Abstract
Cancer stem cells (CSCs) are a promising target for cancer therapy, particularly for metastatic lung cancers, but how CSCs are regulated is largely unknown. We identify two proteins, SLUG (encoded by SNAI2 gene) and SOX9, that are associated with advanced stage lung cancers and are implicated in the regulation of CSCs. Inhibition of either SLUG or SOX9 sufficiently inhibits CSCs in human lung cancer cells and attenuates experimental lung metastasis in a xenograft mouse model. Correlation between SLUG and SOX9 levels was observed remarkably, we therefore sought to explore their mechanistic relationship and regulation. SLUG, beyond its known function as an epithelial-mesenchymal transition transcription factor, was found to regulate SOX9 by controlling its stability via a post-translational modification process. SLUG interacts directly with SOX9 and prevents it from ubiquitin-mediated proteasomal degradation. SLUG expression and binding are necessary for SOX9 promotion of lung CSCs and metastasis in a mouse model. Together, our findings provide a novel mechanistic insight into the regulation of CSCs via SLUG-SOX9 regulatory axis, which represents a potential novel target for CSC therapy that may overcome cancer chemoresistance and relapse.
Keywords: Cancer stem cells, lung cancer metastasis, SLUG, SOX9, ubiquitination
INTRODUCTION
Lung cancer is the leading cause of cancer death that kills more than one million people worldwide each year.1 The poor survival rate of patients is largely attributed to diagnosis at late stages with local or advanced metastasis at distant organs.2,3 While recent chemotherapy and radiotherapy have improved palliation, the treatment outcomes remain poor, as metastasis is largely incurable. In the past decade, subpopulations of cancer stem cells (CSCs; also known as tumor-initiating cells) have been reported in many solid tumors including breast, prostate, colon and lung4–6 which appear to both initiate the bulk of tumors and drive their progression through continuous rounds of self-renewal.4,7,8 CSCs can acquire apoptosis resistance and increased cell migratory and invasive properties, a prerequisite for tumor metastasis.9,10 The presence of CSCs in primary tumors is strongly correlated with an increased incidence of metastasis and poor survival of patients,11–13 suggesting them to be promising target for cancer therapy. Here we compared lung CSCs with their non-CSC counterpart and investigated the regulatory mechanisms that determine their metastatic behavior.
As epithelial-mesenchymal transition (EMT), a trans-differentiation process by which cells undergo a morphological change into a more mesenchymal phenotype, is common occurrence in metastasis of lung and other tumors,14,15 we profiled EMT and identified SLUG (encoded by SNAI2 gene) as significantly upregulated in the tested lung CSCs. SLUG is a member of Snail family with a unique conserve motif near the zinc fingers that is absent in other members.16 A high expression of SNAI2 is found in highly invasive lung cancer cells and tumor specimens, and is associated with poor survival and cancer relapse.17,18 We further observed here that SLUG is not required for EMT activation in lung cancer cells, leading us to the discovery of other pathways that may contribute to the aggressive phenotypes of lung CSCs.
CSCs and normal stem cells share many common characteristics, e.g. self-renewal and differentiation. Correlations between the regulatory pathways critical for normal developmental process and tumor progression have long been hypothesized and are being recognized.20,21 Sex-determining region Y (SRY)-boxes (SOX) family is known to play a pivotal role in the regulation of embryonic development and its members have been used as pluripotent stem cell markers.22 SOX9, in particular, is expressed in lung epithelium and mesenchyme, and is critical in tracheal differentiation and formation.23 Upregulation of SOX9 has been reported in lung adenocarcinoma, supporting its clinical significance in lung cancer.24 We demonstrate here the high-level SOX9 in correlation with high-level SLUG in lung CSCs and advanced stage lung cancers. Thus, we further investigated: (a) the roles of SLUG and SOX9 in lung CSCs and metastasis; (b) the SLUG and SOX9 relationship; and (c) their regulatory mechanisms. Our findings could be important in understanding CSCs and lung metastasis and may have clinical utility for targeted therapy of lung and other cancers whose etiology are dependent on SLUG-SOX9 dysregulation.
RESULTS
CSC phenotypes in human cancer cells
CSCs could self-renew and generate differentiated progeny that constitute the majority of cells in tumors.25,26 To determine whether CSCs could be defined in human non-small cell lung cancer (NSCLC) cell lines, we performed tumor sphere formation assays under CSC-selective conditions in H460 and A549 cells. Indeed, both NSCLC cell lines formed large floating spheres under such detachment and serum-starvation conditions (Supplementary Figure S1A). We isolated and characterized cells bearing CSC properties based on their side population (SP) phenotype, a common feature of CSCs.6,25 Cells were stained with Hoechst 33342 and SP cells which disappear in the presence of fumitremorgin c (FTC), a specific inhibitor of multidrug resistance ABCG2 transporter, were identified by FACS. NSCLC cells contained a distinct fraction of SP cells ranging from approximately 6% (A549) to 11% (H460) (Figure 1a and Supplementary Figure S1B). We verified that the SP cells from NSCLC H460 cells possessed CSC-like properties compared to their non-SP (NSP) counterpart, as assessed by tumor sphere formation, chemoresistance, and cell migration and invasion assays in vitro and tumor formation in vivo (Supplementary Figure S1C–F).
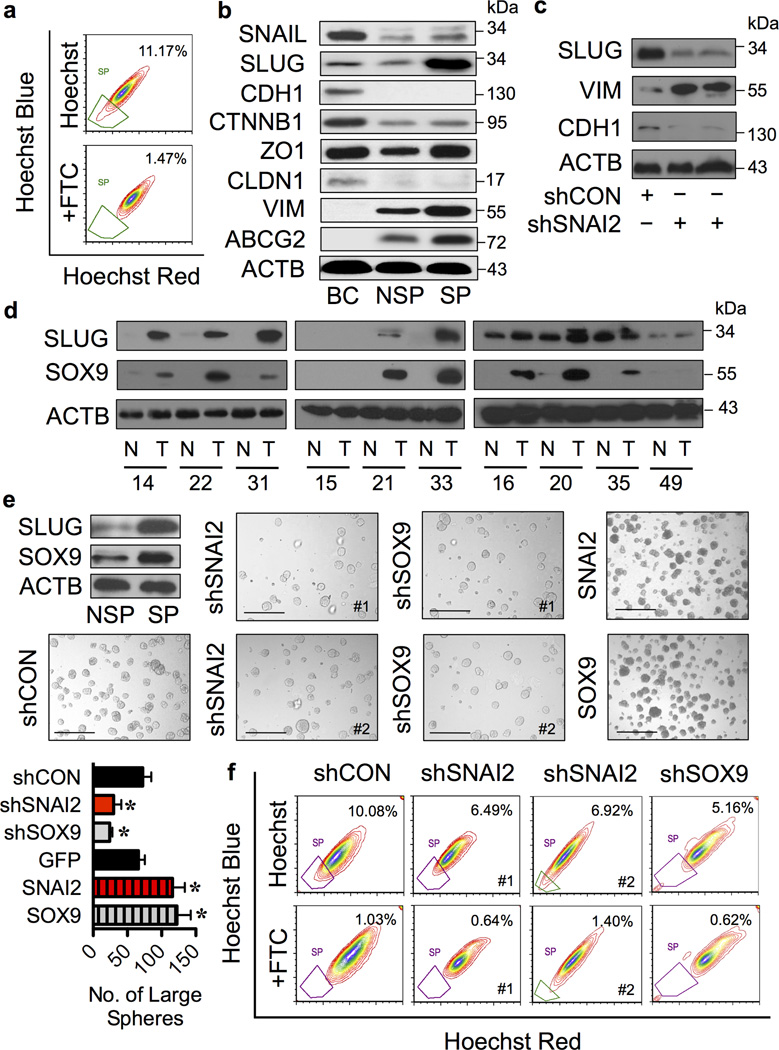
Figure 1. Lung CSCs and clinical lung carcinoma exhibit high levels of SLUG and SOX9.
(a) Analysis of side population (SP) in human lung carcinoma H460 cells in the presence or absence of fumitremorgin C (FTC) using FACS. SP cells (box) were determined by their disappearance in the presence of FTC and were shown as percentage of the pool population. CSCs were isolated based on SP phenotype and their aggressive features were validated in vitro and in vivo as shown in Supplementary Figure S1. (b) Analysis of EMT markers and ABCG2 transporter in human normal lung epithelial BEAS-2B (BC) cells and SP (CSC) and NSP (non-CSC) H460 cells using Western blotting. Immunoblot signals from three-independent experiments (one of which is shown here) were quantified by densitometry, revealing a dominant overexpression of SLUG in SP cells. (c) Western blot analysis of SLUG, VIM and CDH1 in SLUG knockdown (shSNAI2) and control (shCON) H460 cells. (d) Protein expression of SLUG and SOX9 in clinical lung cancer and matched normal lung tissues. Blots were reprobed with anti-β-actin (ACTB) antibody to confirm equal loading of the samples. Quantitative analysis of SLUG and SOX9 levels (Supplementary Figure S2) revealed a striking difference between normal (N) and tumor (T) tissues at the significance level in two-sided Student’s t-test of P < 0.03 and P < 0.003, respectively. (e and f) SLUG and SOX9 knockdown and overexpression experiments were performed using H460 cells treated with lentiviral particles carrying shSNAI2, shSOX9 or shCON and nucleofection of GFP, SNAI2 or SOX9 overexpression plasmids, as described under “Materials and methods”. Analysis of (e) tumor sphere formation and (f) SP in various clones of H460 cells. Scale bar = 200 µm. Data are mean ± S.D. (n = 4). *P < 0.05 vs. shCON cells; two-sided Student’s t-test.
To investigate the potential role of EMT in CSC regulation, we profiled EMT in the SP and NSP cells derived from NSCLC H460 cells by Western blotting. The results revealed a high level of mesenchymal markers (e.g. SLUG and VIM) and a low level of epithelial markers (CDH1 and CLDN1) in the SP cells as compared to NSP cells (Figure 1b), indicating activation of the EMT program in CSC population. The dominant overexpression of SLUG was observed in the SP cells with a > 5-fold increase relative to NSP cells. Additionally, the expression of ABCG2 transporter was strikingly higher in the SP cells compared to NSP cells, thus confirming the SP analysis and sorting by FACS.
To determine whether SLUG is a key transcription factor controlling EMT (EMT-TF) in NSCLC cells, SLUG expression was inhibited by RNA interference using shRNA against SNAI2 (shSNAI2 or shSLUG) and its effect on EMT was examined by analyzing the mesenchymal marker VIM and epithelial marker CDH1. Surprisingly, knockdown of SLUG not only failed to neither suppress the mesenchymal marker nor repress the epithelial marker, but rather upregulated or downregulated it (Figure 1c), suggesting that SLUG level is not critical for EMT in this tested cell system.
Effect of SLUG and SOX9 on CSC regulation
The inability of SLUG knockdown to inhibit EMT, despite the suggested role of EMT in cancer metastasis,14,15 points out that pathways other than EMT may contribute to the aggressive phenotypes of NSCLC cells. We hypothesized that SOX9, a cooperating factor of SLUG in mammary stem cells (MaSCs),27 may be involved, thus we performed an expression analysis of SOX9 and SLUG in human clinical specimens from advanced stage (stage III) lung cancer and matched normal lung tissues using Western blotting. A high level of both proteins was observed in the majority of advanced lung cancer tissues (T) compared to matched normal lung tissues (N) (Figure 1d) as well as in lung CSCs (Figure 1e). To our knowledge, this is the first demonstration of SLUG and SOX9 co-upregulation in advanced stage lung cancer and CSCs.
To test whether SLUG and SOX9 are functionally linked to lung CSCs, which are believed to drive tumor progression and metastasis,7,8 NSCLC H460 cells expressing a high endogenous level of SLUG and SOX9 were stably transduced with shRNA lentiviral particles against SNAI2 (shSNAI2), SOX9 (shSOX9) or control (CON; shCON), and their effects on CSCs were determined using tumor sphere formation and SP assays. Figure 1e and f shows that both shSNAI2 and shSOX9 cells exhibited significantly less tumor sphere formation and SP fraction than shCON cells, indicating the involvement of SLUG and SOX9 in CSC regulation.
SLUG and SOX9 act as regulators of lung metastasis
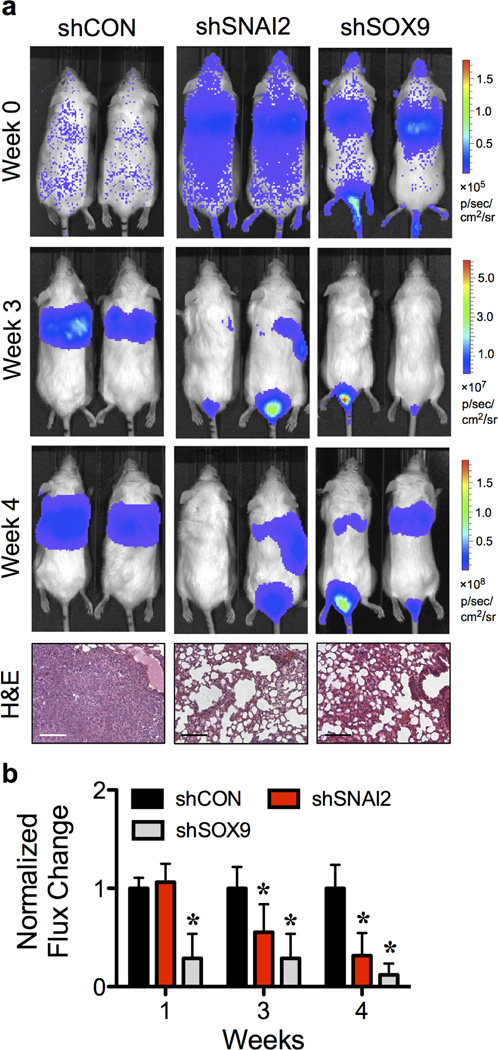
CSCs are critical for dissemination of tumor cells in the circulation to seed metastases at distant sites.10,28 Having demonstrated the role of SLUG and SOX9 in CSCs, we further examined the involvement of SLUG and SOX9 in metastasis using a xenograft mouse model. shSNAI2, shSOX9 and shCON H460 cells were genetically labeled with LUC2 and injected into NOD/SCID gamma (NSG) mice via tail vein at the dose of 1×106 cells/mouse. Tumor growth was monitored weekly by measuring the luciferase activity associated with the growing cells using IVIS® bioluminescence imaging. The results showed that at 4 weeks post-injection, the tumor luminescence was strikingly lower in mice bearing shSNAI2 and shSOX9 cells relative to shCON cells, consistent with the observations of the reduced number of experimental lung metastases and lung volume (Figure 2a and Supplementary Figure S3). For quantitative comparison, tumor luminescence signals were normalized to their initial signals at the time of inoculation (week 0) and relative to shCON cells. Figure 2b shows that the tumor growth was substantially lower in mice bearing shSNAI2 and shSOX9 cells than that of shCON cells at 3 and 4 weeks post-injection, indicating the critical role of SLUG and SOX9 in lung tumor growth and metastasis.
Figure 2. Inhibition of SLUG and SOX9 suppresses experimental lung cancer metastasis in vivo.
LUC2-labeled shSNAI2, shSOX9 or shCON H460 cells were injected into NSG mice via tail vein at the dose of 1×106 cells/mouse. (a) Representative bioluminescence of mice taken at the time of inoculation (week 0) and at 3 and 4 weeks post-injection (top) and H&E micrographs of lung tissues (bottom). Scale bar = 100 µm. (b) Quantitative analysis of bioluminescence signals over time. Tumor luminescence signals were normalized to their initial signals at week 0 and relative to shCON cells. Data are mean ± S.D. (n = 3 or 5). *P < 0.05 vs. shCON cells; two-sided Student’s t-test.
SLUG regulates SOX9 in NSCLC cells
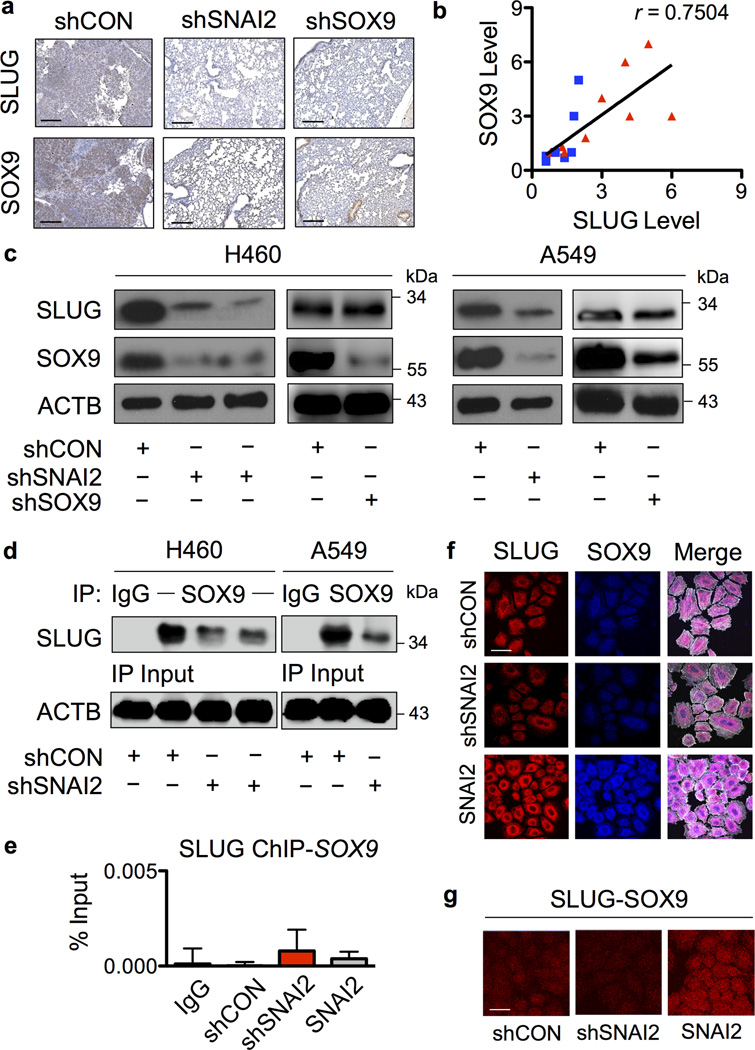
Immunohistochemistry of mouse lung tissues revealed the high levels of SLUG and SOX9 in experimental metastases, consistent with the findings in human clinical specimens (Figure 3a). Such observations and the similar effects of SLUG and SOX9 on lung CSCs and metastasis suggest their potential linkage and shared mechanisms. A correlation plot of SLUG and SOX9 expression generated from normal and tumor clinical specimens reveals their positive correlation with a correlation coefficient of 0.75 (Figure 3b). To experimentally determine their functional linkage, we tested whether knockdown of SLUG affects SOX9 expression in shSNAI2 and shCON cells. Figure 3c shows that knockdown of SLUG in NSCLC H460 cells resulted in a parallel decrease in SOX9 expression as compared to control cells. In contrast, knockdown of SOX9 had minimal effect on SLUG expression, suggesting that SLUG regulates SOX9. To substantiate the effect of SLUG on SOX9, we similarly knocked down SLUG in NSCLC A549 cells and compared its effect on SOX9 expression with nonsilencing control. The results showed that knockdown of SLUG in A549 cells similarly suppressed the expression of SOX9, substantiating the regulatory role of SLUG on SOX9 in NSCLC cells.
Figure 3. SLUG regulates of SOX9 in NSCLC cells.
(a) Immunohistochemistry analysis of SLUG and SOX9 in isolated lungs from mice bearing shSNAI2, shSOX9 and shCON cells. Scale bar = 200 µm. (b) Correlation analysis of the protein expression of SLUG and SOX9 in clinical lung tumor (red triangle) and matched normal (blue square) samples. (c) Western blot analysis of SLUG and SOX9 expression in shSNAI2, shSOX9 and shCon H460 and A549 cells. (d) shSNAI2 and shCON cell lysates were prepared, immunoprecipitated with control IgG or anti-SOX9 antibody, and probed with anti-SLUG antibody. Immunoblots were performed on cell lysates used as input for immunoprecipitation (IP) using anti-β-actin (ACTB) antibody to confirm equal loading of the samples. (e) Chromatin IP (ChIP) analysis of SLUG binding on SOX9 promoter. Sheared chromatin was immunoprecipitated using ChIP-grade anti-SLUG antibody or IgG control, and ChIP DNA was quantified by real-time PCR with primers specific to human SOX9 promoter (see Supplementary Table S1 for primer sequences). Data are mean ± S.D. (n = 3). (f) SLUG overexpressing or knockdown H460 cells were seeded onto type I collagen-coated slides. The cells were immunostained for SLUG (red), SOX9 (blue) and examined under a confocal fluorescence microscope. Colocalization of SLUG and SOX9 is shown in the merged display (purple). Images were taken pairwise with the same instrumental setting. Scale bar = 50 µm. (g) In-cell co-IP using proximity ligation (Duolink®) assay as described under “Materials and methods”. Cells were similarly seeded onto type I collagen-coated slides and in-cell SLUG-SOX9 interaction (red) was determined using the Duolink® assay. Scale bar = 50 µm.
Direct interaction between SLUG and SOX9 was evaluated using immunoprecipitation, immunoblotting and immunocytochemistry techniques. First, cell lysates from shSNAI2 and shCON H460 and A549 cells were prepared and immunoprecipitated with anti-SOX9 antibody. The resulting immune complexes were then analyzed for SLUG-SOX9 interaction using anti-SLUG antibody. Figure 3d shows a high basal level of SLUG-SOX9 complex in the shCON cells as compared to SLUG-knockdown cells, suggesting SLUG and SOX9 protein binding interaction. To investigate the possible binding of SLUG, which is an EMT transcription factor, to SOX9 gene promoter, chromatin immunoprecipitation (ChIP) assay was performed in shSNAI2, shCON and SLUG-overexpressing shCON cells. The results of this study demonstrated that SLUG does not bind to SOX9 promoter (Figure 3e).
Next, immunofluorescence studies were performed to confirm the SLUG and SOX9 protein binding and to evaluate their intracellular localization. A high degree of colocalization of SLUG and SOX9 was observed in the cytoplasm of control cells. A corresponding increase and decrease in the degree of colocalization were observed in SLUG-overexpressing and knockdown cells, respectively (Figure 3f). To confirm SLUG-SOX9 interaction in situ, in-cell co-immunoprecipitation experiments were performed using Duolink proximity ligation assay (PLA). Briefly, cells were fixed and incubated with anti-SLUG and SOX9 antibodies, followed by the addition of plus and minus PLA probes with attached short DNA strands. Two DNA strands in a close proximity were ligased, amplified and visualized by confocal microscopy. Figure 3g shows that the in-cell SLUG-SOX9 complex (red fluorescence) was highest in the SLUG-overexpressing cells, followed by control and SLUG-knockdown cells, respectively. This finding is consistent with the conventional immunofluorescence study and confirms the direct interaction between SLUG and SOX9 in the tested cell systems.
SLUG inhibits SOX9 ubiquitination and degradation
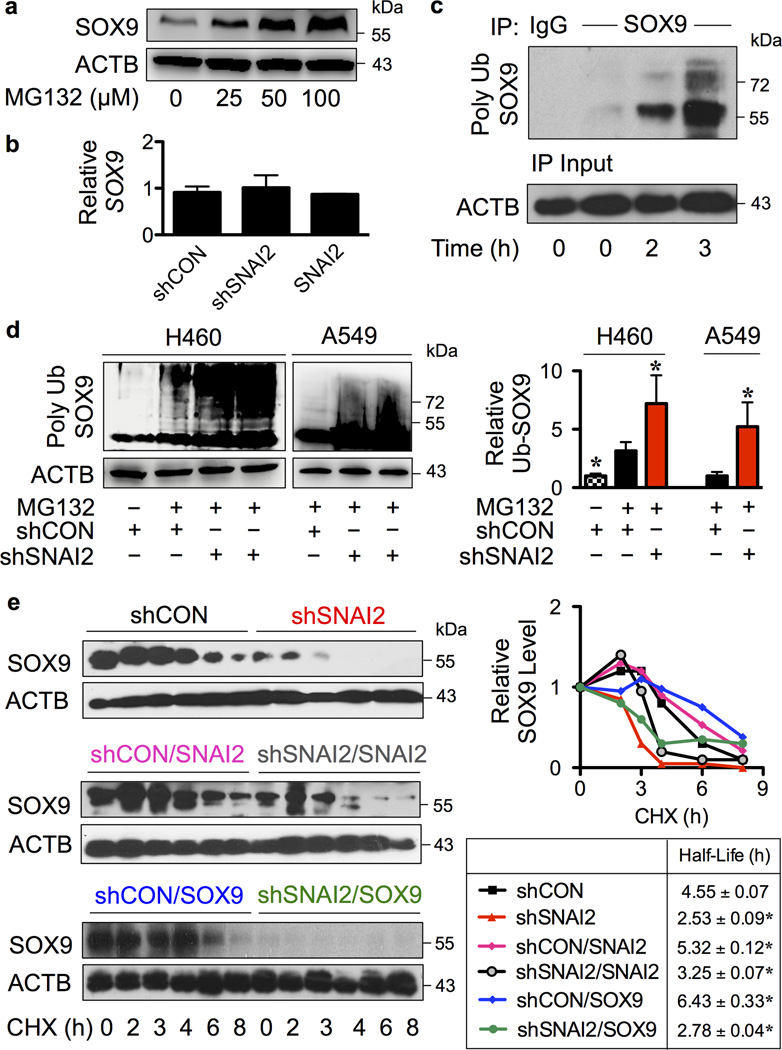
Protein-protein interactions are known to influence protein stability and function.29–31 We further investigated the effect of SLUG binding on SOX9 stability. Proteasomal degradation is a major cellular process that controls protein turnover.32,33 To determine whether SOX9 stability is controlled by proteasomal degradation, NSCLC H460 cells were treated with MG132 for proteasome inhibition. The increase in SOX9 expression by MG132 treatment suggested proteasomal degradation as an important mechanism of SOX9 regulation (Figure 4a). In addition, we performed quantitative real-time PCR and confirmed that SLUG binding had no significant effect on SOX9 transcription (Figure 4b).
Figure 4. SLUG inhibits SOX9 ubiquitination and prolongs its stability.
(a) H460 cells were treated with proteasome inhibitor MG132 (25–100 µM) for 24 hours and SOX9 expression was determined by Western blotting. (b) Quantitative real-time PCR of SOX9 mRNA expression in SLUG overexpressing or knockdown H460 cells. (c) H460 cells were treated with MG132 (50 µM) to prevent proteasomal degradation of SOX9 and cell lysates were prepared and immunoprecipitated using control IgG or anti-SOX9 antibody. The immune complexes were analyzed for ubiquitin at various times (0–3 hours) by Western blotting. Immunoblots were performed on cell lysates used as IP input using anti-β-actin (ACTB) antibody to confirm equal loading of the samples. (d) Analysis of SOX9 ubiquitination (Poly Ub SOX9) in shSNAI2 and shCON H460 and A549 cells in the presence or absence of MG132 at 3 hours, in which ubiquitination was found to be maximal. Equal amounts of protein were loaded in each lane. Data are mean ± S.D. (n = 3). *P < 0.05 vs. shCON cells in the presence of MG132; two-sided Student’s t-test. (e) Various clones of H460 cells, including shCON, shSNAI2, shCON/SNAI2, shSNAI2/SNAI2, shCON/SOX9 and shSNAI2/SOX9 cells were treated with protein translation inhibitor cycloheximide (CHX; 10 µg/mL) for various times to follow the degradation of SOX9 protein. Cell lysates were prepared and SOX9 expression was determined by Western blotting. Representative immunoblots of SOX9 and loading control β-actin (ACTB) are shown (see also Supplementary Figure S4). SOX9 expression time profile was plotted and SOX9 half-life was calculated and shown. Data are mean ± S.D (n = 3). *P < 0.05 vs. shCON cells; two-sided Student’s t-test. No significant differences were observed when compared between shSNAI2 and shSNAI2/SOX9 cells.
Ubiquitination is a post-translational modification (PTM) that triggers proteasomal degradation.34,35 In NSCLC H460 cells, polyubiquitination of SOX9 gradually increased as early as 2 hours and peaked at approximately 3 hours (Figure 4c). To test whether SLUG might regulate SOX9 through ubiquitination, SLUG-knockdown and nonsilencing control NSCLC H460 and A549 cells were compared for SOX9 ubiquitination. Figure 4d shows that knockdown of SLUG resulted in an increase in SOX9 ubiquitination in both cell lines. Such increase could promote proteasomal degradation and thus decreased the expression of SOX9 protein as shown in Figure 3c. To substantiate this finding, SOX9 protein half-life was further analyzed in SLUG knockdown and control cells by cycloheximide-chase assay. shSNAI2 and shCON H460 cells were treated with cycloheximide to inhibit new protein synthesis and SOX9 expression was determined at various times by Western blotting. SOX9 expression-time profile was plotted from non-saturated blots and SOX9 half-life was calculated from the plot. The results showed that SOX9 protein degraded much faster in the shSNAI2 cells than in shCON cells as indicated by its half-life of approximately 2.5 and 4.5 hours, respectively (Figure 4e, top). In addition, overexpression of SLUG in the knockdown (shSNAI2/SNAI2) and control (shCON/SNAI2) cells prolonged the SOX9 half-life to approximately 3.5 and 5.5 hours (Figure 4e, middle), and increased its expression (data not shown), indicating that SOX9 protein stability is a key determinant of its expression.
To validate the role of SLUG in SOX9 stabilization, shSNAI2 and shCON cells were transfected with SOX9 plasmid and analyzed for SOX9 half-life. A substantially lower level of SOX9 was observed in the shSNAI2/SOX9 cells compared to shCON/SOX9 cells (Figure 4e, bottom), indicating its inability to retain SOX9 expression after overexpression. SOX9 half-life was approximately 6.5 and 3 hours in the shCON/SOX9 and shSNAI2/SOX9 cells, respectively. Altogether, these results substantiate the critical role of SLUG in SOX9 stabilization.
SLUG is critical for SOX9-mediated lung CSCs and metastasis
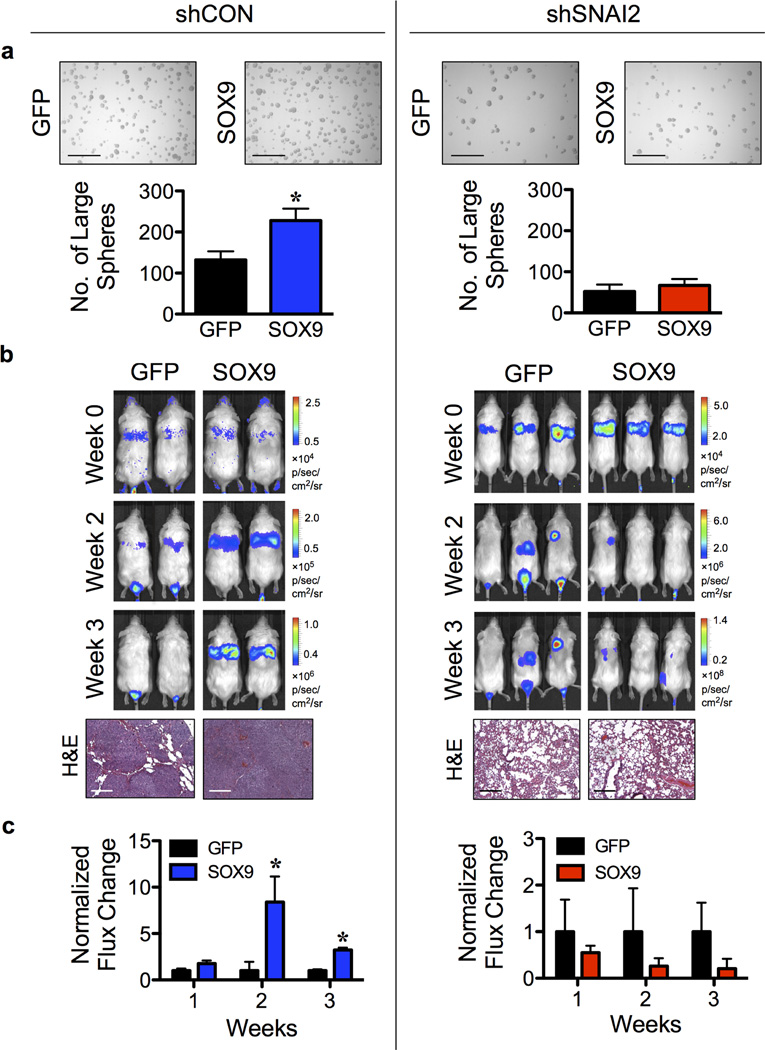
Having demonstrated the role of SOX9 in lung CSCs and experimental metastasis (Figure 1 and 2) and the requirement of SLUG in SOX9 stability (Figure 4), we further tested if SLUG is required for SOX9-mediated lung CSCs and metastasis. LUC2-labeled shCON or shSNAI2 H460 cells were transfected with SOX9 or GFP plasmid and evaluated for tumor formation in vitro and in vivo. Figure 5b shows a much higher rate of tumor growth and experimental lung metastases in mice bearing shCON/SOX9 cells compared to shCON/GFP cells. In contrast, tumor growth in mice bearing shSNAI2/SOX9 cells was comparable to that in mice bearing shSNAI2/GFP cells, in agreement with the in vitro tumor sphere results (Figure 5a), indicating the inability of SOX9 to promote lung CSCs and metastasis in the absence of SLUG. A similar finding was observed when LUC2-labeled A549 cells were used instead of the H460 cells (data not shown). Taken together, our results strongly support the critical role of SLUG in SOX9 stability and tumorigenic function.
Figure 5. SLUG is required in SOX9-mediated lung CSC in vitro and metastasis in vivo.
(a) LUC2-labeled shCON or shSNAI2 H460 cells were transfected with SOX9 or GFP control plasmid and were analyzed for tumor sphere formation. Scale bar = 300 µm. (b,c) LUC2-labeled shCON or shSNAI2 H460 cells were transfected with SOX9 or GFP plasmid and injected into NSG mice via tail vein at the dose of 1×106 cells/mouse. (b) Representative bioluminescence of mice taken at the time of inoculation (week 0) and at 2 and 3 weeks post-injection (top) and H&E micrographs of lung tissues (bottom). Scale bar = 200 µm. (c) Normalization of tumor signals at various times to their initial signal at week 0 and relative to shCON/GFP or shSNAI2/GFP cells. Data are mean ± S.D. (n = 3 or 4). *P < 0.05 vs. shCON/GFP or shSNAI2/GFP cells; two-sided Student’s t-test.
DISCUSSION
Post-translational modifications (PTMs) contribute to the regulation of protein expression and function and have ubiquitous roles in controlling various types of cancer.36,37 We examined the relationship between SLUG and SOX9, which are upregulated in advanced stage lung carcinoma and CSCs, and found that SLUG regulates SOX9 stability as well as tumorigenic and metastatic activities through the PTM of SOX9 ubiquitination. These findings indicate a novel role of SLUG, beyond its known function as an EMT-TF, and its importance in lung carcinoma.
A few studies have suggested the unique functions of SLUG that differ from other EMT-TFs, e.g. it is more relevant to breast CSC regulation than SNAIL (encoded by SNAI1 gene).38 In MaSCs, SLUG and EMT program alone is not sufficient to create fully functional MaSCs from differentiated luminal cells – it needs to work in concert with SOX9.27 In this study, we observed the high levels of SLUG and SOX9 in clinical samples of aggressive lung carcinoma and demonstrated their requirement in lung CSCs and metastasis (Figures 1 and 2). We showed that knockdown of SLUG and SOX9 repressed CSCs, in agreement with the previous report implicating their role in breast tumor initiation,27 and inhibited experimental metastasis, which is likely due to their effect on CSCs and not on the EMT process since EMT remained activated after SLUG knockdown (Figure 1).
SOX9 is a known regulator of cell proliferation and differentiation during lung morphogenesis.23,39 In lung carcinoma, SOX9 has been shown to promote cell growth possibly through the alteration of cell cycle regulators CDK4 and p21.24 Additionally, we demonstrated herein the requirement of SOX9 in the maintenance of lung CSCs. Relatively little is known about the regulation of SOX9. We showed that SOX9 expression is regulated primarily through ubiquitin-proteasomal degradation and that SLUG post-translationally inhibits this process in NSCLC cells (Figure 4). The half-life of SOX9 protein is significantly shortened in the absence of SLUG and prolonged in its presence. In prostate cancer cells, SLUG was shown to regulate the ubiquitin-proteasomal degradation of some other proteins such as cyclin D1.40 To our knowledge, this is the first demonstration of the regulatory role of SLUG on SOX9, in addition to serving as its cooperating partner.
Using correlation and immunoprecipitation analyses, we found that SLUG binds to SOX9 in the cytoplasm of NSCLC cells and the extent of this binding is inversely proportional to SOX9 ubiquitination and stability (Figures 3). Although SLUG and SOX9 are widely known as transcription factors that reside in the nucleus, increasing evidence suggests the significance of their cytosolic expression in cancer progression and metastasis.41–44 For example, cytoplasmic SOX9 was observed in invasive and metastatic breast carcinomas and was linked to indefinite proliferation of breast cancer cells.43,44 It has been shown that covalent attachment of ubiquitin to a target protein, which generates a polyubiquitin chain to guide proteasomal degradation, depends largely on protein conformation.45 Thus, it is likely that the binding of SOX9 by SLUG alters its confirmation, which interferes with its ubiquitination and subsequent degradation by the proteasomes. This stabilization process is consistent with the previous report showing an increase in protein stability by protein-protein interaction and inhibition of protein ubiquitination.31 Thus, the high-level coexpression of SLUG and SOX9 in clinical samples of aggressive lung carcinoma might be attributable to their binding interaction and increased stability. It is worth noting that other mechanisms of lung cancer metastasis are likely involved as the lack of SLUG and SOX9 was observed in some clinical samples of aggressive tumors, e.g. case #15 and 49.
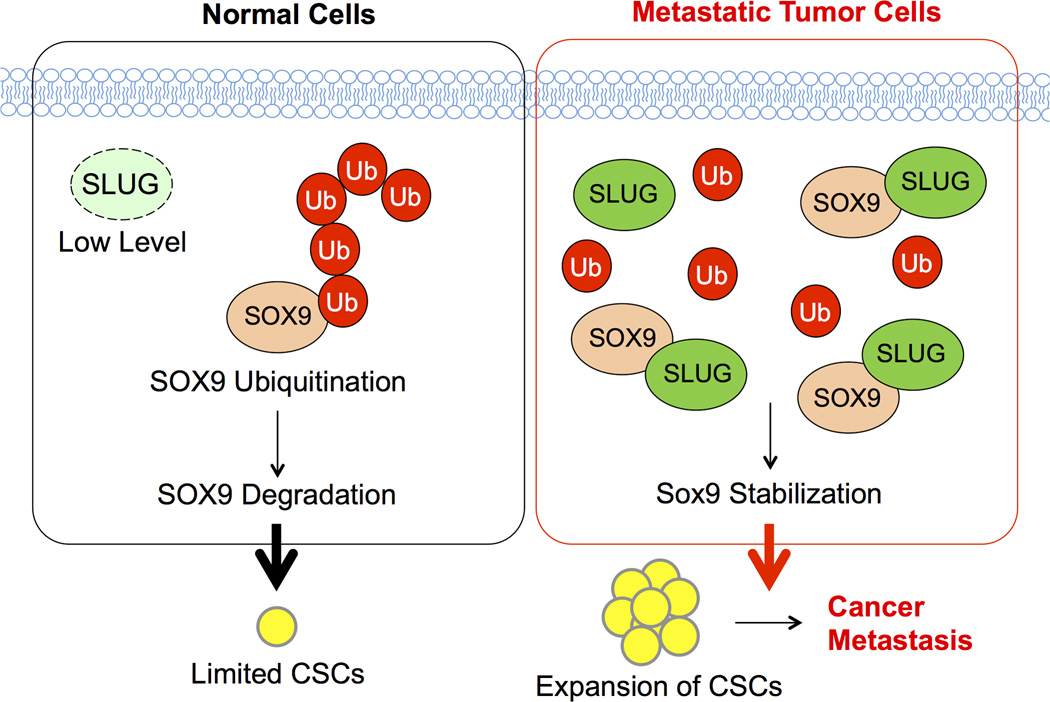
In conclusion, the evidence presented here demonstrated the significance of SLUG-SOX9 axis in regulating lung CSCs and metastasis. It established the essential role of SLUG in regulating SOX9 stability and subsequent functions in NSCLC cells whereby disruption of the SLUG-SOX9 complex could lead to SOX9 ubiquitination and proteasomal degradation. Our novel findings, as schematically summarized in Figure 6, could aid in the understanding of lung cancer progression and metastasis. Because of its important role in lung CSCs, the SLUG-SOX9 regulatory axis could be a promising therapeutic target for advanced and recurrent lung cancers.
Figure 6. A schematic working model for the function of SLUG-SOX9 axis in CSC and metastasis regulation.
In metastatic tumor cells, increased SLUG expression stabilizes SOX9 through their binding interaction, which inhibits SOX9 ubiquitination and proteasomal degradation. SOX9 stabilization promotes the expansion of CSCs and subsequent cancer metastasis.
MATERIALS AND METHODS
Cell culture
NSCLC H460 and A549 cells were obtained from ATCC (Manassas, VA) and cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), while human normal bronchial epithelial BEAS-2B (BC) cells were from ATCC and cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% FBS. All cells were supplemented with 2 mM L-glutamine and 100 U/mL penicillin and 100 µg/mL streptomycin and were maintained in a humidified atmosphere of 5% CO2 environment at 37 °C.
Tumor sphere assay
Tumor sphere assay was performed under non-adherent and serum-free conditions as previously described as stem cell-selective conditions.25,46 Briefly, cells were suspended in 0.8% methylcellulose-based medium (Stem Cell Technologies, Vancouver, Canada) supplemented with 20 ng/mL epidermal growth factor (BD Biosciences, Franklin Lakes, NJ), basic fibroblast growth factor and 4 mg/mL insulin (Sigma-Aldrich, St Louis, MO) and plated at 5×103 cells in ultralow adherent 24-well plates (Corning, Corning, NY). Cells were cultured for two or three weeks and visualized under a light microscope.
Side population analysis and isolation
Cells were labeled with 5 µg/mL Hoechst 33342 in DMEM-F12 medium containing 2% FBS in the presence or absence of 25 µM ABCG2 inhibitor fumitremorgin C (FTC; EMD Biosciences, San Diego, CA) at 37 °C for 90 minutes. SP analysis and sorting were performed using BD FACSAria cell sorter (BD Biosciences) using UV laser and Hoechst Blue (450/20) and Red (675 LP) filters. SP fraction was calculated based on the disappearance of SP cells in the presence of FTC.
Human normal and lung cancer tissue protein lysates
Protein lysates of human lung cancer, including large cell carcinoma (#14–16 and 20–22), squamous cell carcinoma (#31 and 33) and adenocarcinoma (#49), and matched normal lung tissues were obtained from Protein Biotechnologies (Ramona, CA).
Inhibition of SLUG by RNA interference
Lentiviral transduction particles carrying short hairpin RNA (shRNA) sequence against human SNAI2 and control non-target sequence (Sigma-Aldrich; NM_003068/TRCN0000284362 and TRCN0000015389 and SHC002V) were used to knockdown SLUG expression in H460 and A549 cells, according to the manufacturer’s protocol. Briefly, cells were incubated with lentiviral particles in the presence of hexadimethrine bromide (8 µg/mL) for 36 hours. Infected cells were allowed to recover for 48 hours and were cultured for 28 days with puromycin-containing medium (1 µg/mL). The stable knockdown cells were identified by Western blotting using anti-SLUG antibody (Cell Signaling Technology, Beverly, MA; 9585) and were cultured in puromycin-free RPMI 1640 medium.
Lentivirus production and inhibition of SOX9 by RNA interference
Lentiviral plasmids carrying shRNA sequence against human SOX9 were obtained from Addgene (Cambridge, MA; plasmid 40644)27 and Santa Cruz Biotechnology (Dallas, TX; sc-36533-SH) and shSOX9 lentivirus production was performed using HEK293T packaging cells (ATCC) in conjugation with pCMV.dR8.2 dvpr lentiviral packaging and pCMV-VSV-G envelope plasmids (Addgene, plasmids 8454 and 8455).47 H460 and A549 cells were incubated with shSOX9 viral particles in the presence of hexadimethrine bromide for 36 hours. The transfected cells were analyzed prior to use by Western blotting using anti-SOX9 antibody (Millipore, Billerica, MA; AB5535).
Overexpression plasmid and transfection
H460 and A549 cells were transfected with SNAI2 (Addgene, plasmid 31698),48 SOX9 (Origene, Rockville, MD) or GFP (Invitrogen, Carlsbad, MA) plasmid by nucleofection using Nucleofector® (Amexa Biosystems, Cologne, Germany), according to the manufacturer’s instructions, using the device program T020 and X001, respectively. The transfected cells were allowed to recover for 48 hours before each experiment and were cultured for 28 days with G418-containing medium (800 µg/mL). The stable transfectants were identified by Western blotting for SLUG or SOX9 and were cultured in G418-free RPMI 1640 medium.
Western blot analysis
After specific treatments, the cells were incubated in a commercial lysis buffer (Cell Signaling Technology) and a protease inhibitor mixture (Roche Molecular Biochemicals, Indianapolis, IN) at 4 °C for 20 minutes. Protein content was analyzed using the Pierce™ BCA protein assay (Pierce Biotechnology, Rockford, IL) and 50 µg proteins were resolved under denaturing conditions by SDS-PAGE as described previously.31
Cycloheximide-chase assay
A monolayer of cells was treated with cycloheximide (10 µg/mL) to inhibit new protein synthesis for various times (0–8 hours) to follow the degradation of SOX9 protein by Western blot analysis. SOX9 expression profile was plotted and SOX9 half-life was calculated from the unsaturated plot using GraphPad Prism software (La Jolla, CA).
Xenograft mouse model
Animal care and experimental procedures described in this study were performed in accordance with the Guidelines for Animal Experiments at West Virginia University (WVU) with the approval of the Institutional Animal Care and Use Committee (IACUC #12-0502). 1×106 Luc2-labeled cells were injected into male NOD/SCID gamma mice (NSG, strain NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, aged 6–8 weeks; Jackson Laboratory, Bar Harbor, ME) via tail vein and mice were inspected daily for any signs of distress. Tumor growth of LUC2-labeled cells was monitored at the time of inoculation (week 0) and on a weekly basis by using in vivo IVIS® imaging (PerkinElmer, Waltham, MA). At the end of experiments, mice were euthanized and the lungs were dissected and further analyzed for tumor histopathology.
Tumor histopathology
Isolated lung tissues from tumor-bearing mice were formalin-fixed and paraffin-embedded. The specimens were cut into 5-µm sections and stained with hematoxylin and eosin (H&E) to define the tumor morphology and cellular structure within the lungs. All tissue sectioning and H&E staining were performed at the WVU Pathology Laboratory for Translational Medicine.
Immunoprecipitation
Cell lysates (100 µg protein) were immunoprecipitated using Dynabeads® magnetic beads (Invitrogen). Briefly, the beads were conjugated with anti-SOX9 antibody (AB5535) for 10 minutes at room temperature, followed by a BS3 crosslinking step as per manufacturer’s protocol. The conjugated beads were then resuspended with cell lysates for 30 minutes at room temperature. The immune complexes were washed four times and resuspended in 2× Laemmli sample buffer. They were then separated by SDS-PAGE and analyzed for ubiquitination or interaction using anti-ubiquitin (Santa Cruz; FL-76) or anti-SLUG (9585) antibody.
Immunofluorescence
Cells were seeded on rat type I collagen-coated coverslips (5 µg/cm2), fixed with 3.7% paraformaldehyde, incubated in 50 mM glycine, and permeabilized and blocked with 0.5% saponin, 1.5% bovine serum albumin and normal goat serum. SLUG and SOX9 were immunostained with anti-SLUG (Santa Cruz Biotechnology; sc-10437 X) and anti-SOX9 (AB5535) antibodies, followed by Alexa Fluor 488- and 546-conjugated antibodies, phalloidin (Invitrogen) and ProLong® Gold Antifade Mountant with DAPI (Life Technologies). Cells were visualized with a Zeiss LSM 510 confocal on an AxioImager Z1 microscope (Carl Zeiss, Jena, Germany).
In-cell coimmunoprecipitation
In-cell coimmunoprecipitation was performed using Duolink® in situ assay following the manufacturer’s instructions. Briefly, cells were blocked and incubated with anti-SLUG (mouse) and anti-SOX9 (rabbit) antibodies for 2 hours at room temperature, followed by an incubation with anti-rabbit plus and anti-mouse minus PLA® probes for 60 minutes at 37 °C. Ligase was added and incubated for 30 minutes at 37 °C. Fluorescence signal was then amplified by the addition of polymerase for 100 minutes at 37 °C. The in-cell complexes were visualized with a confocal microscopy.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using Zymo-Spin™ ChIP kit (Zymo Research, Irvine, CA), according to the manufacturer’s protocol. Briefly, cells were collected and formaldehyde cross-linked. Chromatin was then mechanically sheared using sonication, followed by immunoprecipitation with ChIP-grade anti-SLUG (sc-10437 X) antibody. Total and immunoprecipitated chromatin were reverse cross-linked and recovered using column purification. The amount of total and ChIP DNA was determined using quantitative real-time PCR, using SYBR® Select Master Mix (Applied Biosystems, Grand Island, NY) and the ChIP primers for human SOX9 promoter.49
Statistical analysis
The data represent means ± SD from three or more independent experiments as indicated to ensure adequate power (> 80%). Statistical analysis was performed by two-sided Student’s t-test at a significance level of P < 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIH; R01-ES022968), National Science Foundation (CBET-1434503) and Mary Babb Randolph Cancer Center (MBRCC) Sara C. Allen Lung and James F. Allen Comp Lung Cancer Research Fund. Flow cytometric analysis was performed in the WVU Flow Cytometry Core Facility, which is supported in part by the NIH Grant P30 GM103488. Animal experiments were performed in the WVU Animal Models and Imaging Facility, which is supported in part by the MBRCC and NIH Grants P20 RR016440, P30 RR032138/GM103488 and S10 RR026378. The authors would like to acknowledge the WVU Pathology Laboratory for Translational Medicine for tissue sectioning and staining services and Drs. Davor Solter and Barbara Knowles for their comments on the manuscript.
Footnotes
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPLEMENTARY INFORMATION
Supplementary Information accompanies the paper on the Oncogene website (http://nature.com/onc).
REFERENCES
- 1.Esposito L, Conti D, Ailavajhala R, Khalil N, Giordano A. Lung cancer: are we up to the challenge? Curr Genomics. 2010;11:513–518. doi: 10.2174/138920210793175903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crino L, Weder W, van Meerbeeck J, Felip E. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21:V103–V115. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 3.Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundkist J, et al. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajk M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumor initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Luanpitpong S, Wang L, Castranova V, Rojanasakul Y. Induction of stem-like cells with malignant properties by chronic exposure of human lung epithelial cells to single-walled carbon nanotubes. Part Fibre Toxicol. 2014;11:22. doi: 10.1186/1743-8977-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 11.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan Y, Chen B, Xu W, Zhao W, Wu J. Clinicopathological significance of CD133 in lung cancer: a meta-analysis. Mol Clin Oncol. 2014;2:111–115. doi: 10.3892/mco.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurwidya F, Takahashi F, Murakami A, Takahashi K. Epithelial mesencymal transition in drug resistance and metastasis of lung cancer. Cancer Res Treat. 2012;44:151–156. doi: 10.4143/crt.2012.44.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 17.Shih JY, Tsai MF, Chang YL, Yuan A, Yu CJ, Lin SB, et al. Transcription repressor Slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res. 2005;11:8070–8078. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- 18.Shih JY, Yang PC. The EMT regulator Slug and lung carcinogenesis. Carcinogenesis. 2011;32:1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 19.Philips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cells Rep. 2014;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronin JC, Watkins-Chow DE, Incao A, Hasskamp JH, Schonewolf N, Aoude LG, et al. SOX10 ablation arrests cell cycle, induces senescence, and suppresses melanomagenesis. Cancer Res. 2013;73:5709–5718. doi: 10.1158/0008-5472.CAN-12-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre V, Dumitrui B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turcatel G, Rubin N, Menke DB, Martin G, Shi W, Warburton D. Lung mesenchymal expression of Sox9 plays a critical role in tracheal development. BMC Biol. 2013;11:117. doi: 10.1186/1741-7007-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res. 2010;16:4363–4373. doi: 10.1158/1078-0432.CCR-10-0138. [DOI] [PubMed] [Google Scholar]
- 25.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Darnell GA, Antallis TM, Johnstone RW, Stringer BW, Ogbourne SM, Harrich D, et al. Inhibition of retinoblastoma protein degradation by interaction with the serpin plasminogen activator inhibitor 2 via a novel consensus motif. Mol Cell Biol. 2003;23:6520–6532. doi: 10.1128/MCB.23.18.6520-6532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Wang B, Jayappa KD, Yao X. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J Biol Chem. 2011;286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luanpitpong S, Chanvorachote P, Stehlik C, Tse W, Callery PS, Wang L, et al. Regulation of apoptosis by Bcl-2 cysteine oxidation in human lung epithelial cells. Mol Cell Biol. 2013;24:858–869. doi: 10.1091/mbc.E12-10-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 35.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 36.Krueger KE, Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics. 2006;5:1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 38.Bhat-Nakshatri P, Appaiah H, Ballas C, Pick-Franke P, Goulet R, Jr, Badve S, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer. 2010;10:411. doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, et al. Sox9 plays multiple role in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci USA. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding GX, Liu J, Feng CC, Jiang HW, Xu JF, Ding Q. Slug regulates cyclin D1 expression by ubiquitin-proteasome pathway in prostate cancer cells. Panminerva Med. 2012;54:219–223. [PubMed] [Google Scholar]
- 41.Hasan MR, Sharma R, Saraya A, Chattopadhyay TK, DattaGupta S, Walfish PG, et al. Slug is a predictor of poor prognosis in esophageal squamous cell carcinoma patients. PLoS ONE. 2013;8:e82846. doi: 10.1371/journal.pone.0082846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cates JM, Byrd RH, Fohn LE, Tatsas AD, Washington MK, Black CC. Epithelial-mesenchymal transition markers in pancreatic ductal adenocarcinoma. Pancreas. 2009;38:e1–e6. doi: 10.1097/MPA.0b013e3181878b7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med. 2011;236:141–155. doi: 10.1258/ebm.2010.010086. [DOI] [PubMed] [Google Scholar]
- 44.Chakravarty G, Rider B, Mondal D. Cytoplasmic compartmentalization of SOX9 abrogates the growth arrest response of breast cancer cells that can be rescued by trichostatin A treatment. Cancer Biol Ther. 2011;11:71–83. doi: 10.4161/cbt.11.1.13952. [DOI] [PubMed] [Google Scholar]
- 45.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains-from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levina V, Marrangnoi A, Wang T, Parikh S, Su Y, Herberman R, et al. Elimination of human lung cancer stem cells through targeting of the stem cell factor–c- kit autocrine signaling loop. Cancer Res. 2010;70:338–346. doi: 10.1158/0008-5472.CAN-09-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell. Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z, Gao X, He Y, Ju J, Zhang M, Liu R, et al. Synergistic effect of SRY and its direct target, WDR5, on Sox9 expression. PLoS One. 2012;7:e34327. doi: 10.1371/journal.pone.0034327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.