 1/50 Lys-C or 1/25 Glu-C (enzyme to protein ratios) digests of Hb modified with 3 molar excess of HONH-AαC. Ion counts of β93Cys-SO-AαC and β93Cys-SO2-AαC of peptide D ( adducts recovered from (C) 1/25 Glu-C and 1/50 Glu-C digests (enzyme to protein ratio) and (D) 1/25 Glu-C digestion at timer intervals of 4, 8,17 and 24 h of Hb modified with 3 molar excess of HONH-AαC. Data are plotted as mean and standard deviation of ion counts (N = 3). The level of adducts were determined from the area of total counts of CID-MS2 spectral data analyzed using front end iontrap MS
Chymotrypsin Digestion
Chymotrypsin generates smaller sized peptides and frequently with one or two missed-cleavages, which restricts its application for this study (Scheme 2). Proteolysis of Hb with a chymotrypsin to Hb ratio of 1:25 or 1:50 protein did not result in the recovery of peptide adducts of AαC, and the level of free AαC was 5-fold higher than those amounts recovered by digestion with trypsin (1:10 trypsin to Hb). Thus, the majority of the sulfinamide adduct was hydrolyzed during digestion with chymotrypsin (data not shown). Multiple missed-cleaved peptides, including, HCDKL, SELHCDKL, and ATLSELHCDKL were also identified in chymotryptic digest of unmodified Hb (data not shown).
Glu-C Digestion
Glu-C is an endoproteinase that cleaves peptide bonds at the carboxy terminus of either Glu or Asp, with higher specificity towards Glu.63 Two Glu residues are situated at positions 91 and 102 of the β-chain of Hb. Both SOAαC and SO2AαC adducts within the peptide sequence 92LH*CDKLHVDPE102 (M.W. 1304.6) were recovered by digestion with Glu-C (Table 2). The optimal amount of Glu-C employed for digestion of Hb used a ratio of Glu-C to Hb of 1:25, and proteolysis was conducted for 17 h There was no evidence of single missed-cleavage peptides, when assayed by MS-tag DDA and screened with the Myrimatch algorithm. The β-Cys93SO2H and β-Cys93SO3H containing modifications also were identified (Table 2, Figure S2 in Supporting Information). The signals of Hb β-Cys93 adducts were greatest with Glu-C, followed by Lys-C, and lastly for trypsin (Figure 4A). Optimal digestion conditions, Glu-C:Hb ratio of 1:25 and a 17 h (Fig. 4C–D) were used for mass spectral characterization of Hb adducts of ABP and ANL described below.
Mass Spectral Characterization of Hb-AαC, Hb-ABP, Hb-ANL Adducts Following Digestion of Hb with Glu-C
Triply charged precursor ion species were the most abundant ions for all the peptide adducts. The reaction of RBC with a 3 mol excess of HONH-AαC, HONH-ABP or HONH-ANL to HbO2 tetramer resulted in formation of both the sulfinamide and sulfonamide adducts at β-Cys93, designated as peptide D, LH*CDKLHVDPE (Table 2). The product ion spectra are described below.
LH*CDKLHVDPE Sulfinamide Adducts of AαC, ABP, and ANL
The [M+3H]3+ precursor ions at m/z 501.5664 (tR:13.26), 496.9029 (tR:14.94) and 471.5591 (tR:12.26) correspond, respectively, to β-Cys93SOAαC, β-Cys93SOABP, and β-Cys93SOANL adducts of Peptide D. The observed m/z values for precursor ions of the adducts were within 3 ppm of the calculated m/z values. The adducted peptides were subjected to CID- or HCD fragmentation and accurate mass measurements were acquired with the Orbitrap. The product ion spectrum of β-Cys93SOAαC ([M+3H]3+ at m/z 501.5664) obtained by CID, displays two fragment ions at m/z 183.0788 and 660.8107, corresponding to AαC•+ and the doubly charged sulfoxide radical LHC[SO]DKLHVDPE (Figure 5A1). The ion at m/z 660.8107 was also the base peak in the product ion spectra of β-Cys93SOABP and β-Cys93SOANL adducts (Figure 5B1, 5C1). In the case of Cys93SOABP, the product ion at m/z 169.0887 (ABP•+) was observed, whereas the radical cation of ANL at m/z 93.0570, arising by bond cleavage between SO and amine group of ANL was not seen due to low mass cutoff of the CID product ion spectra (Figure 5B1, 5C1). HCD product ion spectra were acquired on the triply charged precursor ions of β-Cys93SOAαC (m/z 501.5664), β-Cys93SOAαC (m/z 496.9029), and β-Cys93SOANL (m/z 471.5591). The lower mass range of the spectra show the corresponding product ions of the amines at m/z 183.0787 (AαC•+), 169.0888 (ABP•+), and 93.0570 (ANL•+) (Figure 5A2,B2, and C2), and the –b2 and -y2 -y3 type product ions were present in the HCD product ion spectra. Consecutive reaction monitoring was done by CID at the MS3 scan stage of the product ion at m/z 660.8100 ([M′]2•+), which is in common to all three arylsulfinamde adducts. A complete series of -b2-b9 and -y2-y10 ions confirmed the sulfoxide structure (LHC[SO]DKLHVDPE) and respective LH*CDKLHVPE sulfinamide adducts of AαC, ABP and, ANL (Figure 5D). All the product ions were observed within 3 ppm of the calculated values.
Figure 5.
TIC and MS2 spectra of [M+3H]3+ of peptide D (LH*CDKLHVDPE) adducts (A1) CID, (A2) HCD of β93Cys-SO-AαC at m/z 501.5664 at tR 13.26 min; (B1) CID, (B2) HCD of β93Cys-SO-ABP at m/z 496.9029 at tR 14.94 min; (C1) CID, (C2) HCD of β93Cys-SO-ANL at m/z 471.5591 at tR 12.26 min and (D) CID-MS3 of LHC[SO]DKLHVDPE [M′•]2+ at m/z 471.5591 >660.8101 > recovered from the Glu-C (1/25th enzyme to protein ratio) digest of Hb modified with 3 molar excess of respective HONH-AαC, HONH-ABP or HONH-ANL. MS/MS spectral data were acquired on Orbitrap MS at 30,000 resolving power.
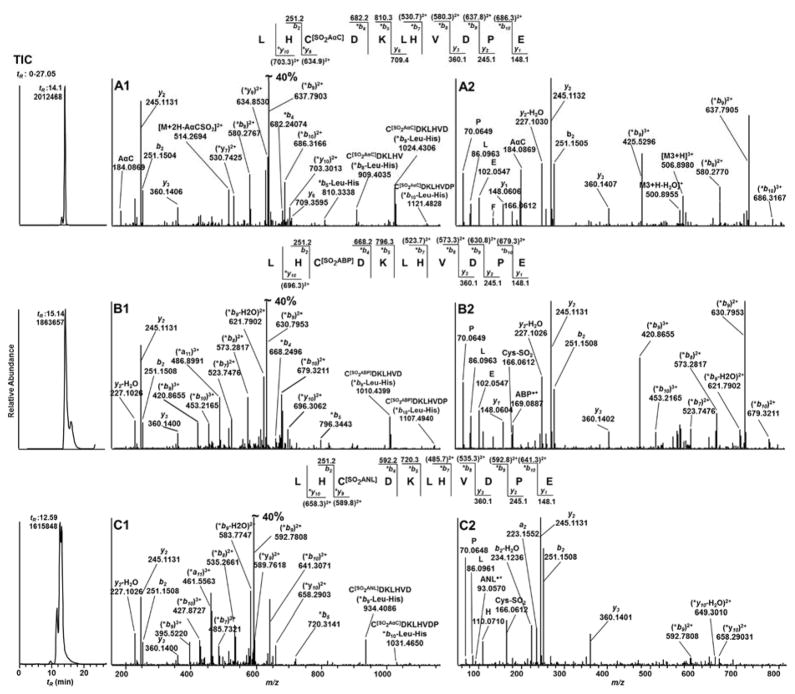
LH*CDKLHVDPE Sulfonamide Adducts of AαC, ABP and ANL
The CID product ion spectra of [M+3H]3+ precursor ions of β-Cys93SO2AαC (m/z 506.8980), β-Cys93SO2ABP (m/z 502.2345), and β-Cys93SO2ANL (m/z 476.8907), show the presence of typical –bn (-b2-b10) and –yn (-y2-y3 and-y9-y10) series of ions, which confirm the structures of the adducts of LH*CDKLHVDPE (Peptide D), (Figure 6A1, B1 and C1). The precursor ion m/z values were observed within 3 ppm of calculated m/z values. The C-terminal of Peptide D is an acidic Glu residue, which resulted in high abundance and sequence coverage of –bn ions. The ions at m/z 909.4035, 1024.4306 and, 1121.4838 in the product ion spectrum of precursor ion at m/z 506.8980 (SO2AαC) were assigned as putative -b type internal ions as a result of loss of N-terminal dipeptide Leu-His from -b8-Leu-His (*CDKLHV), -b9-Leu-His (*C[SO2AαC]DKLHVD), and -b10-Leu-His (*C[SO2AαC]DKLHVDP) ions (Figure 6A1).65 Similarly, -b type internal ions as a result of loss of N-terminal dipeptide Leu-His from -bn ions are observed in the product ion spectra of SO2ABP adduct (m/z 502.2345) and SO2ANL (m/z 476.8907) (Figure 6B1, C1).
Figure 6.
TIC and MS2 spectra of [M+3H]3+ of peptide D (LH*CDKLHVDPE) adducts (A1) CID, (A2) HCD of β93Cys-SO2-AαC at m/z 506.8980 at tR 14.1 min; (B1) CID, (B2) HCD of β93Cys-SO2-ABP at m/z 502.2345 at tR 15.14 min; (C1) CID, (C2) HCD of β93Cys-SO2-ANL at m/z 476.8907 at tR 12.59 min recovered from the Glu-C (1/25th enzyme to protein ratio) digest of Hb modified with 3 molar excess of respective HONH-AαC, HONH-ABP or HONH-ANL.MS/MS spectral data were acquired on Orbitrap MS at 30,000 resolving power.
The HCD product ion spectrum of β-Cys93SO2AαC, ([M+3H]3+ at m/z 506.8980), shows fragment ions at m/z 184.0869 and 183.0791, which are attributed to protonated AαC and the radical cation of AαC•+ (Figure 6A2). Similarly, the HCD spectra of β-Cys93SO2ABP and β-Cys93SO2ANL ([M+3H]3+ at m/z 502.2345, and m/z 476.8907, respectively) display product ions at m/z 169.0888 and 93.0570, corresponding to ABP•+ and ANL•+ (Figures 6B2, 6C2).
Concentration-dependent Adduct Formation of AαC and ABP in RBC
The reactivity of HONH-AαC and HONH-ABP with Hb was carried out over a 300-fold concentration range (0, 3, 0.1 and 0.01 mol of HONH substrate to HbO2 tetramer). The Hb adducts were monitored by measuring ion counts of sulfinamide and sulfonamide of Peptide D adducts. The adduct formation and recovery of AαC and ABP showed an approximate linear-concentration response relationship (Figure 7A,B).
Figure 7.
Ion counts of peptide D (LH*CDKLHVDPE) adducts, (A) β93Cys-SO-AαC and β93Cys-SO2-AαC adducts and, (B) β93Cys-SO-ABP and β93Cys-SO2-ABP adducts recovered from the Glu-C digests of 1 mole Hb modified with 3, 0.1 and 0.01 mol equivalents of HONH-AαC or HONH-ABP. The level of adducts were determined from the area of total counts of CID-MS2 spectral data analyzed using orbitrap MS analyzer at 30,000 resolving power. (C) Ion counts of peptide D (LH*CDKLHVDPE) of β93Cys-SO2H and β93Cys-SO3H recovered from the Glu-C digests of 1molar Hb modified with 3, 0.1 and 0.01molar excess of HONH-AαC or HONH-ABP. Data are plotted as mean and standard deviation of ion counts (N = 3).
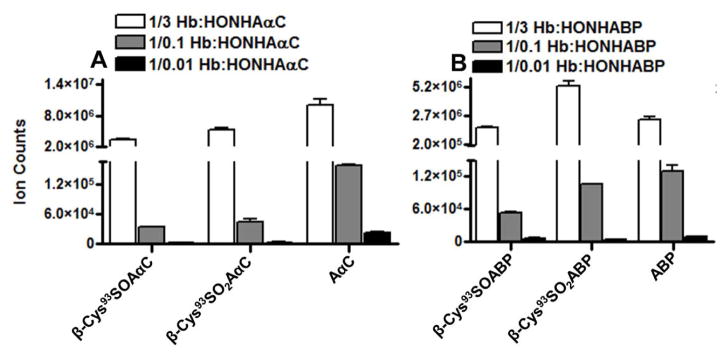
ABP-Hb Adduct Formation in Mouse
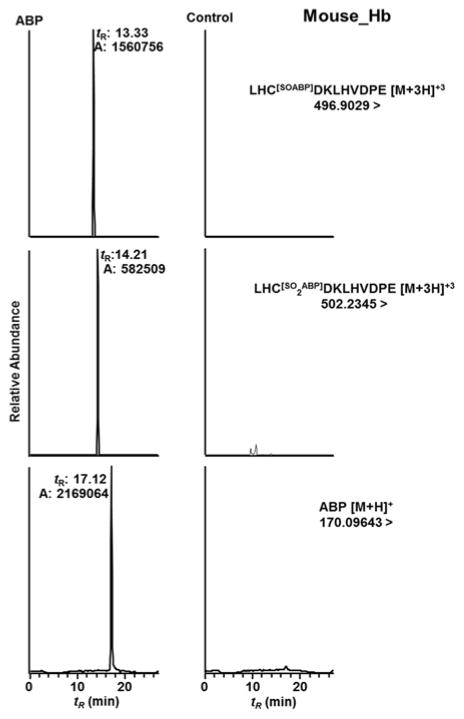
The β-chain of mouse and human Hb share 80.3% sequence homology, and the amino acid sequence is identical in both species from residues 74–108 containing the β-Cys93.66 Thus, we sought to determine if the intact sulfinamide and/or sulfonamide adduct of ABP could be recovered in vivo from mice dose with ABP (40 mg/kg body weight). Hb from ABP-treated and untreated mice was collected and digested with Glu-C as described above. Mouse Hb digests (200 ng Hb) were used for targeted UPLC-MS/MS analysis in the Orbitrap, by assaying for β-Cys93SOABP ([M+3H]3+ at m/z 496.9029) and β-Cys93SO2ABP ([M+3H]3+ at m/z 502.2345) adducts of Peptide D (LH*CDKLHVDPE), and free ABP ([M+H]+ at m/z 170.0964) released during proteolysis of mouse Hb-ABP. The HbCys93β-ABP adducts and free ABP are present in the ion chromatograms of Hb obtained from ABP-treated mice (Figure 8), but the signals are absent in the chromatograms of Hb from the control mice. The CID product ion spectra of β-Cys93SOABP and β-Cys93SO2ABP adducts of Peptide D (LH*CDKLHVDPE) from mouse Hb (Figure S3A and S3B in Supporting Information) are identical to those observed for human Hb treated with HONH-ABP (Figure 5B1 and 6B1). The levels of β-Cys93SOABP and β-Cys93SO2ABP and, ABP in ABP treated mice are summarized in Figure 9.
Figure 8.
Ion chromatograms of peptide D (LH*CDKLHVDPE) β93Cys-SO-ABP, β93Cys-SO2-ABP adducts and free ABP recovered from Glu-C digest of Hb from mouse treated with ABP (40 mg/kg body weight) and untreated mouse.
Figure 9.
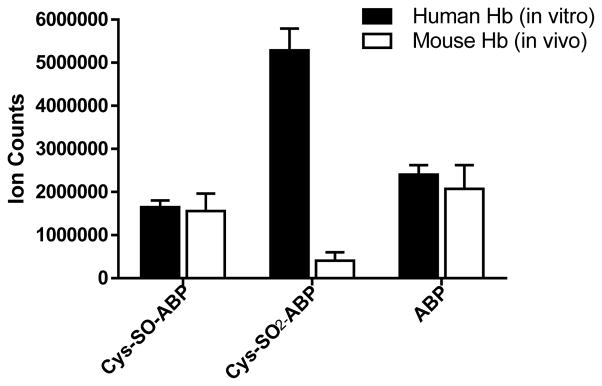
Ion counts of β93Cys-SO-ABP, β93Cys-SO2-ABP adducts and free ABP recovered from Glu-C digests of Hb from mouse treated with ABP (40 mg/kg body weight) and human Hb (2nmol) in vitro modified with 3 molar excess of HONH-ABP (6 nmol). The Glu-C digestion was carried out with Hb (5 μg of Hb from in vivo mice treated with ABP and Hb of modified in vitro with HONH-ABP). The protein digest (100 ng) was used for the LC-MS/MS based measurement of ABP-Hb adducts and free ABP. Data are plotted as mean and standard deviation of ion counts of adducts recovered from five ABP-treated mice and three independent experiments conducted in vitro. The level of adducts were determined from the area of total counts of CID-MS2 spectral data analyzed using orbitrap MS analyzer at 30,000 resolving power.
The relative abundance of mouse Hb-ABP sulfinamide adduct and free ABP in treated mice was comparable to those obtained from the incubation of a 3-fold molar excess of HONH-ABP with Hb in human RBC; however, the level of the Hb-ABP sulfonamide adduct relative to the Hb-ABP sulfinamide in mouse was 13-fold lower than level obtained from the in vitro studies with human RBC (Figure 9). Considering the average mouse Hb content (~268 mg mouse Hb/1.8 mL of total blood volume/30 g body weight, 4.1 μmol),67 the dose of 40 mg ABP/kg body weight (1.2 mg/30 g body weight, 7.1 μmol) was equivalent to ~2-fold molar excess of ABP/mol mouse Hb tetramer. Although, the metabolism of ABP in vivo is a far more complex biological process than the in vitro reaction of HbO2 with HONH-ABP, the amount of the ABP-Hb adduct formed in vivo is comparable to that obtained from the in vitro study.
Molecular Modeling
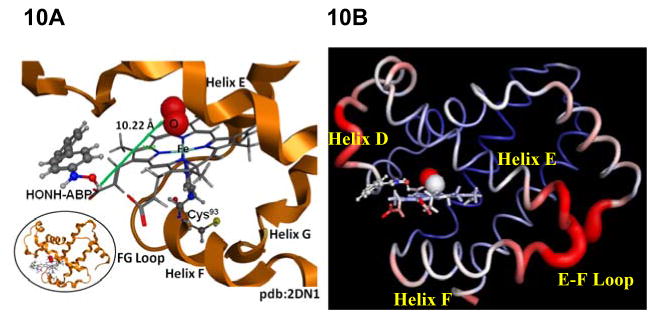
We sought to determine if variations in met-Hb formation among different HONH-AA and HONH-HAA substrates could be attributed to differences in their proximity of binding to the oxy-heme complex. The β chains of Hb from four crystal structures: 2DN1,68 4NI0,69 1YZI,70 and 1GZX71 were examined to identify a suitable overall hemoglobin structure into which to dock HONH-ANL, HONH-ABP, HONH-AαC, HONH-PhIP, and HONH-MeIQx. All four β chains were close to identical in configuration, including the proximity of their heme pockets; RMSD values between the four B chains were 0.77–0.79Å. Given these slight variations, 2DN1.pdb was chosen as the docking target for all five AAs and HAAs, chiefly because 2DN1.pdb exhibits the HbO2 in its relaxed state.26,68 The heme pocket is narrow and there is very little room for the compounds addressed here to interact with the heme oxygen, a finding which is supported by our docking results. Figure 10A illustrates the highest-scoring docked pose of HONH-ABP with 2DN1.pdb. The measured distance between H-ONH-ABP and the oxy-heme moiety of 2DN1.pdb was predicted to be located far from the heme oxygen (10.22 Å). The measured distances (Table 3) between the H-ONH-substrates of other AA and HAAs were also predicted to be located too far from the heme oxygen to undergo co-oxidation and induce methemoglobinemia.72 To explore the flexibility of the heme pocket, the secondary structure of human hemoglobin (2DN1.pdb) was color coded according to a crystallographic temperature factor (B-factor) spectrum using PyMOL (Schrödinger, LLC), ranging from red (high B-factor) to blue (low B-factor), with white representing average values over the range of B-factors in this system. As shown in Figure 10B, helix D, the end of helix E, E-F loop, and the initial residues of helix F are more flexible regions according to experimental temperature factors. Therefore, we hypothesize that significant conformational changes are likely to take place in the flexible helix D near the heme pocket. Such changes in configuration would expose the heme pocket to interactions between the heme oxygen and small molecules.
Figure 10.
(A) The highest-scoring docked pose of HONH-ABP with HbO2 β chain (2DN1.pdb). The H-ONH-ABP substrate and the oxy-heme moiety of 2DN1.pdb shows the predicted bond length between the H-ON-ABP substrate and the heme oxygen is 10.22 Å. (B) 2DN1.pdb colored according to crystallographic temperature factors (B-factors). Areas with high B-factors are colored red (hot) while areas with low B-factors are colored blue (cold). Flexible helix D is near the heme pocket.
Table 3.
Measured distances between the H-ONH-AA and H-ONH-HAA substrates and oxy-heme moiety of 2DN1.pdb in the highest-scoring docked poses of the five AAs and HAAs
| HONH-AA or HONH-HAA | Measured distance between the H-ON-substrate and oxy-heme moiety of 2DN1.pdb in the highest-scoring docked pose |
|---|---|
| HONH-ANL | 7.15 Å |
| HONH-ABP | 10.22 Å |
| HONH-AαC | 10.01 Å |
| HONH-PhIP | 7.52 Å |
| HONH-MeIQx | 9.38 Å |
DISCUSSION
Hb adducts of AAs have been used in human biomonitoring studies.39,73,74 The adducts have been measured indirectly, following acid or base treatment of Hb in vitro, to hydrolyze the presumed sulfinamide adducts formed at the β-Cys93 of Hb, and liberate the parent amines, which are assayed by MS.34,37 Because of their purported instability during proteolysis, mass spectral structural characterization of Hb arylsulfinamide adducts have not been reported,35,36 although we recently characterized, by UPLC/MS2, sulfinamide and sulfonamide peptide adducts of ABP, PhIP, and AαC formed at the Cys34 of human albumin, following proteolysis with trypsin/chymotrypsin.53,75 We have shown that the primary structure of Hb-peptide containing β-Cys93 adducts of ABP, ANL, and AαC formed in vitro can be characterized by UPLC-MS/MS, employing bottom-up proteomic approaches following proteolytic digestion of the adducted Hb with trypsin, chymotrypsin, Lys-C or Glu-C. The Glu-C was an ideal protease to recover the peptide, 92LH*CDKLHVDPE102 with minimal hydrolysis of the arylsulfinamide linkage. The pattern of CID fragmentation of arylsufinamide and arylsulfonamide adducts at β-Cys93 of Hb was similar to the previously characterized sulfur-nitrogen linked albumin adducts of ABP and AαC.53,75 As a proof of principle, we demonstrate that Hb sulfinamide and sulfonamide adducts of ABP formed in mice dosed with ABP (40 mg/kg) also can be detected by bottom-up proteomic approaches. At this dose, the levels of ABP adducts of Hb were comparable to the in vitro studies using a 3-fold molar excess of ABP/Hb tetramer. The identification of arylsulfinamide and arylsulfonamide adducts at 92LH*CDKLHVDPE102 suggests that application of Glu-C and an UPLC-MS/MS based bottom-up proteomic approach may be achieved for the characterization of other reactive electrophiles that adduct to the β-Cys93 of Hb. Indeed, data-independent-acquisition mass spectrometry has been employed to identify peptide site-specific modifications at the β-Cys93 of Hb.76 However, because of the challenges in the enrichment of the peptide adduct from the vast excess of non-modified protein digest, the biomonitoring of AαC in humans, like other arylamine Hb adducts, is expected to be more facile by acid or base hydrolysis of the sulfinamide linked adduct followed by the direct measurement of AαC.
We confirmed that the N-hydroxylated intermediates of the primary arylamines ANL and ABP induce methemoglobinemia and form sulfinamide adducts with Hb in RBC. Unexpectedly, the N-hydroxylated metabolite of AαC, a larger, linear tricyclic ring structure, was discovered to be more potent than HONH-ABP and HONH-ANL in inducing methemoglobinemia, and HONH-AαC also formed a sulfinamide adduct with Hb. However, HONH-PhIP and HONH-MeIQx, HAAs with angular tricyclic ring structures, did not interact with the oxy-heme complex and failed to induce met-Hb formation. Furthermore, the titratable β-Cys93 residues were relatively unchanged following treatment of HbO2 with HONH-PhIP or HONH-MeIQx. The binding proclivities of N-hydroxylated AAs to Hb are thought to depend upon their ability to undergo co-oxidation of HbO2 to form met-Hb and the arylnitroso intermediates, which selectively bind to the β-Cys93 residue of Hb (Scheme 1).35,36 N-Hydroxy-AA and N-hydroxy-HAA undergo facile aerial oxidation to form their nitroso intermediates.77,78 Moreover, we showed that HONH-ABP, HONH-PhIP, and HONH-AαC incubated with human serum albumin under similar buffer and pH conditions to those employed for HbO2 resulted in the formation of AA and HAA sulfinamide adducts at the Cys34 residue of albumin.75 These findings signify that the nitroso intermediates were produced by aerial oxidation of the HONH-AA and HONH-HAA substrates. Thus, in addition to the poor reactivity of HONH-PhIP and HOHN-MeIQx with the oxy-heme complex of Hb, we surmise that the nitroso intermediates of PhIP and MeIQx may not reach and efficiently react with the β-Cys93 Hb residue, which is on the distal side of the oxy-heme complex.
A study on the structure-activity relationship of arylamine binding to Hb conducted by Sabbioni reported that the binding of ortho-substituted alkyl or halogen derivatives of ANL to Hb was 5-fold or lower than the binding of ANL in the rat;79 however, the reactivity of the N-hydroxylated ANL derivatives with HbO2 was not examined. The decrease in the level of binding of ortho-substituted ANL derivatives to HbO2 may have been attributed to lower amounts of the N-hydroxylated metabolites produced in vivo, or by hindered reactivity of the N-hydroxylated metabolites with the oxy-heme complex, resulting in diminished levels of met-Hb and the arylnitroso intermediates.79 Another study reported that the binding of 2-aminofluorene, 2-napthylamine, and 2-aminopyrene to Hb in the rat were, respectively, 2.7%, 0.1%, and < 0.005% of the level of binding of 4-ABP.80 Thus, certain arylamines with fused bicyclic or tricyclic ring structures can form sulfinamide adducts with Hb. The level of Hb adduct formation is dependent upon the extent of metabolism the arylamines to their N-hydroxy intermediates, the ability of the N-hydroxy metabolites to react with oxy-heme complex to form the arylnitroso species, and their ensuing reactivity with β-Cys93 residue of Hb. The N-methyl substituents ortho to the hydroxylamine groups of HONH-PhIP and HONH-MeIQx may contribute to the poor reactivity of these HAA with HbO2 in vitro. The low level of binding of PhIP and MeIQx to Hb in vitro is consistent with that observed in vivo in rodents and humans.47,49
The β-Cys93 is located between heme-linked proximal His92(F8) and Asp94(FG1), and near the functionally important α1 and β2 interface. In the native form of tetrameric Hb, the two β-Cys93 residues are accessible to solvent and can react with low molecular weight electrophiles,27 whereas the α-Cys104 and β-Cys112 are shielded and their reactivity with electrophiles is minor.60,81 However, under high concentrations of PMB and prolonged incubation, a partial disassembly of the Hb subunits occurs, and the α-Cys104 and β-Cys112 become more accessible to solvent and can react with PMB and other electrophiles.29,82,83 The reactivity of β-Cys93 with sulfhydryl agents has been employed to separate the Hb tetramer into separate monomer chains.84 The β-Cys93 also binds low molecular weight alkylating agents such as styrene oxide,85 benzene oxide,25 nitric oxide,86 and acrylate contact allergens present in paint.87 The induced-fit docking program of MOE 2013.08 (Chemical Computing Group, Inc.) allowed only the active site residue side chains to be included in the final minimization stage of docking.58,59 Based on these docking results (Figure 10 and Table 3), variations in sidechain configuration were too small to allow any of the five AA and HAA substrates to approach the oxy-heme complex closely enough to engage in hydrogen bonding and/or electron donation. Therefore, significant conformational changes are likely to occur in the flexible helical and loop regions near the heme pocket upon ligand binding, resulting in enlargement of the cavity to permit interactions between the heme oxygen and HONH-AA or HONH-AαC substrates. With this in mind, our future work will incorporate additional experimental studies, including redox potential determination, other electronic properties and chemical descriptors, to elucidate the chemical reactivity of the various N-hydroxy AA and HAA substrates with HbO2 and the role that protein conformational changes may play in reactivity of these genotoxicants with HbO2.79,88
We had reported the crystal structure of the single stable Hb adduct of ABP, which confirmed that adduct formation had occurred at the β-Cys93 chain of Hb.28 The biphenyl group was wedged between the F and H helices close to the section where the two helices cross. The predominant interactions between adduct and protein occurred along the backbone of residues Leu141(H19) Ala142(H20) and the side chain of Lys144 (HC1) along one side of the phenyl rings, and residues Ala86(F2) - Cys93(F9) along the other side. The second phenyl ring was exposed at the surface of the heme pocket. It has been largely inferred that Hb adducts of other primary arylamines exist as sulfinamide linkages formed at the β-Cys93 chain.34,37 Our findings demonstrate that AαC also binds to the β-Cys93 to form the sulfinamide linkage. This results reinforces the findings that certain HONH-substrates with considerable diversity of shape can induce structural changes in Hb which allow them to interact with the oxy-heme complex and burrow into mobile regions surrounding the heme pocket and adduct to the distal the β-Cys93 group (Fig. 10). The high reactivity of HONH-AαC with HbO2 suggests that the Hb sulfinamide adduct of AαC may be a promising biomarker of exposure to this HAA in humans.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH grants RO1CA134700 and R01CA134700-03S1 of the Family Smoking Prevention and Tobacco Control Act, and Cancer Center Support grant no. CA-077598 (R.J.T.).
The authors thank the Mass Spectrometry core facility, a subsidiary of “The Analytical Biochemistry shared resource” at Masonic Cancer Center, University of Minnesota, Minneapolis and Dr. Peter Villalta, core facility coordinator for helpful discussion on MS instrument operation. We also thank the Dr. David Tabb Laboratory at Department of Biomedical informatics (Nashville, TN), for providing the MyriMatch software program. The authors acknowledge Mrs. Lihua Yao for providing blood samples of mice treated with ABP. Grateful acknowledgment is also given to the Minnesota Supercomputing Institute for Advanced Computational Research (MSI).
ABBREVIATIONS
- AA
arylamines
- AαC
2-Amino-9H-pyrido[2,3-b]indole
- HONH-AαC
2-hydroxyamino-9H-pyrido[2,3-b]indole
- NO-AαC
2-nitroso-9H-pyrido[2,3-b]indole
- NO2-AαC
2-nitro-9H-pyrido[2,3-b]indole
- ABP
4-aminobiphenyl
- HONH-AA
N-hydroxylated derivatives of arylamines
- HONH-ABP
4-hydroxyaminobiphenyl
- HONH-ANL
phenylhydroxylamine
- HAA
heterocyclic aromatic amines
- Hb
hemoglobin
- HbO2
Oxyhemoglobin
- RBC
red blood cell
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HONH-PhIP
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NO2-PhIP
2-nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine
- MeIQx
2-amino-3,8-dimethylimidaxo[4,5-f]quinoxaline (MeIQx)
- HONH-MeIQx
2-hydroxyamino-3,8-dimethylimidaxo[4,5-f]quinoxaline
- NO2-MeIQx
2-nitro-3,8-dimethylimidaxo[4,5-f]quinoxaline
- IAM
iodoacetamide
- SPE
solid phase extraction
- THF
tetrahydrofuran
- DDA
data dependent acquisition
- MS/MS
tandem mass spectrometry
- CID
collision induced dissociation
- HCD
high-energy collision dissociation
- RMSD
root-mean-square deviation
- UPLC
ultraperformance liquid chromatography
Footnotes
Notes
Authors declare no competing financial interest
The determination of free Cys and met-Hb Formation by N-Hydroxy Metabolites of AαC, ABP, MeIQx, and PhIP with purified Hb; Recovery of β93Cys-SO-AαC and β93Cys-SO2-AαC adducts of as a function of the amount of trypsin; CID-MS2 spectra of [M+3H]3+ of β93Cys-SO2H and β93Cys-SO3H of (LH*CDKLHVDPE); and TIC and CID-MS2 spectra of [M+3H]3+ of LH*CDKLHVDPE adducts of β93Cys-SO-ABP and β93Cys-SO2-ABP recovered from mice dosed with 4-ABP. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turesky RJ, Le Marchand L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem Res Toxicol. 2011;24:1169–1214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Agency for Research on Cancer. IARC Monographs: Some aromatic amines, organic dyes, and related exposures. Vol. 99. International Agency for Research on Cancer; Lyon, France: 2010. [Google Scholar]
- 4.Cioni F, Bartolucci G, Pieraccini G, Meloni S, Moneti G. Development of a solid phase microextraction method for detection of the use of banned azo dyes in coloured textiles and leather. Rapid Commun Mass Spectrom. 1999;13:1833–1837. doi: 10.1002/(SICI)1097-0231(19990930)13:18<1833::AID-RCM725>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 5.Turesky RJ, Freeman JP, Holland RD, Nestorick DM, Miller DW, Ratnasinghe DL, Kadlubar FF. Identification of aminobiphenyl derivatives in commercial hair dyes. Chem Res Toxicol. 2003;16:1162–1173. doi: 10.1021/tx030029r. [DOI] [PubMed] [Google Scholar]
- 6.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoke and involuntary smoking. Vol. 83. International Agency for Research on Cancer; Lyon, France: 2002. [PMC free article] [PubMed] [Google Scholar]
- 7.Manabe S, Izumikawa S, Asakuno K, Wada O, Kanai Y. Detection of carcinogenic amino-alpha-carbolines and amino-gamma-carbolines in diesel-exhaust particles. Environ Pollut. 1991;70:255–265. doi: 10.1016/0269-7491(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida D, Matsumoto T. Amino-alpha-carbolines as mutagenic agents in cigarette smoke condensate. Cancer Lett. 1980;10:141–149. doi: 10.1016/0304-3835(80)90037-3. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Ashley DL, Watson CH. Quantitative analysis of six heterocyclic aromatic amines in mainstream cigarette smoke condensate using isotope dilution liquid chromatography-electrospray ionization tandem mass spectrometry. Nicotine Tob Res. 2011;13:120–126. doi: 10.1093/ntr/ntq219. [DOI] [PubMed] [Google Scholar]
- 10.Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K. Contents in foods, beverages and tobacco. In: Nagao M, Sugimura T, editors. Food Borne Carcinogens Heterocyclic Amines. John Wiley & Sons Ltd; Chichester, England: 2000. pp. 31–71. [Google Scholar]
- 11.Shimada T, Guengerich FP. Activation of amino-α-carboline, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and a copper phthalocyanine cellulose extract of cigarette smoke condensate by cytochrome P-450 enzymes in rat and human liver microsomes. Cancer Res. 1991;51:5284–5291. [PubMed] [Google Scholar]
- 12.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem Res Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 13.Yamazoe Y, Nagata K, Ozawa S, Gong DW, Kato R. Activation and detoxication of carcinogenic arylamines by sulfation. Princess Takamatsu Symp. 1995;23:154–162. [PubMed] [Google Scholar]
- 14.Windmill KF, McKinnon RA, Zhu X, Gaedigk A, Grant DM, McManus ME. The role of xenobiotic metabolizing enzymes in arylamine toxicity and carcinogenesis: functional and localization studies. Mutat Res. 1997;376:153–160. doi: 10.1016/s0027-5107(97)00038-9. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen TM, Novak M. Synthesis and decomposition of an ester derivative of the procarcinogen and promutagen, PhIP, 2-amino-1-methyl-6-phenyl-1H-imidazo[4,5-b]pyridine: unusual nitrenium ion chemistry. J Org Chem. 2007;72:4698–4706. doi: 10.1021/jo070306p. [DOI] [PubMed] [Google Scholar]
- 16.Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 17.Tretyakova N, Goggin M, Sangaraju D, Janis G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem Res Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller JA. Carcinogenesis by chemicals: an overview--G. H. A. Clowes memorial lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 19.Rubino FM, Pitton M, Di Fabio D, Colombi A. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 20.Skipper PL, Tannenbaum SR. Protein adducts in the molecular dosimetry of chemical carcinogens. Carcinogenesis. 1990;11:507–518. doi: 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- 21.Tornqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 22.Skipper PL, Peng X, SooHoo CK, Tannenbaum SR. Protein adducts as biomarkers of human carcinogen exposure. Drug Metab Rev. 1994;26:111–124. doi: 10.3109/03602539409029787. [DOI] [PubMed] [Google Scholar]
- 23.Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ Health Perspect. 2002;110(Suppl 1):3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldini G, Regazzoni L, Orioli M, Rimoldi I, Facino RM, Carini M. A tandem MS precursor-ion scan approach to identify variable covalent modification of albumin Cys34: a new tool for studying vascular carbonylation. J Mass Spectrom. 2008;43:1470–1481. doi: 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- 25.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perutz MF. Hemoglobin structure and respiratory transport. Sci Am. 1978;239:92–125. doi: 10.1038/scientificamerican1278-92. [DOI] [PubMed] [Google Scholar]
- 27.Boyer PD. Spectrophotometric Study of the Reaction of Protein Sulfhydryl Groups with Organic Mercurials. J Am Chem Soc. 1954;76:4331–4337. [Google Scholar]
- 28.Ringe D, Turesky RJ, Skipper PL, Tannenbaum SR. Structure of the single stable hemoglobin adduct formed by 4-aminobiphenyl in vivo. Chem Res Toxicol. 1988;1:22–24. doi: 10.1021/tx00001a003. [DOI] [PubMed] [Google Scholar]
- 29.Kan HI, Chen IY, Zulfajri M, Wang CC. Subunit disassembly pathway of human hemoglobin revealing the site-specific role of its cysteine residues. J Phys Chem B. 2013;117:9831–9839. doi: 10.1021/jp402292b. [DOI] [PubMed] [Google Scholar]
- 30.Kiese M. The biochemical production of ferrihemoglobin-forming derivatives from aromatic amines, and mechanisms of ferrihemoglobin formation. Pharmacol Rev. 1966;18:1091–1161. [PubMed] [Google Scholar]
- 31.Kiese M, Taeger K. The fate of phenylhydroxylamine in human red cells. Naunyn Schmiedebergs Arch Pharmacol. 1976;292:59–66. doi: 10.1007/BF00506490. [DOI] [PubMed] [Google Scholar]
- 32.Butler MA, Guengerich FP, Kadlubar FF. Metabolic oxidation of the carcinogens 4-aminobiphenyl and 4,4′-methylenebis(2-chloroaniline) by human hepatic microsomes and by purified rat hepatic cytochrome P-450 monooxygenases. Cancer Res. 1989;49:25–31. [PubMed] [Google Scholar]
- 33.Butler MA, Iwasaki M, Guengerich FP, Kadlubar FF. Human cytochrome P-450 PA (P450IA2), the phenacetin O -deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc Natl Acad Sci USA. 1989;86:7696–7700. doi: 10.1073/pnas.86.20.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann HG. Analysis of hemoglobin as a dose monitor for alkylating and arylating agents. Arch Toxicol. 1984;56:1–6. doi: 10.1007/BF00316343. [DOI] [PubMed] [Google Scholar]
- 35.Green LC, Skipper PL, Turesky RJ, Bryant MS, Tannenbaum SR. In vivo dosimetry of 4-aminobiphenyl in rats via a cysteine adduct in hemoglobin. Cancer Res. 1984;44:4254–4259. [PubMed] [Google Scholar]
- 36.Neumann HG. Biomonitoring of aromatic amines and alkylating agents by measuring hemoglobin adducts. Int Arch Occup Environ Health. 1988;60:151–155. doi: 10.1007/BF00378690. [DOI] [PubMed] [Google Scholar]
- 37.Skipper PL, Stillwell WG. Aromatic amine-hemoglobin adducts. Methods Enzymol. 1994;231:643–649. doi: 10.1016/0076-6879(94)31044-0. [DOI] [PubMed] [Google Scholar]
- 38.Bryant MS, Skipper PL, Tannenbaum SR, Maclure M. Hemoglobin adducts of 4-aminobiphenyl in smokers and nonsmokers. Cancer Res. 1987;47:602–608. [PubMed] [Google Scholar]
- 39.Bryant MS, Vineis P, Skipper PL, Tannenbaum SR. Hemoglobin adducts of aromatic amines: associations with smoking status and type of tobacco. Proc Natl Acad Sci USA. 1988;85:9788–9791. doi: 10.1073/pnas.85.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu MC, Skipper PL, Tannenbaum SR, Chan KK, Ross RK. Arylamine exposures and bladder cancer risk. Mutat Res. 2002;506–507:21–28. doi: 10.1016/s0027-5107(02)00148-3. [DOI] [PubMed] [Google Scholar]
- 41.Gan J, Skipper PL, Gago-Dominguez M, Arakawa K, Ross RK, Yu MC, Tannenbaum SR. Alkylaniline-hemoglobin adducts and risk of non-smoking-related bladder cancer. J Natl Cancer Inst. 2004;96:1425–1431. doi: 10.1093/jnci/djh274. [DOI] [PubMed] [Google Scholar]
- 42.Malfatti MA, Dingley KH, Nowell-Kadlubar S, Ubick EA, Mulakken N, Nelson D, Lang NP, Felton JS, Turteltaub KW. The urinary metabolite profile of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine is predictive of colon DNA adducts after a low-dose exposure in humans. Cancer Res. 2006;66:10541–10547. doi: 10.1158/0008-5472.CAN-06-1573. [DOI] [PubMed] [Google Scholar]
- 43.Stillwell WG, Sinha R, Tannenbaum SR. Excretion of the N 2 -glucuronide conjugate of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in urine and its relationship to CYP1A2 and NAT2 activity levels in humans. Carcinogenesis. 2002;23:831–838. doi: 10.1093/carcin/23.5.831. [DOI] [PubMed] [Google Scholar]
- 44.Walters DG, Young PJ, Agus C, Knize MG, Boobis AR, Gooderham NJ, Lake BG. Cruciferous vegetable consumption alters the metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Carcinogenesis. 2004;25:1659–1669. doi: 10.1093/carcin/bgh164. [DOI] [PubMed] [Google Scholar]
- 45.Lynch AM, Murray S, Boobis AR, Davies DS, Gooderham NJ. The measurement of MeIQx adducts with mouse haemoglobin in vitro and in vivo: implications for human dosimetry. Carcinogenesis. 1991;12:1067–1072. doi: 10.1093/carcin/12.6.1067. [DOI] [PubMed] [Google Scholar]
- 46.Turesky RJ, Garner RC, Welti DH, Richoz J, Leveson SH, Dingley KH, Turteltaub KW, Fay LB. Metabolism of the food-borne mutagen 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in humans. Chem Res Toxicol. 1998;11:217–225. doi: 10.1021/tx9701891. [DOI] [PubMed] [Google Scholar]
- 47.Dingley KH, Freeman SP, Nelson DO, Garner RC, Turteltaub KW. Covalent binding of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline to albumin and hemoglobin at environmentally relevant doses. Comparison of human subjects and F344 rats. Drug Metab Dispos. 1998;26:825–828. [PubMed] [Google Scholar]
- 48.Turteltaub KW, Dingley KH, Curtis KD, Malfatti MA, Turesky RJ, Garner RC, Felton JS, Lang NP. Macromolecular adduct formation and metabolism of heterocyclic amines in humans and rodents at low doses. Cancer Lett. 1999;143:149–155. doi: 10.1016/s0304-3835(99)00116-0. [DOI] [PubMed] [Google Scholar]
- 49.Dingley KH, Curtis KD, Nowell S, Felton JS, Lang NP, Turteltaub KW. DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol Biomarkers Prev. 1999;8:507–512. [PubMed] [Google Scholar]
- 50.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 51.King RS, Teitel CH, Kadlubar FF. In vitro bioactivation of N-hydroxy-2-amino-alpha-carboline. Carcinogenesis. 2000;21:1347–1354. [PubMed] [Google Scholar]
- 52.Zijlstra WG, Buursma A, Meeuwsen-van der Roest WP. Absorption spectra of human fetal and adult oxyhemoglobin, de-oxyhemoglobin, carboxyhemoglobin, and methemoglobin. Clin Chem. 1991;37:1633–1638. [PubMed] [Google Scholar]
- 53.Pathak KV, Bellamri M, Wang Y, Langouët S, Turesky RJ. 2-Amino-9H-pyrido[2,3-b]indole (AalphaC) adducts and thiol oxidation of serum albumin as potential biomarkers of tobacco smoke. J Biol Chem. 2015;290:16304–16318. doi: 10.1074/jbc.M115.646539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng L, Dasari S, Tabb DL, Turesky RJ. Mapping serum albumin adducts of the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by data-dependent tandem mass spectrometry. Chem Res Toxicol. 2012;25:2179–2193. doi: 10.1021/tx300253j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shapiro A, Botha JD, Pastore A, Lesk AM. A method for multiple superposition of structures. Acta Crystallogr A. 1992;48(Pt 1):11–14. doi: 10.1107/s010876739100867x. [DOI] [PubMed] [Google Scholar]
- 57.Damm KL, Carlson HA. Gaussian-weighted RMSD superposition of proteins: a structural comparison for flexible proteins and predicted protein structures. Biophys J. 2006;90:4558–4573. doi: 10.1529/biophysj.105.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corbeil CR, Williams CI, Labute P. Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des. 2012;26:775–786. doi: 10.1007/s10822-012-9570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29:1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 60.Birchmeier W, Glatthaar BE, Winterhalter KH, Bradshaw RA. The thiol groups of human hemoglobin carrying heme only on the -chains. Eur J Biochem. 1972;28:533–537. doi: 10.1111/j.1432-1033.1972.tb01941.x. [DOI] [PubMed] [Google Scholar]
- 61.Kiese M. Methemoglobinemia: a Comprehensive Treatise: Causes, Consequences, and Correction of Increased Contents of Ferrihemoglobin in Blood. CRC Press; 1974. [Google Scholar]
- 62.Raijmakers R, Neerincx P, Mohammed S, Heck AJ. Cleavage specificities of the brother and sister proteases Lys-C and Lys-N. Chem Commun (Camb) 2010;46:8827–8829. doi: 10.1039/c0cc02523b. [DOI] [PubMed] [Google Scholar]
- 63.Breddam K, Meldal M. Substrate preferences of glutamic-acid-specific endopeptidases assessed by synthetic peptide substrates based on intramolecular fluorescence quenching. Eur J Biochem. 1992;206:103–107. doi: 10.1111/j.1432-1033.1992.tb16906.x. [DOI] [PubMed] [Google Scholar]
- 64.Graumann J, Scheltema RA, Zhang Y, Cox J, Mann M. A framework for intelligent data acquisition and real-time database searching for shotgun proteomics. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.013185. 013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salek M, Lehmann WD. Neutral loss of amino acid residues from protonated peptides in collision-induced dissociation generates N- or C-terminal sequence ladders. J Mass Spectrom. 2003;38:1143–1149. doi: 10.1002/jms.531. [DOI] [PubMed] [Google Scholar]
- 66.Hempe JM, Ory-Ascani J, Hsia D. Genetic variation in mouse beta globin cysteine content modifies glutathione metabolism: implications for the use of mouse models. Exp Biol Med (Maywood) 2007;232:437–444. [PubMed] [Google Scholar]
- 67.Mitruka BM, Rawnsley HM, Mitruka BM. Clinical biochemical and hematological reference values in normal experimental animals and normal humans. Masson Pub; New York: 1981. [Google Scholar]
- 68.Park SY, Yokoyama T, Shibayama N, Shiro Y, Tame JR. 1.25 A resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J Mol Biol. 2006;360:690–701. doi: 10.1016/j.jmb.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 69.Nakagawa A, Lui FE, Wassaf D, Yefidoff-Freedman R, Casalena D, Palmer MA, Meadows J, Mozzarelli A, Ronda L, Abdulmalik O, Bloch KD, Safo MK, Zapol WM. Identification of a small molecule that increases hemoglobin oxygen affinity and reduces SS erythrocyte sickling. ACS Chem Biol. 2014;9:2318–2325. doi: 10.1021/cb500230b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Safo MK, Abraham DJ. The enigma of the liganded hemoglobin end state: a novel quaternary structure of human carbonmonoxy hemoglobin. Biochemistry. 2005;44:8347–8359. doi: 10.1021/bi050412q. [DOI] [PubMed] [Google Scholar]
- 71.Paoli M, Liddington R, Tame J, Wilkinson A, Dodson G. Crystal structure of T state haemoglobin with oxygen bound at all four haems. J Mol Biol. 1996;256:775–792. doi: 10.1006/jmbi.1996.0124. [DOI] [PubMed] [Google Scholar]
- 72.Harris TK, Mildvan AS. High-precision measurement of hydrogen bond lengths in proteins by nuclear magnetic resonance methods. Proteins. 1999;35:275–282. doi: 10.1002/(sici)1097-0134(19990515)35:3<275::aid-prot1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 73.Castelao JE, Yuan JM, Skipper PL, Tannenbaum SR, Gago-Dominguez M, Crowder JS, Ross RK, Yu MC. Gender- and smoking-related bladder cancer risk. J Natl Cancer Inst. 2001;93:538–545. doi: 10.1093/jnci/93.7.538. [DOI] [PubMed] [Google Scholar]
- 74.Skipper PL, Tannenbaum SR, Ross RK, Yu MC. Nonsmoking-related arylamine exposure and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:503–507. [PubMed] [Google Scholar]
- 75.Peng L, Turesky RJ. Capturing labile sulfenamide and sulfinamide serum albumin adducts of carcinogenic arylamines by chemical oxidation. Anal Chem. 2013;85:1065–1072. doi: 10.1021/ac3028273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porter CJ, Bereman MS. Data-independent-acquisition mass spectrometry for identification of targeted-peptide site-specific modifications. Anal Bioanal Chem. 2015;407:6627–6635. doi: 10.1007/s00216-015-8819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Becker AR, Sternson LA. Oxidation of phenylhydroxylamine in aqueous solution: a model for study of the carcinogenic effect of primary aromatic amines. Proc Natl Acad Sci U S A. 1981;78:2003–2007. doi: 10.1073/pnas.78.4.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hiramoto K, Negishi K, Namba T, Katsu T, Hayatsu H. Superoxide dismutase-mediated reversible conversion of 3-hydroxyamino-1-methyl-5H-pyrido[4,3-b]indole, the N-hydroxy derivative of Trp-P-2, into its nitroso derivative. Carcinogenesis. 1988;9:2003–2008. doi: 10.1093/carcin/9.11.2003. [DOI] [PubMed] [Google Scholar]
- 79.Sabbioni G. Hemoglobin binding of aromatic amines: molecular dosimetry and quantitative structure-activity relationships for N-oxidation. Environ Health Perspect. 1993;99:213–216. doi: 10.1289/ehp.9399213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki J, Meguro S, Morita O, Hirayama S, Suzuki S. Comparison of in vivo binding of aromatic nitro and amino compounds to rat hemoglobin. Biochem Pharmacol. 1989;38:3511–3519. doi: 10.1016/0006-2952(89)90122-6. [DOI] [PubMed] [Google Scholar]
- 81.Benesch RE, Benesch R. The influence of oxygenation on the reactivity of the--SH groups of hemoglobin. Biochemistry. 1962;1:735–738. doi: 10.1021/bi00911a002. [DOI] [PubMed] [Google Scholar]
- 82.Guidotti G, Konigsberg W. The Characterization of Modified Human Hemoglobin. I Reaction with Iodoacetamide and N-Ethylmaleimide. J Biol Chem. 1964;239:1474–1484. [PubMed] [Google Scholar]
- 83.Knee KM, Roden CK, Flory MR, Mukerji I. The role of beta93 Cys in the inhibition of Hb S fiber formation. Biophys Chem. 2007;127:181–193. doi: 10.1016/j.bpc.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bucci E, Fronticelli C. A New Method for the Preparation of Alpha and Beta Subunits of Human Hemoglobin. J Biol Chem. 1965;240:PC551–552. [PubMed] [Google Scholar]
- 85.Basile A, Ferranti P, Mamone G, Manco I, Pocsfalvi G, Malorni A, Acampora A, Sannolo N. Structural analysis of styrene oxide/haemoglobin adducts by mass spectrometry: identification of suitable biomarkers for human exposure evaluation. Rapid Commun Mass Spectrom. 2002;16:871–878. doi: 10.1002/rcm.655. [DOI] [PubMed] [Google Scholar]
- 86.Ferranti P, Malorni A, Mamone G, Sannolo N, Marino G. Characterisation of S-nitrosohaemoglobin by mass spectrometry. FEBS Lett. 1997;400:19–24. doi: 10.1016/s0014-5793(96)01258-6. [DOI] [PubMed] [Google Scholar]
- 87.Jeppsson MC, Mortstedt H, Ferrari G, Jonsson BA, Lindh CH. Identification of covalent binding sites of ethyl 2-cyanoacrylate, methyl methacrylate and 2-hydroxyethyl methacrylate in human hemoglobin using LC/MS/MS techniques. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2474–2482. doi: 10.1016/j.jchromb.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 88.Sabbioni G. Hemoglobin binding of monocyclic aromatic amines: molecular dosimetry and quantitative structure activity relationships for the N-oxidation. Chem Biol Interact. 1992;81:91–117. doi: 10.1016/0009-2797(92)90029-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.