Abstract
Background
With advances in cell capture, gene expression can now be studied in neuronal subtypes and single cells; however, studying epigenetic mechanisms that underlie these changes presents challenges. Moreover, chromatin immunoprecipitation (ChIP) protocols optimized for low cell number do not adequately address technical issues and cell loss while preparing tissue for fluorescence activated cell sorting (FACS). Developing a reliable FACS-ChIP protocol without the need for pooling tissue from multiple animals would enable study of epigenetic mechanisms in neuronal subtypes.
Methods
FACS was used to isolate dopamine 1 receptor (D1R) expressing cells from the nucleus accumbens (NAc) of a commercially available BAC transgenic mouse strain. D1R+ cells were used to study gene expression as well as histone modifications at gene promoters using a novel native ChIP protocol.
Results
Isolated cells had enrichment of the dopamine 1 receptor (D1R) mRNA and nearly undetectable levels of GFAP and D2R mRNA. ChIP analysis demonstrated the association of activating or repressive histone modifications with highly expressed or silent gene promoters, respectively. Comparison with existing methods: The ChIP protocol developed in this paper enables characterization of histone modifications from ~30,000 FAC-sorted neurons.
Conclusions
We describe a one day FACS-ChIP protocol that can be applied to epigenetic studies of neuronal subtypes without pooling tissue.
1. Introduction
Regulation of gene expression is critical for neuronal adaptation to the environment and is enacted by epigenetic mechanisms. These mechanisms are induced by a range of chromatin modifying enzymes and include covalent modifications to histone subunit N-terminal tails and methylation of cytosine residues on DNA (1). Changes in histone modifications and DNA methylation across the brain have been implicated in regulation of neuropsychiatric and neurological disorders (2–4); therefore, characterizing epigenetic processes that control gene expression may reveal new targets for treatment.
While discovery of epigenetic modifications in the brain has greatly accelerated over the past two decades, technical issues remain in studying histone modifications in this complex tissue type. One issue relates to correlating changes in RNA transcript levels to changes in histone modifications at gene promoters using chromatin immunoprecipitation (ChIP). For instance, while a cell may produce hundreds of RNA molecules of the same transcript, it only contains two loci for those RNA sequences in the genome (excluding the potential contribution of copy number variation). Therefore, it may be difficult to detect changes in histone modifications at a specific gene promoter using chromatin made from millions of cells if only a few thousand cells are contributing to a detectable increase in RNA transcript number. This issue may be particularly relevant in the brain, which is composed of a plethora of neuronal subtypes, glia, and other cell types with distinct transcriptional profiles (5). Additionally, substantial loss of chromatin occurs during formaldehyde cross-linking, multiple cell lysis steps, and sonication that are components of nearly all standard ChIP protocols, so that most protocols recommend using tens of millions of cells as input (6, 7). Some of these limitations are overcome using cell culture systems where histone modifications within a distinct cell type can be studied; however, this lacks complexity relative to whole animal models.
Recent advances in fluorescence activated cell sorting (FACS) and laser capture microdissection using brain tissue have made it possible to isolate large numbers of neuronal subtypes. In particular, FACS allows for automated capture of a large number of cells within a matter of hours based on antibody-mediated or endogenous cell fluorescence. FACS studies also benefited by the completion of the Gene Expression Nervous System Atlas project, which developed transgenic mice with promoter-specific fluorescent proteins integrated into the genome using bacterial artificial chromosomes (BAC) (8). Studies characterizing neuronal subtypes using BAC transgenic strains indicate fluorescent reporter expression is highly cell-type specific and these animals have been used in anatomical, electrophysiological, and optogenetic studies (8–10).
Transcriptional differences between neuronal subtypes are also emerging. An early study utilized BAC transgenic mouse strains and FACS to identify transcriptional differences between dopamine D1 receptor (D1R) and dopamine D2 receptor (D2R) medium spiny neurons (MSN) in the striatum (11). Application of this technique has been particularly important for studying drug-induced regulation of gene expression. Eric Nestler’s group identified differences in cocaine-induced gene expression in D1R versus D2R MSNs (12) and showed that epigenetic mechanisms intrinsic to these neuronal subtypes underlie the observed transcriptional changes (13). A recent study utilized BAC transgenic strains along with antibody-mediated tagging of histone modifications to examine how cocaine alters histone modifications in D1R and D2R MSNs (14). Other groups have used antibody-mediated FACS to study gene regulation induced by cocaine (15, 16), opioids (17, 18), and methamphetamine (19) in cellular subtypes. However, while antibody-based approaches allow for selection of a wider array of cell types compared to BAC transgenic strains, they also require fixing cells and have issues with off-target binding by antibodies that may reduce their specificity.
While the use of FACS to study neuronal subtype specific gene expression is well-established, interrogating neuronal subtype-specific epigenetic mechanisms would benefit by studying histone modifications at gene promoters using ChIP. Several papers have reported methods for ChIP from small numbers of cells (20, 21); however, they have not adequately addressed technical issues of handling FAC-sorted neurons. One study reported a ChIP protocol after isolating olfactory epithelial cells using FACS (22); however, they required an average of ten animals per ChIP to achieve reliable results. Additionally, isolation of neuronal nuclei using FACS is associated with increased cell yield and can be used to study epigenetic modifications in neuronal subtypes (23, 24); however, this technique precludes simultaneous study of cytosolic RNA in these neurons.
This study utilized a commercially available BAC transgenic strain where the dopamine D1 receptor (D1R) promoter drives expression of tdTomato, a variant of the red fluorescent protein. This strain has been well-characterized with tdTomato expression restricted to D1R MSNs and no apparent behavioral differences relative to wild type mice (25). We describe a one day protocol to isolate D1R+ medium spiny neurons (MSN) from the nucleus accumbens (NAc) and complete ChIP and RNA-specific studies using FAC-sorted neurons from a single animal.
2. Materials and methods
2.1. Animals
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh and conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Two heterozygous transgenic males containing a gene for the tdTomato fluorescent protein driven by the D1R promoter were purchased from the Jackson Laboratory (Stock #016204). These males were bred to isogenic C57BL/6J females and offspring were checked for the presence of the transgene with PCR after weaning. Male offspring that possessed the transgene were housed with their wild type littermates and used for all experiments at 8–16 weeks of age. Mice were housed under 12 hour light/dark cycles and had ad libitum access to food and water. Separate animals were used for chromatin and RNA extraction described below.
2.2. PCR genotyping
Tail snips (< 0.5 cm) were taken at the time of weaning and DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instruction. DNA was used for a PCR assay containing GoTaq HotStart Polymerase (Promega, Madison, WI), primers for the transgene and positive control region (20 µM), dNTPs (2.5 mM), 5× reaction buffer, and MgCl2 (2.5 mM). Primer sequences used were Transgene, F: 5’-CTT CTG AGG CGG AAA GAA CC-3’ and R: 5’-TTT CTG ATT GAG AGC ATT CG-3’, Positive control region, F: 5’-CTA GGC CAC AGA ATT GAA AGA TCT-3’ and 5’-GTA GGT GGA AAT TCT AGC ATC ATC C-3’. PCR conditions were 3 min at 94° C, then 30 s at 94° C, 1 min at 59° C, 1 min at 72° C repeated 35 times, then 2 min at 72° C. PCR products were run on a 1.5% agarose gel. Transgenic animals were identified by the presence of two bands: a 750 bp band for the transgene and 324 bp band for the positive control region.
2.3. Dissociation of the nucleus accumbens into a single cell suspension
Tissue dissociation was adapted from a previous report of FACS using BAC transgenic mouse strains (26). The following reagents were made fresh before the start of each experiment. HABG: 200 µl of 50× B27 supplement (Life Technologies) and 25 µl of 100× Glutamax (Life Technologies) into 9.8 ml of Hibernate A cell culture media (Life Technologies). Papain dissociation buffer: 14 µl of 100× Glutamax (Life Technologies) into 5.5 ml Hibernate E cell culture media (BrainBits LLC, Springfield, IL). Papain: 5 ml of papain dissociation buffer into one vial containing 100 U of papain (Worthington Biochem, Lakewood, NJ). DNase: 500 µl of papain dissociation buffer into one vial containing 1000 U of DNase (Worthington Biochem).
Adult animals were sacrificed by cervical dislocation and the brain was extracted and placed into an ice cold adult mouse brain slicer matrix with 1 mm coronal section slice intervals (Zivic Instruments). The NAc was identified under a dissecting microscope as the region surrounding the anterior commissure and manually dissected using a scalpel; it was differentiated from the overlying dorsal striatum by its homogenous appearance relative to the striated appearance of the dorsal striatum. The NAc was placed into a small culture dish containing 1 mL HABG supplemented with 100 U/ml of RiboLock RNase inhibitor (Thermo Scientific, Waltham, MA) and 5 mM sodium butyrate (Sigma-Aldrich, St. Lous, MO). Tissue was cut into ~1 mm3 pieces using a scalpel. Tissue pieces were gently aspirated using a cut 1 ml pipet tip and allowed to fall to the edge of the tip. The pipet tip was placed into 2 ml of Papain supplemented with 110 µl DNAse and 5 mM sodium butyrate that was pre-warmed to 37° C and the tissue pieces were allowed to gently fall into the Papain so that there was minimal transfer of HABG into Papain. Tissue pieces in Papain were covered in aluminum foil and incubated at 32° C with gentle rotation for 15 minutes.
After incubation in Papain and DNase, tissue pieces were collected with a cut 1 ml pipet tip and allowed to fall to the edge of the tip. The pipet tip was placed into 2 ml of HABG supplemented with 100 U/ml of RiboLock RNase inhibitor, 1× cOmplete protease inhibitor (Roche, Basel, Switzerland), and 5 mM sodium butyrate, and the tissue pieces were allowed to gently fall into the HABG so that there was minimal transfer of Papain. The tissue pieces in HABG were covered with aluminum foil and incubated for 5 minutes at room temperature. Tissue pieces were triturated using a latex pipette bulb and autoclaved, cotton-plugged Pasteur pipette that was fire-polished to a ~0.5 mm tip. For trituration, tissue pieces in HABG were sucked into the pipette and ejected over ~5 seconds until large tissue pieces were no longer visible, which usually took ~10 cycles of trituration. Care was taken to avoid air bubbles in the cell suspension.
The dissociated cell suspension was passed through a 70 µm cell strainer (Fisher Scientific, Pittsburgh, PA) by aspirating 800 µl of solution at a time through a cut 1 ml pipet tip and gently placing the tip over the cell strainer. After the cell suspension was strained, 5 ml of HABG supplemented with 100 U/ml of RiboLock RNase inhibitor, 1× cOmplete protease inhibitor, and 5 mM sodium butyrate was passed over the same cell strainer. Therefore, the total volume of the cell suspension was ~7 ml. The cell suspension was covered in aluminum foil and immediately transported for cell sorting. It is important to note that this process takes ~45 minutes from the time the animal is sacrificed to cell sorting.
2.4. Fluorescence activated cell sorting of tdTomato+ cells
Neurons were sorted in collaboration with the Rheumatology Flow Cytometry Core Facility at the University of Pittsburgh. Briefly, the dissociated cell suspension was sorted on a FACS ARIA II (Becton Dickinson Inc.) using an 85 µm nozzle with a pressure of 45 p.s.i. Debris and doublets in the sample were excluded from sorting by gating for intact cells using forward and side scatter profiles. tdTomato was excited by an argon-ion laser (488 nm) and detected using an emission spectra between 564 nm and 606 nm. Fluorescence gating thresholds were established by sorting wild-type animals that do not express tdTomato and isolating events that were above the highest detectable limit in wild type animals. tdTomato+ and tdTomato− cells were separated using fluorescence gating and collected into two separate tubes.
To visualize fluorescence of live FAC-sorted neurons, cells were collected in PBS and applied to a glass slide using a Cytospin centrifuge (Thermo Scientific). Cells were visualized on a Nikon fluorescent microscope and photographed with a Photometrics CoolSNAP digital camera.
2.5. RNA extraction
For RNA studies, cells were sorted directly into 750 µl of Trizol LS (Invitrogen). After sorting, volume of Trizol LS with sorted cells was measured and nuclease free water was added to bring the volume up to 1 ml. Total RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol without DNase digestion and eluted in 30 µl of nuclease free water. RNA concentration and RNA integrity numbers (RIN) were determined on an Agilent Bioanalyzer using the Eukaryotic Total RNA Nano Series II kit (Agilent Technologies, Santa Clara, CA).
2.6. RT-qPCR
23 ul of RNA (section 2.5) was converted into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) in a 30 ul reaction with 6 ul 5× reaction buffer and 1 ul reverse transcriptase. Reaction conditions were performed according to the manufacturer’s protocol: 5 min at 25°C, 30 min at 40 °C, 5 min at 85°C, then a hold at 4 °C.. Reactions were carried out in duplicate for each gene. SYBR green fluorescent master mix (Bio-Rad) was added to each well and visualized using a Bio-Rad iCycler. All primers were optimized for 90% to 110% efficiency at the following conditions: 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 1 min at 60°C, and 30 s at 72°C. Primer sequences used were β-actin, F: 5’-TCA TGA AGT GTG ACG TTG ACA TCC GT-3’ and R: 5’-CCT AGA AGC ATT TGC GGT GCA CGA TG-3’, D1R, F: 5’-GAA CCC AGA AGA CAG GTG GA-3’ and R: 5’-GCT TAG CCC TCA CGT TCT TG-3’, dopamine D2 receptor (D2R), F: 5’-ATC TCT TGC CCA CTG CTC TTT GGA-3’ and R: 5’-ATA GAC CAG CAG GGT GAC GAT GAA-3’; glial fibrillary acidic protein (GFAP), F: 5’-AGA AAA CCG CAT CAC CAT TC-3’ and R: 5’-TCA CAT CAC CAC GTC CTT GT. Threshold cycle (Ct) values were calculated for each well and duplicate values averaged. The difference between specific genes and β-actin (ΔCt) was calculated for each animal and normalized to the average of tdTomato- or whole NAc (ΔΔCt). Fold change over tdTomato× or whole NAc was calculated for each animal using the following formula: 2−ΔΔCt.
2.7. Native FACS-ChIP
Reaction buffers were adapted from a previous report of low cell number ChIP (21). To prevent histone deacetylation and protease activation during tissue dissociation for ChIP studies, all media and buffers were supplemented with 5 mM sodium butyrate and, beginning after papain treatment, all media and buffers were supplemented with cOmplete protease inhibitor cocktail (Roche, Basel, Switzerland). Sodium butyrate was used to inactive histone deacetylases (HDACs) that are active during chromatin preparation and is commonly used during native ChIP protocols that lack formaldehyde fixation (27). Protease inhibitors prevent histone degradation during chromatin preparation and are also commonly used in ChIP protocols (28). Cells were sorted directly into 250 µl of 2× micrococcal nuclease (MNase) buffer (100 mM Tris, pH 8.0, 2 mM CaCl2, 0.4% Triton X-100) and volume was measured and brought up to 500 µl by adding nuclease free water. MNase (New England Biolabs, Ipswich, MA) was diluted by adding 1 µl of MNase (2,000 gel units) to 500 µl 1× MNase buffer (50 mM Tris, pH 8.0, 1 mM CaCl2, 0.2% Triton X-100). For MNase digestion of chromatin, 15 µl of diluted MNase was added to sorted cells and cells were incubated in a 37° C heat block for 5 minutes. To stop the MNase digestion, 50 µl of 10× MNase stop buffer (110 mM Tris, pH 8.0, 55mM EDTA) was added to the cells. From this point, chromatin was kept on ice or in a 4° C room through immunoprecipitation. To lyse cells, 550 µl of ice cold 2× RIPA cell lysis buffer (280 mM NaCl, 1.8% Triton X-100, 0.2% SDS, 0.2% Na Deoxycholate, and 5 mM EGTA) was added. The solution was mixed by inverting the tube and the tube was left on ice for 15 minutes. To remove cell debris, the tube was centrifuged at 4° C at 13,000 rpm for 10 minutes. Chromatin was decanted into a new 1.5 ml Eppendorf tube and placed on ice.
Chromatin was divided into two 400 µl aliquots for immunoprecipitation and one 200 µl aliquot used as input. For immunopreciptiation reactions, 20 µl of MagnaChIP Protein A/G magnetic beads (Millipore, Billerica, MA) were added to each tube. To one tube, 5 µl of an anti-histone H3 tri-methylated at lysine 27 (H3K27me3) (#17-622, Millipore) was added and to the other tube 9 µl of an anti-histone H3 acetylated at lysine 18 (H3K18ac) (#9675, Cell Signaling Technology, Danvers, MA) was added. We also attempted to optimize anti-histone H3 tri-methylated at lysine 4 (H3K4me3) (#9727, Cell Signaling Techology) and anti-histone H3 acetylated at lysines 9 and 14 (H3K9,14ac) (#06-559b, Millipore) but these antibodies did not produce a reliable signal above input. As a negative control for optimization experiments, nonspecific IgG antibody (#PP64B, Millipore) was also used. The immunoprecipitation tubes were incubated at 4° C with gentle end-over-end rotation for 4 hours. After incubation, tubes were placed onto a magnetic rack and the supernatant was removed using a pipet. Magnetic beads were washed 3 times with 500 µl RIPA buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% Na Deoxycholate) and 1 time with 500 µl TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA); between washes, the beads were incubated for 5 minutes at 4° C with gentle end-over-end rotation and the supernatant was removed using a pipet.
Following washes, magnetic beads were eluted in 200 µl elution buffer (50 mM NaCl, 20 mM Tris, pH 8.0, 5 mM EDTA, 1% SDS, 50 µg/ml proteinase K); for the input, proteinase K was added to a concentration of 50 µg/ml. The magnetic beads in elution buffer and input were incubated at 65° C with gentle rotation for 45 minutes. Following proteinase K digestion, tubes were briefly spun on a tabletop centrifuge and placed into a magnetic rack. The supernatant (200 µl) was transferred to a tube containing 10 µl 3 M Na Acetate and 1 ml buffer PB (Qiagen). DNA was purified using the Qiagen MinElute Reaction Cleanup Kit according to the manufacturer’s protocol and eluted in 14 µl nuclease free water.
ChIP DNA samples were quantified using a Qubit fluorometer (Life Technologies). For optimization of FACS-ChIP experiments using tdTomato+ neurons, 0.5 ng of DNA were loaded into each well and analyzed by qPCR in duplicate for each primer set. qPCR conditions were the same as reported in the RT-qPCR section. Primer sequences used were: D1R promoter, F: 5’-GCC TCT GGT TTC CTA CAC CC-3’ and R: 5’-AGG GAA AAG CAT GGT CGA GG-3’, GFAP promoter, F: 5’-ACA AAA GGC CTG GGT TGA CA-3’ and R: 5’-CTC TGG ATC TGG AAC TCG CC-3’, LINE1 5’ UTR, F: 5’-CCG GGA CTC CAA GGA ACT TA-3’ and R: 5’-CCT CCT GGC CGA AGA AGA-3’. Ct values for antibodies were normalized to the input Ct value for each primer set. Data are present as fold enrichment over input DNA.
2.8. Statistics
For RT-qPCR and ChIP-qPCR experiments, a Student’s t-test was used to compare groups. Statistical significance was defined by a p-value < 0.05. All data are presented as mean +/− SEM.
3. Results
3.1. FACS Validation
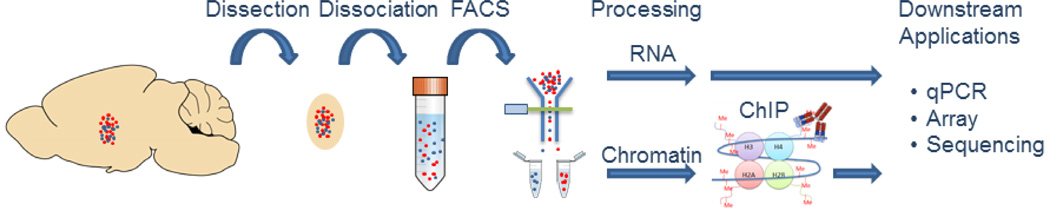
We developed a one day protocol for performing ChIP using FAC-sorted neurons (Fig. 1). Notably, cells were sorted within one hour of collecting brain tissue and RNA and chromatin were available within four hours, providing a one-day protocol for analyzing RNA and histone modifications at a gene promoter in a specific neuronal subtype.
Figure 1. Experimental Overview.
The nucleus accumbens (NAc) from a D1R::tdTomato BAC transgenic mouse was dissociated into a single cell suspension and cells expressing tdTomato were isolated using FACS. RNA and histone-associated DNA from chromatin immunoprecipitation were available within one day for downstream applications.
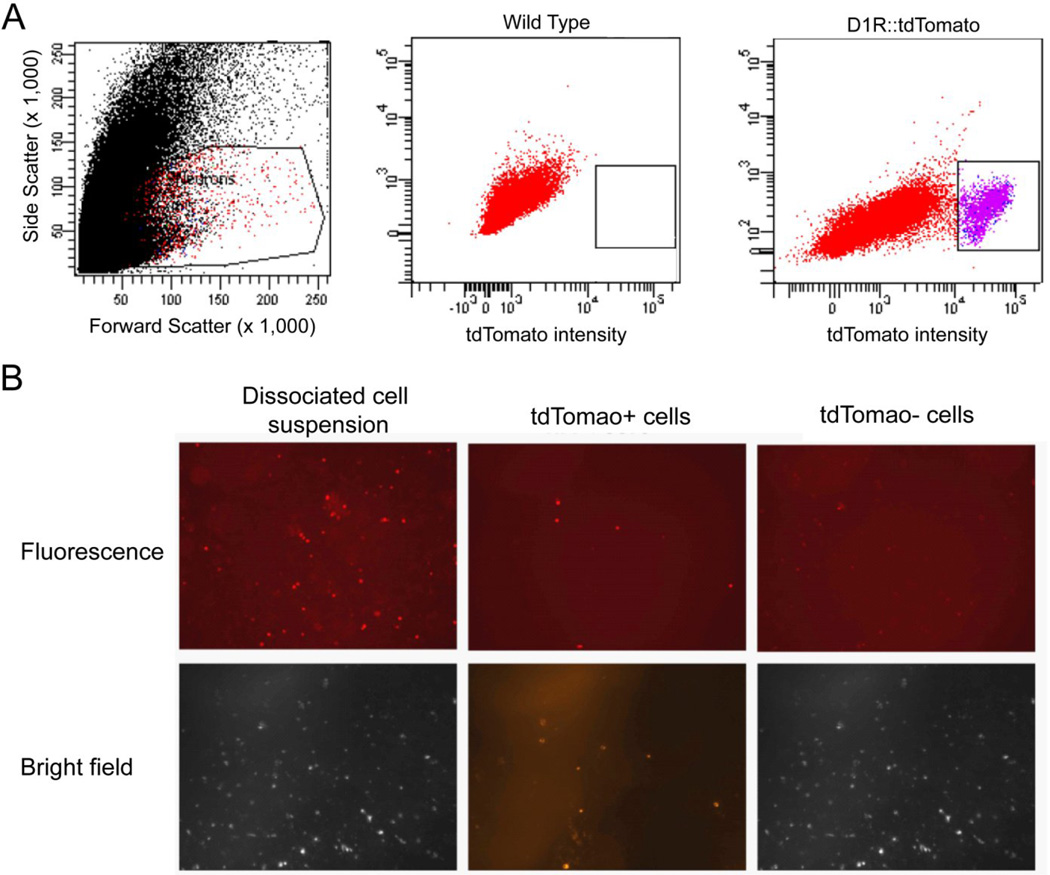
NAc was dissociated into a single cell suspension and cells were sorted by their size based on forward and side scatter profiles to exclude debris and doublets. A distinct group of cells with high tdTomato expression were clearly discriminated using FACS (Fig. 2A). These cells showed high endogenous tdTomato fluorescence under microscopy compared to cells with low tdTomato fluorescence (Fig. 2B).
Figure 2. Fluorescence activated cell sorting is selective for cells with high tdTomato expression.
A dissociated neuronal suspension derived from the NAc of a BAC D1R::tdTomato transgenic mouse was sorted on a FACS Aria II. (A) Neurons were selected by forward and side scatter profiles. Fluorescence gates were established by comparing tdTomato intensity in wild type and D1R::tdTomato mice. Purple cells were denoted as tdTomato+ and collected for analysis. (B) Using fluorescent microscopy, tdTomato fluorescence could be visualized before sorting and in tdTomato+ neurons but rarely cells with low tdTomato expression.
3.2. RNA analysis
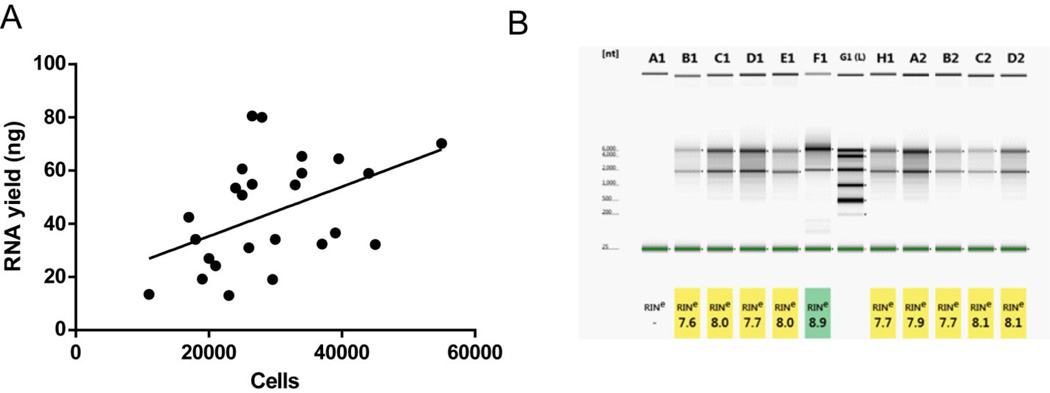
Cells were prepared in buffer containing RNase inhibitor and sorted directly intro Trizol, ensuring RNase inhibition throughout the experiment. We collected ~30,000 tdTomato positive cells per animal from NAc, which is higher than previously reported yields from the striatum (29). RNA was quantified and assessed for quality using the Agilent Bioanalyzer. RNA recovery averaged 43.5 +/− 4.0 ng (mean +/− SEM) (range: 13.0 ng – 80.1 ng) and there was a significant correlation between cells recovered and RNA yield (n = 26, r = 0.46, p < 0.05) (Fig. 3A). RNA integrity numbers (RIN) were used to determine RNA quality based on height of 28s and 18s ribosomal subunit peaks [33]. RIN values below 7 are associated with poor performance on gene expression microarrays [34]; importantly, all samples analyzed had RIN values greater than 7 (Fig. 3B).
Figure 3. RNA integrity of tdTomato+ neurons is suitable for microarray studies.
(A) There is a significant correlation between cells collected and RNA extracted (r = 0.46, p < 0.05), indicating efficient RNA extraction. (B) RNA integrity numbers (RIN) measured using the Agilent Bioanalyzer are all greater than 7, indicating suitability of microarray experiments.
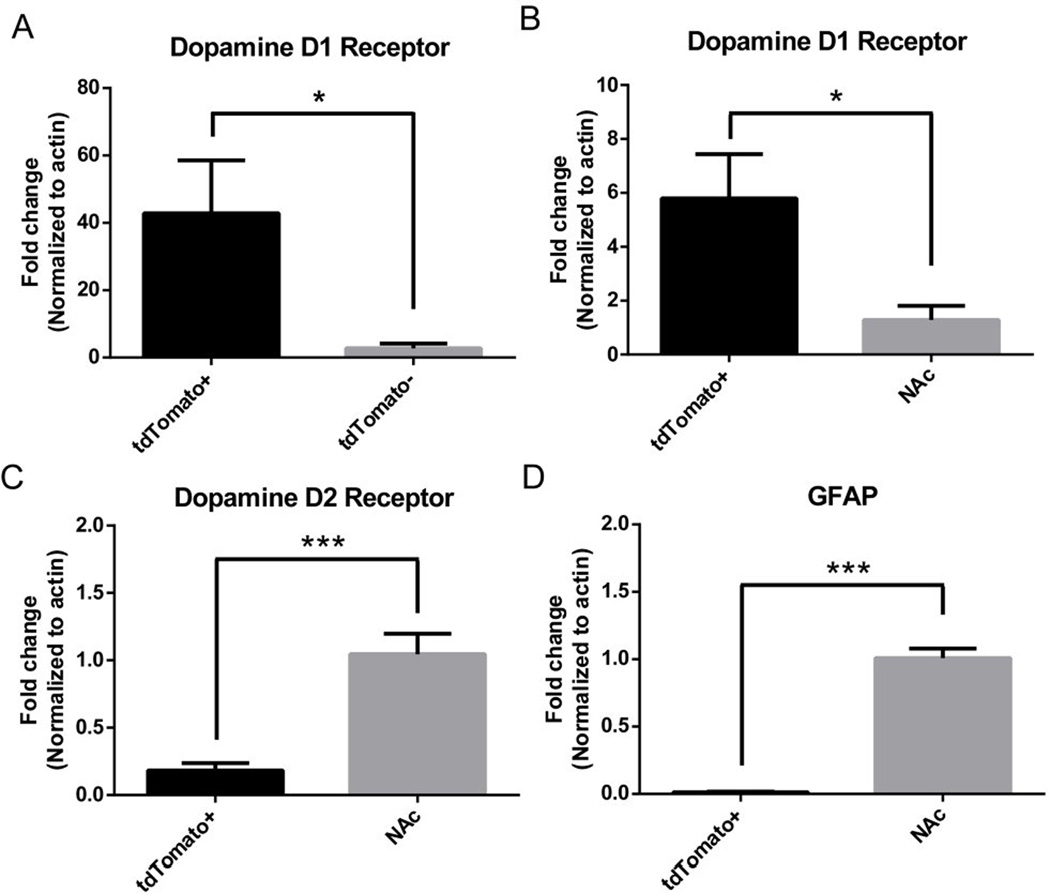
Using RT-qPCR, we showed that tdTomato positive cells have a 40-fold enrichment of D1R compared to cells with low tdTomato expression after FACS (Fig. 4A). Additionally, there was significant enrichment of D1R with significant down-regulation of GFAP and the D2 dopamine receptor (D2R) in tdTomato positive cells compared to whole NAc (Fig. 4B–D). Based on gene expression analysis, tdTomato+ cells represent an enriched population of D1R MSNs.
Figure 4. Gene expression in tdTomato+ cells.
Using RT-qPCR, tdTomato+ were found to have (A) 40-fold enrichment of the Dopamine D1 receptor relative to cells with low tdTomato expression sorted by FACS and (B–D) significant enrichment of the dopamine D1 receptor and nearly undetectable expression of the dopamine D2 receptor and GFAP relative to whole nucleus accumbens. Data presented as mean +/− SEM. n = 5–8/group, *p < 0.05. ***p < 0.01
3.3. Chromatin Immunoprecipitation
Chromatin was collected from sorted tdTomato+ cells and digested using MNase. Chromatin was left in its native state and immunoprecipitated immediately, so that there were no freeze-thaw cycles. MNase concentration was optimized to generate DNA fragments between 200–1000 bp (Fig. 5) Optimization using several antibodies revealed that anti-H3K18ac and anti-H3K27me3 antibodies could be used to discriminate active and repressed gene promoters. Notably, H3K18ac is exclusively found near the transcriptional start site of actively transcribed gene promoters and highly correlated with other marks of transcriptional activation like H3K9ac, H3K27ac, and H3K4me1/2/3 (30), so that its presence at a gene promoter is specific for transcriptional activation. H3K27me3 is a component of the polycomb repressive complex and largely associated with transcriptional repression of gene promoters and repetitive DNA sequences (30, 31). To test enrichment of D1R MSNs, H3K18ac and H3K27me3 levels were measured relative to input at the D1R and GFAP promoters, which were enriched and repressed, respectively, based on RT-qPCR analysis of this cell type. Therefore, the D1R promoter should have high levels of H3K18ac and low levels of H3K27me3 while the GFAP promoter should have the opposite pattern. Because this study used a low cell number for ChIP and only two copies for each gene promoter are available at each cell, the long interspersed nuclear element (LINE1) 5’ UTR was used as a control that could be easily quantified. There are ~500,000 copies for this retrotransposable element in the murine genome and it is associated with H3K27me3 and exclusion of H3K18ac (31, 32).
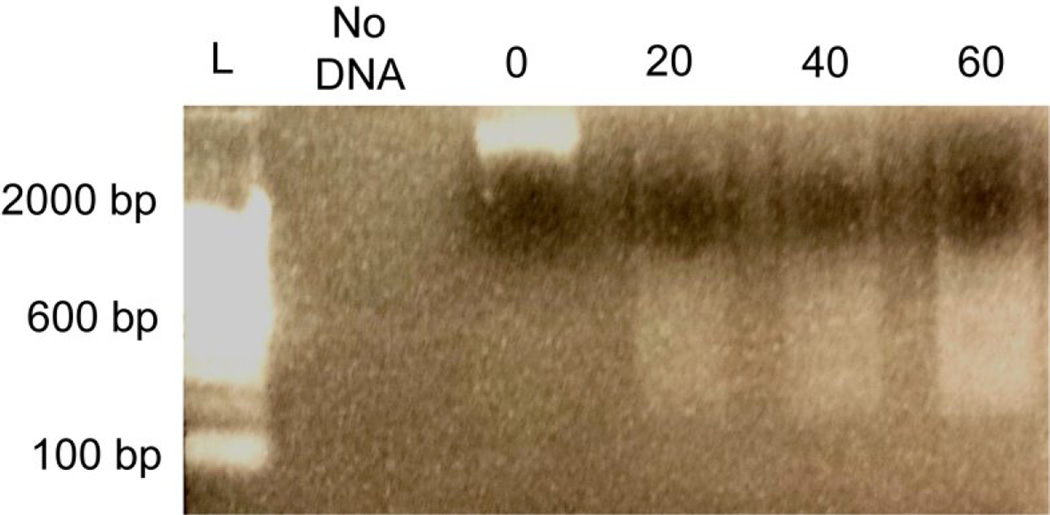
Figure 5. Optimization of micrococcal nuclease concentration.
Micrococcal nuclease (MNase) digestion for native ChIP was optimized by dividing 100,000 FAC-sorted neurons into 4 aliquots (25,000 cells/digestion). Increasing concentration of MNase resulted in greater intensity of 200 – 1000 bp DNA fragments. L = Invitrogen 100 bp ladder; 0, 20, 40, 60 = units of MNase used per 500 ul reaction.
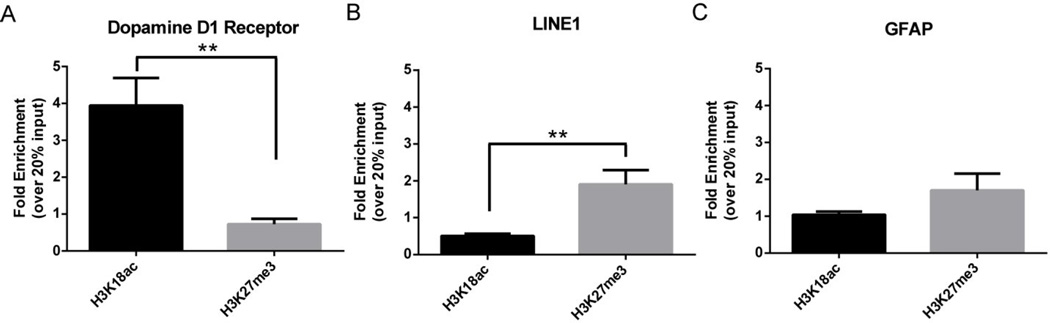
Using H3K18ac, H3K27me3, and IgG antibodies, immunoprecipitated DNA was quantified relative to input using qPCR. Prior to qPCR, DNA was quantified using a Qubit fluorometer and could be detected in the input as well as anti-H3K18ac and anti-H3K27me3 immunoprecipitations; no DNA was detected using the IgG antibody. We averaged a total mass of ~10 ng for H3K27me3, ~10 ng for 20% input, and ~2 ng for H3K18ac per 30,000 cells following immunoprecipitation. At the D1R promoter, there was a significant increase in H3K18ac occupancy relative to H3K27me3 (p < 0.01) (Fig. 6A). At LINE1, there was a significant decrease in H3K18ac occupancy relative to H3K27me3 (Fig. 6B). There was no significant difference in H3K18ac and H3K27me3 occupancy at the GFAP promoter (Fig. 6C). Additionally, there were nearly undetectable levels of DNA relative to input using a nonspecific IgG antibody (data not shown), indicating specificity for the histone modifications studied. Our data indicate that ChIP can be used to discriminate active and repressed genomic regions in FAC-sorted neuronal subtypes with low variability.
Figure 6. Chromatin immunoprecipitation identifies histone acetylation and methylation across gene promoters in tdTomato+ cells.
tdTomato+ cells were collected from D1R::tdTomato BAC transgenic mice and used to analyze histone occupancy at gene promoters. (A) There was a significant increase in H3K18ac levels relative to H3K27me3 at the D1R promoter. (B) There was a significant increase in H3K27me3 levels relative to H3K18ac levels at the repressed retrotransposable element LINE1. (C) There was no difference between H3K18ac and H3K27me3 at the GFAP promoter. Data presented as mean +/− SEM. n = 6–8/group, **p < 0.01.
4. Discussion
We describe a novel protocol for analyzing chromatin as well as efficient extraction of RNA from neuronal subtypes collected from a BAC transgenic mouse strain. Using FACS, ~30,000 tdTomato+ cells were collected from NAc with minimal tissue processing time (~45 min), providing sufficient RNA and chromatin for downstream analysis. This one day ChIP protocol is an improvement over previous protocols that describe pooling multiple animals (16, 22, 33) and is expected to facilitate epigenetic analysis of neuronal subtypes in a variety of experimental settings.
Our experimental validation shows that tdTomato positive cells have 40-fold enrichment of D1R relative to those with low tdTomato expression as well as significantly increased D1R and significant decreased D2R and GFAP relative to whole NAc, indicating cell type specificity for D1R MSNs. Despite low input for traditional chromatin studies, the ChIP protocol optimized in this study discriminated transcriptionally active and repressed regions on the basis of H3K18ac and H3K27me3 levels at gene promoters. Importantly, it showed elevated H3K18ac at the D1R promoter and elevated H3K27me3 at the LINE1 5’ UTR in tdTomato positive cells, indicating cell type specificity at the level of chromatin. There was no significant difference between H3K27me3 and H3K18ac enrichment at the GFAP promoter despite transcriptional repression. This may indicate that transcriptional repression of GFAP is achieved by mechanisms other than H3K27me3-mediated repression at the promoter. Additionally, while we designed our qPCR primers to cover regions of histone modification enrichment noted in the UCSC genome browser, it is possible that the primers did not cover the portion of the GFAP promoter where H3K27me3 accumulates to induce repression. Promoter tiling using qPCR primers would likely reveal the critical region of the GFAP promoter where these changes occur.
Another important feature of the ChIP protocol was ability to detect differences in histone occupancy with a relatively small sample size (n = 6–8), indicating reproducibility of the assay across mice. One feature that may contribute to reproducibility is use of native ChIP, which does not use formaldehyde crosslinking that can be associated with over-fixation of chromatin known as “formaldehyde artifact” (reviewed in (34)). While traditionally confined to studies of histone modifications, native ChIP can also be used to study transcription factor binding to DNA (35). The potential use of native ChIP and formaldehyde crosslinking to study transcription factor binding in FAC-sorted neurons will be interesting for further study.
While we were able to successfully quantify gene expression and histone occupancy following ChIP using qPCR, our RNA and DNA recovery were low for traditional whole genome studies. Single-cell whole exome (36) and whole genome sequencing (37) using neurons have recently been reported, suggesting that DNA and RNA analysis are possible using starting material reported in this study. Additionally, RNA yield for the majority of the samples in this study is sufficient (>25 ng) for Illumina microarrays using TargetAmp kits available from EpiCentre (Illumina). Low input ChIP-seq protocols are also being developed from as few as 1000 cells (38), so it may be possible to perform genomewide analysis of histone occupancy using the protocol described in this study or pool animals as done in previous studies. While these recently reported approaches are possible based on the reported DNA and RNA yields in this study, substantial optimization would need to be performed prior to initiation whole genome studies.
FAC-sorting of NAc from D1R::tdTomato BAC transgenic mice was efficient, with a distinct population of cells with high tdTomato expression. Gene expression and chromatin analysis of the D1R promoter in these cells confirmed that these are highly enriched D1R MSNs. While collection of ~30,000 tdTomato+ cells per sort is a substantial improvement over previous studies using BAC transgenic stains (11, 12, 26), this amount is much smaller than the total population of D1R MSNs in the NAc and suggests considerable loss of neurons during tissue processing. Improving yields will likely require optimization of tissue trituration, which is associated with considerable mechanical stress on cells and is likely the greatest source of cell loss. Unfortunately, neuronal FACS studies are still limited, so there are no reports of how changing experimental parameters affects yield. Gentler trituration over a longer time period, decreasing papain concentration, and modifying incubation temperatures are all potential ways to improve downstream analysis by providing more input for RNA and chromatin studies.
In conclusion, we describe a ChIP and RNA extraction protocol that is applicable to any cell type that can be discriminated on the basis of gene expression by a fluorescent reporter. Currently, GENSAT has 1386 commercially available mouse lines with promoter driven fluorescent reporters (39), allowing for a wide array of studies. We also provide optimization for two antibodies, anti-H3K18ac and anti-H3K27me3, that can be used to discriminate transcriptionally active and repressed gene promoters. While we do not provide a direct application of this protocol in studying neuropsychiatric disease, we hope sharing it will improve detection of epigenetic mechanisms involved in cell-type specific disease processes.
Highlights.
This paper describes a one day chromatin immunoprecipitation protocol for neuronal subtypes isolated using fluorescence activated cell sorting
The protocol achieves increased yield of cells per animal relative to previously reported FACS studies
ChIP is able to discriminate differences in histone acetylation and methylation at multiple gene promoters that correspond to gene expression
Acknowledgments
We are greatly appreciative to Erich Wilkerson and Dr. Marc Levesque for use of their flow cytometry core, Elizabeth Osterndorff-Kahanek and Dr. R. Adron Harris for their assistance with RNA analysis, and Carolyn Ferguson for technical assistance. This project is supported by NIH/NIAAA grants AA10422, AA020889, and AA021632.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12(11):623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the Depressed Brain: Role of Histone Acetylation and Methylation. Neuropsychopharmacology. 2013;38(1):124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancent Neurol. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 5.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 6.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- 7.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (ChIP) Nat Protoc. 2008;3(6):1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 8.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 9.Valjent E, Bertran-Gonzalez J, Hervé D, Fisone G, Girault J-A. Looking BAC at striatal signaling: cell-specific analysis in new transgenic mice. Trends Neurosci. 2009;32(10):538–547. doi: 10.1016/j.tins.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Durieux PF, Schiffmann SN, de Kerchove d’Exaerde A. Targeting Neuronal Populations of the Striatum. Front Neuroanat. 2011;5:40. doi: 10.3389/fnana.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 12.Lobo MK, et al. Cell Type–Specific Loss of BDNF Signaling Mimics Optogenetic Control of Cocaine Reward. Science. 2010;330(6002):385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maze I, et al. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17(4):533–539. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordi E, et al. Differential effects of cocaine on histone posttranslational modifications in identified populations of striatal neurons. Proc Nat Acad Sci. 2013;110(23):9511–9516. doi: 10.1073/pnas.1307116110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guez-Barber D, et al. FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci. 2011;31(11):4251–4259. doi: 10.1523/JNEUROSCI.6195-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozburn AR, et al. Direct regulation of diurnal Drd3 expression and cocaine reward by NPAS2. Biol Psychiatry. 2015;77(5):425–433. doi: 10.1016/j.biopsych.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fanous S, et al. Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. J Neurochem. 2013;124(1):100–108. doi: 10.1111/jnc.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz JM, Smith SH, Bilbo SD. FACS analysis of neuronal-glial interactions in the nucleus accumbens following morphine administration. Psychopharmacology (Berl) 2013;230(4):525–535. doi: 10.1007/s00213-013-3180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu QR, et al. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS) J Neurochem. 2014;128(1):173–185. doi: 10.1111/jnc.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acevedo LG, et al. Genome-scale ChIP-chip analysis using 10,000 human cells. BioTechniques. 2007;43(6):791–797. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilfillan GD, et al. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 2012;13:645. doi: 10.1186/1471-2164-13-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magklara A, et al. An Epigenetic Signature for Monoallelic Olfactory Receptor Expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo A, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86(6):1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Matevossian A, Huang H-S, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neuroscience. 2008;9(1):1–9. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ade KK, Wan Y, Chen M, Gloss B, Calakos N. An Improved BAC Transgenic Fluorescent Reporter Line for Sensitive and Specific Identification of Striatonigral Medium Spiny Neurons. Front Syst Neurosci. 2011;5:32. doi: 10.3389/fnsys.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crook ZR, Housman DE. Dysregulation of dopamine receptor D2 as a sensitive measure for Huntington disease pathology in model mice. Proc Nat Acad Sci. 2012;109(19):7487–7492. doi: 10.1073/pnas.1204542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorne A, Myers F, Hebbes T. Native Chromatin Immunoprecipitation. Epigenetics Protocols. In: Tollefsbol T, editor. Methods in Molecular Biology™. Vol. 287. Humana Press; 2004. pp. 21–44. [DOI] [PubMed] [Google Scholar]
- 28.Schoppee Bortz PD, Wamhoff BR. Chromatin Immunoprecipitation (ChIP): Revisiting the Efficacy of Sample Preparation, Sonication, Quantification of Sheared DNA, and Analysis via PCR. PLoS ONE. 2011;6(10):e26015. doi: 10.1371/journal.pone.0026015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nature neuroscience. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunami E, Vu AT, Nguyen SL, Giuliano AE, Hoon DS. Quantification of LINE1 in circulating DNA as a molecular biomarker of breast cancer. Ann N Y Acad Sci. 2008;1137(1):171–174. doi: 10.1196/annals.1448.011. [DOI] [PubMed] [Google Scholar]
- 33.Lobo MK, et al. DeltaFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 2013;33(47):18381–18395. doi: 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nat Rev Genet. 2014;15(12):814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 35.Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods. 2014;11(2):203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 37.Cai X, et al. Single-Cell, Genome-wide Sequencing Identifies Clonal Somatic Copy-Number Variation in the Human Brain. Cell Reports. 2014;8(5):1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brind’Amour J, et al. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 39.Heintz N. Gene expression nervous system atlas (GENSAT) Nature neuroscience. 2004;7(5):483–483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]