Abstract
Heterogeneity of obesity within a population of inbred mice fed an obesogenic high-fat diet (HFD) is associated with changes of gene expression in white adipose tissue (WAT). One gene in particular with large variations among mice, mesoderm-specific transcript (Mest), has been shown to be highly inducible after being fed a short-term HFD, and its expression in WAT before HFD feeding is predictive for susceptibility to the development of obesity. To gain further insight on the association of Mest with rapid changes in body composition, 96 individually housed C57BL/6J mice were fed an HFD for only 2 weeks, resultin in a 12-fold and 90-fold variation in Mest mRNA in visceral epididymal and subcutaneous inguinal WAT, respectively. WAT Mest mRNA was positively associated with interindividual variation of fat mass. Surprisingly, there was only a slight association of WAT Mest with food intake when normalized by body weight or lean mass. In addition, WAT Mest expression coincided highly with the expression of the transcription factor Kruppel-like factor 14 (Klf14), an imprinted gene that regulates lipid metabolism in WAT. Our data suggest that KLF14 transcriptional activity may partially mediate, or act in concert with, MEST as part of an epigenetic mechanism that promotes fat mass accumulation in mice fed an obesogenic diet.
Keywords: Mest, Klf14, adiposity, obesity, epigenetics
Introduction
Complex metabolic phenotypes, such as obesity and type 2 diabetes (T2D), have been shown to be highly variable among individuals within genetically identical populations of mice.1–3 Our studies have also demonstrated that variation of fat mass expansion in inbred mice fed an obesogenic diet is highly stable and distinctive among individual animals and suggests that an epigenetic etiology, possibly established via developmental programming, could be involved in these phenotypic differences between mice.3,4 Analyses of gene expression in subcutaneous white adipose tissue (WAT) of male C57BL/6J (B6) mice fed a high-fat diet (HFD) for 4 weeks identified a subset of genes that correlate highly with fat mass accretion, including imprinted genes and genes involved in the Wnt and Tgf-β signaling pathways.3,5 Mesoderm-specific transcript (Mest), a maternally imprinted gene belonging to the α/β hydrolase family, showed the largest variation (almost 80-fold) among mice, and its expression was highly and positively associated with secreted frizzled-related sequence protein 5 (Sfrp5) and bone morphogenetic protein 3 (Bmp3). In addition, mRNA expression of Mest, Sfrp5, and Bmp3 in WAT biopsies of mice before exposure to an obesogenic diet was predictive for the future development of adiposity.3
Although evidence for a role of Mest in facilitating fat accumulation in cell-based in vitro models and in WAT3,5–8 is compelling, little is known about its catalytic function and how it is transcriptionally regulated. Evidence suggests that Mest is localized within the endoplasmic reticulum/Golgi apparatus of the adipocyte where it may act to facilitate fat storage in lipid droplets.5,6 We have also demonstrated that increased WAT Sfrp5 expression is associated with enhanced adiposity in B6 mice after an HFD, which is consistent with findings on the role of these soluble inhibitors of Wnt signaling in the development of adiposity in mice.3,9 Recent evidence suggests that Sfrp5 may act to stimulate adipocyte hypertrophy via inhibition of oxidative metabolism and/or act as an anti-inflammatory adipokine that regulates metabolic dysfunction and inflammation.10–12 Bmps have been shown to be involved in a wide variety of morphogenetic processes during development.13–17 However, Bmp3, unlike other Bmps, antagonizes osteogenesis by activation of the Tgf-β/activin pathway.18,19 This antagonistic effect of Bmp3 on osteogenesis is consistent with a morphogenetic role for Bmp3 in adipogenesis.19–21
The recent identification of Kruppel-like factor 14 (Klf14), a maternally-expressed transcription factor, as a regulator of lipid metabolism and lipid signaling22,23 suggests that KLF14 may potentially play a role in mediating fat mass expansion in populations of genetically identical inbred mice. Subsequent analyses of global gene expression data of inguinal (ING) fat from male C57BL/6J mice fed an obesogenic diet for 4 weeks revealed significantly elevated Klf14 expression (1.52-fold; P = 0.0054) in mice with high versus low weight gain (deposited in NCBI's Gene Expression Omnibus (GEO), with accession number GSE4692).3 In this study, a cohort of 96 male C57BL/6J mice were subjected to short-term (2 week) feedings of dietary fat to determine the effects on WAT Mest mRNA expression and the association of Mest with indices of fat mass expansion, caloric intake, and WAT gene expression. As expected, our results showed strong associations between adipose Mest mRNA with indices of adiposity, but, surprisingly, little association was observed for caloric intake during the 2-week dietary-fat feeding regimen. In addition, adipose Mest expression correlated very highly with Klf14 expression in both subcutaneous and visceral WAT, suggesting that KLF14 transcriptional activity may at least partially mediate WAT MEST to promote fat mass accumulation in this model designed to investigate the epigenetic basis for the development of obesity.
Materials and methods
Animals and phenotyping
All in vivo experiments were carried out in age-matched male C57BL/6J (B6) mice (n = 96) purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained in a temperature-controlled room (23 °C) with a 12-h light/12-h dark cycle. They were reared under conventional conditions and fed PicoLab Rodent Diet 20 (LabDiet, 13% kcal fat) until 8 weeks of age. Mice were singly housed at 7 weeks of age and then fed an HFD (Surwit Diet D12331, Research Diets; 58% kcal fat; 23.26 kJ/g) for 2 weeks starting at 8 weeks of age. Body composition was analyzed at 8 and 10 weeks of age with a nuclear magnetic resonance (NMR) minispec (Bruker), which uses the contrasting hydrogen density and/or hydrogen spin properties from fat and lean mass to estimate body composition. A quality-control check of NMR parameters using a standard provided by the manufacturer was performed at the beginning of each day of testing. Food intake was measured for all mice during the 2 weeks of HFD feeding. All animal experiments were approved by the Pennington Biomedical Research Center and Maine Medical Center Research Institute Institutional Animal Care and Use Committees and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Gene expression analyses
Epididymal (EPI) and ING WAT of mice were isolated from euthanized mice and quickly frozen in liquid nitrogen. RNA was extracted from tissue homogenized in TriReagent (Molecular Research Center, Inc.) and then purified using RNeasy Mini Kit and RNase-free DNAse (Qiagen). Isolated RNA was protected from RNAse contamination with SUPERase-In RNase inhibitor (Life Technologies). RNA quantity and quality was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed using total RNA, with specific primers and probes designed using Primer Express Software v3.0.1 (Life Technologies) as previously described5 except that a BioRad CFX Real-Time System was used for analyses. Gene expression data were normalized to cyclophilin B (Ppib).
Statistics
Statistical calculations were performed using GraphPad Prism 6 and Microsoft Excel 2010. Differences between two groups were calculated using a two-tailed unpaired parametric t-test with confidence level at 95%. Significance among multiple groups was calculated with a one-way ANOVA followed by Tukey’s multiple comparison test with an alpha of 0.5. Pearson correlation coefficients (two-tailed; assuming Gaussian distribution) were calculated with a confidence interval of 95%. Data are presented as mean ± SEM. A P value of < 0.05 was considered to be significant.
Results
Body weight and composition of male C57BL/6J mice fed HFD for 2 weeks
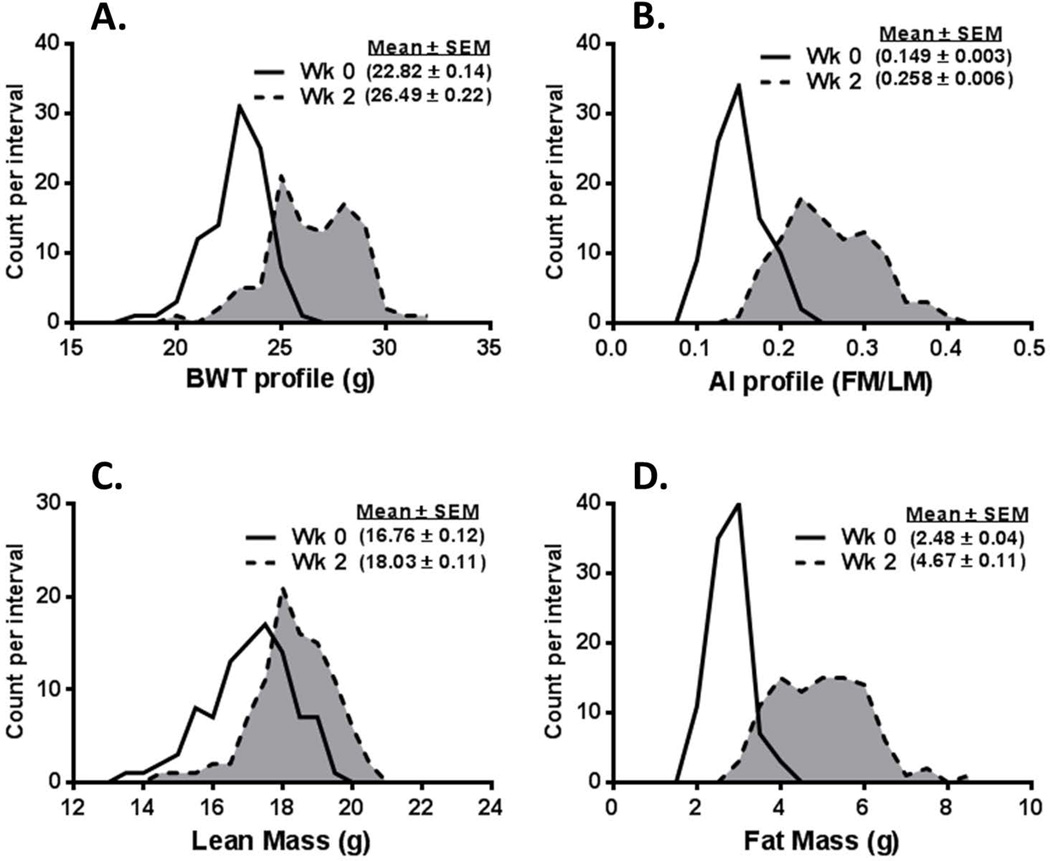
Frequency distribution profiles for body weight (BWT) of 96 male B6 mice (Fig. 1A) fed an HFD from 8 (Wk 0) to 10 weeks of age (Wk 2) showed an average weight gain of approximately 3.7 g, with a range of 1.5–7.4 grams. Surprisingly, after feeding mice an HFD for only 2 weeks, a broad and somewhat bimodal distribution of BWTs became evident, with peaks at about 25 and 28 grams. Indices of adiposity as measured by the adiposity index (AI; fat mass/lean mass ratio; Fig.1B) or fat mass (Fig. 1D) showed frequency distribution patterns that are consistent with the observed change in BWT and exhibit a modest bimodal distribution. The average increase in fat mass was 2.19 g/mouse, with a range of 0.2–5.5 g, whereas lean mass increased by an average of only 1.27 g/mouse, with a range of 0.1–2.7 g (Fig. 1C), indicating a significantly greater contribution of adiposity to changes in BWT during the 2-week obesogenic diet. Results from a regression analysis comparing fat mass (R = 0.83; P = 1.8E-25) and lean mass (R = 0.46; P = 3.0E-06) gain versus the change in BWT are consistent with the significance of the contribution of fat mass to overall BWT gain.
Figure 1.
Data represent the frequency distribution for (A) body weight (BWT), (B) adiposity index (AI) represented as fat mass (FM)/lean mass (LM), (C) LM, and (D) FM of 96 male C57BL/6J mice before (solid line, unshaded) and after (dashed line, shaded) being fed a high-fat diet for 2 weeks.
Association of Mest with BWT, body composition, and food intake
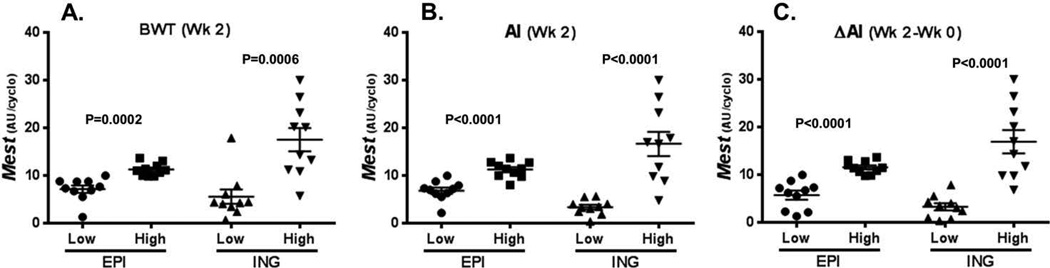
Data in Figure 2 show the level of Mest expression in both EPI and ING WAT of mice within the lowest and highest 10th percentile (10 mice per group) for BWT (Fig. 2A), AI (Fig. 2B), and the change of AI during the 2 weeks of feeding an obesogenic diet (Fig. 2C). Although differences in WAT Mest mRNA in mice can be observed 2–7 days after the initiation of an HFD,5 it was somewhat unexpected that changes in both BWT and adiposity indices occurring after feeding mice an obesogenic diet for only 2 weeks would show significant associations with Mest mRNA expression in both EPI and ING WAT.
Figure 2.
The expression of Mest mRNA was measured in epididymal (EPI) and inguinal (ING) adipose tissue of mice within the lowest and highest 10th percentile (10 mice/group) for (A) body weight (BWT), (B) adiposity index (AI), and (C) change in adiposity index (ΔAI) after 2 weeks of feeding an obesogenic diet. Gene expression measured by TaqMan quantitative RT-PCR is represented as arbitrary units (AU) normalized to cyclophilin B. Significance between groups measured by two-tailed unpaired parametric t-tests is indicated on each figure.
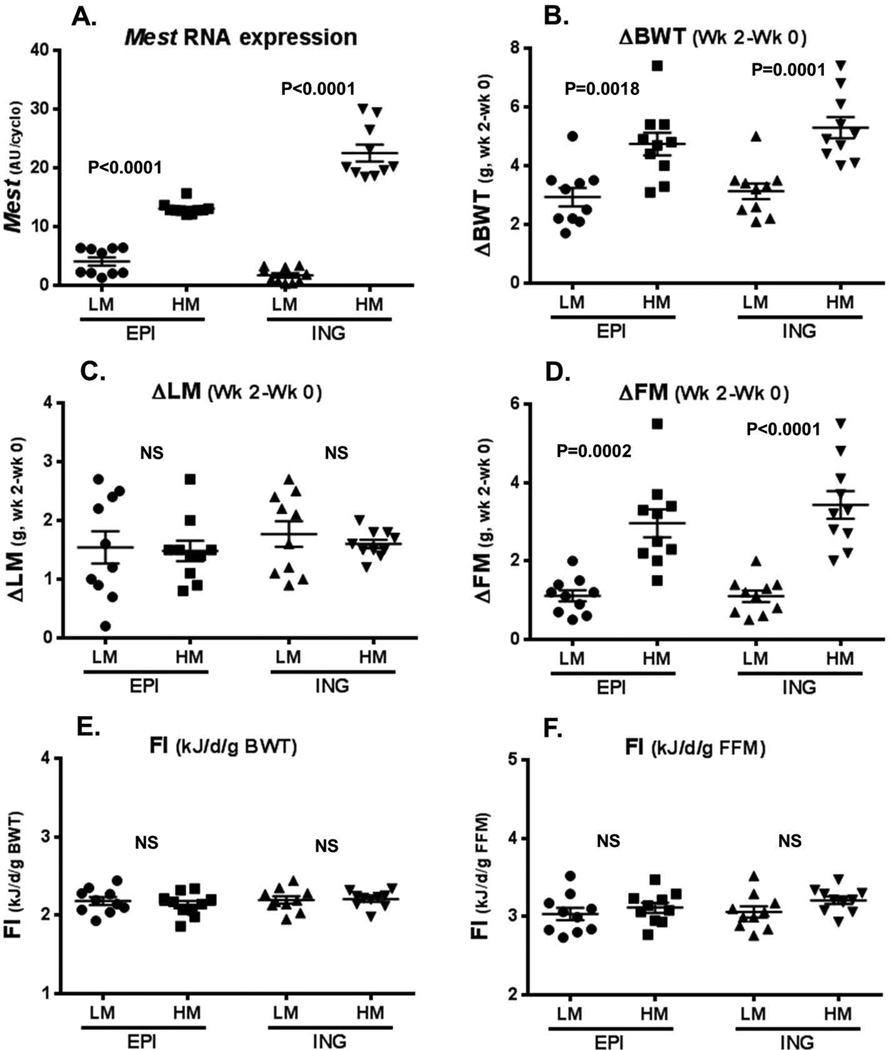
A comparison of the lowest and highest 10th percentiles of Mest expression in both EPI and ING fat showed highly significant differences between subsets of mice, as indicated by a lack of overlap in gene expression values (Fig. 3A). Although the range for Mest expression in ING fat was large compared with EPI fat after 2 weeks of an HFD, the average for Mest expression among the cohort of 96 mice was not significantly different between the two WAT depots (EPI, 9.23 ± 0.26 AU/cyclo; ING 9.67 ± 0.62 AU/cyclo; P = 0.52). In addition, regression analysis showed a strong correlation (R = 0.69; P = 1.2E-14) between EPI and ING fat Mest expression in individual mice.
Figure 3.
Data represent a comparison of phenotypes of mice within the lowest (low Mest; LM) and highest (high Mest; HM) 10th percentile groups (10 mice/group) for epididymal (EPI) and inguinal (ING) adipose tissue Mest mRNA after the 2 weeks of feeding an obesogenic diet. A represents Mest mRNA levels of the LM and HM cohorts in EPI and ING adipose tissue; and B–D show the change in body weight (ΔBWT), lean mass (ΔLM), and fat mass (ΔFM) in the LM and HM mice. Panels (E–F) represent food intake in mice with LM and HM normalized to BWT or fat-free mass (FFM), respectively. Gene expression measured by TaqMan quantitative RT-PCR is represented as arbitrary units (AU) normalized to cyclophilin B. Significance between groups measured by two-tailed unpaired parametric t-tests is indicated on each figure.
The analyses of the changes in BWT, fat mass, and lean mass in mice with the lowest and highest 10th percentiles of EPI and ING fat Mest expression after two weeks on an HFD showed highly significant increases in BWT (ΔBWT; Fig. 3B) and fat mass (ΔFM; Fig. 3D), but not lean mass (ΔLM; Fig. 3C), in mice in the highest 10th percentile of Mest expression, regardless of fat depot. These results are consistent with the data in Figure 2 showing EPI and ING WAT Mest expression in mice with the lowest and highest 10th percentiles of BWT and adiposity. Surprisingly, although the association of WAT Mest with increased BWT and fat mass was highly significant, measurements of food intake, adjusted to either BWT (Fig. 3E) or fat-free mass (FFM; Fig. 3F) for each mouse, showed no association with WAT Mest expression.
Molecular correlates of Mest gene expression and adipose tissue expansion
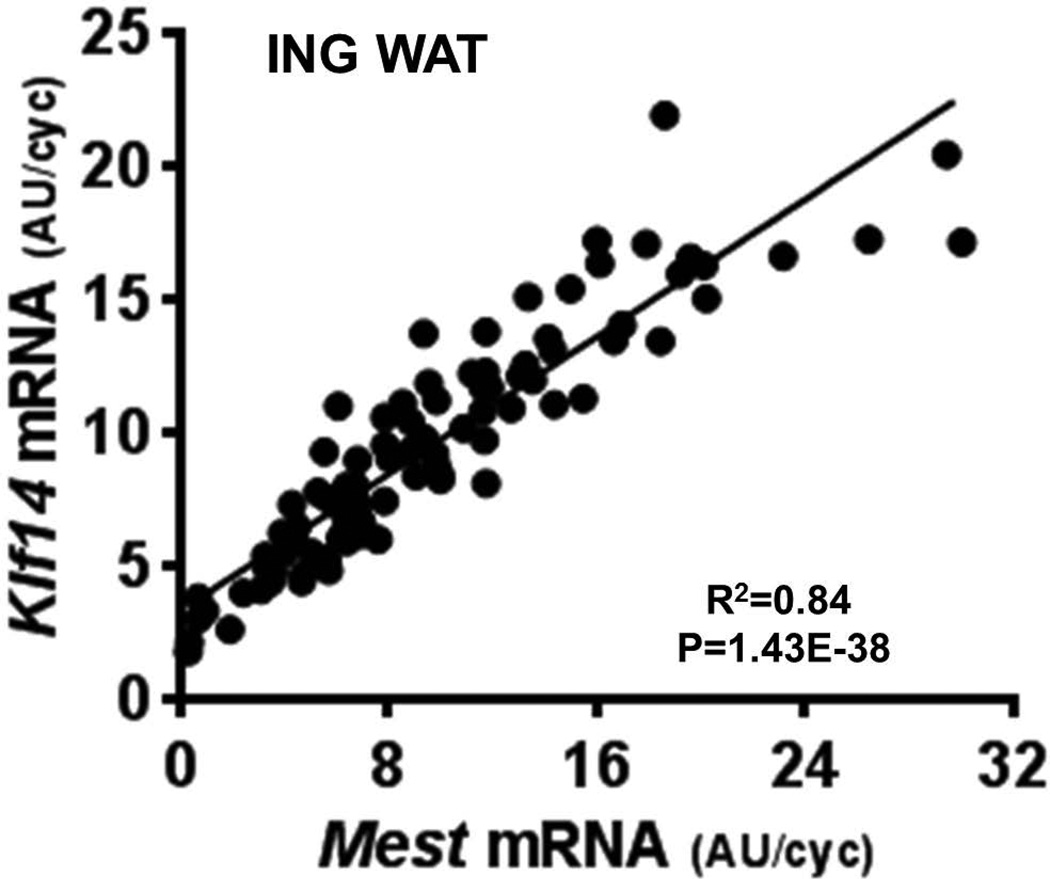
Previous studies showed a strong association between adipose tissue Mest expression and genes associated with Tgf-β (i.e., Bmp3) and Wnt signaling (i.e., Sfrp5 and Naked 1 (Nkd1) in mice fed an HFD for 4 weeks.3,5 In the present study, expression of Bmp3, Sfrp5, and Nkd1 were measured in EPI and ING fat of mice with the lowest and highest 10th percentiles of Mest expression in order to determine whether coexpression of these genes emerges after a short 2-week HFD feeding regimen. Data in Table 1 show significantly upregulated expression for Bmp3, Sfrp5, and Nkd1 in the highest 10th percentile of Mest for both fat depots, consistent with previous observations following long-term exposure to obesogenic diets. Leptin (Lep) mRNA expression was also shown to be more highly expressed in both WAT depots of the mice with the highest 10th percentile of Mest expression, possibly reflecting the increased adiposity in these groups (Fig. 3D). Because of recent studies suggesting a role for Klf14 as a master regulator of gene expression in WAT,22 and its possible link to obesity and type 2 diabetes,24,25 Klf14 mRNA was measured in WAT of mice with the lowest and highest 10th percentiles Mest expression. Results in Table 1 show significantly higher Klf14 expression in the highest-percentile groups for both WAT depots. Furthermore, regression analyses with the entire cohort of 96 mice showed a similar range of expression (Mest, 0.32–30.03 AU/cyclo; Klf14, 1.78–21.91 AU/cyclo) and an extremely strong correlation between Mest and Klf14 (Fig. 4) in ING WAT (R = 0.91; P = 1.4E-38). Measurements of Pparg and Cebpa, genes involved in adipogenesis, were not significantly different in EPI WAT of mice with the lowest versus highest 10th percentile of Mest expression; and only Cebpa showed a modest, albeit significant, increase in expression in ING WAT of the highest-percentile mice (Table 1). However, analyses of the complete cohort of 96 mice showed no association of EPI or ING fat Pparg or Cebpa with fat mass accumulation (Table 2), whereas Mest, Nkd1, and Lep all showed significant associations with fat mass accumulation during the 2 weeks of HFD feeding, regardless of the WAT type used for gene expression analyses. Correlation analyses of genes previously identified as being associated with nongenetic variation of obesity in the cohort of 96 mice showed Mest to be most robustly associated with Klf14 (Table 3) in both ING and EPI fat.3 Strong correlations of Mest with Bmp3, Sfrp5, Nkd1 and Lep were also observed and are consistent with previous observations.3
Table 1.
Analyses of gene expression in mice within the lowest and highest 10th percentile for adipose tissue Mest mRNA
| EPI fat | ING fat | |||||
|---|---|---|---|---|---|---|
| Gene | LM | HM | P value | LM | HM | P value |
| Mest | 4.06 ± 0.71 | 13.06 ± 0.33 | 3.28E-08 | 1.70 ± 0.39 | 22.52 ± 1.42 | 6.40E-08 |
| Bmp3 | 6.99 ± 0.64 | 14.44 ± 0.87 | 2.78E-06 | 5.99 ± 0.61 | 13.68 ± 0.69 | 1.31E-07 |
| Nkd1 | 5.87 ± 0.28 | 9.90 ± 0.49 | 5.41E-06 | 5.14 ± 0.30 | 14.61 ± 1.16 | 1.31E-05 |
| Sfrp5 | 3.02 ± 0.49 | 7.44 ± 1.27 | 0.0070 | 4.52 ± 0.52 | 17.47 ± 1.59 | 8.82E-06 |
| Lep | 6.00 ± 0.43 | 11.53 ± 0.74 | 1.52E-05 | 5.95 ± 0.85 | 15.68 ± 1.60 | 9.79E-05 |
| Klf14 | 3.27 ± 0.52 | 10.14 ± 1.19 | 0.00019 | 3.50 ± 0.36 | 17.05 ± 0.78 | 7.38E-10 |
| Pparg | 10.52 ± 0.66 | 10.90 ± 0.53 | 0.67 | 8.47 ± 1.52 | 12.14 ± 0.95 | 0.058 |
| Cebpa | 9.92 ± 0.38 | 9.00 ± 0.27 | 0.064 | 7.63 ± 0.77 | 10.36 ± 0.33 | 0.0068 |
LM, lowest 10th percentile Mest mRNA expression; HM, highest 10th percentile Mest mRNA expression
Figure 4.
A highly significant correlation between inguinal (ING) fat mRNA expression for Mest and Klf14 is represented in a scatterplot. Gene expression measured by TaqMan quantitative RT-PCR is represented as arbitrary units (AU) normalized to cyclophilin B.
Table 2.
Correlation between adipose tissue gene expression and fat deposition in mice fed an obesogenic diet for 2 weeks
| EPI GenExp vs. ΔFM (wk 2–wk 0) | ING GenExp vs. ΔFM (wk 2–wk 0) | |||
|---|---|---|---|---|
| Gene | R | P value | R | P value |
| Mest | 0.57 | 1.9E-09 | 0.66 | 3.7E-13 |
| Bmp3 | 0.39 | 7.2E-05 | 0.46 | 2.2E-06 |
| Nkd1 | 0.68 | 3.1E-14 | 0.65 | 5.6E-13 |
| Sfrp5 | 0.25 | 0.015 | 0.40 | 4.8E-05 |
| Lep | 0.68 | 2.3E-14 | 0.59 | 2.3E-10 |
| Klf14 | 0.31 | 0.0022 | 0.50 | 1.6E-07 |
| Pparg | 0.01 | 0.91 | 0.07 | 0.51 |
| Cebpa | 0.01 | 0.90 | 0.05 | 0.63 |
Table 3.
Pearson (R) correlation matrix of ING (unshaded) and EPI (shaded) fat gene expression in mice fed an obesogenic diet for 2 weeks
| Gene expression |
Mest | Bmp3 | Sfrp5 | Klf14 | Nkd1 | Lep | Pparg | Cebpa |
|---|---|---|---|---|---|---|---|---|
| Mest | 0.79 | 0.83 | 0.91 | 0.86 | 0.76 | 0.30 | 0.36 | |
| Bmp3 | 0.48 | 0.82 | 0.85 | 0.80 | 0.77 | 0.52 | 0.57 | |
| Sfrp5 | 0.41 | 0.62 | 0.88 | 0.70 | 0.67 | 0.30 | 0.37 | |
| Klf14 | 0.71 | 0.62 | 0.78 | 0.76 | 0.74 | 0.41 | 0.43 | |
| Nkd1 | 0.62 | 0.30 | 0.23 | 0.40 | 0.86 | 0.41 | 0.48 | |
| Lep | 0.51 | 0.48 | 0.41 | 0.44 | 0.66 | 0.62 | 0.56 | |
| Pparg | −0.05 | 0.19 | 0.12 | 0.10 | −0.20 | 0.20 | 0.81 | |
| Cebpa | −0.03 | −0.17 | −0.005 | −0.08 | 0.07 | 0.01 | 0.02 |
Discussion
In previous studies aimed to ascertain epigenetic mechanisms underlying the development of obesity within an inbred, genetically identical population of mice, we identified several genes including the imprinted gene Mest, inhibitors of Wnt signaling (i.e., Sfrp5 and Nkd1), and genes associated with Tgfβ/activin signaling (i.e., Bmp3) that are associated with heterogeneity in fat accretion and storage. Because prior studies examined WAT gene expression in mice fed an HFD for at least 4 weeks, when WAT has already undergone robust development, the aim of the current study was to determine whether these same gene targets continue to emerge as correlative indicators of fat mass expansion after a short-term 2-week exposure of mice to an obesogenic diet. It was anticipated that short-term dietary studies will provide a greater likelihood for the identification of genes linked with epigenetic mechanism(s) involved in the regulation fat mass accumulation, rather than genes that show changes in expression as a result of the development of obesity.
Feeding mice an HFD for 2 weeks resulted in an approximate 3.7-g average increase in BWT, with fat mass accounting for more than 60% of the weight gained. In addition, a bimodal pattern of BWT emerged that paralleled the pattern for indices of adiposity (Fig. 1). Although a similar distribution of BWT was also observed in a previous study of male B6 mice fed an HFD for 4 weeks,3 this was originally regarded as a possible artifact caused by the combination of two independent cohorts of mice for this dataset and thus was interpreted as a bell-shaped curve. The consistent replication of a bimodal distribution in BWT and corresponding indices of adiposity may be indicative of a metabolic difference in fat mass accumulation between these two subsets of mice. Since all mice in our study are genetically homogeneous, it is unlikely that genetic differences contribute to this BWT and adiposity pattern; however, it is possible that epigenetic variation could at least be partly responsible for the emergence of this phenotypic profile. A recent study determined that differences in intestinal microbial profiles in 129/SvEv mice acquired from different vendors resulted in mice that were susceptible or resistant to the development of obesity.26 Subsequent analyses with 129/SvEv and C57BL/6J mice determined that environmental history, diet, and genetic background can strongly affect gut microbiota content.26 Since the mice in our study are reared in the same environment and fed the same diet, the likelihood for significant differences in gut microbiota between individuals, at least initially, is relatively small. However, as mice begin to metabolically diverge when fed an obesogenic diet, it is possible that differences in bacterial taxa profiles in intestinal microbiota may begin to emerge in animals with low or high weight gain, which could exacerbate adipose inflammation and insulin resistance. Another potential nongenomic developmental variable that could mediate behavior and metabolic status of mice is the intrauterine position (IUP) of the fetus and resulting prenatal exposure to androgens.27 An early study by Kinsley et al. suggested that the IUP of male and female fetuses between two male fetuses could establish a metabolic set point that promotes increased BWT and fat storage during postnatal growth.28
We have shown in previous studies that WAT Mest gene expression can be rapidly, but variably, induced in mice fed an HFD for only 2–5 days.5 However, variability in the induction of WAT Mest gene expression among individual mice after this short duration of HFD feeding does not reflect differences in adiposity. Although it is unclear whether mice susceptible to rapid induction of WAT Mest are more susceptible to the development of dietary obesity, we have shown that Mest expression in WAT biopsies in mice prior to exposure to an obesogenic environment is predictive for the development of obesity as they age.3 A recent study by Voigt et al.29 has also determined that WAT Mest expression is highly predictive for adipose tissue expansion after a 5-day HFD-feeding trial with C57BL/6J mice and can be used to test efficacy of anti-obesity treatments. In addition, this same study showed that Mest mRNA in whole blood correlated strongly with adiposity, leading to the possibility of developing a minimally invasive approach to identify mice that differ in their susceptibility to the development of obesity. After two weeks on an HFD, we determined WAT Mest to be highly associated with mice within the lowest and highest 10th percentiles for both BWT and measurements of adiposity (Fig. 2). When examined from the perspective of how WAT Mest expression affects phenotypic traits, analyses of changes in BWT, body composition, and food intake in mice with the lowest and highest 10th percentiles of EPI and ING WAT Mest mRNA showed a positive association with changes in BWT and fat mass, but not with lean mass or food intake (Fig. 3). This strong association of WAT Mest with indices of adiposity after a short 2-week dietary fat regimen is consistent with past studies of mice fed an HFD for 4 weeks.3 Food intake during the 2-week dietary study normalized to either BWT or fat-free mass showed no association with mice in the lowest and highest 10th percentile WAT Mest expression groups (Fig. 3) and only marginal significance (R = 0.25; P = 0.015) to ING WAT Mest mRNA in a regression analysis of the entire cohort of 96 mice. These data can be interpreted to suggest that heterogeneity in the development of adiposity observed among individual mice is not caused by increased energy intake, but rather by differences in the utilization and compartmentalization of energy, metabolic efficiency, or capacity for fat storage in adipocytes.
Analyses of WAT gene expression within the lowest and highest 10th percentiles for EPI and ING WAT Mest expression (Table 1) show similar strong associations between Mest, Bmp3, Nkd1, and Sfrp5 after HFD feeding for 2 weeks, as observed in studies with mice fed an HFD for 4 weeks.3 Both Lep and Klf14, previously shown to be upregulated in ING WAT in a global gene expression study (GEO accession number GSE4692) of mice with low and high weight gain when fed an HFD for 4 weeks,3 were also strongly associated with Mest in this 2-week dietary study. In addition, regression analyses of EPI and ING WAT gene expression in the entire cohort of 96 mice versus the change of fat mass during the 2 weeks of HFD feeding (Table 2) show strong associations for Mest, Bmp3, Nkd1, Sfrp5, Lep, and Klf14. WAT expression of the well-characterized adipogenic genes Pparg or Cebpa was not associated with indices of fat mass expansion, which is consistent with observations in previous studies.3
Our interest in the potential contribution of Klf14 in the heterogeneous development of obesity within a population of genetically identical mice stems from recent studies suggesting that Klf14, an imprinted maternally expressed transcriptional regulator, is an important mediator of the regulation of lipid metabolism and signaling in adipose tissue.22,23,30 In addition, since Mest and Klf14 are located in close proximity on the same genomic locus in mice (Chr 6; 30.73–30.96 mb) and humans (CHR 7; 130.49–130.73 mb), it is possible that a common mechanism (e.g., imprinting control region (ICR) could be involved in their regulation.30 The observation of consistent patterns and range for gene expression for Mest and Klf14 in ING WAT among mice was confirmed by regression analyses (Fig. 4), which showed an almost linear relationship (R = 0.91; P < 10–37) between the two genes. Correlation matrices of gene expression in Table 3 showed that the strongest association for Mest in both fat depots was with Klf14. Although this association was not as strong in EPI WAT (R = 0.70; P < 10–14) as in ING WAT, these data were consistent with a possible interaction and contribution of the imprinted genes Klf14 and Mest, and genes previously shown to co-regulate with Mest (i.e., Sfrp5, Nkd1, and Bmp3),3 in an epigenetic model for the regulation of fat mass expansion in mice fed an obesogenic diet.
The identification of a strong association of Mest and Klf14 in WAT with indices of adiposity in male C57BL/6J mice fed an HFD for only 2 weeks establishes an exciting opportunity for further study. Although loss of imprinting (LOI) of Mest has been shown to increase its expression in fat of interspecies hybrids and results in increased body weight,31 our studies and those of others have shown that variation in Mest mRNA in WAT does not show loss of imprinting, and Mest mRNA is derived exclusively from the paternal allele.32–34 Although it is possible that epigenetic modifications of distal enhancers and regulatory elements could lead to variation in the expression of Mest mRNA from the paternal allele, it is equally plausible that the epigenetic regulation of a gene, or a transcription factor that regulates Mest transcription from the paternal allele, could lead to a similar result. On the basis of the data presented here, studies are now underway to investigate Klf14-mediated transcriptional regulation of Mest, as well as global changes in histone modifications in DNA from WAT with low and high Mest and Klf14 expression. We anticipate that the results of this current study will begin to delineate a novel epigenetic mechanism for the regulation of fat mass accumulation in an obesogenic environment.
Acknowledgements
We would like to thank David C. Higgins for assistance in generating some of the data for this study. These studies were supported by the Molecular Phenotyping Core Facility (funded by NIGM 8P30-GM106391) at the Maine Medical Center Research Institute and by NIH RO1DK090361 (R.A.K).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Burcelin R, Crivelli V, Dacosta A, Roy-Tirelli A, Thorens B. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am J Physiol Endocrinol Metab. 2002;282:E834–E842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 2.Koza RA, et al. Contributions of dysregulated energy metabolism to type 2 diabetes development in NZO/H1Lt mice with polygenic obesity. Metabolism. 2004;53:799–808. doi: 10.1016/j.metabol.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Koza RA, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak LP, Newman S, Chao PM, Mendoza T, Koza RA. The early nutritional environment of mice determines the capacity for adipose tissue expansion by modulating genes of caveolae structure. PLoS One. 2010;5:e11015. doi: 10.1371/journal.pone.0011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikonova L, et al. Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:3925–3937. doi: 10.1096/fj.08-108266. doi:fj.08-108266 [pii] 10.1096/fj.08-108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi M, Kamei Y, Ezaki O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am J Physiol Endocrinol Metab. 2005;288:E117–E124. doi: 10.1152/ajpendo.00244.2004. [DOI] [PubMed] [Google Scholar]
- 7.Moraes RC, et al. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144:4773–4782. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 8.Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes & development. 2000;14:963–980. [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett CN, et al. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 10.Mori H, et al. Secreted frizzled-related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. The Journal of clinical investigation. 2012;122:2405–2416. doi: 10.1172/JCI63604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulte DM, et al. Pro-inflammatory wnt5a and anti-inflammatory sFRP5 are differentially regulated by nutritional factors in obese human subjects. PLoS One. 2012;7:e32437. doi: 10.1371/journal.pone.0032437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouchi N, et al. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329:454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes & development. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 14.Wang EA, et al. Purification and characterization of other distinct bone-inducing factors. Proc Natl Acad Sci U S A. 1988;85:9484–9488. doi: 10.1073/pnas.85.24.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang EA, et al. Recombinant human bone morphogenetic protein induces bone formation. Proc Natl Acad Sci U S A. 1990;87:2220–2224. doi: 10.1073/pnas.87.6.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & development. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 17.Wozney JM, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 18.Bahamonde ME, Lyons KM. BMP3: to be or not to be a BMP. The Journal of bone and joint surgery. American volume. 2001;83-A(Suppl 1):S56–S62. [PubMed] [Google Scholar]
- 19.Daluiski A, et al. Bone morphogenetic protein-3 is a negative regulator of bone density. Nature genetics. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 20.Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. Journal of cell science. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 21.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 22.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nature genetics. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Assuncao TM, et al. New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids. J Biol Chem. 2014;289:15798–15809. doi: 10.1074/jbc.M113.544346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimas AS, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohshige T, et al. Association of new loci identified in European genome-wide association studies with susceptibility to type 2 diabetes in the Japanese. PLoS One. 2011;6:e26911. doi: 10.1371/journal.pone.0026911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ussar S, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell metabolism. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan BC, Vandenbergh JG. Intrauterine position effects. Neuroscience and biobehavioral reviews. 2002;26:665–678. doi: 10.1016/s0149-7634(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 28.Kinsley C, et al. Prior intrauterine position influences body weight in male and female mice. Hormones and behavior. 1986;20:201–211. doi: 10.1016/0018-506x(86)90018-8. [DOI] [PubMed] [Google Scholar]
- 29.Voigt A, et al. Identification of Mest/Peg1 gene expression as a predictive biomarker of adipose tissue expansion sensitive to dietary anti-obesity interventions. Genes & nutrition. 2015;10:477. doi: 10.1007/s12263-015-0477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker-Katiraee L, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, et al. Loss-of-imprinting of Peg1 in mouse interspecies hybrids is correlated with altered growth. Genesis. 2004;39:65–72. doi: 10.1002/gene.20027. [DOI] [PubMed] [Google Scholar]
- 32.Kamei Y, et al. Peg1/Mest in obese adipose tissue is expressed from the paternal allele in an isoform-specific manner. FEBS letters. 2007;581:91–96. doi: 10.1016/j.febslet.2006.12.002. doi:S0014-5793(06)01432-3 [pii] 10.1016/j.febslet.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Koza RA, Rogers P, Kozak LP. Inter-individual variation of dietary fat-induced mesoderm specific transcript in adipose tissue within inbred mice is not caused by altered promoter methylation. Epigenetics. 2009;4:512–518. doi: 10.4161/epi.4.7.10031. doi:10031 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada Y, Sakaue H, Nagare T, Kasuga M. Diet-induced up-regulation of gene expression in adipocytes without changes in DNA methylation. Kobe J Med Sci. 2009;54:E241–E249. [PubMed] [Google Scholar]