Abstract
T lymphocytes are essential contributors to the adaptive immune system and consist of multiple lineages that serve various effector and regulatory roles. As such, precise control of gene expression is essential to the proper development and function of these cells. Previously, we identified Snai2 and Snai3 as being essential regulators of immune tolerance partly due to the impaired function of CD4+ regulatory T cells in Snai2/3 conditional double knockout mice. Here we extend those previous findings using a bone marrow transplantation model to provide an environmentally unbiased view of the molecular changes imparted onto various T lymphocyte populations once Snai2 and Snai3 are deleted. The data presented here demonstrate that Snai2 and Snai3 transcriptionally regulate the cellular fitness and functionality of not only CD4+ regulatory T cells but effector CD8α+ and CD4+ conventional T cells as well. This is achieved through the modulation of gene sets unique to each cell type and includes transcriptional targets relevant to the survival and function of each T cell lineage. As such, Snai2 and Snai3 are essential regulators of T cell immunobiology.
Keywords: Snail transcription factors, autoimmunity, genetic deficiency, T lymphocyte, T cell receptor
1. Introduction
Cellular immunity is regulated through the interactions of multiple cell types that constitute the innate and adaptive arms of the immune system. Within the adaptive immune system, T lymphocytes play a key role not only as immune effectors but also as immune regulators. To this end, the T lymphocyte “family” consists of multiple lineages. Such examples include CD8α+, CD4+ CD25− conventional (TConv) and CD4+ CD25+ regulatory (TReg) cells. Within the thymus, various transcriptional regulators precisely control the decisions regarding lineage fate. Following the CD4+ CD8α+ double positive T cell stage, Runx3 and ThPOK drive CD8α+ and CD4+ T cell differentiation, respectively, while also antagonizing the alternative differentiation program[1, 2]. Once the CD4+ fate choice has been made, selective expression of Foxp3 “turns on” the gene program required for TReg specification[3]. While these transcriptional regulators are key to the development and function of the above mentioned cell types, the totality of transcriptional regulatory networks involved is much more complicated. This is perhaps best illustrated in the TReg lineage which modifies nuanced regulatory responses through the use of an evolving list of transcriptional regulatory mechanisms[4]. Not surprisingly, multiple layers of redundancy have developed to ensure the proper expression of these gene programs[5, 6]. One such example includes the Id family of transcriptional regulators. These proteins negatively regulate the function of E-box DNA-binding factors such E2A and Snai1[7, 8]. Upon deletion of Id2 or Id3, the other family member is able to functionally compensate and ensure the ability of TRegs to suppress T helper (TH2)-mediated inflammation. This specific TReg function is subsequently lost upon the deletion of both Id2 and Id3[5].
The Snail family consists of 3 evolutionarily conserved transcriptional regulators: Snai1 (Snail), Snai2 (Slug) and Snai3 (Smuc)[9]. These proteins share the same basic structure inclusive of an N-terminal SNAG (Snail/Gfi-1) domain and multiple C2H2-type zinc finger DNA-binding domains (DBDs) within the C-terminus[10]. Using their DBDs, Snail factors recognize consensus E-box DNA elements (CANNTG) with a preference for G/C-rich central dinucleotides[11]. Once bound to target genes, Snail family members augment transcription through the recruitment of various chromatin modifiers via the SNAG domain[12, 13]. Although classically known as transcriptional repressors, a growing body of data supports the ability of Snail proteins to positively regulate their targets upon interaction with other transcription factors[14, 15].
The founding member, Snai1, was discovered in Drosophila melanogaster where it was first characterized as a key factor involved in embryonic patterning[16, 17]. Since then, Snail family members have been most well characterized in the areas of embryonic and developmental biology[18-20]. However, this family also plays a number of roles in the development and function of the immune system (summarized in [21]). Recently, we demonstrated a previously unappreciated functional redundancy for Snai2 and Snai3 in lymphoid development[22]. Furthermore, the conditional deletion of Snai2 and Snai3 (cDKO) resulted in fatal autoimmunity that could be corrected by the transplantation of wildtype (WT) TRegs [23]. While high levels of autoantibodies characterized this disease, these animals lacked the T cell proliferation commonly associated with many autoimmune diseases[24, 25]. This led us to hypothesize that the deletion of Snai2 and Snai3 not only affected TRegs but also diminished the fitness of CD8α+ and TConv cells as well. Using competitive reconstitution, we demonstrated that cDKO CD8α+, TConv and TReg cells were compromised in their ability to compete with their WT counterparts. Additionally, a reduced amount of cDKO T cells was able to enter the activated, effector/memory-like pool. RNA sequencing (RNA-seq) analysis showed that Snai2 and Snai3 regulated genes essential for the cellular fitness and function of all 3 lineages. Importantly, Snai2 and Snai3 accomplished this via modulation of transcriptional targets almost completely exclusive to each individual cell type. Thus, Snai2 and Snai3 are key transcriptional regulators of T cell biology.
2. Materials and Methods
2.1 Animal strains and care
Animals were housed in the Animal Resource Center (University of Utah Health Science Center, Salt Lake City, UT) according to the guidelines of the National Institute of Health for the care and use of laboratory animals. All animal protocols were reviewed and approved by the University of Utah Institutional Animal Use and Care Committee. Vav-Cre (Stock #: 008610), Rag2−/− (Stock #: 008449) and UBC-GFP (Stock #: 004353) mice were purchased from The Jackson Laboratory and bred in house. Snai2/Snai3 conditional double knockout (cDKO) mice were derived from Snai2+/− Snai3Fl/Fl Vav-Cre+/− breeding pairs. Snai3Fl/Fl have been made available from the Jackson Laboratory (Stock #: 027276). Animal numbers used per experiment are noted in the figure legends.
2.2 DNA isolation and genomic DNA PCR
Approximately 5 mm portions of tail were boiled in 50 mM NaOH until fully dissolved. 1 M Tris was added to neutralize the NaOH. Following centrifugation to remove insoluble material, DNA was precipitated from supernatants following standard ethanol precipitation guidelines. Snai2, Snai3 and Vav-Cre genotyping was performed with Thermo Scientific Taq DNA Polymerase (Cat. #: FEREP0402) using 2 μL of DNA per reaction. Products were electrophoresed in 2% agarose gels. Cycling parameters are available upon request. Primer sequences are provided in Supplementary Table 1.
2.3 RNA isolation and RNA sequencing (RNA-seq)
Total RNA was isolated from cells using the Qiagen miRNeasy Micro Kit (Cat. #: 217084) according to the manufacturer’s instructions. Isolated RNA was utilized for RNA-seq library preparation using the Illumina TruSeq Stranded Total RNA Sample Preparation Kit with Ribo-Zero Gold treatment to eliminate ribosomal RNAs. Libraries were subjected to HiSeq2000 50 Cycle Single Read Sequencing. Greater than 2.5 × 107 reads per sample (quality score, Q ≥ 20) were obtained and aligned to the mm10 (Ensembl build 75) transcriptome index using Novoalign. Aligned reads were further processed for splicing and expression variance using the Useq 8.7.4 software package. The data has been submitted to the NCBI GEO database (GSE74467). 4 replicates were performed for wildtype (WT) and cDKO CD8α+ and CD4+ CD25− conventional (TConv) T cells. For CD4+ CD25+ regulatory (TReg) T cells, 4 and 3 replicates were performed for WT and cDKO genotypes, respectively. For mathematical purposes, a value of 0.0001 was added to all gene fragments per kilobase per million mapped reads (FPKM) values as to avoid “zero” values. Mean fold changes for each gene were calculated by dividing mean WT by mean cDKO FPKM values for a given T cell lineage. Significantly altered genes for CD8α+, TConv and TReg cells are listed in Supplementary Tables 3-5. Analysis for all detectable CD8α+, TConv and TReg genes can be found in Supplementary Tables 6-8. Data tracks were visualized with the University of California-Santa Cruz (UCSC) genome browser. Venn diagrams for gene expression analysis were created using the online program Venny 2.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html)[26, 27]. Gene ontology analysis was performed using Gorilla online software. The Benjamini and Hochberg method was used to correct p-values for multiple testing (FDR q-value). Gene Set Enrichment Analysis (GSEA) was performed using software made publicly available by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp).
2.4 Fluorescence-activated cell sorting (FACS) analysis and isolation of T cell populations
Upon dissection, the plunger of a 5 mL syringe was used to dissociate thymus, spleen and mesenteric lymph node (mLN) tissues. Cells were strained through a 100 μm filter and collected in 10 mL of FACS buffer (1× PBS + 0.1% BSA). Peripheral blood was isolated from the retro-orbital (r.o.) sinus via heparin-lined capillary tubes and complete blood counts were obtained using a Hemavet 950 FS (Drew Scientific). Remaining contents were then collected in 5 mL of FACS buffer. After centrifugation, erythrocytes were lysed on ice for 10 minutes using ammonium-chloride-potassium (ACK) buffer. Following lysis, cells were respun, resuspended in FACS buffer and counted using a Hemoctyometer. Cells were stained on ice for 30 minutes using the appropriate antibody cocktail. Samples were washed with FACS buffer, centrifuged and resuspended in FACS buffer. To discriminate between live and dead cells, 4’6-diamidino-2-phenylindole (DAPI) was added at a final concentration of 3 μM. The antibodies utilized with their indicated dilutions are available in Supplementary Table 2. Population analysis was performed on the FACS Canto II (BD Biosciences) and results for a given cell type are graphically represented as mean values ± standard error measurement (SEM). Cell sorting of select populations was performed on the FacsAria Cell Sorter (BD Biosciences) at the University of Utah Flow Cytometry Core.
2.5 Bone marrow transplantation
One day prior to transplantation, 8-12 weeks old UBC-GFP recipient mice were lethally irradiated with 2 doses of 500 cGy split approximately 4 to 6 hours apart. WT and cDKO bone marrow was isolated from pooled tibias and femurs. Samples were lineage depleted using the mouse-specific Lineage Depletion Kit from Miltenyi (Cat. #: 130-090-858). Isolated progenitors were washed and resuspended in 1× PBS at a concentration of 7.5 × 105 cells/mL. A total of 7.5 × 104 cells (100 μL) were delivered r.o. to each recipient animal. Animals were maintained on Sulfamethoxazole and Trimethoprim antibiotic-treated water for 21 days post-transplant.
2.6 Statistical analysis
Using Prism GraphPad software, the unpaired Student’s t-Test was applied for all FACS-based analyses. For RNA-Seq analysis, two-tailed Student’s t-Tests using two-sample equal variance were performed using Microsoft Excel software[23]. Comparisons of up or downregulated gene percentages were performed with the Fisher’s Exact Test in Prism. Statistical cutoffs are described in the figure legends.
3. Results
3.1. Deletion of Snai2 and Snai3 impairs T cell competitive fitness
Through the combined deletion of Snai2 in the germline with the hematopoietic-specific deletion of Snai3, we previously identified Snai2 and Snai3 as redundant transcription factors necessary for the preservation of immunological tolerance[23]. Snai2/Snai3 conditional double knockout (cDKO) mice were reminiscent of Scurfy (i.e. Foxp3 functional mutant) mice in that they developed lethal autoimmunity, which could be attributed to the absence of wildtype (WT) regulatory (TReg) T cells[28]. However, unlike Scurfy mice, Snai2/Snai3 cDKO animals did not develop a T cell-driven lymphoproliferative disease[24, 29]. This led us to hypothesize that the deletion of Snai2 and Snai3 compromised the fidelity of not only TRegs but that of effector T cells as well. It has previously been shown that T cells, in particular TRegs, are highly radioresistant and provide a source of competition for WT or cDKO T cells generated from transplanted bone marrow progenitors[30]. Additionally, via a series of bone marrow transplantation experiments we have previously demonstrated that autoimmunity derived from cDKO progenitors is nullified in the presence of WT radioresistant TRegs[23]. As such, the use of a bone marrow transplantation system using WT hosts allows for the examination of WT and cDKO T cells on an even playing field as these cells: 1) would be generated from an equivalent stromal environment and 2) never exposed to a proinflammatory autoimmune milieu.
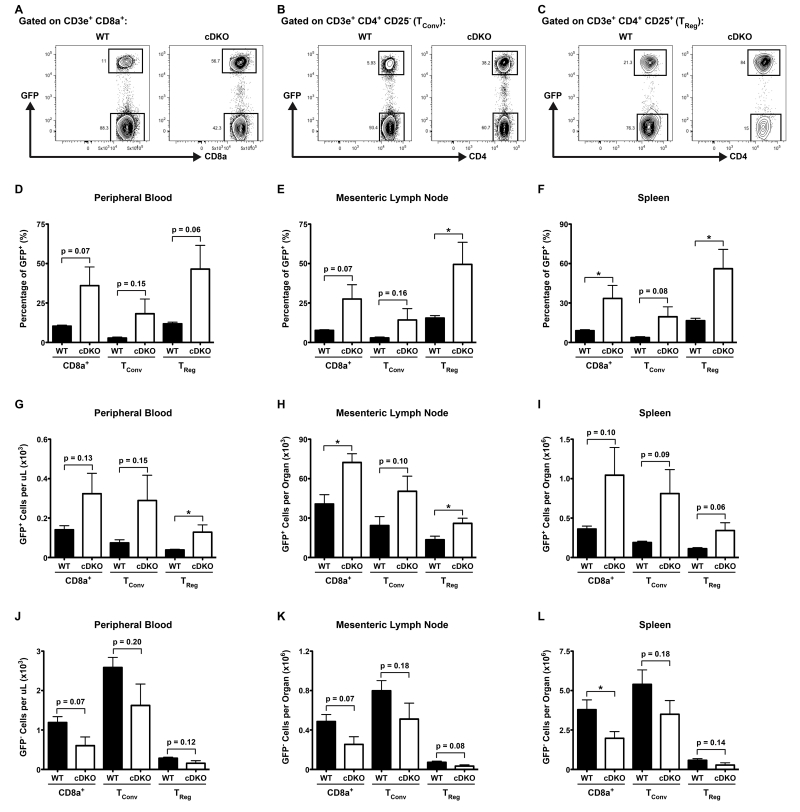
To test the overall fitness of various cDKO T cell lineages in comparison to their WT counterparts, we transplanted lineage-depleted bone marrow progenitors from either genotype into lethally irradiated mice expressing green fluorescent protein (GFP) under the control of the human ubiquitin C (UBC) promoter (UBC-GFP). In this system, donor-derived hematopoietic cells would be GFP− while all host-derived cells would be GFP+. 9 weeks later, we analyzed donor versus host chimerism within T cell lineages via fluorescence-activated cell sorting (FACS). Representative gating for splenic CD8α+, CD4+ conventional (TConv) and TReg cells is shown in Supplementary Figure 1. Within each lineage, donor-derived and host-derived radioresistant T cells were identifiable by the absence or presence of GFP, respectively (Figure 1A-C).
Figure 1.
Deletion of Snai2 and Snai3 impairs T cell competitive fitness. (A-C) Representative FACS plots from spleens of UBC-GFP mice nine weeks post-transplantation with either WT or cDKO GFP− bone marrow progenitors. GFP+ (host-derived) versus GFP− (donor-derived) cells are shown for (A) CD8α+, (B) TConv and (C) TReg cells. (D-F) Quantification of the percentage of GFP+ cells per T cell lineage within the (D) PBCs, (E) mLNs and (F) spleens from all animals analyzed. (G-I) Absolute numbers of GFP+ recipient cells T cells per lineage within the (G) PBCs, (H) mLNs and (I) spleens from all animals analyzed. (J-L) Absolute numbers of GFP− donor cells T cells per lineage within the (J) PBCs, (K) mLNs and (L) spleens from all animals analyzed. (D-L) Bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 4. Student’s t-Test: * p ≤ 0.05.
To minimize the potential caveat of trafficking differences among genotypes, we analyzed 3 tissues inclusive of the peripheral blood (PBC), mesenteric lymph node (mLN) and spleen of each animal. Overall, mice that were reconstituted with cDKO bone marrow had a higher ratio of GFP+ host-derived radioresistant T cells 9 weeks post-transplantation (Figure 1D-F). Analysis of the log2-transformed fold changes for percentages of GFP+ cells from cDKO and WT (i.e. cDKO / WT) animals indicated a uniform effect for all organs analyzed for a given T cell lineage (Table 1, compare “a” for Peripheral Blood, Mesenteric Lymph Node and Spleen within each individual lineage). Interestingly, CD8α+ and TReg lineages shared a similar gap between WT and cDKO animals while TConv lymphocytes displayed a significantly larger discrepancy between the two genotypes (Table 1, Combined log2 fold change for “a”: CD8α+ = 1.86 ± 0.03, TConv = 2.49 ± 0.12, TReg = 1.81 ± 0.09). This indicated a larger effect was imparted upon cDKO TConv cells following Snai2/Snai3 deletion. To further assess the origin of this difference we analyzed absolute numbers of GFP+ host-derived (Figure 1G-I) and GFP− donor-derived (Figure J-L) T cells. What is most readily apparent is that both expanded GFP+ and reduced GFP− compartments contributed to the altered GFP+ frequencies seen in the recipients of cDKO bone marrow (Figure 1, Table 1). Interestingly, the mLNs displayed the least expanded GFP+ compartment for all lineages analyzed (Table 1, compare “b” for Peripheral Blood, Mesenteric Lymph Node and Spleen within each individual lineage). This was independent of the loss of GFP− cDKO donor-derived T cells and may just reflect kinetic limitations of cellular expansion within the mLN. In regards to the expansion of GFP+ radio-resistant cells, the TConv and TReg compartments trended towards having the largest increases relative to CD8α+ T cells (Table 1, Combined log2 fold change for “b”: CD8α+ = 1.19 ± 0.20, TConv = 1.70 ± 0.33, TReg = 1.43 ± 0.25). In contrast, CD8α+ and T regulatory cells demonstrated a significantly greater loss of cDKO GFP- cells when compared to the TConv lineage (Table 1, Combined log2 fold change for “c”: CD8α+ = −0.95 ± 0.02, TConv = −0.64 ± 0.01, TReg = −1.00 ± 0.08). The relative consistency in which cDKO T cells were lost on an organ-to-organ basis within a particular lineage was more consistent with an all-encompassing cell intrinsic defect rather than an impairment driven by a particular extracellular niche.
Table 1.
Mean log2 fold change comparisons of the percentage of GFP+ host-deriveda, number of GFP+ host-derivedb and number of GFP− donor-derivedc CD8α+, TConv and TReg cells in animals receiving cDKO or WT bone marrow (i.e. cDKO / WT). Combined represents mean log2 fold change ± SEM for all 3 organs analyzed. Number of animals per group: N = 4 WT, N= 4 cDKO. Student’s t-Test: For “a” ** p ≤ 0.01 CD8α+ versus TConv, * p ≤ 0.05 TReg versus TConv. For “c” *** p ≤ 0.001 CD8α+ versus Tconv, * p ≤ 0.05 TReg versus Tconv.
| Organ | CD8α+ | TConv | TReg |
|---|---|---|---|
|
| |||
| Peripheral Blood | 1.80a | 2.72a | 1.98a |
| 1.20b | 1.96b | 1.75b | |
| −0.98c | −0.67c | −0.84c | |
|
| |||
| Mesenteric Lymph | 1.86a | 2.33a | 1.68a |
| Node | 0.83b | 1.05b | 0.94b |
| −0.93c | −0.64c | −1.08c | |
|
| |||
| Spleen | 1.90a | 2.41a | 1.76a |
| 1.53b | 2.10b | 1.61b | |
| −0.94c | −0.62c | −1.08c | |
|
| |||
| Combined | 1.86 ± 0.03a ** | 2.49 ± 0.12a | 1.81 ± 0.09a * |
| 1.19 ± 0.20b | 1.70 ± 0.33b | 1.43 ± 0.25b | |
| −0.95 ± 0.02c *** | −0.64 ± 0.01c | −1.00 ± 0.08a * | |
The reduced number of peripheral cDKO T cells may have been a result of diminished thymic output rather than a defect in peripheral “maintenance”. To test this, we analyzed thymi from recipients of both WT and cDKO bone marrow. The gating strategy used to delineate donor- and host-derived thymic T cells is depicted in Supplementary Figure 2A-E. Interestingly, we saw an enhanced presence of GFP+ cells in thymi of animals reconstituted with cDKO bone marrow as indicated by both increased cell percentage and number (Supplementary Figure 2F,G). However, there were no observable differences between WT and cDKO animals in the number of GFP− donor-derived T cells suggesting that the enhanced presence of GFP+ T cells in cDKO thymi may have been due to a migratory effect or a local expansion of these cells (Supplementary Figure 2H)[23]. Thus, the lack of cDKO donor-derived peripheral T lymphocytes was not due to a thymic deficiency but was most likely rooted in an impaired ability to successfully compete in peripheral survival niches.
3.2 Snai2/Snai3 cDKO T cells display an impaired activation/memory phenotype
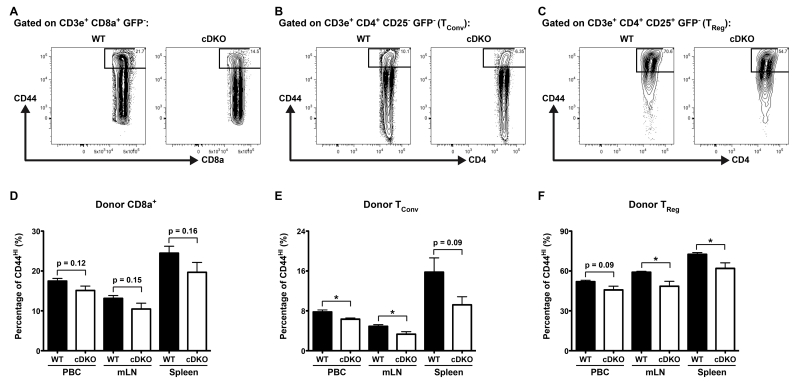
There are multiple potential explanations for the observed loss of donor-derived T cells in the recipients of cDKO bone marrow. One possibility may be due to the decreased activation and expansion of naïve cells. In unchallenged hosts such as these mice (i.e. pathogen free), maintenance of peripheral T cell populations inclusive of naïve and memory phenotypes is modulated via homeostatic proliferation[31]. The generation and maintenance of these cells have requirements for cytokines and peptide:MHC interactions that vary upon context[32]. To determine the activation state of WT and cDKO donor-derived T cells, we performed FACS analysis as described in Supplementary Figure 1.
However, we also included the activation marker CD44 to bifurcate resting (i.e. naïve) and activated, or effector/memory-like, T cells[33]. Figure 2A-C shows representative gating for the surface expression of CD44 on WT and cDKO CD8α+, TConv and TReg cells from spleens of recipient animals. Quantification of data derived from the analysis of PBCs, mLNs and spleens is shown in Figure 2D-F. Overall, the data demonstrated trends towards a lower ratio of activated cDKO CD8α+ T cells (PBC = 13% decrease [p = 0.12], mLN = 20% decrease [p = 0.15] and spleen = 20% [p = 0.16]). Furthermore, similar analysis of both TConv and TReg cells revealed significant decreases in the activation state of cDKO donor-derived cells over multiple organs. Similar to cDKO CD8α+ T cells, cDKO TReg cells possessed decreased ratios of CD44HI cells that were relatively similar among all organs analyzed (PBC = 12% decrease [p = 0.09], mLN = 18% decrease [* p ≤ 0.05] and spleen = 15% [p = 0.16]). In contrast, TConv cells from cDKO donors displayed much larger decreases at sites of T cell priming (PBC = 15% decrease [* p ≤ 0.05] versus mLN = 32% decrease [* p ≤ 0.05], spleen = 42% decrease [p = 0.09]). In total, this data is suggestive of a lower level of basal activation among T cells derived from cDKO bone marrow. However, further experiments are required to clarify the origin of this defect (e.g. increased cell death or higher activation thresholds).
Figure 2.
Snai2/Snai3 cDKO peripheral T cells display an impaired activation, effector/memory-like phenotype. (A-C) Representative FACS plots from spleens of UBC-GFP mice 9 weeks post-transplantation with either WT or cDKO GFP− bone marrow progenitors. CD44+ activated, effector/memory-like T cells are gated within GFP− (donor-derived) populations of (A) CD8α+, (B) TConv and (C) TReg cells. (D-F) Quantification of the percentage of CD44+ activated, effector/memory-like donor (D) CD8α+, (E) TConv and (F) TReg cells from the PBCs, mLNs and spleens of recipient animals. (D-F) Bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 4. Student’s t-Test: * p ≤ 0.05.
3.3 Snai2 and Snai3 regulate unique gene programs among T cell lineages
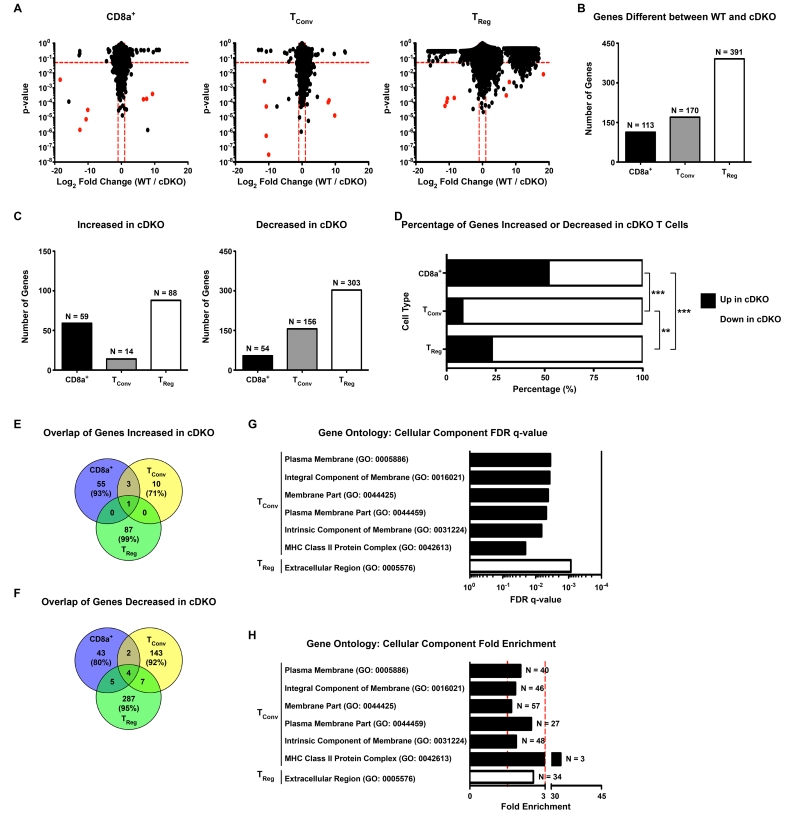
The above data suggested that Snai2 and Snai3 are important regulators of competitive fitness among T cell lineages. As mentioned above, Snai2 and Snai3 are transcriptional regulators most classically associated with gene repression. To gain global and unbiased insight into how these factors may transcriptionally regulate T lymphocytes, we performed RNA sequencing (RNA-seq) on WT and cDKO donor-derived CD8α+, TConv and TReg cells sorted from the spleens of recipient mice. T cell lineages were identified and FACS-sorted as previously shown in Supplementary Figure 1 and Figure 1. To validate the populations analyzed, we confirmed the expression, or lack thereof, of Cd8a, Cd4 and Foxp3 in each cell type. Only CD8α+ T cells expressed Cd8a and Foxp3 transcripts were restricted to TRegs as depicted by University of California-Santa Cruz (UCSC) genome browser tracks (Supplementary Figure 3). Graphical comparisons of the p-values and log2 fold changes (WT / cDKO) for all three cell types are shown in Figure 3A (left = CD8α+, middle = TConv, right = TReg). For these experiments, WT bone marrow was derived from female mice while male mice supplied cDKO bone marrow. As such, Y-linked and X chromosome inactivation-associated genes are highlighted in red and excluded from further analyses. Upon examination of the data, it was most readily apparent that the greatest level of gene augmentation occurred in TRegs. Excluding the sex-linked genes referenced above, all 3 lineages displayed similar magnitudes of increase for genes that were upregulated in T cells derived from cDKO bone marrow (mean WT / cDKO log2 fold change: CD8α+ = −1.96, TConv = −1.99, TReg = −1.96). However, TRegs displayed greater levels of differential expression for those genes that were downregulated in the cDKO samples (mean WT / cDKO log2 fold change: CD8α+ = 1.63, TConv = 1.50, TReg = 4.18). Quantification of the number of significantly different genes among genotypes (p-value ≤ 0.05, WT / cDKO log2 fold change ≥ |1|, dotted red lines equal significance cutoffs in Figure 3A) yielded a total of 391 genes dysregulated in Snai2/Snai3 cDKO TRegs versus only 113 and 170 genes for CD8α+ and TConv cells, respectively (Figure 3B and Supplementary Tables 3-5). When we compared the pattern of gene expression changes (i.e. increased versus decreased), there were clear differences among the cell types (Figure 3C,D). CD8α+ T cells had a relatively even distribution between up and downregulated genes (59 genes up (52%) versus 54 genes down (48%)). In stark contrast, TConv and TReg cells were heavily biased towards the downregulation of genes upon the deletion of Snai2 and Snai3 (TConv: 14 genes up (8%) versus 156 genes down (92%), TReg: 88 genes up (23%) versus 303 genes down (77%)). Furthermore, there was minimal conservation of dysregulated genes among the 3 cells types (Figure 3E,F). This was most exemplified in the TReg gene set as 99% of upregulated and 95% of downregulated genes were unique to that particular cell type. CD8α+ (93% of increased, 80% of decreased) and TConv (71% of increased, 92% of decreased) cells displayed a similar phenomenon albeit to a slightly lesser degree. Table 2 lists genes up and downregulated in the cDKO common to multiple T cell lineages. Of the “shared” upregulated genes, none have been previously annotated as having a direct immune-related function (PubMed). In contrast, multiple “shared” genes downregulated in cDKO T cell lineages have known roles in immunological function, albeit bot necessarily in a T cell-specific fashion. This was most evident for TConv and TReg cells in which Serpinc1, Themis2, Mlkl and March1 all had reduced expression upon the deletion of Snai2 and Snai3 (Supplementary Tables 4 and 5)[34-39]. To summarize, the data suggested that the deletion of Snai2 and Snai3 resulted in transcriptional alterations that were mostly specific to a given T cell lineage. Additionally, Snai2 and Snai3 appeared to play the largest role in TRegs mainly through the preservation of gene activation.
Figure 3.
Snai2 and Snai3 regulate unique gene programs among T cell lineages. (A) Graphical depiction of RNA-seq data from WT and cDKO (Left) CD8α+, (Middle) TConv and (Right) TReg cells. The y-axis represents p-value and the x-axis shows log2 fold chance (WT / cDKO). Dotted red lines mark cut-offs for significantly altered genes (p-value ≤ 0.05, WT / cDKO log2 fold change ≥ |1|). Red dots indicate Y-linked and X-chromosome inactivation-associated genes as WT donors were female and cDKO donors were male. (B) Quantification of the number of significantly altered genes upon comparison of transcriptomes from WT and cDKO CD8α+, TConv and TReg cells. Bars represent the number of genes with the exact number (N=) indicated above each bar. (C) The number of genes significantly (Left) increased or (Right) decreased in cDKO CD8α+, TConv and TReg cells. Bars represent the number of genes with the exact number (N=) indicated above each bar. (D) Percentage of genes significantly increased or decreased in cDKO CD8α+, TConv and TReg cells. Black bars represent the percentage of genes increased while white bars indicate the percentage of genes decreased. (E,F) Venn diagrams depicting the overlap of genes significantly (E) increased or (F) decreased in cDKO CD8α+, TConv and TReg cells. Number of genes and percentage of total genes (in parentheses) are indicated for each cell type. (G, H) Gene ontology analysis of cDKO dysregulated genes showing enrichment for various Cellular Component categories represented within TConv and TReg significantly altered gene sets. (G) Bars represent FDR q-values for each associated category. (H) Bars represent fold enrichment for each associated category. Vertical dotted red lines demarcate 1.5- and 3-fold enrichment, respectively. The number of contributing number of genes (N=) for each category is indicated next to each bar. (A-H) Number of animals: N = 4 for all cell types except for cDKO TRegs (N = 3). (D) Fisher’s Exact Test: ** p ≤ 0.01, *** p ≤ 0.001.
Table 2.
Genes significantly up and downregulated in multiple T cell lineages upon the deletion of Snai2 and Snai3. Mean ± SEM FPKM values for each analyzed gene are indicated for both WT and cDKO versions of all 3 cell types. Grey shaded boxes indicate statistically significant differences (p < 0.05) between WT and cDKO samples for a given gene and lineage. Bolded gene names indicate those with a previously published immunological association. N/A = not applicable; N.D. = not detectable
| Category | Lineages | Gene | WT CD8α+ |
cDKO CD8α+ |
WT TConv | cDKO TConv | WT TReg | cDKO TReg |
|---|---|---|---|---|---|---|---|---|
|
Upregulated
in cDKO |
CD8α+ and TConv | Sva | 0.001 ±0.000 |
5.568 ±0.631 |
0.031 ±0.030 |
5.578 ±0.548 |
0.001 ±0.000 |
3.049 ±1.524 |
| Spaca1 | 0.653 ±0.285 |
1.893 ±0.420 |
0.816 ±0.348 |
2.130 ±0.231 |
0.473 ±0.279 |
1.308 ±0.537 |
||
| Srp54b | 0.297 ±0.020 |
0.658 ±0.070 |
0.396 ±0.062 |
0.841 ±0.122 |
0.698 ±0.129 |
0.065 ±0.034 |
||
| CD8α+ and TReg |
N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| TConv and TReg | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| CD8α+, TConv and TReg |
Gm9522 | 1.826 ±0.499 |
6.032 ±1.220 |
2.265 ±0.244 |
7.323 ±1.213 |
3.442 ±0.987 |
9.873 ±1.122 |
|
|
Downregulated in
cDKO |
CD8α+ and TConv |
Gm15819 | 3.737 ±0.085 |
1.301 ±0.206 |
3.236 ±0.168 |
1.351 ±0.269 |
2.362 ±0.813 |
1.147 ±0.573 |
| Rapgefl1 | 0.141 ±0.041 |
0.019 ±0.015 |
0.112 ±0.019 |
0.043 ±0.018 |
N.D. | N.D. | ||
| CD8α+ and TReg |
Cables1 | 0.347 ±0.043 |
0.172 ±0.014 |
N.D. | N.D. | 1.039 ±0.244 |
0.032 ±0.022 |
|
| Ube2l6 | 1.042 ±0.154 |
0.509 ±0.112 |
2.004 ±0.394 |
1.100 ±0.499 |
9.394 ±0.664 |
4.128 ±1.202 |
||
| Zfp69 | 3.988 ±0.785 |
1.751 ±0.049 |
3.824 ±0.887 |
1.809 ±0.205 |
4.061 ±0.565 |
0.977 ±0.565 |
||
| Gp49a | 2.829 ±0.572 |
1.138 ±0.248 |
1.551 ±0.717 |
0.649 ±0.352 |
10.575 ±1.775 |
2.340 ±0.671 |
||
| Map2 | 0.084 ±0.018 |
0.029 ±0.007 |
0.243 ±0.077 |
0.068 ±0.029 |
0.811 ±0.153 |
0.259 ±0.112 |
||
| TConv and TReg | Mir5107 | N.D. | N.D. | 29.253 ±4.359 |
14.401 ±2.203 |
39.095 ±4.858 |
2.185 ±2.184 |
|
| Serpinc1 | 0.309 ±0.097 |
0.336 ±0.091 |
0.718 ±0.100 |
0.341 ±0.035 |
3.624 ±0.116 |
1.697 ±0.796 |
||
| Themis2 | 4.456 ±0.441 |
3.146 ±0.722 |
2.311 ±0.165 |
1.017 ±0.066 |
1.768 ±0.194 |
0.656 ±0.350 |
||
| Phox2a | 0.119 ±0.083 |
0.015 ±0.014 |
0.343 ±0.055 |
0.139 ±0.053 |
0.786 ±0.075 |
0.292 ±0.162 |
||
| Mlkl | 0.804 ±0.201 |
0.609 ±0.198 |
0.571 ±0.109 |
0.215 ±0.057 |
4.278 ±0.586 |
1.924 ±0.638 |
||
| March1 | 0.171 ±0.039 |
0.145 ±0.031 |
0.417 ±0.068 |
0.153 ±0.021 |
0.179 ±0.042 |
0.0130 ±0.012 |
||
| Penk | 0.489 ±0.266 |
0.074 ±0.062 |
1.951 ±0.552 |
0.396 ±0.111 |
45.828 ±2.334 |
19.140 ±4.292 |
||
| CD8α+, TConv and TReg |
Rell1 | 3.731 ±0.281 |
1.797 ±0.164 |
4.062 ±0.353 |
1.988 ±0.139 |
3.969 ±0.334 |
1.711 ±0.169 |
|
| Col27a1 | 0.085 ±0.015 |
0.035 ±0.006 |
0.144 ±0.022 |
0.058 ±0.006 |
0.454 ±0.037 |
0.088 ±0.073 |
||
| Gm11212 | 1.452 ±0.082 |
0.582 ±0.100 |
1.740 ±0.446 |
0.546 ±0.193 |
0.686 ±0.198 |
0.001 ±0.000 |
||
| Snai3 | 7.005 ±0.370 |
0.030 ±0.018 |
0.787 ±0.095 |
0.034 ±0.021 |
0.733 ±0.118 |
0.041 ±0.040 |
3.4 Snai2/Snai3 cDKO T lymphocytes have impaired expression of key modulators of cellular fitness and function
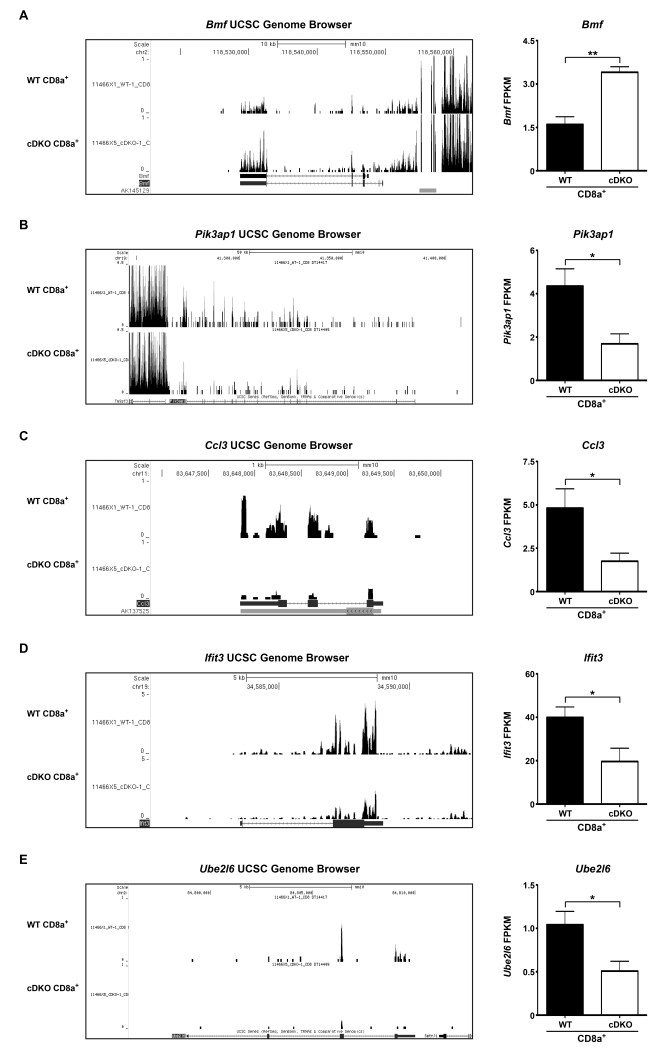
To better understand the global gene changes taking place, we performed gene ontology (GO) analysis using Gorilla software (see Materials and Methods). Our lists of significantly altered genes only demonstrated significant enrichments in categories related to Cellular Components, rather than Biological Processes and/or Molecular Functions (Figure 3G,H). CD8α+ T lymphocytes displayed no enrichment for any GO terms. However, a closer examination of the data yielded a multitude of dysregulated genes that may directly impact the survival and/or function of CD8α+ T cells. These genes included the pro-apoptotic Bmf, which displayed a 113% increase in expression in the cDKO samples when compared to WT (Supplementary Table 3, Figure 4A shown are (left) UCSC genome browser tracks and (right) quantification of fragments per kilobase per million mapped reads (FPKM))[40]. Additionally, Pik3ap1, also known as BCAP, was significantly reduced by 61% in CD8α+ T cells derived from cDKO bone marrow (Supplementary Table 3, Figure 4B shown are (left) UCSC genome browser tracks and (right) quantification of FPKM). In B cells, BCAP is a key potentiator of the PI3K-AKT survival pathway[41, 42]. Furthermore, AKT signaling has been shown to suppress BMF expression in human breast cancer cell lines[43]. Thus, it is plausible that Pik3ap1, or BCAP, may play a role in the survival of CD8α+ T cells. These gene changes, among others, would be expected to in appropriately prime, or sensitize, CD8α+ T lymphocytes to apoptotic stimuli thus preventing normal functionality. Accordingly, Ccl3, a cytokine secreted by memory CD8α+ T cells was decreased by 64% in cDKO samples (Supplementary Table 3, Figure 4C shown are (left) UCSC genome browser tracks and (right) quantification of FPKM).[44]. In addition, cDKO CD8α+ T cells displayed significantly lower basal expression of Ifit3 and Ube2l6, genes associated with the interferon response (Supplementary Table 3, Figure 4D,E shown are (left) UCSC genome browser tracks and (right) quantification of FPKM)[45, 46]. Aside from the interferon response, Ube2l6 is a key regulator of the cell cycle by targeting the cell cycle inhibitors, p21 and Cdt1, for proteolytic degradation[47].
Figure 4.
Snai2 and Snai3 are important regulators of CD8α+ T cell genes involved in cellular expansion and function. (Left) Representative UCSC genome browser tracks and (Right) FPKM for (A) Bmf, (B) Pik3ap1, (C) Ccl3, (D) Ifit3 and (E) Ube2l6 from WT and cDKO CD8α+ T cells. FPKM bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 4. Student’s t-Test: * p ≤ 0.05, ** p ≤ 0.01.
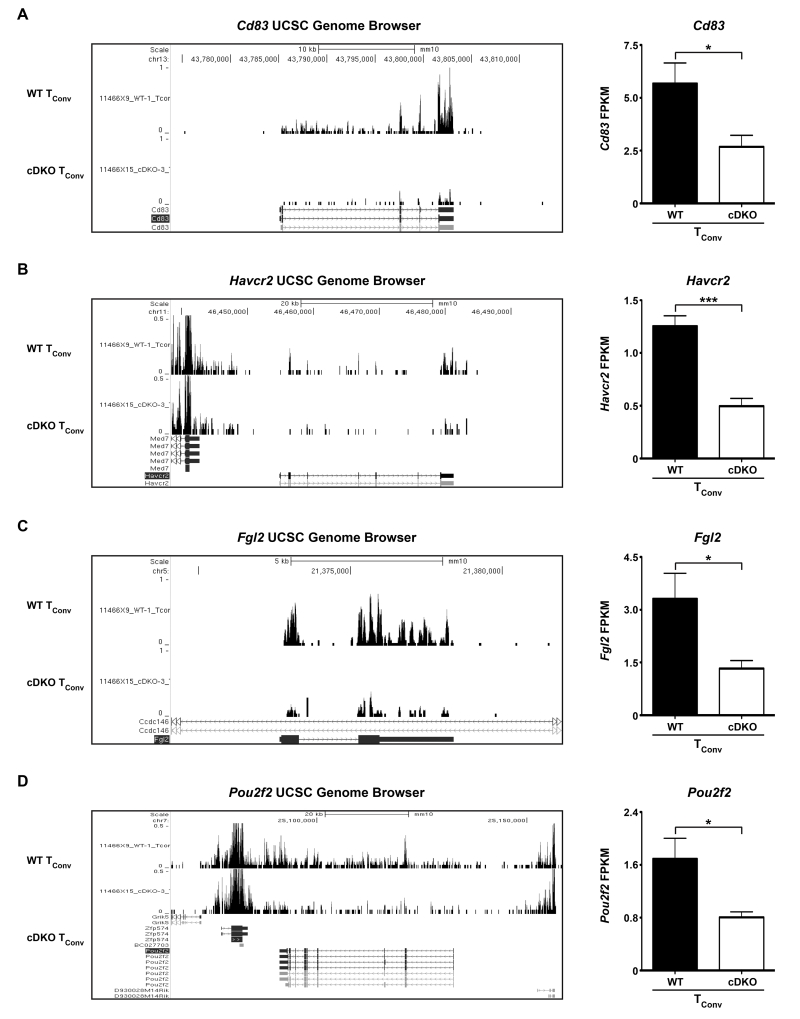
In contrast to CD8α+ T cells, gene changes associated with cDKO TConv cells displayed enrichment for a multitude of Cellular Components such as: Plasma Membrane (2.03-fold), Integral Component of Membrane (1.82-fold), Membrane Part (1.66-fold), Plasma Membrane Part (2.45-fold), Intrinsic Component of Membrane (1.85-fold) and MHC Class II Protein Complex (32.87-fold) (Figure 3G,H). Not surprisingly, these categories included a plethora of genes encoding for cell surface receptors/molecules. Examples being Cd83 and Havcr2 (TIM-3) which were downregulated by 53% and 60%, respectively, in cDKO TConv cells (Supplementary Table 4, Figure 5A,B shown are (left) UCSC genome browser tracks and (right) quantification of FPKM). Cd83 is a marker of B and T cell activation and is required for sustained survival of both cell types[48]. In a similar fashion, TIM-3 is upregulated by activated human CD4+ T cells[49]. However, in the case of TIM-3, this protein has been shown to be a negative regulator of T cell cytokine production. We also observed a 60% decrease in expression of Fgl2 by cDKO TConv cells (Supplementary Table 4, Figure 5C shown are (left) UCSC genome browser tracks and (right) quantification of FPKM). Previous data has demonstrated a requirement for Fgl2 in TReg-mediated immunosuppression[50]. While a role for Fgl2 in TConv biology has not been described, splenic memory CD4+ T cells and TRegs express similar amounts of Fgl2 transcripts perhaps suggesting an important role for this gene product in T cell memory formation/function (Immunological Genome Project database). Not falling within the GO categories but potentially meaningful, cDKO-derived TConv cells possessed 52% lower levels of the transcription factor, Pou2f2 (Oct-2) (Supplementary Table 4, Figure 5D shown are (left) UCSC genome browser tracks and (right) quantification of FPKM). Oct-2 is known to be upregulated post-T cell activation and subsequently transactivate the Il2 gene, a critical T cell survival factor[51, 52]. In this case, we did not see reduced Il2 gene expression most likely due to our cells being isolated from naïve mice perhaps indicative of a more complex role for Oct-2 in TConv cell biology.
Figure 5.
Snai2 and Snai3 transcriptionally control important modulators of TConv lymphocyte fitness and function. (Left) Representative UCSC genome browser tracks and (Right) quantification of FPKM for (A) Cd83, (B) Havcr2, (C) Fgl2 and (D) Pou2f2 from WT and cDKO TConv cells. FPKM bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 4. Student’s t-Test: * p ≤ 0.05, *** p ≤ 0.001.
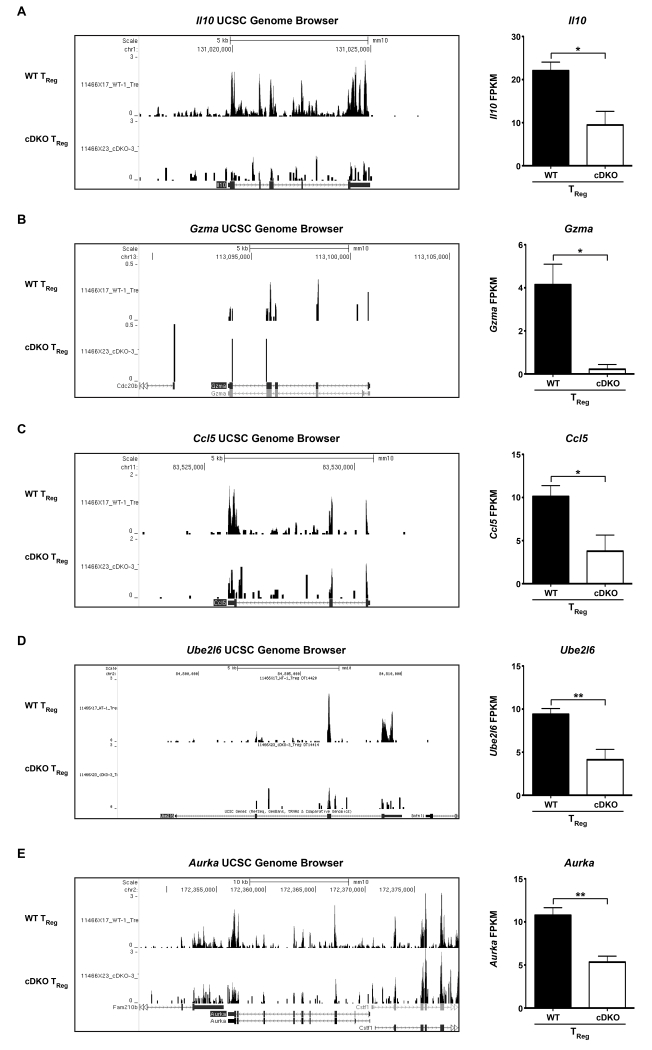
Since cDKO TRegs possessed the greatest number of gene alterations, we expected that this cell type to have the largest number of gene ontology associations. However, this was not the case as the TReg gene set was only enriched for the Cellular Compartment category of Extracellular Region (2.53-fold) that consisted of 34 genes, or approximately 9% of the total gene set (Figure 3G,H). However, this small cohort of targets included known TReg effector molecules such as Il10 and Gzma[53, 54]. Il10 and Gzma were significantly reduced by 57% and 95%, respectively (Supplementary Table 5, Figure 6A,B shown are (left) UCSC genome browser tracks and (right) quantification of FPKM). Aside from a deficiency in the expression of key suppressive molecules, cDKO TRegs may have also been unable to efficiently recruit their regulatory targets due to reduced expression of Ccl5 (63% reduced compared to WT), a well characterized chemoattractant of antigen presenting cells and activated T cells. (Supplementary Table 5, Figure 6C shown are (left) UCSC genome browser tracks and (right) quantification of FPKM)[55, 56]. As with CD8α+ T cells, cDKO TRegs also displayed a significant decrease in Ube2l6 potentially associating Snai2/Snai3 with the regulation of cell cycle dynamics in both T cell lineages (Supplementary Table 5, Figure 6D shown are (left) UCSC genome browser tracks and (right) quantification of FPKM)[15]. This was further supported by the 50% decrease in expression of Aurka in cDKO TRegs (Supplementary Table 5, Figure 6E shown are (left) UCSC genome browser tracks and (right) quantification of FPKM)[57].
Figure 6.
Snai2 and Snai3 transcriptionally augment key TReg cellular fitness and effector genes. (Left) Representative UCSC genome browser tracks and (Right) quantification of FPKM for (A) Il10, (B) Gzma, (C) Ccl5, (D) Ube2l6 and (E) Aurka from WT and cDKO TRegs. FPKM bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 3. Student’s t-Test: * p ≤ 0.05, ** p ≤ 0.01.
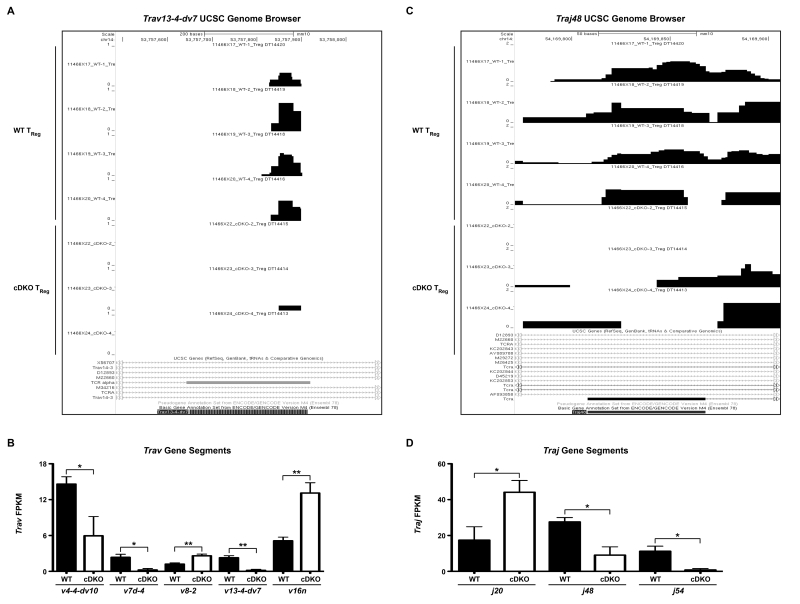
On a curious note, we found multiple T cell receptor (TCR) gene loci whose representation was significantly different between WT and cDKO TReg samples. Figure 7A shows representative UCSC genome browser tracks for all WT and cDKO TReg samples sequenced with a specific focus on the TCRα variable region 13-4-dv7 (Trav13-4-dv7). As can be seen, expression of this locus was almost completely lost in the 3 cDKO samples compared to the 4 WT samples. In total, usage of 5 TCRα variable regions (Trav4-4-dv10, Trav7d-4, Trav8-2, Trav13-4-dv7 and Trav16n) was significantly altered between WT and cDKO TRegs (Figure 7B). This represented approximately 8%, or 5 out of 62, of the total detectable variable region sequences. In a similar fashion, we saw fluctuations in the prevalence of multiple TCRα joining loci. Figure 7C shows representative UCSC genome browser tracks for TCRα joining region 48 (Traj48) with quantification of Traj20, Traj48 and Traj54 depicted in Figure 7D. Overall, 3 of 56 (approximately 8%) joining regions were altered in cDKO TRegs. This effect was not seen in the TCRβ chain. However, it should be noted that only 21 variable, 2 diversity and 13 joining regions for the TCRβ chain were detected and any differences may have been below the limit of detection using standard RNA-seq methodology. The observed variation in TCRα usage was restricted to TRegs as both CD8α+ and TConv cells, with equivalent sequencing depth, showed no apparent alterations in variable and joining region usage. Overall, these data imply a role for Snai2 and Snai3 in the “selection” of TCRα gene loci represented within the peripheral TReg pool.
Figure 7.
T cell receptor α variable and joining region usage is altered in Snai2/Snai3 cDKO regulatory T cells. (A) UCSC genome browser tracks showing Trav13-4-dv7 expression in WT and cDKO TRegs. (B) Quantification of FPKM for multiple variable regions from WT and cDKO TRegs. (C) UCSC genome browser tracks showing Traj48 expression in WT and cDKO TRegs. (D) Quantification of FPKM for multiple joining regions from WT and cDKO TRegs. (B,D) FPKM bars represent mean ± SEM. Number of animals per group: WT = 4, cDKO = 3. Student’s t-Test: ** p ≤ 0.01, * p ≤ 0.05.
3.5 Snai2/Snai3 regulate a Foxp3-independent TReg transcriptional program
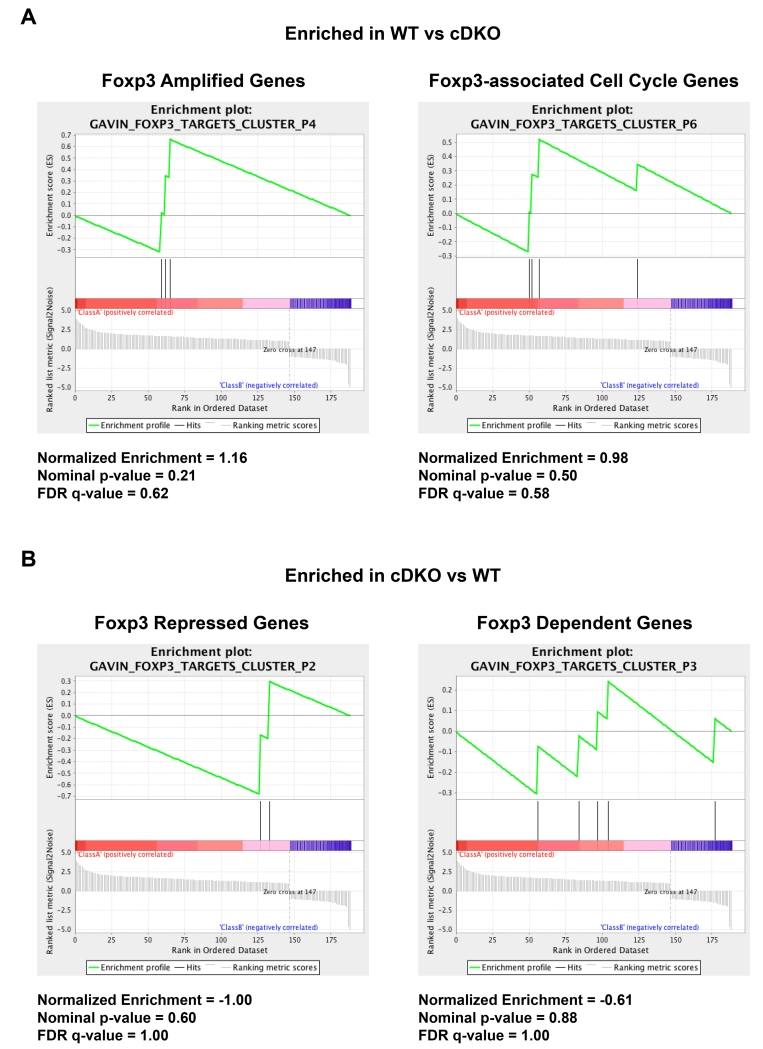
Finally, we were interested to assess the similarity between Snai2/Snai3- and Foxp3-regulated genes, as this data would indicate a Foxp3-independent versus Foxp3-dependent role for Snai2/Snai3 in augmenting TReg biology. To this end, we performed Gene Set Enrichment Analysis (GSEA) to compare significantly altered genes in cDKO TRegs versus Foxp3 target genes identified by the Rudensky lab (Figure 8)[3]. A small number of Foxp3-regulated genes such as Atp6vod2 (P3) and Aurka (P6) were also regulated by Snai2/Snai3. However, the vast majority of Snai2/Snai3 transcriptional targets (≥95%) were not present in any of the Foxp3-regulated categories analyzed. Therefore, the data imply that Snai2/Snai3 regulate a Foxp3-independent TReg transcriptional program.
Figure 8.
Snai2/Snai3 regulate a Foxp3-independent regulatory T cell transcriptional program. (A,B) GSEA of Snai2/Snai3 regulated gene targets as compared to Foxp3-dependent genes. Normalized enrichment scores, nominal p-values and FDR q-values are shown for each category analyzed.
Discussion
This study was initiated based on the observation that Snai2/Snai3 cDKO animals while developing a substantial autoantibody response did not also possess a T cell lymphoproliferative disease, a common facet of many autoimmune syndromes[23, 25]. This led to the hypothesis that Snai2 and Snai3 were also essential for the proper “function” of effector T cells and not just their regulatory counterparts (TRegs). Using a combination of in vivo competition and global transcriptome analysis (RNA-seq), we have definitively shown that Snai2 and Snai3 are key transcriptional regulators of not only TRegs but of CD8α+ and TConv cells as well. While the transcriptional regulatory profile of Snai2/Snai3 was unique to each cell type, the overall theme of augmenting cellular fitness (i.e. survival and proliferation) and function was conserved.
Phenotypically, Snai2/Snai3 cDKO T cells as a whole were less competent in their ability to replace radioresistant T cells retained in lethally irradiated host animals. While it has been previously demonstrated that both TConv and TReg cells are radioresistant, to our knowledge, this is the first account of CD8α+ T cells also sharing this property[30]. Interestingly, the differences between WT and cDKO T cells also included a diminished amount of “activation” within each cDKO T cell lineage. This could arise from multiple sources such as increased activation thresholds for cDKO naïve T cells or the impaired ability of cDKO T cells to be promoted into and subsist within the memory pool (i.e. impaired downstream integration of TCR signals). Given the relatively normal expression levels of key TCR signaling components (e.g. Lat, Zap70, Cd3e), we would expect that proximal TCR signaling would be intact in cDKO T cells and that any potential defects would lie downstream. That is, Snai2/Snai3 would be key in integrating a portion of the transcriptional response downstream of T cell activation. Previous studies of Snail transcription factors in hematopoiesis have defined a clear role for these factors in responses to cellular stress such as irradiation or cytokine withdrawal[58-60]. In those contexts, Puma, or Bbc3, was a key transcriptional target of Snai2. While dysregulation of multiple genes involved in survival (e.g. Bmf) and proliferation (e.g. Aurka) were apparent in cDKO T cells, none included known Snail targets such as Puma, Mmac1 (PTEN), Cdkn3b (p15ink4b) or Cdkn1a (p21WAF/CIP1) indicative of novel regulatory mechanisms for these factors in T cell biology[15, 61, 62]. However, an in depth comparison of WT and cDKO T cell activation responses as a whole will be needed to better understand the apparent deficit of cDKO T cell “activation”.
Perhaps most surprising were the different routes these factors took in each cell type in essentially accomplishing the same goals (i.e. lowered fitness and/or functionality). The minimal number of genes disrupted in CD8α+ T cells upon the deletion of Snai2 and Snai3 was striking. Given that these cells possessed the highest levels of Snai3 transcripts, it would have been logical to expect the largest gene changes within these cells[22, 23]. The unexpected results may be due to a nonlinear relationship between the amount of Snai3 transcript and protein (and Snai2 as well?) within a given CD8α+ T cell. A well-known mediator of Snai1 protein stability is Mdm2 which targets Snai1 for proteosomal degradation upon p53 activation[63]. Interestingly, the Notch1 pathway, intact in peripheral CD8α+ T cells, can recruit Mdm2 to Snai1 promoting subsequent degradation[64, 65]. Our attempts to analyze Snai2 and Snai3 protein levels in CD8α+, TConv and TReg cells via FACS suggested that both Snai2 and Snai3 proteins were enriched for in TConv and TReg cells, in particular in the CD44HI, activated, effector/memory-like, compartment (PDP, unpublished data). However, due to the non-specific nature of these antibodies when comparing cell types from WT and Snai2 or Snai3 single knockout (KO) animals, these results were more qualitative than quantitative (i.e. variable levels of background on an experiment-to-experiment basis). In addition, multiple regulators of Snai1 DNA-binding and transcriptional repressor activity have been described[66-68]. Included within this group is Lats2, a kinase that phosphorylates Snai1 at threonine 203 leading to Snai1 nuclear retention and enhanced stability[68]. Interestingly enough, TConv and TReg cells express much higher levels of Lats2 compared to CD8α+ T lymphocytes, which could potentially explain the higher levels of Snail proteins in these cell types (Immunological Genome Project database). However, proposed mechanisms of Snai3, and Snai2, protein stability based off of Snai1 studies require further assessment to determine how well these regulatory circuits are conserved among family members.
Examination of the gene expression patterns in TConv and TReg cells from WT and cDKO animals suggested that the presence of Snai2 and Snai3 was required to maintain gene activation rather than repression. As mentioned before, except for a limited number of circumstances, Snail proteins mainly function as transcriptional repressors[14, 15]. As such, our data leaves us with 2 basic hypotheses as to how this may be occurring in TConv and TReg cells. First, Snai2 and Snai3 are not directly binding genes that lose expression upon Snai2/Snai3 deletion. Rather, Snai2/Snai3 may directly repress a repressor(s) of the aforementioned gene targets. As a result, enhanced expression of this repressor(s) would lead to further downregulation of its gene targets. Overall, there was a paucity of upregulated transcription factors in cDKO TConv and TReg cells. However, cDKO TConv cells had increased expression of Zfp618 (Supplementary Table 4) and TRegs had higher levels of Zfp608 (Supplementary Table 5). While little information exists as to the function of Zfp618, Zfp608 has been described as a regulator of thymocyte development and a repressor of Rag gene expression[69, 70]. Unlike peripheral CD8α+ and TConv cells that have low Zfp608 expression upon thymic egress, TRegs maintain a relatively high level of the gene transcript (Immunological Genome Project database). The specific deletion of Zfp608 via Foxp3-Cre would aid in the clarification of any roles this factor may play in TReg development and/or function. An alternative hypothesis to the above stated is that Snai2/Snai3 interact with a differential set of “cofactors” in a cell type specific manner “flipping the switch” from repressors to activators. This idea would be inclusive of both chromatin modifiers and DNA-binding transcription factors and would further explain the unique set of gene targets per each T cell lineage examined. However, to fully evaluate these possibilities would require Snai2/Snai3 chromatin immunoprecipitation sequencing (ChIP-seq) coupled with an unbiased protein-protein interactome analysis (e.g. co-immunoprecipitation coupled with mass spectrometry) in different T lymphocyte populations.
Upon examination of the RNA-seq data, it was not surprising to see a strong association with Cellular Component GO terms that related to plasma membrane and extracellular dynamics. After all, perhaps the best know function for all Snail family members is the regulation of epithelial-to-mesenchymal transition (EMT), which directly affects how cells interact with one another[71]. Rather interestingly, there was an overall lack of association with immune-related GO terms. Ultimately, this may indicate that Snai2/Snai3 play a larger role in correctly localizing TRegs. This would allow for the proper regulation of TReg target cells and in addition, the reception of extracellular signals (i.e. IL-2) that would further induce the TReg immuno-suppressive program[72].
The analysis of transcriptomes from Snai2/Snai3 cDKO TRegs formally demonstrated the importance of this family in the transcriptional regulation of key TReg effector genes (e.g. Il10, Gzma), as proposed in our previous study[23]. However, given that Snai2/Snai3 were not global regulators of immune-related processes, we were curious to see as to what level Snai2/Snai3 function autonomously of Foxp3, the “master transcription factor” of TRegs. To assess this, we used GSEA to compare previous data generated from the Rudensky lab versus the list of significantly altered genes in Snai2/Snai3 cDKO TRegs (Figure 8). Some Foxp3 target genes such as Atp6v0d2, Aurka, Ccl5, Gpr15, Ifitm3, Igfbp7, Igf2bp3, Lgals3, Selp and Zfp608 were also regulated by Snai2/Snai3 (Supplementary Table 5). While deletion of Foxp3 and Snai2/Snai3 had a parallel effect on most of these common targets, genes such as Ifitm3, Igfbp7 and Zfp608 exhibited an anti-parallel effect (e.g. Igfbp7: increased in Foxp3 KO, decreased in Snai2/Snai3 cDKO) suggesting that there is a complex dynamic for the few Foxp3 and Snai2/Snai3 co-regulated genes. However, the vast majority of genes (≥95%) from our cDKO TReg data set did not appear to also fall under Foxp3-mediated regulation (e.g. Il10) indicative of a Foxp3-independent role for Snai2/Snai3. So how then, would Snai2/Snai3 function to regulate the TReg transcriptional landscape? Recently, the Sakaguchi group defined the independent roles that Foxp3 and epigenetic changes (in general) play in forming the TReg transcriptional landscape[73]. In particular, they were interested in genes that were upregulated in TReg versus TConv cells under both resting and TCR-stimulated conditions. Transcription factor motif analysis focused on the transcriptional start sites (TSS) of genes upregulated in TRegs (potentially downregulated in Snai2/Snai3 cDKO TRegs) did not demonstrate enrichment for E-box binding sites, which would be consistent with the aforementioned hypothesis of unique interacting partners recruiting Snai2/Snai3 to cell type-specific targets ultimately leading to the conversion of Snai2/Snai3 to activators of gene expression. Potential Snai2/Snai3 interactors may include Id family members, Id2 and Id3. Albeit not a DNA-binding protein, Id2 has been previously shown to block the transcriptionally repressive nature of Snai1 by blocking the SNAG domain-mediated recruitment of LSD1[7]. Interestingly, Il10 gene expression was enhanced in regulatory T cells upon the deletion of both Id2 and Id3[5]. In this context, we would hypothesize that Id2 and/or Id3 would potentially block the interaction of Snai2/Snai3 with putative transcriptional activators. Thus, removal of the Ids would boost the ability of Snai2/Snai3 to induce transcription in a locus specific manner.
Within the TReg data set, we also noticed that a modest percentage of TCRα variable and joining regions displayed aberrant expression in cDKO TRegs, an effect that was specific to this cell type. Given the relatively limited nature of this phenotype (5 TCRα variable and 3 TCRα joining regions), we do not believe this reflects the Snai2/Snai3-mediated regulation of TCR recombination during thymocyte development or necessarily massive shifts in positive and/or negative selection thresholds within the TReg lineage. Rather, this most likely represents the results of selective pressures exerted on WT and cDKO TRegs in the peripheral environment. Evaluating the cellular responses of cDKO TRegs to TCR- and cytokine-mediated stimulation in combination with the sequencing of TCR repertoires in both thymic and peripheral TRegs will be informative in further understanding this phenomenon.
Finally, it should be mentioned that our analysis has been in the context of the deletion of both Snai2 and Snai3 in the aforementioned T cell lineages. However, our previous work has suggested impaired TH1 antibody generation upon the deletion of only Snai3[23]. Whether this was due to a B or TConv cell defect remains unknown. However, it provides the impetus to further understand the single gene contributions of Snail family members in lymphocyte develop and function. To summarize, we have demonstrated the novel finding that Snai2 and Snai3 are critical regulators of the transcriptional landscape not only of TRegs but of CD8α+ and TConv lymphocytes as well. Clearly, these factors play specific roles among different T cell lineages, which require further investigation. To fully understand the phenotypic outcomes of Snai2/Snai3 deletion in a particular immune cell will require nuanced approaches (e.g. cell type-specific deletion, single lineage adoptive transfers, etc…) as the interplay between multiple defective cell types may dampen the true magnitude of any observable phenotypes.
Supplementary Material
Highlights.
Snai2/Snai3 regulate the competitive fitness of CD8α+, TConv and TReg cells.
Snai2/Snai3 transcriptionally target not only survival/proliferation genes but also genes essential to the function of particular T cell lineages (e.g. Il10 and T Regs).
Snai2/Snai3 regulate unique transcriptional programs among CD8α+, TConv and TReg cells suggesting a cell type specific function for these factors.
Acknowledgments
The authors would like to thank the University of Utah Flow Cytometry and Bioinformatics Core Facilities. We would like to thank the Weis labs for their critique of this work and many intellectual contributions. PDP would specifically like to thank JHW, a mentor and a friend, for the past few years…it has been an honor and a privilege.
The Department of Pathology and the Weber Presidential Endowed Chair in Immunology provided research funding. Usage of the FacsAria flow cytometer was supported by the National Center for Research Resources of the National Institutes of Health under award #1S10RR026802-01. JJW was supported by NIH AR043521 and AI032223. JHW was supported by NIH AI114462. PDP was supported as a pre-doctoral trainee on the NIH Hematology T32 training grant, #T-32DK007115-40.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest
The authors have no competing financial interests.
References
- [1].He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annual review of immunology. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- [2].Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7731–6. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- [4].Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nature reviews Immunology. 2011;11:119–30. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu LF, et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nature immunology. 2014;15:767–76. doi: 10.1038/ni.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nature immunology. 2010;11:618–27. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- [7].Chang C, Yang X, Pursell B, Mercurio AM. Id2 complexes with the SNAG domain of Snai1 inhibiting Snai1-mediated repression of integrin beta4. Molecular and cellular biology. 2013;33:3795–804. doi: 10.1128/MCB.00434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20:8308–16. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- [9].Manzanares M, Locascio A, Nieto MA. The increasing complexity of the Snail gene superfamily in metazoan evolution. Trends in genetics : TIG. 2001;17:178–81. doi: 10.1016/s0168-9525(01)02232-6. [DOI] [PubMed] [Google Scholar]
- [10].Kataoka H, Murayama T, Yokode M, Mori S, Sano H, Ozaki H, et al. A novel snail-related transcription factor Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic acids research. 2000;28:626–33. doi: 10.1093/nar/28.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, et al. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Molecular cell. 2012;47:457–68. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S, et al. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer research. 2013;73:235–45. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Molecular and cellular biology. 2004;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gingold JA, Fidalgo M, Guallar D, Lau Z, Sun Z, Zhou H, et al. A genome-wide RNAi screen identifies opposing functions of Snai1 and Snai2 on the Nanog dependency in reprogramming. Molecular cell. 2014;56:140–52. doi: 10.1016/j.molcel.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu CT, Chang TY, Cheng CC, Liu CS, Wu JR, Li MC, et al. Snail associates with EGR-1 and SP-1 to upregulate transcriptional activation of p15INK4b. The FEBS journal. 2010;277:1202–18. doi: 10.1111/j.1742-4658.2009.07553.x. [DOI] [PubMed] [Google Scholar]
- [16].Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–92. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- [17].Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–8. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- [18].Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC research notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10300–4. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–97. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- [21].Pioli PD, Weis JH. Snail transcription factors in hematopoietic cell development: a model of functional redundancy. Experimental hematology. 2014;42:425–30. doi: 10.1016/j.exphem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pioli PD, Dahlem TJ, Weis JJ, Weis JH. Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency. PloS one. 2013;8:e69216. doi: 10.1371/journal.pone.0069216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pioli PD, Chen X, Weis JJ, Weis JH. Fatal autoimmunity results from the conditional deletion of Snai2 and Snai3. Cellular immunology. 2015;295:1–18. doi: 10.1016/j.cellimm.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nature genetics. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- [25].Oliveira JB. The expanding spectrum of the autoimmune lymphoproliferative syndromes. Current opinion in pediatrics. 2013;25:722–9. doi: 10.1097/MOP.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS computational biology. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lyon MF, Peters J, Glenister PH, Ball S, Wright E. The scurfy mouse mutant has previously unrecognized hematological abnormalities and resembles Wiskott-Aldrich syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:2433–7. doi: 10.1073/pnas.87.7.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Clark LB, Appleby MW, Brunkow ME, Wilkinson JE, Ziegler SF, Ramsdell F. Cellular and molecular characterization of the scurfy mouse mutant. Journal of immunology. 1999;162:2546–54. [PubMed] [Google Scholar]
- [30].Komatsu N, Hori S. Full restoration of peripheral Foxp3+ regulatory T cell pool by radioresistant host cells in scurfy bone marrow chimeras. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8959–64. doi: 10.1073/pnas.0702004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Almeida AR, Rocha B, Freitas AA, Tanchot C. Homeostasis of T cell numbers: from thymus production to peripheral compartmentalization and the indexation of regulatory T cells. Seminars in immunology. 2005;17:239–49. doi: 10.1016/j.smim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- [32].Le Campion A, Pommier A, Delpoux A, Stouvenel L, Auffray C, Martin B, et al. IL-2 and IL-7 determine the homeostatic balance between the regulatory and conventional CD4+ T cell compartments during peripheral T cell reconstitution. Journal of immunology. 2012;189:3339–46. doi: 10.4049/jimmunol.1103152. [DOI] [PubMed] [Google Scholar]
- [33].Chang X, Zheng P, Liu Y. Homeostatic proliferation in the mice with germline FoxP3 mutation and its contribution to fatal autoimmunity. Journal of immunology. 2008;181:2399–406. doi: 10.4049/jimmunol.181.4.2399. [DOI] [PubMed] [Google Scholar]
- [34].Kang S, Fernandes-Alnemri T, Rogers C, Mayes L, Wang Y, Dillon C, et al. Caspase-8 scaffolding function and MLKL regulate NLRP3 inflammasome activation downstream of TLR3. Nature communications. 2015;6:7515. doi: 10.1038/ncomms8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lesourne R, Zvezdova E, Song KD, El-Khoury D, Uehara S, Barr VA, et al. Interchangeability of Themis1 and Themis2 in thymocyte development reveals two related proteins with conserved molecular function. Journal of immunology. 2012;189:1154–61. doi: 10.4049/jimmunol.1200123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Oh J, Wu N, Baravalle G, Cohn B, Ma J, Lo B, et al. MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. The Journal of experimental medicine. 2013;210:1069–77. doi: 10.1084/jem.20122695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Peirce MJ, Brook M, Morrice N, Snelgrove R, Begum S, Lanfrancotti A, et al. Themis2/ICB1 is a signaling scaffold that selectively regulates macrophage Toll-like receptor signaling and cytokine production. PloS one. 2010;5:e11465. doi: 10.1371/journal.pone.0011465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zuo XJ, Nicolaidou E, Okada Y, Toyoda M, Jordan SC. Antithrombin III inhibits lymphocyte proliferation, immunoglobulin production and mRNA expression of lymphocyte growth factors (IL-2, gamma-IFN and IL-4) in vitro. Transplant immunology. 2001;9:1–6. doi: 10.1016/s0966-3274(01)00042-9. [DOI] [PubMed] [Google Scholar]
- [39].Zuo XJ, Okada Y, Nicolaidou E, Toyoda T, Marchevsky A, Matloff JM, et al. Antithrombin III inhibits T and B lymphocyte activation in vitro and improves parameters of inflammation in a rat model of acute lung allograft rejection. Transplantation proceedings. 1999;31:847–8. doi: 10.1016/s0041-1345(98)01800-4. [DOI] [PubMed] [Google Scholar]
- [40].Labi V, Woess C, Tuzlak S, Erlacher M, Bouillet P, Strasser A, et al. Deregulated cell death and lymphocyte homeostasis cause premature lethality in mice lacking the BH3-only proteins Bim and Bmf. Blood. 2014;123:2652–62. doi: 10.1182/blood-2013-11-537217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- [42].Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13:817–27. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- [43].Tan BS, Tiong KH, Choo HL, Fei-Lei Chung F, Hii LW, Tan SH, et al. Mutant p53-R273H mediates cancer cell survival and anoikis resistance through AKT-dependent suppression of BCL2-modifying factor (BMF) Cell death & disease. 2015;6:e1826. doi: 10.1038/cddis.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Narni-Mancinelli E, Campisi L, Bassand D, Cazareth J, Gounon P, Glaichenhaus N, et al. Memory CD8+ T cells mediate antibacterial immunity via CCL3 activation of TNF/ROI+ phagocytes. The Journal of experimental medicine. 2007;204:2075–87. doi: 10.1084/jem.20070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].D’Cunha J, Knight E, Jr., Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:211–5. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7578–82. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shibata E, Abbas T, Huang X, Wohlschlegel JA, Dutta A. Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin-conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Molecular and cellular biology. 2011;31:3136–45. doi: 10.1128/MCB.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Prazma CM, Yazawa N, Fujimoto Y, Fujimoto M, Tedder TF. CD83 expression is a sensitive marker of activation required for B cell and CD4+ T cell longevity in vivo. Journal of immunology. 2007;179:4550–62. doi: 10.4049/jimmunol.179.7.4550. [DOI] [PubMed] [Google Scholar]
- [49].Hastings WD, Anderson DE, Kassam N, Koguchi K, Greenfield EA, Kent SC, et al. TIM-3 is expressed on activated human CD4+ T cells and regulates Th1 and Th17 cytokines. European journal of immunology. 2009;39:2492–501. doi: 10.1002/eji.200939274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM, et al. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. Journal of immunology. 2008;180:249–60. doi: 10.4049/jimmunol.180.1.249. [DOI] [PubMed] [Google Scholar]
- [51].Kang SM, Tsang W, Doll S, Scherle P, Ko HS, Tran AC, et al. Induction of the POU domain transcription factor Oct-2 during T-cell activation by cognate antigen. Molecular and cellular biology. 1992;12:3149–54. doi: 10.1128/mcb.12.7.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].de Grazia U, Felli MP, Vacca A, Farina AR, Maroder M, Cappabianca L, et al. Positive and negative regulation of the composite octamer motif of the interleukin 2 enhancer by AP-1, Oct-2, and retinoic acid receptor. The Journal of experimental medicine. 1994;180:1485–97. doi: 10.1084/jem.180.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chattopadhyay G, Shevach EM. Antigen-specific induced T regulatory cells impair dendritic cell function via an IL-10/MARCH1-dependent mechanism. Journal of immunology. 2013;191:5875–84. doi: 10.4049/jimmunol.1301693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Velaga S, Ukena SN, Dringenberg U, Alter C, Pardo J, Kershaw O, et al. Granzyme A Is Required for Regulatory T-Cell Mediated Prevention of Gastrointestinal Graft-versus-Host Disease. PloS one. 2015;10:e0124927. doi: 10.1371/journal.pone.0124927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Choi SW, Hildebrandt GC, Olkiewicz KM, Hanauer DA, Chaudhary MN, Silva IA, et al. CCR1/CCL5 (RANTES) receptor-ligand interactions modulate allogeneic T-cell responses and graft-versus-host disease following stem-cell transplantation. Blood. 2007;110:3447–55. doi: 10.1182/blood-2007-05-087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dal Secco V, Soldani C, Debrat C, Asperti-Boursin F, Donnadieu E, Viola A, et al. Tunable chemokine production by antigen presenting dendritic cells in response to changes in regulatory T cell frequency in mouse reactive lymph nodes. PloS one. 2009;4:e7696. doi: 10.1371/journal.pone.0007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Jr., Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cellular and molecular life sciences : CMLS. 2013;70:661–87. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer cell. 2002;2:279–88. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- [59].Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Molecular cell. 1999;4:343–52. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- [60].Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–53. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- [61].Lin Y, Kang T, Zhou BP. Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1-dependent manner. Cell cycle. 2014;13:1708–16. doi: 10.4161/cc.28619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Takahashi E, Funato N, Higashihori N, Hata Y, Gridley T, Nakamura M. Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist. Biochemical and biophysical research communications. 2004;325:1136–44. doi: 10.1016/j.bbrc.2004.10.148. [DOI] [PubMed] [Google Scholar]
- [63].Lim SO, Kim H, Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS letters. 2010;584:2231–6. doi: 10.1016/j.febslet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- [64].Lim SO, Kim HS, Quan X, Ahn SM, Kim H, Hsieh D, et al. Notch1 binds and induces degradation of Snail in hepatocellular carcinoma. BMC biology. 2011;9:83. doi: 10.1186/1741-7007-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mathieu M, Cotta-Grand N, Daudelin JF, Thebault P, Labrecque N. Notch signaling regulates PD-1 expression during CD8(+) T-cell activation. Immunology and cell biology. 2013;91:82–8. doi: 10.1038/icb.2012.53. [DOI] [PubMed] [Google Scholar]
- [66].Bastea LI, Doppler H, Balogun B, Storz P. Protein kinase D1 maintains the epithelial phenotype by inducing a DNA-bound, inactive SNAI1 transcriptional repressor complex. PloS one. 2012;7:e30459. doi: 10.1371/journal.pone.0030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mingot JM, Vega S, Cano A, Portillo F, Nieto MA. eEF1A mediates the nuclear export of SNAG-containing proteins via the Exportin5-aminoacyl-tRNA complex. Cell reports. 2013;5:727–37. doi: 10.1016/j.celrep.2013.09.030. [DOI] [PubMed] [Google Scholar]
- [68].Zhang K, Rodriguez-Aznar E, Yabuta N, Owen RJ, Mingot JM, Nojima H, et al. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. The EMBO journal. 2012;31:29–43. doi: 10.1038/emboj.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Reed NP, Henderson MA, Oltz EM, Aune TM. Reciprocal regulation of Rag expression in thymocytes by the zinc-finger proteins, Zfp608 and Zfp609. Genes and immunity. 2013;14:7–12. doi: 10.1038/gene.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang F, Thomas LR, Oltz EM, Aune TM. Control of thymocyte development and recombination-activating gene expression by the zinc finger protein Zfp608. Nature immunology. 2006;7:1309–16. doi: 10.1038/ni1397. [DOI] [PubMed] [Google Scholar]
- [71].Gras B, Jacqueroud L, Wierinckx A, Lamblot C, Fauvet F, Lachuer J, et al. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PloS one. 2014;9:e92254. doi: 10.1371/journal.pone.0092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wuest TY, Willette-Brown J, Durum SK, Hurwitz AA. The influence of IL-2 family cytokines on activation and function of naturally occurring regulatory T cells. Journal of leukocyte biology. 2008;84:973–80. doi: 10.1189/jlb.1107778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Morikawa H, Ohkura N, Vandenbon A, Itoh M, Nagao-Sato S, Kawaji H, et al. Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5289–94. doi: 10.1073/pnas.1312717110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.