Abstract
Objectives
Copy number variants (CNVs) are duplications or deletions of genomic regions. Large CNVs are potentially pathogenic and over-represented in children with congenital heart disease (CHD). We sought to determine the frequency of large CNVs in children with isolated CHD and evaluate the relationship of these potentially pathogenic CNVs with transplant-free survival.
Methods
These cases are derived from a prospective cohort of non-syndromic CHD patients (n=422) ascertained prior to their first surgery. Healthy pediatric controls (n=500) were obtained from the electronic Medical Records and Genetic Epidemiology (eMERGE) Network and CNV frequency was contrasted for CHD cases and controls. CNVs were algorithmically determined, subsequently screened for >95% overlap between two methods, size (>300kb), quality score, overlap with a gene, and novelty (absent from databases of known, benign CNVs), and separately validated with quantitative-PCR. Survival likelihoods were calculated for cases using Cox proportional hazards modeling to evaluate the joint effect of CNV burden and known confounders on transplant-free survival.
Results
Children with nonsyndromic CHD had a higher burden of potentially pathogenic CNVs compared to pediatric controls (12.1% vs. 5.0%, P=0.00016). Presence of a CNV was associated with significantly decreased transplant-free survival after surgery (HR=3.42, 95% CI: 1.66-7.09, P=0.00090) with confounder adjustment.
Conclusions
We confirm that children with isolated CHD have a greater burden of rare/large CNVs. We report a novel finding that these CNVs are associated with an adjusted 2.55-fold increased risk of death or transplant. These data suggest that CNV burden is an important modifier of survival after surgery for CHD.
INTRODUCTION
Congenital heart disease (CHDs) represents the most common human birth defect, often requiring surgical intervention with cardiopulmonary bypass (CPB) or circulatory arrest soon after birth. Survival after surgery has improved, but long-term mortality remains considerable, particularly for more severe CHD including single-ventricle lesions1.
Genetic factors, particularly those that are rare and alter proteins, are hypothesized to be major contributors to human disease2. Copy number variants (CNVs) are duplicated or deleted regions of the genome, and have been reported as potential causes of sporadic CHDs3,4. CNVs have been reported to be more frequent in children with CHD as compared to controls5. In addition, large CNVs greater than 300 kilobases (kb) in size and overlapping a gene have been reported to be more frequent in CHD cases and associated with poorer growth and cognitive outcomes6. Notably, studies reporting the prevalence of CNVs in children with CHD have included syndromic patients (e.g., those with DiGeorge 22q11.2 microdeletions).
We previously have used data from this cohort of children with isolated CHD to demonstrate the strong protective effects of VEGFA and SOD2 genetic variants on transplant-free survival7. Given the protein disrupting potential of these large, gene-overlapping CNVs, we hypothesize that such CNVs are also likely to affect survival. Thus, in this study we sought to determine the frequency of these potentially pathogenic CNVs among children with CHD compared to healthy pediatric controls and whether these large CNVs affect transplant-free survival in the first three years of follow-up after surgical correction of CHD.
METHODS
Ethics Statement
Subjects were enrolled at the Children's Hospital of Philadelphia (CHOP) on a protocol approved by the Institutional Review Boards of CHOP and the University of Washington from 10/1998 – 04/2003. Informed, written consent was obtained from parents or guardians of all the subjects.
CHD Patient Population
This is an analysis of a previously described prospective cohort of 550 participants enrolled in a prospective study at the Children's Hospital of Philadelphia (CHOP) to study neurodevelopmental dysfunction after surgical correction for CHD (hereafter referred to as the “CHD cases”)8-10. Patients 6 months of age or younger who underwent CPB with or without deep hypothermic circulatory arrest (DHCA) for repair of CHD were eligible for enrollment. Exclusion criteria included (1) multiple distinct congenital anomalies, (2) a recognizable genetic or phenotypic syndrome, and (3) a language other than English spoken in the home. However, recognition of dysmorphic features can be difficult in neonates and genetic syndromes were identified in some patients at subsequent evaluation and removed from the dataset prior to analyses. This study examined a subset of the cohort with genetic data (n=422) to establish the prevalence of large, gene-overlapping CNVs as compared to healthy pediatric controls, and determine whether these potentially pathogenic CNVs were associated with differential transplant-free survival. We note that no genome-wide association analyses have been attempted with this CNV data; this is solely a study of the global burden of large, gene-overlapping CNVs and how they affect transplant-free survival in the first three years after surgical correction of CHD.
Of the original 550 CHD cases, 73 were excluded due to a lack of high quality genotype data, leaving a total of 477 participants for analysis. An additional 55 participants were excluded due to presence of DiGeorge syndrome or other chromosomal/genetic abnormalities, which would be expected to bias both the estimation of CNV prevalence and the effects of CNVs on survival for CHD, as patients with genetic syndromes generally have worse survival11. There were 422 patients considered after these exclusions. Information on data collection (including further information on inclusion/exclusion criteria) operative management, and genotyping have been previously reported in detail7.
Control Population
Healthy controls from the same site (CHOP) for comparison of CNV prevalence were obtained from the Electronic Medical Records and Genetic Epidemiology (eMERGE) consortium (hereafter referred to as “Controls”)12. In total, a total of 500 healthy controls without CHD or other conditions associated with increased CNV prevalence (autism and schizophrenia) were analyzed for determination of large, gene-overlapping CNV presence.
Genetic Evaluation to Exclude Syndromic CHD Subjects
CHD participants were evaluated by a genetic dysmorphologist at the 1-year and/or 4-year examinations. Patients were classified as either: having no indication of genetic syndrome or chromosomal abnormality (normal, isolated CHD), suspected genetic syndrome (suspect), or a definite genetic syndrome or chromosomal abnormality (genetic). Following this classification, each CHD patient's genetics records were individually reviewed by a second senior board-certified medical geneticist, blinded to the genetic data, to determine whether subjects were to be included or excluded from the current analysis, which focuses on non-syndromic subjects. Due to this review, 55 CHD participants with known or suspected genetic syndromes were excluded from analysis due to the potential for genetic confounding effects on CNV prevalence and their effects on transplant-free survival within the first 3 years of follow-up after surgery.
Genotyping
Whole blood or buccal swab samples were obtained before surgery and were stored at 4°C for CHD case subjects. Similarly, control samples were obtained at study enrollment (as there was no surgery in this cohort) and stored at 4°C. Genomic DNA was isolated from white blood cells and genotyping was performed at the Center for Applied Genomics of the Children's Hospital of Philadelphia.
Copy Number Variation Determination and Validation
CNVs were determined algorithmically from Illumina HumanHap 550k BeadChip (for the CHD cases) and Illumina 610k-Quad BeadChip data (for the controls) using the programs PennCNV13 and GWASTools14. In brief, CNVs were considered potentially pathogenic after filtering for size (>300kb), PennCNV quality score >100, overlap with genes, and novelty, as previously reported by Carey et al6. Novelty was determined by comparing the called CNVs to the Database of Genomic Variants (http://projects.tcag.ca/variation/). CNVs were filtered from the data if greater than 50% of the CNV overlapped with another non-pathogenic CNV already catalogued in the Database of Genomic Variants. If CNVs were noted to be pathogenic and present in the Database of Genomic Variants, they were not filtered from the data. To prevent false positive CNV calls from algorithmic methods, we filtered all CNVs for a minimum of 95% overlap in calls from the two programs, PennCNV and GWASTools. The above methods were performed separately for the CHD cases and controls. A full list of the CNVs algorithmically determined in both the CHD cases and controls is presented in Supplemental Table S1.
CNVs were validated at the Center for Applied Genomics at CHOP using quantitative polymerase chain reaction (qPCR). Of the 38 tested CNVs from the CHD cases for whom we had DNA available, 2 samples’ CNVs failed to validate (i.e. showed the reference copy number), while 1 sample failed to undergo DNA amplification (likely due to poor DNA quality) and therefore could not be tested for CNV. Excluding the DNA-quality related qPCR failure, 35/37 predicted CNVs were successfully validated for a validation rate of 94.6% (see Supplemental Table S1). Primer information for qPCR on the 38 tested CNVs is separately available as a Microsoft Excel Worksheet in Supplemental Table S2.
Statistical Analyses
All analyses and graphics were performed in R (http://www.r-project.org/) using standard regression packages. A chi-square test was used to test the significance of the difference in frequency of large, gene-overlapping CNVs between the CHD cases and controls.
Genetic ancestry was determined using previously described methods15. Due to the mixed ancestry of the cohort, the first three principal component eigenvectors were used as covariates in Cox proportional hazards regression models to adjust for potential population stratification16.
Time to long-term mortality of cases was calculated from the date of initial surgery to the date of death. A Cox proportional hazards model was used to evaluate the joint effect of the global burden of potentially pathogenic CNVs and covariates affecting survival. Output from Cox proportional hazards model was used for plotting of survival curves. Survival analyses were adjusted for the previously reported confounding variables: the first three principal component eigenvectors for race, gestational age, birth weight, diagnostic class (coded as a dummy variable with diagnostic class 1 as the reference group)17, total surgical support time (total minutes on either cardiopulmonary bypass or DHCA), and extracorporeal membrane oxygenation (ECMO) use. Diagnosis class was assigned based on preoperative diagnosis according to a previously proposed scheme17: class I, two-ventricle heart without arch obstruction; class II, two-ventricle heart with arch obstruction; class III single-ventricle heart without arch obstruction; and class IV, single-ventricle heart with arch obstruction. Confidence intervals and two-sided p-values were calculated using an asymptotic normal distribution of the estimated hazard ratio (HR) using Wald statistics.
RESULTS
Baseline characteristics of the studied subset of the CHD cases stratified by presence or absence of a potentially pathogenic CNV are presented in Table 1. CHD cases with a potentially pathogenic CNV had a significantly higher proportion of males (73% vs. 56%, P=0.040) and nominally lower gestational age (38.0 vs. 38.5 weeks, P=0.062). Birth weight was not significantly different (3.03 kg in CHD cases with a qualifying CNV vs. 3.16 kg in those without, P=0.13). No significant differences were seen between for distribution of race/ethnicity (where European followed by African ancestry individuals composed the majority of the cases), diagnostic class, preoperative length of stay (LOS), incidence of preoperative intubation, total surgical support time, incidence of delayed sternal closure, ECMO use, and postoperative LOS.
Table 1.
Baseline characteristics of the CHD cases, stratified by presence or absence of potentially pathogenic CNV.
| No CNV (n=371) | CNV (n=51) | Combined (n=422) | P-Value | |
|---|---|---|---|---|
| Sex, male (%) | 208 (56%) | 37 (73%) | 245 (58%) | P = 0.040a |
| Ethnicity | P = 0.49a | |||
| Caucasian | 240 (65%) | 33 (65%) | 273 (65%) | |
| African | 85 (23%) | 14 (27%) | 99 (23%) | |
| Hispanic | 18 (5%) | 1 (2%) | 19 (5%) | |
| Asian | 12 (3%) | 0 (0%) | 12 (3%) | |
| Native American | 7 (2%) | 2 (4%) | 9 (2%) | |
| Other | 9 (2%) | 1 (2%) | 10 (2%) | |
| Gestational age, weeks | 38.5 ± 2.0 | 38.0 ± 2.1 | 38.5 ± 2.1 | P = 0.062b |
| Birth weight, kg | 3.16 ± 0.60 | 3.03 ± 0.72 | 3.15 ± 0.62 | P = 0.13b |
| Diagnostic class (%) | P = 0.88a | |||
| Class 1 | 179 (48%) | 25 (49%) | 204 (48%) | |
| Class 2 | 36 (10%) | 5 (10%) | 41 (10%) | |
| Class 3 | 39 (11%) | 7 (14%) | 46 (11%) | |
| Class 4 | 117 (32%) | 14 (27%) | 131 (31%) | |
| Preoperative LOS, days | 2.2 ± 2.6 | 2.1 ± 2.7 | 2.1 ± 2.6 | P = 0.57b |
| Intubation (%) | 104 (28%) | 15 (29%) | 119 (28%) | P = 0.97a |
| Total surgical support time, min | 30 ± 50 | 20 ± 35 | 29 ± 48 | P = 0.22b |
| Delayed sternal closure (%) | 56 (15%) | 5 (10%) | 61 (14%) | P = 0.48a |
| ECMO use (%) | 22 (6%) | 1 (2%) | 23 (5%) | P = 0.40a |
| Postoperative LOS, days | 16 ± 23 | 18 ± 29 | 16 ± 24 | P = 0.62b |
| Mortality (%) | 34 (9%) | 13 (25%) | 47 (11%) | - |
Abbreviations used: CHD = congenital heart defect; CNV = potentially pathogenic copy number variant; ECMO = extracorporeal membrane oxygenation; HLHS = hypoplastic left heart syndrome; LOS = length of stay; TGA = transposition of the great arteries; VSD = ventricular septal defect.
Tests used:
Chi-square test
Wilcoxon rank sum test
Mean ± Standard Deviation are presented for continuous variables
Fifty-one of the 422 CHD cases considered (12.1%) had CNVs that were >300kb in size, overlapped with a gene, and were novel (potentially pathogenic). In comparison, 25 such potentially pathogenic CNVs were identified in 500 (5.0%) of the healthy control cohort (see Supplemental Table S1 for a list of identified CNVs). A chi-square test found a significant difference in the proportion of participants with potentially pathogenic CNVs among those with isolated CHD vs. controls (12.1% vs. 5.0%, P=0.00016).
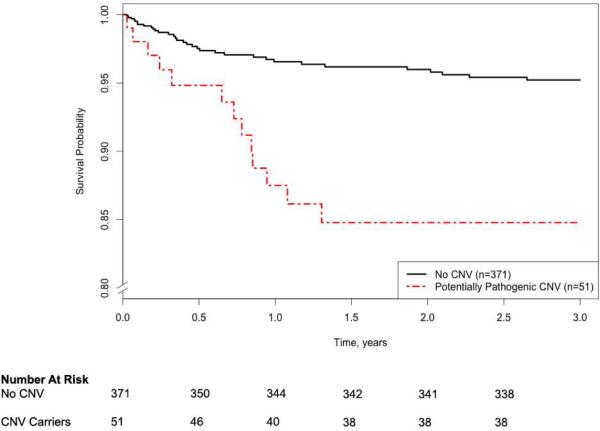
To determine whether the presence of a large, gene-overlapping, novel CNV was associated with transplant-free survival, a Cox proportional hazards model was applied. Survival analyses demonstrated a strong increased risk of death is associated with potentially pathogenic CNVs in a model jointly adjusting for the covariates: the first three principal component eigenvectors for race, gestational age, birth weight, sex, diagnostic class, total surgical support time, and ECMO use (HR = 3.43, 95% CI 1.66-7.09, P=0.0009, see Table 2 and Figure 1). From Cox proportional hazard model r2 comparison, the effect of CNV burden on transplant-free survival is estimated to be 2.1%. Sensitivity analyses of the effect of covariate-adjusted CNV burden on transplant-free survival across diagnostic classes are presented in Supplemental Table S3.
Table 2.
Association of potentially pathogenic CNV on transplant-free survival in the CHD cases, adjusting for confounders with cox proportional hazards regression (n=422 with 47 deaths or heart transplants observed).
| Covariate | HR (95% CI) | P-Value |
|---|---|---|
| Gestational Age | 0.90 (0.80-1.02) | 0.93 |
| Birth weight | 0.99 (0.99-1.02) | 0.19 |
| Male gender | 0.84 (0.44-1.58) | 0.64 |
| Diagnostic class 2 | 1.19 (0.25-5.71) | 0.82 |
| Diagnostic class 3 | 2.77 (0.91-8.41) | 0.073 |
| Diagnostic class 4 | 9.70 (3.89-24.22) | 1.13×10−6 |
| Total surgical support time | 1.01 (1.00-1.02) | 0.037 |
| ECMO use | 14.44 (6.73-31.00) | 7.42×10−12 |
| Genetic ancestry PC1 | - | 0.40 |
| Genetic ancestry PC2 | - | 0.24 |
| Genetic ancestry PC3 | - | 0.0086 |
| Potentially Pathogenic CNV* | 3.43 (1.66-7.09) | 0.0009 |
Abbreviations: CNV = copy number variant; ECMO = extracorporeal membrane oxygenation; HR = hazard ratio; PC = principal component eigenvector (for genetic ancestry adjustment).
Presence of a potentially pathogenic CNV is associated with a 2.1% increased risk of transplant or death.
Figure 1. Covariate-adjusted long-term survival by presence of potentially pathogenic CNV in CHD cases (n=422, with 47 observed events).
Please note the discontinuous y-axis, which begins at an adjusted survival probability of 0.8. The 95% confidence intervals (CIs) for covariate-adjusted survival probability for the no CNV group are 0.960-0.990, 0.947-0.985, 0.941-0.982, 0.939-0.981, and 0.931-0.978 at 0.5, 1, 1.5, 2, and 2.5 years of follow-up, respectively. The 95% CIs for covariate-adjusted survival probability for the CNV group are 0.901-0.997, 0.798-0.959, 0.762-0.943, 0.762-0.943, and 0.762-0.943 at 0.5, 1, 1.5, 2, and 2.5 years of follow-up, respectively.
DISCUSSION
We confirm prior published reports5,6 that large, gene-overlapping, novel CNVs are present at higher frequency in children with isolated, non-syndromic CHD as compared to controls (12.1% vs. 5.0%). CNVs present in a catalogue of non-pathogenic CNVs were excluded, as they were less likely to contribute to survival given their known benign status. We note that we compute CNV frequency specifically for participants with isolated CHD not attributed to any known genetic or chromosomal anomaly that may bias our results. In addition, we present the first finding that large, gene-overlapping, novel CNVs are associated with an estimated 3.43-fold increased risk of death as compared to patients without such CNVs (p=0.00009 and 95% CI 1.66-7.09) Overall, this novel association of large, gene-overlapping CNVs and survival further emphasizes the importance of genetic factors in explaining complex phenotype variation and outcomes.
Copy number variants (CNVs), which can result in the duplication or deletion of entire genes (i.e., a change in the “copy number” of a particular gene), are one of several types of mutations that are currently thought to account for the “missing heritability” of complex genetic traits not yet adequately explained by common genetic variants studied in genome-wide association scans2. In children, CNVs have previously been implicated in the pathogenesis of numerous diseases, such as schizophrenia18 and autism19. Moreover, CNVs have also been reported as potential causes of specific CHD diagnoses3,4. In this work we expand upon prior reports and present evidence that global burden of large, gene-overlapping CNVs is likely pathogenic, with a significantly increased risk of heart transplant or death after surgery in CNV carriers. Notably, analyses in adult cohorts have similarly found that burden of large CNVs is associated with mortality20.
Some limitations of this study should be considered. First, statistical power was limited owing to the size of the CHD case study and lack of comparable cohorts. We addressed the rarity of each individual CNV by focusing on the global burden of these large, gene-overlapping, and novel CNVs. However, due to this pooling approach to analysis, we were unable to determine whether CNVs in a given region were more responsible for the effect on survival as compared to others. In addition, due to this analytic method we cannot specifically identify the reason for the impact of CNVs on survival. As noted in Supplemental Table S1, multiple chromosomal regions are affected and it is unlikely that a singular, common pathway is acting to affect survival in these patients. Finally, because we lack genotype data on parents of the affected CHD participants we are unable to infer whether these rare CNV events are de novo or inherited. De novo variants in affected children of unaffected parents are considered more likely to be pathogenic.
In conclusion, the results confirm that large, gene-overlapping, novel CNVs are enriched in children with isolated CHD as compared to healthy children, and a provide new evidence that these CNVs are associated with poorer survival. Further follow-up of the pathogenic effects of these potentially pathogenic CNVs in a similar prospective cohort lacking survivor bias is imperative. Given the approximate 3.5-fold enrichment of these pathogenic CNVs in children with isolated CHD, validation of these results could lead to novel preventative and risk assessment strategies to decrease the morbidity and mortality of CHD.
Supplementary Material
Acknowledgements
We would like to thank all the children and families for their participation. Genotyping was performed by the Center for Applied Genomics at the Children's Hospital of Philadelphia.
Sources of Funding: This work was supported by a grant from the Fannie E. Rippel Foundation, an American Heart Association National Grant-in-Aid (9950480N), NIH HL071834, and a Washington State Life Sciences Discovery Award to the Northwest Institute for Genetic Medicine. The CHOP site of the eMERGE network was supported by U01HG006830. DSK was supported by NIH 1F31MH101905-01 and T32HL007312. JHK was supported by NCRR Grant KL2 TR000421.
Abbreviations
- CHD
congenital heart disease
- CHOP
Children's Hospital of Philadelphia
- CNV
copy number variant
- CPB
cardiopulmonary bypass
- DHCA
deep hypothermic circulatory arrest
- ECMO
extracorporeal membrane oxygenation
- eMERGE
electronic Medical Records and Genetic Epidemiology
- LOS
length of stay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented at the 95th Annual Meeting of the American Association for Thoracic Surgery in Seattle, WA.
Conflicts of Interest: The authors declare no conflicts of interest.
REFERENCES
- 1.Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, et al. Hypoplastic left heart syndrome: current considerations and expectations. Journal of the American College of Cardiology. (3) 2012 Jan;59(1 Suppl):S1–42. doi: 10.1016/j.jacc.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010 Jun;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hitz M-P, Lemieux-Perreault L-P, Marshall C, Feroz-Zada Y, Davies R, Yang SW, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012 Sep;8(9):e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soemedi R, Wilson IJ, Bentham J, Darlay R, Töpf A, Zelenika D, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012 Sep 7;91(3):489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, et al. Increased Frequency of De Novo Copy Number Variants in Congenital Heart Disease by Integrative Analysis of Single Nucleotide Polymorphism Array and Exome Sequence Data. Circulation Research. 2014;115(10):884–96. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey AS, Liang L, Edwards J, Brandt T, Mei H, Sharp AJ, et al. Effect of Copy Number Variants on Outcomes for Infants With Single Ventricle Heart Defects. Circulation: Cardiovascular Genetics. 2013 Oct 15;6(5):444–51. doi: 10.1161/CIRCGENETICS.113.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DS, Kim JH, Burt AA, Crosslin DR, Burnham N, McDonald-McGinn DM, et al. Patient genotypes impact survival after surgery for isolated congenital heart disease. The Annals of Thoracic Surgery. 2014 Jul;98(1):104–10–discussion110–1. doi: 10.1016/j.athoracsur.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaynor JW, Gerdes M, Zackai EH, Bernbaum J, Wernovsky G, Clancy RR, et al. Apolipoprotein E genotype and neurodevelopmental sequelae of infant cardiac surgery. The Journal of Thoracic and Cardiovascular Surgery. 2003 Dec;126(6):1736–45. doi: 10.1016/s0022-5223(03)01188-7. [DOI] [PubMed] [Google Scholar]
- 9.Gaynor JW, Nord AS, Wernovsky G, Bernbaum J, Solot CB, Burnham N, et al. Apolipoprotein E genotype modifies the risk of behavior problems after infant cardiac surgery. PEDIATRICS. 2009 Jul;124(1):241–50. doi: 10.1542/peds.2008-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DS, Stanaway IB, Rajagopalan R, Bernbaum JC, Solot CB, Burnham N, et al. Results of genome-wide analyses on neurodevelopmental phenotypes at four-year follow-up following cardiac surgery in infancy. PLoS ONE. 2012;7(9):e45936. doi: 10.1371/journal.pone.0045936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyburz A, Bauersfeld U, Schinzel A, Riegel M, Hug M, Tomaske M, et al. The Fate of Children with Microdeletion 22q11.2 Syndrome and Congenital Heart Defect: Clinical Course and Cardiac Outcome. Pediatr Cardiol. 2007 Sep 29;29(1):76–83. doi: 10.1007/s00246-007-9074-2. [DOI] [PubMed] [Google Scholar]
- 12.Sleiman P, Bradfield J, Mentch F, Almoguera B, Connolly J, Hakonarson H. Assessing the functional consequence of loss of function variants using electronic medical record and large-scale genomics consortium efforts. Front Genet. 2014;5:105. doi: 10.3389/fgene.2014.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research. 2007 Nov 1;17(11):1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012 Dec 7;28(24):3329–31. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009 Jan;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Clancy RR, McGaurn SA, Wernovsky G, Spray TL, Norwood WI, Jacobs ML, et al. Preoperative risk-of-death prediction model in heart surgery with deep hypothermic circulatory arrest in the neonate. The Journal of Thoracic and Cardiovascular Surgery. 2000 Feb;119(2):347–57. doi: 10.1016/S0022-5223(00)70191-7. [DOI] [PubMed] [Google Scholar]
- 18.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008 Apr 25;320(5875):539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 19.Girirajan S, Johnson RL, Tassone F, Balciuniene J, Katiyar N, Fox K, et al. Global increases in both common and rare copy number load associated with autism. Human Molecular Genetics. 2013 Jul 15;22(14):2870–80. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuningas M, Estrada K, Hsu Y-H, Nandakumar K, Uitterlinden AG, Lunetta KL, et al. Human Molecular Genetics. 21. Vol. 20. Oxford University Press; Nov 1, 2011. Large common deletions associate with mortality at old age. pp. 4290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.