Abstract
Objectives
Given the sparse evidence for selection of first-line therapy for acute atrial fibrillation (AF) based on clinical factors alone, incorporation of genotype data may improve the effectiveness of treatment algorithms and advance the understanding of inter-patient heterogeneity. We tested whether candidate nucleotide polymorphisms (SNPs) related to AF physiologic responses are associated with ventricular rate control after intravenous diltiazem in the emergency department (ED).
Methods
We conducted an analysis within a prospective observational cohort of ED patients with acute symptomatic AF, ventricular rate > 110 beats per minute within the first 2 hours, initially treated with intravenous diltiazem, and who had DNA available for analysis. We evaluated 24 candidate SNPs that were grouped into 3 categories based on their phenotype response (atrioventricular nodal (AVN) conduction, resting heart rate, disease susceptibility) and calculated 3 genetic scores for each patient. Our primary outcome was maximum heart rate reduction within 4 hours of diltiazem administration. Multivariable regression was used to identify associations with the outcome while adjusting for age, sex, baseline heart rate, and diltiazem dose.
Results
Of the 142 patients, 127 had complete data for the primary outcome. None of the genetic scores for AVN conduction, resting heart rate, or AF susceptibility showed a significant association with maximal heart rate response.
Conclusion
Using a candidate SNP approach, screening for genetic variants associated with AVN conduction, resting heart rate, or AF susceptibility failed to provide significant data for predicting successful rate control response to intravenous diltiazem for treating acute AF in the ED.
Keywords: atrial fibrillation, pharmacogenetics, emergency department
1. Introduction
Atrial fibrillation (AF), the most common sustained cardiac arrhythmia, affects 33 million individuals worldwide.1 Emergency department (ED) evaluations and hospitalizations for primary AF have progressively increased with 463,000 ED evaluations and 422,000 hospitalizations in 2009.2,3 The administration of rate control medications, specifically atrioventricular nodal (AVN) blocking medications, is the predominant acute treatment strategy in the United States.2,4 Diltiazem, a non-dihydropyridine calcium channel blocker, is used most commonly in the United States and Canada.5 The decision to treat an ED patient with an AVN blocking agent or cardioversion depends on factors including the treating physician preference, duration of symptoms, patient hemodynamic stability, and the patient’s reported response to prior cardioversion procedures.6,7 To our knowledge, no investigations have studied whether genetic data might identify individuals who are adequate responders to acute rate control treatment when they have symptomatic AF with rapid ventricular rate. Given the sparse evidence for selection of first-line therapy for acute AF based on clinical factors alone, incorporation of genetic information may improve the effectiveness of treatment algorithms and provide a better understanding of inter-patient heterogeneity. We tested whether several nucleotide polymorphisms (SNPs) related to AF physiologic responses are associated with ventricular rate control after intravenous diltiazem in the ED with AF and rapid ventricular rates.
2. Methods
This investigation was conducted in patients who were enrolled in the Atrial Fibrillation and Flutter Outcomes and Risk Determination (AFFORD) study (Clinicaltrials.gov identifier NCT01138644), a prospective observational cohort of patients presenting with AF to the ED of a single large academic medical center. Details on the AFFORD methodology and cohort have been previously described.8,9 Briefly, the research team enrolled a convenience sample of adult (≥ 18 years old) ED patients who had an electrocardiogram (ECG) demonstrating AF or atrial flutter, and signs (e.g., tachycardia, dyspnea) or symptoms (e.g., palpitations, chest pain, shortness of breath, weakness, pre-syncope, or syncope) consistent with symptomatic AF or atrial flutter. Patients provided written informed consent to participate in the study and a supplementary informed consent was obtained for the collection and storage of genetic material. Our medical center Institutional Review Board approved this study.
We identified a subgroup of individuals from the AFFORD cohort who met the following inclusion criteria: ECG documented AF at time of presentation to the ED, ventricular rate was > 110 beats per minute (bpm) within the first two hours of their ED evaluation, the first rate control medication administered was IV diltiazem, genetic material was available for analysis, and Caucasian race. Although individuals of all ethnicities were enrolled in AFFORD, we limited this analysis only to patients who identified themselves as Caucasian to reduce the genetic heterogeneity.
Clinical data were obtained through patient interviews and review of electronic health records (EHR). Vital signs, medication administration, laboratory and radiographic results with time stamps were collected from the EHR. The baseline ventricular rate measurement was defined as the measured ventricular rate documented at the closest time point before the administration of the first dose of IV diltiazem in the ED. Our institution requires vital signs reassessment at a minimum of every 2 hours with more frequent recordings and documentation in hemodynamically unstable individuals. Data were compiled and stored in a web-based REDCap database.10 The investigators were blinded to genotype while creating the database.
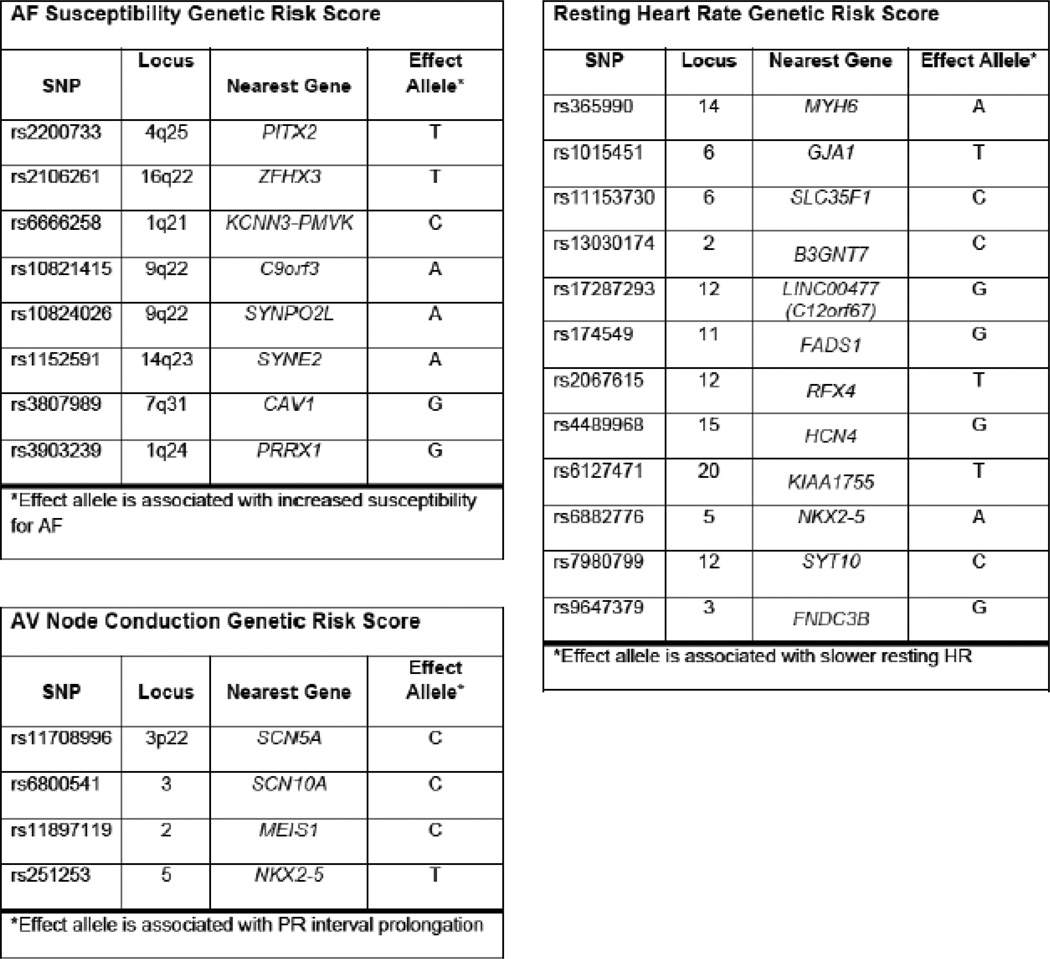
The primary exposures were genotypes of the patients. Genotypes were classified as having 0, 1 or 2 copies of the effect allele for the 24 SNPs associated with AF treatment response. SNPs were grouped into 3 categories (Figure 1) based on their phenotype response in prior studies evaluating chronic AF:11–18 (1) AF Disease Susceptibility; 2) Atrioventricular Nodal (AVN) Conduction; and 3) Resting Heart Rate. Genotyping was conducted by the Vanderbilt DNA Resources Core with the use of the Sequenom genotyping platform, based on a single-base primer extension reaction coupled with mass spectrometry. Quality-control procedures included examination of marker and sample genotyping efficiency, allele-frequency calculations, and tests of Hardy-Weinberg equilibrium (HWE). Laboratory personnel performing the genetic sequencing were blinded to patient’s response to acute rate-control therapy.
Figure 1. Categorization of SNPs associated with AF treatment response.
Detailed listing of the components of the 3 genetic scores, a score for each of the phenotype responses: 1) AF Disease Susceptibility; 2) Atrioventricular Nodal (AVN) Conduction; and 3) Resting Heart Rate for each patient
The primary outcome was maximum decrease in heart rate, defined as the largest difference between the baseline heart rate prior to diltiazem administration and lowest heart rate recorded within 4 hours following diltiazem administration. A secondary outcome included a dichotomous assessment of ventricular heart rate < 110 bpm within 4 hours of diltiazem administration (termed “rate controlled”) vs. ventricular heart rate remaining > 110 bpm after diltiazem (“rate not controlled”). A ventricular rate <110 bpm to define rate control was chosen based on data suggesting this target goal is non-inferior to strict rate control in the outpatient setting.19
All data are reported as number (%) or median (interquartile range) as applicable. We calculated 3 genetic scores, a score for each of the phenotype responses: 1) AF Disease Susceptibility; 2) Atrioventricular Nodal (AVN) Conduction; and 3) Resting Heart Rate for each patient. We assigned 0 points if the patient had neither effect allele for the SNP, 1 point for heterozygotes, and 2 points for homozygotes. Patients who were missing genetic data on 1 or more of the SNPs in each score category were excluded from the analysis.
The primary statistical analysis was pre-specified as a linear regression model with maximum heart rate reduction within the first 4 hours after IV diltiazem administration as the primary outcome. The following predictor variables were specified a priori for the model: age, sex, baseline ventricular rate, total weight-based diltiazem dose (mg/kg) received in the first 4 hours of treatment, and AF susceptibility genetic risk score. Patients were censored at the time of type I or III anti-arrhythmic medication administration and electrical cardioversion. No patients were lost to follow-up as all were followed for at least 4 hours after diltiazem administration.
In a secondary analysis, we used binary logistic regression to evaluate the association between the risk alleles and the dichotomous outcome, ventricular rate control at 4 hours after IV diltiazem. The model adjusted for the same clinical covariates as in the primary analysis. Statistical analyses were done with SPSS software (Version 21, IBM, Armonk, NY) and R.20
3. Results
Between June 8, 2010 and March 30, 2013, 4444 patients were screened for the study, 1045 met all eligibility criteria, and 532 were enrolled. Of the 513 patients not enrolled, 411 refused participation, 63 were not consentable, and 39 were missed. Of the 532 patients enrolled in the cohort, 439 consented for collection of specimens for genetic testing. Forty-one subjects were non-Caucasians (37 African Americans, 3 Asian Americans, 1 multiracial) and were excluded from the analysis. A total of 142 patients met all inclusion criteria for this genetic analysis. The baseline demographic and clinical characteristics for the study population are presented in Table 1. The detailed genotype results for the 142 patients are presented in Appendix Table 1. The minor allele frequency (MAF) was 0.19. The call rate was greater than 99%. The allelic distribution did not differ from Hardy-Weinberg equilibrium.
Table 1.
Study Cohort Demographics (n = 142)
| Age (yr) | 62 (54, 72) |
| Female | 46 (32%) |
| CHA2DS2-VASc | 2 (1 – 3) |
| Duration of Symptoms | |
| Less than 12 hours | 68 (48%) |
| 12 to 24 hours | 12 (9%) |
| 24 to 36 hours | 9 (6%) |
| 36 to 48 hours | 4 (3%) |
| Greater than 48 hours | 35 (25%) |
| Unknown duration | 14 (9%) |
| Atrial Fibrillation, Type | |
| Newly detected | 67 (47%) |
| Paroxysmal | 52 (37%) |
| Persistent | 16 (11%) |
| Permanent | 7 (5%) |
| Atrial Flutter | 25 (18%) |
| Comorbidities | |
| Heart Failure | 27 (19%) |
| Myocardial infarction | 18 (13%) |
| Coronary artery disease | 33 (23%) |
| Hypertension | 97 (68%) |
| Chronic kidney disease | 15 (11%) |
| Chronic obstructive pulmonary disease | 20 (14%) |
| Diabetes mellitus | 34 (24%) |
| Peripheral vascular disease | 15 (11%) |
| Cerebrovascular accident or transient ischemic attack | 20 (14%) |
| Hyperthyroidism | 14 (10%) |
| Hypothyroidism | 5 (4%) |
| Baseline ventricular rate, beats/min | 140 (127, 154) |
| Weight (kg) | 90 (77, 102) |
| Diltiazem First Dose (mg/kg) | 0.13 (0.1, 0.19) |
| Ventricular Rate Control at 2 hours (<110 bpm) | 62 (44%) |
| Ventricular Rate Control at 4 hours (<110 bpm) | 102 (72%) |
| Maximum Ventricular Rate Reduction within 4 hours (bpm) | 42 (23, 59) |
| Ejection Fraction, measured during this ED visit or subsequent inpatient hospitalization (percentage)* | 55 (50, 60) |
Continuous variables expressed as median (interquartile range) and dichotomous as n (percent).
72 (54%) patients had their ejection fraction measured during the index ED visit or subsequent inpatient hospitalization
Complete genetic data was available for 141 patients for the AVN conduction score. The scores ranged from 0 to 6 with a median (IQR) of 3 (2, 4). Among those in the cohort with available genetic data, 39 (28%) scored 2 and 33 (23%) scored 3.
The AF susceptibility score was calculated for the 127 patients with complete genetic data. The distribution of scores ranged from 1 to 9 with a median (IQR) of 6 (5, 7).
Complete genetic data was available for 132 patients to calculate the baseline resting heart rate score. The resting heart rate scores ranged from 6 to 19 with a median (IQR) of 11 (10, 12).
Of the 142 patients in the cohort, the median (interquartile range) maximum heart rate reduction within 4 hours was 42 (23 to 59) beats per minute. After adjusting for the clinical covariates, none of the scores were associated with predicting heart rate response to diltiazem (Tables 2 a–c).
Table 2.
a–c: Multivariable Analysis of Genetic Risk Scores and Clinical Covariates Predicting Maximum Heart Rate Reduction Within 4 hours Following Intravenous Diltiazem Administration
| a. AF Disease Susceptibility Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Standardized Beta | 95% CI | P-value |
| Age, year | 0.05 | −0.2, 0.45 | 0.49 |
| Female | −0.06 | −11.9, 5.4 | 0.46 |
| Baseline ventricular rate | 0.57 | 0.5, 0.9 | 0.00 |
| Diltiazem Dose (mg/kg) | 0.10 | −21.1, 107.3 | 0.19 |
| AF Susceptibility Genetic Risk Score | −0.04 | −2.9, 1.6 | 0.57 |
| b. Atrioventricular Nodal (AVN) Conduction Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Standardized Beta | 95% CI | P-value |
| Age, year | 0.65 | −0.14, 0.37 | 0.38 |
| Female | −0.03 | −9.4, 6.5 | 0.72 |
| Baseline ventricular rate | 0.55 | 0.5, 0.9 | 0.00 |
| Diltiazem Dose (mg/kg) | 0.08 | −29.3, 94.5 | 0.30 |
| AVN Conduction Genetic Risk Score | −0.02 | −3.0, 2.4 | 0.81 |
| c. Resting Heart Rate Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Standardized Beta | 95% CI | P-value |
| Age, year | 0.07 | −0.14, 0.38 | 0.35 |
| Female | −0.06 | −11.5, 4.9 | 0.42 |
| Baseline ventricular rate | 0.55 | 0.52, 0.90 | 0.00 |
| Diltiazem Dose (mg/kg) | 0.05 | −43.9, 84.7 | 0.53 |
| Resting Heart Rate Genetic Risk Score | 0.04 | −1.2, 2.2 | 0.58 |
Successful ventricular rate control (heart rate < 110 beats/minute) was achieved in 102 (72%) of the cohort within 4 hours of diltiazem administration. The secondary analysis found no significant association between these genetic scores and successful ventricular rate control within 4 hours (Tables 3 a–c).
Table 3.
a–c: Multivariable Analysis of Genetic Risk Scores and Clinical Covariates Predicting Successful Ventricular Rate Control (Rate Less than 110 beats per minute) Within 4 hours Following Intravenous Diltiazem Administration
| a. AF Disease Susceptibility Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Odds Ratio | 95% CI | P-value |
| Age, year | 1.0 | 0.97, 1.03 | 0.9 |
| Female | 0.73 | 0.3, 1.9 | 0.7 |
| Baseline ventricular rate | 1.02 | 1.0, 1.38 | 0.07 |
| Diltiazem Dose (mg/kg) | 0.01 | 0, 8.55 | 0.17 |
| AF Susceptibility Genetic Risk Score | 1.09 | 0.86, 1.38 | 0.5 |
| b. Atrioventricular Nodal (AVN) Conduction Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Odds Ratio | 95% CI | P-value |
| Age, year | 1.0 | 0.97, 1.02 | 0.8 |
| Female | 0.57 | 0.2, 1.4 | 0.6 |
| Baseline ventricular rate | 1.02 | 1.0, 1.04 | 0.03 |
| Diltiazem Dose (mg/kg) | 0.008 | 0, 7.99 | 0.17 |
| AVN Conduction Genetic Risk Score | 1.02 | 0.77, 1.35 | 0.9 |
| c. Resting Heart Rate Genetic Risk Score | |||
|---|---|---|---|
| Covariate | Odds Ratio | 95% CI | P-value |
| Age, year | 1.0 | 0.97, 1.03 | 0.86 |
| Female | 0.63 | 0.26, 1.5 | 0.32 |
| Baseline ventricular rate | 1.03 | 1, 1.05 | 0.02 |
| Diltiazem Dose (mg/kg) | .012 | 0, 16.1 | 0.23 |
| Resting Heart Rate Genetic Risk Score | 0.96 | 0.8, 1.15 | 0.11 |
4. Discussion
The current understanding and ability to accurately risk stratify which patients with AF are most likely to develop stroke, heart failure, and permanent AF is insufficient.21,22 Van Wagoner et al reported in their summary of the Heart Rhythm Society Research Forum on the Treatment and Prevention of Atrial Fibrillation that key knowledge gaps include whether genetic markers can predict response to therapy and can genetic data be successfully incorporated into AF/stroke risk prediction tools.21 Even the universally accepted CHA2DS2-VASc score for predicting strokes has modest c-statistics (0.59 – 0.79) reported in the literature.23 The potential that a patient’s genotype and genetic scoring may provide a more accurate individualized risk score is an area of ongoing research and an AF treatment goal.21, 22 In this paper, we aimed to identify whether an individual’s genotype and specific genetic scores may identify patients who are likely to respond to the most commonly used rate control agent in the US. The accurate identification of such patients might reduce unnecessary adverse effects related to administering rate control medications to patients who are genetically predisposed to fail these rate control drugs.
To date, studies investigating the acute heart rate response to AV nodal blocking medications in patients with AF have been unable to account for the significant inter-individual variability using clinical characteristics alone. Incorporating genotype data into clinical decision-making, facilitating a “precision medicine” approach to care, has been a recent emphasis of clinical and translational research. Therefore, we designed this analysis as a first-step towards exploring the association between common genetic variants and acute rate control in the ED. We used a candidate SNP approach to select SNPs with known associations at the genome-wide significance level with phenotypes potentially related to acute rate control (resting heart rate, AVN conduction, and susceptibility to AF). Given the effect size of a single common genetic variant is usually small for a complex, polygenetic trait such as rate control in AF, our primary analysis consisted of calculating three separate genetic risk scores to test for an association with our primary outcome. A significant association was not detected between our primary outcome-maximum heart rate reduction following IV diltiazem, or our secondary outcome of rate control < 110 beats/minute. Our findings suggest that genetic variants associated with sinus and AV node function at rest may not affect pathways important for ventricular response during AF, and/or have little relationship with drug response to diltiazem. Our findings may shape future genomic research exploring acute rate control in AF by suggesting the need to: 1) focus on other candidate pathways such as the metabolism of diltiazem, 2) utilize a more comprehensive genetic risk score with inclusion of sub-threshold SNPs, or 3) employ an agnostic genome wide association study (GWAS) approach.
To select SNPs for this study, we included only those that were found to reach the genome wide significance threshold in published genome-wide association studies of phenotypes that we believed may be important for modulating ventricular response to AF. To avoid including SNPs that may be in linkage disequilibrium, only one SNP per risk locus was included to prevent over-representation of a single risk signal in our genetic risk scores. For example, in the case of the 4q25 risk locus, only the rs2200733 SNP was included and it was selected based on the strongest previously published association with AF among other SNPs at that locus.17 Using these selection criteria for candidate SNPs, some individual SNPs may have been omitted from our analysis that could otherwise have been of interest. For example, the β1-adrenergic receptor polymorphisms that were previously found to associate with AF rate control during chronic therapy14 were not included because they were not signals detected in the genome-wide association studies for any of our 3 phenotypes of interest (resting heart rate, AVN conduction, AF susceptibility).
Although the AFFORD study enrolled the largest prospective cohort of ED patients with AF, our sample size with complete genetic data is relatively small. There is the potential that a larger study might detect an association. There is a possibility of bias in our analysis. This is mitigated by a number of factors. First, the investigators were blinded to genotype until after the database was curated. Second, since genotype was unknown to the clinician, treatment bias based on the genetic determinant is highly unlikely. The predominant treatment strategy was rate control with diltiazem, and this is consistent with usual practice in the US.2 Further prospective studies are needed to determine the difference in efficacy of other treatments between risk allele carriers. Our analysis was limited to Caucasians with AF to reduce the genetic heterogeneity and extrapolation of these results to non-Caucasians should be done with caution. Ongoing studies examining the role of common variants in other ethnic groups may provide insight into their role in AF.
In conclusion, screening for presence of AF susceptibility risk alleles failed to provide significant data for predicting successful rate control response to intravenous diltiazem for the management of acute AF in the emergency department.
Acknowledgments
No industry financial support or compensation has been or will be received for conducting this study. Barrett: Consultant, Red Bull GmbH, Fuschl am See, Salzburg and Boehringer Ingelheim Pharmaceuticals, Inc. Ridgefield, Connecticut. Others: None
Funding Sources: Dr. Barrett and this study are funded by NIH grant K23 HL102069 from the National Heart, Lung and Blood Institute, Bethesda, MD. Dr. Self is supported by NIH K23GM110469. Dr. Darbar is supported by the NIH (U19 HL65962, HL092217 and R01HL124935). Dr. Shoemaker is supported by NIH K23HL127704 from the National Heart, Lung and Blood Institute, Bethesda, MD. The project described was supported by The Clinical and Translational Science Award (CTSA) from the National Center for Research Resources, UL1 RR024975 and, UL1 TR000445. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Role of the Sponsors: The funding organizations had no role in the design and conduct of the study; the collection, analysis, and interpretation of the data; the preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Appendix
Table 1.
Results from the logistic regression model investigating the association between heart rate control (< 110 bpm) and the SNPs.
| Continuous univariate |
Continuous multivariable |
Categorical univariate |
Categorical multivariable |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P |
| rs10033464 | 0.98 | (0.45, 2.15) | 0.96 | 1.03 | (0.46, 2.32) | 0.94 | 1.08 | (0.46, 2.53) | 0.85 | 1.18 | (0.49, 2.85) | 0.72 |
| rs2200733 | 1.34 | (0.75, 2.41) | 0.32 | 1.22 | (0.66, 2.23) | 0.53 | 1.31 | (0.61, 2.82) | 0.49 | 1.16 | (0.52, 2.58) | 0.72 |
| rs6817105 | 1.34 | (0.75, 2.41) | 0.32 | 1.22 | (0.66, 2.23) | 0.53 | 1.31 | (0.61, 2.82) | 0.49 | 1.16 | (0.52, 2.58) | 0.72 |
| rs2106261 | 0.53 | (0.26, 1.11) | 0.09 | 0.47 | (0.22, 1.01) | 0.05 | 0.56 | (0.25, 1.25) | 0.16 | 0.50 | (0.22, 1.16) | 0.11 |
| rs11708996 | 1.27 | (0.64, 2.52) | 0.50 | 1.57 | (0.75, 3.29) | 0.23 | 1.19 | (0.53, 2.67) | 0.67 | 1.47 | (0.63, 3.45) | 0.37 |
| rs6795970 | 1.29 | (0.76, 2.21) | 0.35 | 1.32 | (0.76, 2.31) | 0.33 | 2.08 | (0.94, 4.62) | 0.07 | 2.22 | (0.97, 5.07) | 0.06 |
| rs6800541 | 1.17 | (0.69, 1.99) | 0.56 | 1.21 | (0.7, 2.09) | 0.50 | 1.77 | (0.81, 3.87) | 0.15 | 1.93 | (0.86, 4.36) | 0.11 |
| rs12632942 | 0.87 | (0.47, 1.6) | 0.65 | 0.92 | (0.49, 1.73) | 0.80 | 0.75 | (0.34, 1.61) | 0.46 | 0.81 | (0.36, 1.82) | 0.62 |
| rs6666258 | 1.29 | (0.73, 2.27) | 0.39 | 1.54 | (0.84, 2.84) | 0.16 | 1.63 | (0.75, 3.57) | 0.22 | 1.91 | (0.84, 4.34) | 0.12 |
| rs365990 | 0.89 | (0.53, 1.47) | 0.64 | 0.93 | (0.55, 1.59) | 0.80 | 0.53 | (0.25, 1.12) | 0.09 | 0.53 | (0.24, 1.15) | 0.11 |
| rs1015451 | 0.76 | (0.32, 1.8) | 0.53 | 0.79 | (0.32, 1.93) | 0.61 | 0.76 | (0.32, 1.8) | 0.53 | 0.79 | (0.32, 1.93) | 0.61 |
| rs10821415 | 1.48 | (0.88, 2.5) | 0.14 | 1.64 | (0.94, 2.85) | 0.08 | 1.23 | (0.57, 2.67) | 0.60 | 1.43 | (0.64, 3.23) | 0.38 |
| rs10824026 | 0.68 | (0.32, 1.44) | 0.31 | 0.63 | (0.29, 1.37) | 0.24 | 0.64 | (0.26, 1.59) | 0.34 | 0.59 | (0.23, 1.5) | 0.27 |
| rs11153730 | 1.07 | (0.65, 1.77) | 0.78 | 0.98 | (0.58, 1.68) | 0.95 | 1.50 | (0.67, 3.35) | 0.32 | 1.27 | (0.55, 2.95) | 0.57 |
| rs1152591 | 0.77 | (0.46, 1.31) | 0.33 | 0.77 | (0.44, 1.34) | 0.36 | 0.78 | (0.37, 1.66) | 0.53 | 0.74 | (0.33, 1.62) | 0.45 |
| rs11897119 | 0.93 | (0.53, 1.61) | 0.78 | 0.86 | (0.48, 1.53) | 0.61 | 0.91 | (0.43, 1.94) | 0.81 | 0.87 | (0.4, 1.93) | 0.74 |
| rs13030174 | 0.90 | (0.47, 1.73) | 0.76 | 0.89 | (0.45, 1.76) | 0.73 | 0.97 | (0.46, 2.06) | 0.93 | 0.95 | (0.43, 2.09) | 0.89 |
| rs17287293 | 1.37 | (0.57, 3.25) | 0.48 | 1.66 | (0.66, 4.15) | 0.28 | 1.37 | (0.57, 3.25) | 0.48 | 1.66 | (0.66, 4.15) | 0.28 |
| rs1733724 | 1.30 | (0.74, 2.27) | 0.36 | 1.22 | (0.68, 2.19) | 0.49 | 1.59 | (0.76, 3.33) | 0.22 | 1.54 | (0.71, 3.33) | 0.27 |
| rs174549 | 1.25 | (0.74, 2.12) | 0.40 | 1.33 | (0.76, 2.31) | 0.32 | 1.08 | (0.52, 2.25) | 0.83 | 1.12 | (0.52, 2.41) | 0.77 |
| rs1801252 | 0.77 | (0.36, 1.62) | 0.49 | 0.69 | (0.32, 1.48) | 0.34 | 0.72 | (0.31, 1.7) | 0.45 | 0.66 | (0.27, 1.63) | 0.37 |
| rs1801253 | 0.89 | (0.52, 1.52) | 0.68 | 0.85 | (0.49, 1.49) | 0.58 | 1.04 | (0.5, 2.16) | 0.92 | 0.94 | (0.43, 2.03) | 0.87 |
| rs2067615 | 1.00 | (0.59, 1.69) | 0.99 | 1.01 | (0.58, 1.75) | 0.97 | 0.70 | (0.32, 1.54) | 0.38 | 0.70 | (0.31, 1.59) | 0.40 |
| rs251253 | 0.84 | (0.47, 1.49) | 0.55 | 0.76 | (0.42, 1.38) | 0.37 | 0.58 | (0.26, 1.26) | 0.17 | 0.55 | (0.24, 1.24) | 0.15 |
| rs3807989 | 0.61 | (0.33, 1.11) | 0.11 | 0.60 | (0.32, 1.12) | 0.11 | 0.66 | (0.32, 1.37) | 0.27 | 0.66 | (0.3, 1.43) | 0.29 |
| rs3903239 | 1.48 | (0.86, 2.58) | 0.16 | 1.46 | (0.82, 2.61) | 0.20 | 2.46 | (0.99, 6.13) | 0.05 | 2.98 | (1.07, 8.29) | 0.04 |
| rs4489968 | 1.24 | (0.68, 2.26) | 0.48 | 1.34 | (0.72, 2.5) | 0.36 | 1.08 | (0.5, 2.32) | 0.85 | 1.16 | (0.52, 2.6) | 0.72 |
| rs6127471 | 1.19 | (0.7, 2.04) | 0.52 | 1.13 | (0.65, 1.99) | 0.66 | 1.27 | (0.58, 2.8) | 0.55 | 1.19 | (0.52, 2.69) | 0.68 |
| rs6882776 | 0.51 | (0.27, 0.97) | 0.04 | 0.52 | (0.27, 0.99) | 0.04 | 0.43 | (0.2, 0.92) | 0.03 | 0.44 | (0.2, 0.98) | 0.05 |
| rs7980799 | 1.00 | (0.6, 1.68) | 0.99 | 0.91 | (0.53, 1.57) | 0.74 | 1.38 | (0.62, 3.08) | 0.44 | 1.15 | (0.5, 2.65) | 0.75 |
| rs9647379 | 1.14 | (0.68, 1.91) | 0.62 | 1.25 | (0.72, 2.17) | 0.44 | 1.17 | (0.55, 2.47) | 0.69 | 1.30 | (0.58, 2.91) | 0.52 |
Table 2.
Results from the linear regression model investigating the association between maximum heart rate reduction in the first 4 hours after administering diltiazem and the SNPs
| Continuous univariate | Continuous multivariable |
Categorical univariate | Categorical multivariable |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Beta | 95% CI | P | Beta | 95% CI | P | Beta | 95% CI | P | Beta | 95% CI | P |
| rs10033464 | −4.19 | (−13.17, 4.8) | 0.36 | −3.32 | (−10.94, 4.31) | 0.40 | −6.86 | (−16.64, 2.93) | 0.17 | −4.51 | (−12.85, 3.82) | 0.29 |
| rs2200733 | −0.11 | (−7.06, 6.84) | 0.98 | −1.72 | (−7.64, 4.21) | 0.57 | 1.52 | (−7.42, 10.45) | 0.74 | 0.60 | (−7.04, 8.24) | 0.88 |
| rs6817105 | −0.11 | (−7.06, 6.84) | 0.98 | −1.72 | (−7.64, 4.21) | 0.57 | 1.52 | (−7.42,10.45) | 0.74 | 0.60 | (−7.04, 8.24) | 0.88 |
| rs2106261 | 8.28 | (0.87, 15.7) | 0.03 | 5.54 | (−0.83, 11.9) | 0.09 | 8.13 | (−0.49, 16.75) | 0.07 | 5.55 | (−1.8, 12.9) | 0.14 |
| rs11708996 | −4.70 | (−12.8, 3.39) | 0.26 | −2.29 | (−9.26, 4.68) | 0.52 | −3.04 | (−12.44, 6.37) | 0.53 | −0.17 | (−8.23, 7.9) | 0.97 |
| rs6795970 | −1.13 | (−7.32, 5.06) | 0.72 | −1.99 | (−7.24, 3.26) | 0.46 | −4.82 | (−13.38, 3.75) | 0.27 | −4.82 | (−12.08, 2.44) | 0.20 |
| rs6800541 | 0.09 | (−6.03, 6.22) | 0.98 | −0.86 | (−6.06, 4.34) | 0.75 | −3.81 | (−12.39, 4.77) | 0.39 | −3.81 | (−11.09, 3.47) | 0.31 |
| rs12632942 | −2.40 | (−9.28, 4.47) | 0.49 | 0.06 | (−5.79, 5.92) | 0.98 | −2.04 | (−10.73, 6.64) | 0.65 | 2.51 | (−4.92, 9.94) | 0.51 |
| rs6666258 | −8.30 | (−14.72, −1.89) | 0.01 | −5.51 | (−11.06, 0.05) | 0.05 | −8.58 | (−17.12, −0.04) | 0.05 | −7.56 | (−14.82, −0.31) | 0.04 |
| rs365990 | −3.69 | (−9.45, 2.08) | 0.21 | −1.92 | (−6.84, 3.01) | 0.45 | 0.11 | (−8.63, 8.85) | 0.98 | 4.03 | (−3.41, 11.47) | 0.29 |
| rs1015451 | −3.75 | (−13.36, 5.85) | 0.45 | −2.72 | (−10.85, 5.42) | 0.51 | −3.75 | (−13.36, 5.85) | 0.45 | −2.72 | (−10.85, 5.42) | 0.51 |
| rs10821415 | −2.89 | (−8.87, 3.08) | 0.34 | −4.12 | (−9.28, 1.04) | 0.12 | −1.71 | (−10.47, 7.05) | 0.70 | −2.19 | (−9.74, 5.37) | 0.57 |
| rs10824026 | 4.38 | (−3.53, 12.3) | 0.28 | 3.39 | (−3.18, 9.96) | 0.31 | 5.56 | (−4.31, 15.43) | 0.27 | 4.02 | (−4.19, 12.24) | 0.34 |
| rs11153730 | 2.72 | (−3.02, 8.45) | 0.36 | 0.57 | (−4.4, 5.54) | 0.82 | 1.20 | (−7.68, 10.09) | 0.79 | −2.29 | (−9.94, 5.37) | 0.56 |
| rs1152591 | 5.53 | (−0.33, 11.39) | 0.07 | 4.20 | (−0.81, 9.2) | 0.10 | 8.63 | (−0.02, 17.28) | 0.05 | 5.04 | (−2.41, 12.49) | 0.19 |
| rs11897119 | 2.09 | (−4.22, 8.41) | 0.52 | −0.51 | (−5.9, 4.89) | 0.85 | 3.92 | (−4.82, 12.66) | 0.38 | 0.11 | (−7.39, 7.62) | 0.98 |
| rs13030174 | 5.55 | (−1.82, 12.92) | 0.14 | 6.59 | (0.37, 12.8) | 0.04 | 5.55 | (−3.14, 14.24) | 0.21 | 7.39 | (0.09, 14.69) | 0.05 |
| rs17287293 | −2.01 | (−12.31, 8.28) | 0.70 | −2.32 | (−11.15, 6.51) | 0.61 | −2.01 | (−12.31, 8.28) | 0.70 | −2.32 | (−11.15, 6.51) | 0.61 |
| rs1733724 | −3.92 | (−10.5, 2.66) | 0.25 | −4.67 | (−10.27, 0.94) | 0.11 | −3.41 | (−11.99, 5.17) | 0.44 | −5.83 | (−13.1, 1.44) | 0.12 |
| rs174549 | −2.89 | (−9.04, 3.25) | 0.36 | −0.42 | (−5.71, 4.86) | 0.88 | −0.70 | (−9.11, 7.72) | 0.87 | 1.77 | (−5.42, 8.96) | 0.63 |
| rs1801252 | 6.71 | (−1.41, 14.83) | 0.11 | 3.44 | (−3.5, 10.37) | 0.33 | 6.26 | (−3.21, 15.73) | 0.20 | 3.35 | (−4.71, 11.41) | 0.42 |
| rs1801253 | 0.87 | (−5.16, 6.91) | 0.78 | −0.46 | (−5.58, 4.65) | 0.86 | 0.48 | (−7.98, 8.94) | 0.91 | −2.34 | (−9.53, 4.85) | 0.52 |
| rs2067615 | 0.12 | (−5.93, 6.16) | 0.97 | 1.16 | (−3.96, 6.28) | 0.66 | 2.63 | (−6.57, 11.83) | 0.58 | 4.34 | (−3.44, 12.12) | 0.28 |
| rs251253 | 1.71 | (−4.88, 8.31) | 0.61 | 1.71 | (−3.92, 7.33) | 0.55 | 2.80 | (−6.51, 12.1) | 0.56 | 4.91 | (−2.95, 12.77) | 0.22 |
| rs3807989 | 1.48 | (−5.05, 8.02) | 0.66 | 3.17 | (−2.41, 8.75) | 0.27 | 1.34 | (−7.11, 9.78) | 0.76 | 3.20 | (−4.03, 10.42) | 0.39 |
| rs3903239 | −0.74 | (−7.02, 5.53) | 0.82 | −2.16 | (−7.59, 3.28) | 0.44 | −3.93 | (−13.12, 5.26) | 0.40 | −5.50 | (−13.65, 2.66) | 0.19 |
| rs4489968 | −6.08 | (−13.1, 0.94) | 0.09 | −6.00 | (−11.96, −0.04) | 0.05 | −6.59 | (−15.41, 2.23) | 0.15 | −5.89 | (−13.46, 1.68) | 0.13 |
| rs6127471 | 1.85 | (−4.34, 8.05) | 0.56 | −0.10 | (−5.43, 5.23) | 0.97 | 2.24 | (−6.64, 11.12) | 0.62 | 0.40 | (−7.2, 8) | 0.92 |
| rs6882776 | 3.66 | (−2.83, 10.15) | 0.27 | 5.09 | (−0.41, 10.6) | 0.07 | 3.14 | (−5.26, 11.55) | 0.46 | 7.24 | (0.1, 14.38) | 0.05 |
| rs7980799 | 4.28 | (−1.6, 10.15) | 0.16 | 3.44 | (−1.59, 8.47) | 0.18 | 6.19 | (−2.74, 15.11) | 0.18 | 2.62 | (−5.07, 10.31) | 0.51 |
| rs9647379 | −5.21 | (−11.12, 0.7) | 0.09 | −3.22 | (−8.35, 1.91) | 0.22 | −6.99 | (−15.49, 1.51) | 0.11 | −4.25 | (−11.61, 3.11) | 0.26 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Clinical Trial Registration: ClinicalTrials.gov, NCT01138644, Available at: http://clinicaltrials.gov/ct2/show/NCT01138644?term=AFFORD&rank=1
Conflicts of Interests/Disclosures: There are no conflicts of interest in connection with this submission or are there any copyright constraints.
References
- 1.Chugh SS, Havmoeller R, Narayanan K, et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation. 2014;129:837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett TW, Self WH, Jenkins CA, et al. Predictors of regional variations in hospitalizations following emergency department visits for atrial fibrillation. Am J Cardiol. 2013;112:1410–1416. doi: 10.1016/j.amjcard.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel NJ, Deshmukh A, Pant S, et al. Contemporary trends of hospitalization for atrial fibrillation in the United States, 2000 through 2010: implications for healthcare planning. Circulation. 2014;129:2371–2379. doi: 10.1161/CIRCULATIONAHA.114.008201. [DOI] [PubMed] [Google Scholar]
- 4.McDonald AJ, Pelletier AJ, Ellinor PT, Camargo CA., Jr Increasing US emergency department visit rates and subsequent hospital admissions for atrial fibrillation from 1993 to 2004. Ann Emerg Med. 2008;51:58–65. doi: 10.1016/j.annemergmed.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Rogenstein C, Kelly A-M, Mason S, et al. An international view of how recent-onset atrial fibrillation is treated in the emergency department. 2012;19:1255–1260. doi: 10.1111/acem.12016. [DOI] [PubMed] [Google Scholar]
- 6.Atzema CL, Barrett TW. Managing atrial fibrillation. Ann Emerg Med. 2015;65:532–539. doi: 10.1016/j.annemergmed.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.January CT, Wann S, Alpert JS, et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. Circulation. 2014 doi: 10.1161/CIR.0000000000000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett TW, Storrow AB, Jenkins CA, et al. The AFFORD clinical decision aid to identify emergency department patients with atrial fibrillation at low risk for 30-day adverse events. Am J Cardiol. 2015;115:763–770. doi: 10.1016/j.amjcard.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett TW, Storrow AB, Jenkins CA, et al. Atrial fibrillation and flutter outcomes and risk determination (AFFORD): design and rationale. J Cardiol. 2011;58:124–130. doi: 10.1016/j.jjcc.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker MB, Bollmann A, Lubitz SA, et al. Common genetic variants and response to atrial fibrillation ablation. Circ Arrhythm Electrophysiol. 2015;8:296–302. doi: 10.1161/CIRCEP.114.001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolek MJ, Edwards TL, Muhammad R, et al. A genome-wide association study to identify genomic modulators of rate control therapy in patients with atrial fibrillation. Am J Cardiol. 2014;114:593–600. doi: 10.1016/j.amjcard.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolek MJ, Parvez B, Muhammad R, et al. A common variant on chromosome 4q25 is associated with prolonged PR interval in subjects with and without atrial fibrillation. Am J Cardiol. 2014;113:309–313. doi: 10.1016/j.amjcard.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvez B, Shoemaker MB, Muhammad R, et al. Common genetic polymorphism at 4q25 locus predicts atrial fibrillation recurrence after successful cardioversion. Heart Rhythm. 2013;10:849–855. doi: 10.1016/j.hrthm.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–573. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darbar D. Genetics of atrial fibrillation: rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5:483–486. doi: 10.1016/j.hrthm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubitz SA, Ozcan C, Magnani JW, Kaab S, Benjamin EJ, Ellinor PT. Genetics of atrial fibrillation: implications for future research directions and personalized medicine. Circulation Arrhythm Electrophysiol. 2010;3:291–299. doi: 10.1161/CIRCEP.110.942441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Gelder IC, Groenveld HF, Crijns HJ, et al. Lenient versus strict rate control in patients with atrial fibrillation. N Engl J Med. 2010;362:1363–1373. doi: 10.1056/NEJMoa1001337. [DOI] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 21.Van Wagoner DR, Piccini JP, Albert CM, et al. Progress toward the prevention and treatment of atrial fibrillation: A summary of the Heart Rhythm Society Research Forum on the Treatment and Prevention of Atrial Fibrillation, Washington, DC, December 9–10, 2013. Heart Rhythm. 2015;12:e5–e29. doi: 10.1016/j.hrthm.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christophersen IE, Ellinor PT. Genetics of atrial fibrillation: from families to genomes. J Hum Genet. 2015 doi: 10.1038/jhg.2015.44. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Chen JY, Zhang AD, Lu HY, Guo J, Wang FF, Li ZC. CHADS2 versus CHA2DS2-VASc score in assessing the stroke and thromboembolism risk stratification in patients with atrial fibrillation: a systematic review and meta-analysis. J Geriatr Cardiol. 2013;10:258–266. doi: 10.3969/j.issn.1671-5411.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]