Abstract
A crosstalk between commensals, gut immune cells, and colonic epithelia is required for a proper function of intestinal mucosal barrier. Here we investigated the importance of two distinct intestinal dendritic cell (DC) subsets in controlling intestinal inflammation. We show that Clec9A–diphtheria toxin receptor (DTR) mice after depletion of CD103+CD11b− DCs developed severe, low-dose dextran sodium sulfate (DSS)-induced colitis, whereas the lack of CD103+CD11b+ DCs in Clec4a4-DTR mice did not exacerbate intestinal inflammation. The CD103+CD11b− DC subset has gained a functional specialization that able them to repress inflammation via several epithelial interferon-γ (IFN-γ)-induced proteins. Among others, we identified that epithelial IDO1 and interleukin-18-binding protein (IL-18bp) were strongly modulated by CD103+CD11b− DCs. Through its preferential property to express IL-12 and IL-15, this particular DC subset can induce lymphocytes in colonic lamina propria and in epithelia to secrete IFN-γ that then can trigger a reversible early anti-inflammatory response in intestinal epithelial cells.
INTRODUCTION
Intestinal mucosal surfaces, as the major interface between the body tissues and a potentially hostile outer environment, have evolved as a well-structured barrier against physical, chemical, and microbial insults. The epithelial layer, mucus, antimicrobial peptides, secreted immunoglobulin A, and innate and adaptive immune cells together form efficient and complex mucosal barrier (reviewed in ref. 1). All these components help to establish a beneficial “ecosystem” where a diverse and dense commensal community is tolerated, without immune attack for the benefit of the host. Perturbation of this mutualistic relationship can result in severe inflammatory bowel diseases (IBD) such as Crohn's disease and ulcerative colitis, and even more insidiously intestinal malignancies or metabolic syndromes.
The role of intestinal epithelial cells (IECs) in maintaining barrier function and in the pathogenesis of several common intestinal diseases such as IBD has been extensively studied.2, 3 In fact, enhanced gut epithelial permeability and leakage of luminal microbes across the barrier has been linked with IBD in humans and animals.4, 5
The contribution of myeloid cells such as dendritic cells (DCs) in regulating the mucosal barrier, such as epithelial integrity, production of bactericidal enzymes, or antimicrobial peptides, has not been established. It is possible that gut DCs, sentinels of mucosal immune responses, can also contribute in supporting intestinal barrier homeostasis.
Major myeloid cell subsets have been described in the colon lamina propria (LP) that differ in their ontogeny as well as in function.6, 7 There are at least two subsets of bona fide LP DCs that both express the mucosal integrin αE (CD103) β7 but differ in their expression of CD11b.8 One subset, the CD103+CD11b−DCs, the main population in the colonic LP, is dependent on the transcription factors BATF3/IRF8/Id2 for its development9, 10 and is expressing the lectin Clec9A also termed DNGR1.11 The second subset, the migratory CD103+CD11b+ DCs, is IRF4 dependent and expresses the lectin Clec4a4 (also known as DCIR2)12 and signal regulatory protein-α (SIRPα).13 Lineage affiliation of the third CD11c+CD103−CD11b+ myeloid subpopulation is still controversial, i.e., do they belong to DC or macrophage lineage.14, 15, 16
Although different DC subpopulations have been described in the gut, their exact roles in controlling gut inflammatory responses or in protection against potential infections are still elusive.17 To assess their importance in the context of intestinal damage and inflammation, we exploited two diphtheria toxin receptor (DTR) transgenic mouse lines, Clec9A-DTR and Clec4a4-DTR, enabling us to in vivo ablate both bona fide DC subsets (CD103+CD11b− and CD103+CD11b+ respectively) and test these mouse strains in a dextran sodium sulfate (DSS)-induced acute colitis model.18 Our findings show clearly that only mice lacking CD103+CD11b− DCs were highly susceptible to intestinal inflammation, whereas the lack of CD103+CD11b+ DCs did not exacerbate intestinal inflammation. Here we propose a novel pathway mediated by CD103+CD11b− DCs that controls the expression of a series of interferon-γ (IFN-γ)-inducible proteins in intestinal epithelial cells including the anti-inflammatory indoleamine 2,3 dioxygenase (IDO1) enzyme and the decoy protein interleukin-18-binding protein (IL-18bp). Our results underscore the unique role of CD103+CD11b− DCs as major intestinal immune regulators and reveal an efficient cellular network between specific intestinal DC subsets, lymphocytes, and epithelial cells to control colonic inflammation.
RESULTS
Characterization of colon CD11chighMHCII+ myeloid cell subsets: Clec9A and Clec4a4 lectins are differentially expressed on distinct colon bona fide DC subsets
Mouse large intestine contains three distinct CD11chighMHCII+ myeloid cell subsets that express CD103+CD11b−, CD103+CD11b+, or CD103−CD11b+, respectively, as shown in Figure 1a. To further characterize and classify them, we generated genome-wide transcriptional profiles of sorted colon CD11chighMHCII+ cells (Figure 1a,b) isolated from control (steady state) or DSS-treated mice (day 4). A hierarchical clustering of the differentially expressed genes using Pearson's correlation and complete linkage showed a clear clustering of CD103+CD11b−, CD103+CD11b+, and CD103−CD11b+ cells as visible in the principal component analysis plot (Figure 1c). CD103+CD11b− cells were delineated as bona fide DCs because of expression of elevated levels of transcription factors Irf8, Irf5, and Id2 and other markers such as Clec9A, Cd24, Flt3, Xcr1, and Itga2 (Figure 1d lower part, in red). Furthermore, our analysis clearly suggested the macrophage identity for CD103−CD11b+ cells that differentially expressed the macrophage transcription factor MafB as well as other macrophage-related markers such as F4/80 (Emr1), Cd68, Cd14, Tlr4, Lamp1, mannose receptor (Mrc1), MP scavenger receptor (Msr1), chemokine receptor Cx3Cr1, matrix metalloproteinase (Mmp13, Mmp14), and complement receptors (C5ar1 and C3ar1) (Figure 1d middle part, in red). The third subset expressing both CD103 and CD11b markers displayed the highest levels of Irf4 and Clec4a4 and co-shared molecules, although at lower levels, such as Flt3, Irf5, and Id2 with the bona fide CD103+CD11b− DC subset, and some myeloid-related markers with CD103−CD11b+ cells such as granulocyte-macrophage colony stimulating factor 2 receptor (Csf2rb2), triggering receptor expressed on myeloid cells 1 (Trem-1), macrophage galactose N-acetyl-galactosamine-specific lectin 2 (Mgl2), SIRP-α and -β (Sirpa, Sirpb1a, Sirpb1b), different lectins (Clec4a1, Clec4d, Clec4d, Clec10a), and Mmp12. Taken together, our results strongly suggest that colon CD11chighMHCII+ myeloid cells can be subdivided into two distinct bona fide DC subsets and into a distinct macrophage-related cell subpopulation. Interestingly, our microarray analysis did not show any major significant changes between distinct DC subset collected at steady state or under DSS treatment, most likely because of the early time point of chemical treatment (4 days).
Figure 1.
Transcriptome of colon dendritic cell (DC) subsets. (a) Sorting strategy for colon DC isolation. Large intestines obtained from 8 mice, either control (steady state (SS)) or dextran sodium sulfate (DSS) treated (day 4 DSS), were pooled and lamina propria (LP) cells were isolated. Procedure was repeated three times independently. Following CD11chighMHCII+ DC subsets were sorted and analyzed in a gene expression microarray: (1) CD103+CD11b−, (2) CD103+CD11b+, and (3) CD103−CD11b+. (b) Transcript heat map of the ∼640 genes that are at least twofold differentially expressed in one comparison (red: upregulated; green: downregulated). Clustering was performed using Pearson's correlation and complete linkage. Heat map was z-score normalized by row. (c) XY plot of the first two components of a principal component analysis (PCA) of all six groups (SS 1–3 and DSS 1–3). (d) Heat map showing differential expression of selected genes involved in DC development and function; heat map was generated as described in b.
We next validated whether DC subpopulations defined above express Clec9A and Clec4a4 by flow cytometry. After gating on CD11c+MHCII+ cells, Clec9A-expressing cells were confirmed to be located in the CD103+CD11b− fraction, whereas Clec4a4 expression was detectable only in CD103+CD11b+ DC subset (Supplementary Figure S1 online, upper panels). CD11cintMHCII+ macrophages did not express any of the Clec9A and Clec4a4 lectins (Supplementary Figure S1, lower panels). Therefore, Clec9A- and Clec4a4-DTR mice can be used to specifically ablate different subsets of LP DCs.
Efficient and specific in vivo ablation of gut DC subsets
CX3CR1GFP/Clec9A- and CX3CR1GFP/Clec4a4-DTR mice were then tested to see whether they could be used to ablate intestinal DC subsets. Both transgenic mouse strains were injected twice with 20 ng g−1 body weight DT (days −2 and −1) and subsequently analyzed for the presence of different colon and mesenteric lymph node (MLN) DC subsets. As shown in Figure 2a, DT-treated CX3CR1GFP/Clec9A-DTR mice efficiently ablated the CD11chighMHCII+CD103+CD11b− DC subset in colon. In the MLN, both classical lymphoid organ-resident CD11chighMHCII+CD8+CD11b− and LP-derived migratory CD11cintMHCII++CD103+CD11b− disappeared upon DT treatment (Figure 2b). On the contrary, DT-treated CX3CR1GFP/Clec4a4-DTR mice reduced the CD11chighCD103+CD11b+DC fraction by 70% in the colon and by 50% in the MLN. Classical lymphoid organ-resident CD11chighMHCII+CD8−CD11b+DC fraction was effectively diminished by 80% (Figure 2b). Interestingly, for unknown reasons, DT-treated CX3CR1GFP/Clec4a4-DTR mice, but not DT-treated CX3CR1GFP/Clec9A-DTR mice, also partially ablated the CD11cintMHCII+CX3CR1high macrophage fraction as shown in Figure 2a, whereas the CD11cintMHCII+CX3CR1int monocyte-derived macrophage fraction was unaffected. This unexpected ablation, however, had no functional consequences (see below). As Clec9A is also expressed in common DC progenitors and pre-dendritic cells (DCs) in the bone marrow,19 the repetitive DT injections could possibly affect all DC subsets. To exclude this, we analyzed spleen and colon 15 days after the first DT injection (followed by further DT injections at days 4 and 8) and could confirm the spleen CD11b+ DC subset as well as the CD103+CD11b+ DCs in the colon were not affected in our Clec9A-DTR mouse. On the contrary, CD8+ DCs and CD103+CD11b− stayed efficiently ablated over the observation period (data not shown).
Figure 2.
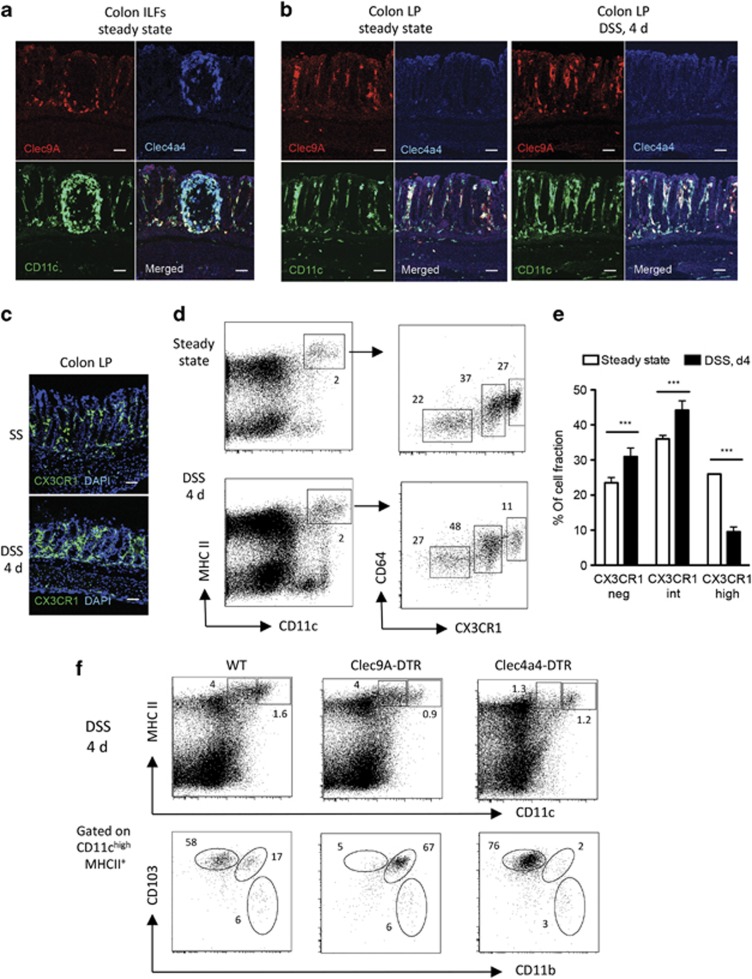
Distinct intestinal myeloid cells are ablated in Clec9A- and Clec4a4-–diphtheria toxin receptor (DTR) mice. Colon cells were obtained from DT-treated CX3CR1GFP wild-type (WT) controls, CX3CR1GFP/Clec9A-DTR, and CX3CR1GFP/Clec4a4-DTR mice. (a) Colon lamina propria (LP) cells were analyzed for CD103 and CD11b expression by gating on CD11chighMHC II+ cells (gate 1) and for CX3CR1 and CD64 expression by gating CD11cintMHC II+ cells (gate 2). (b) Mesenteric lymph nodes (MLNs) were obtained from the same mice and analyzed for CD103 and CD11b expression by gating on CD11cintMHCII++ migratory dendritic cells (DCs; gate 3) and classical lymphoid CD11chighMHC II+ DCs (gate 4). Representative dot plots of colons and MLNs isolated from three different mice are shown. Indicated numbers show the percentage of each gated cell subset.
Clec9A+CD103+CD11b− and Clec4a4+CD103+CD11b+ DCs localize differently in colon LP
We analyzed the localization of both DC populations in the colon LP during steady state as well as during early events of DSS-mediated colitis before any obvious onset of disease (day 4). To achieve this, proximal colon cryosections were costained with anti-CD11c together with anti-Clec9A or anti-Clec4a4 antibodies. As shown in Figure 3 both Clec9A+ and Clec4a4+ DC subsets are colocalized in different areas of colonic innate lymphoid follicles (ILFs). Some CX3CR1+ macrophages were also found in ILFs (data not shown). However, only Clec9A+ DCs together with the CX3CR1+ macrophages could be visualized abundantly in the LP under steady-state conditions, whereas the Clec4a4+ DC subset was absent (Figure 3b,c). The Clec4a4+ DC fraction did not become detectable in the LP even upon DSS treatment (Figure 3b, right panel), whereas a clear shift from CX3CR1high to CX3CR1int cells, presumably inflammatory monocytes,20 could be observed in the LP (Figure 3d,e). The ablation of targeted colonic LP DC subpopulations was also confirmed during DSS treatment (day 4). In fact, LP of Clec9A-DTR mice lacked the CD103+CD11b− DCs and accumulated CD103+CD11b+ DCs, whereas, vice versa, in Clec4a4-DTR mice, CD103+CD11b+ DCs were efficiently ablated whereas CD103+CD11b− DCs remained (Figure 3f). Confocal analysis of Clec9A+ and Clec4a4+ DC subsets in colon sections of DT-treated Clec9A- and Clec4a4-DTR mice also confirmed their ablation at steady state and after 4 days of DSS treatment (Supplementary Figure S2). In summary, our data indicate that Clec9A+ DCs and CX3CR1+ macrophages/monocytes are present in both ILFs and LP compartment, whereas Clec4a4+ DC are restricted to ILFs and no major changes in DC anatomical redistribution (ILF vs. LP) could be detected during DSS treatment or in absence of one of the DC subset.
Figure 3.
Differential localization of distinct colon lamina propria (LP) dendritic cell (DC) subsets and CX3CR1+ macrophages at steady state and during dextran sodium sulfate (DSS)-mediated colitis. (a) Representative confocal images of Clec9A+ and Clec4a4+ DC subsets in the colonic innate lymphoid follicles (ILFs). Sections were stained with phycoerythrin (PE)-labeled anti-Clec9A antibody (red), allophycocyanin (APC)-labeled anti-Clec4a4 (blue), and fluorescein isothiocyanate (FITC)-labeled anti-CD11c (green). Original magnification × 20. Bars=50 μm. (b) Representative confocal images of Clec9A+ and Clec4a4+ DC subsets in the colonic LP under steady-state conditions and 4 days (4 d) after DSS treatment. Cryosections were stained as above in (a). Original magnification × 20. Bars=50 μm. (c) Representative confocal images of CX3CR1GFP+ cells (green) in the colonic LP under steady-state (SS) conditions and 4 days after DSS treatment. Sections were stained with 4',6-diamidino-2-phenylindole (DAPI; blue, nuclei). Original magnification × 20. Bars=45 μm. (d) Flow cytometry of colon LP cells of CX3CR1GFP mice analyzed at steady state and after 4 days of DSS. CD11c/major histocompatibility complex (MHC class II)-positive cells were analyzed for CD64/CX3CR1GFP. Indicated numbers show the percentage of each gated cell subset. (e) Percentage of differentially expressing CX3CR1 populations in the colon during steady state and upon 4 days of DSS treatment. N=5 mice, Student's t-test ***P>0.001. (f) DC ablation profiles during DSS treatment. LP cells were collected at day 4 from the distal part of colon of DT-treated wild-type (WT), Clec9A–diphtheria toxin receptor (DTR), and Clec4a4-DTR (n=3 each group). Representative fluorescence-activated cell sorting (FACS) dot plots showing the staining profile of CD11c and MHC class II as well as CD103 and CD11b.
Absence of Clec9A+CD103+CD11b− DCs, but not of Clec4a4+CD103+CD11b+ DCs, aggravates DSS-induced colitis
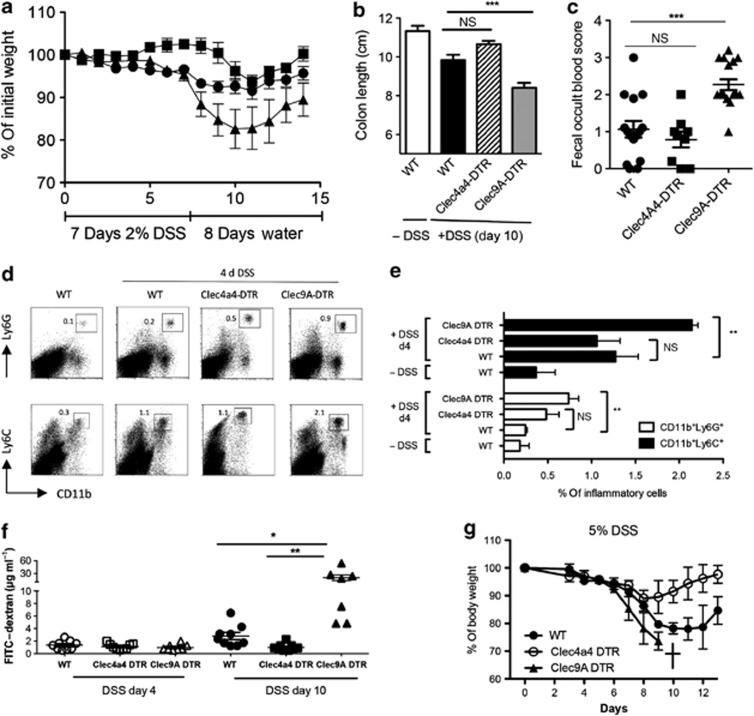
As changes of the mucosal barrier composition have long been identified in gastrointestinal pathologies, we analyzed both Clec9A- and Clec4a4-DTR strains in a DSS-mediated acute experimental colitis and compared the intensity of inflammation in mice with unperturbed intestinal DC repertoire. Therefore, we challenged 8-week-old DT-treated wild-type (WT), Clec9A-DTR, and Clec4a4-DTR mice with 2% DSS in drinking water for 7 days followed by water without DSS (for simplicity, DT-treated WT and Clec9A- and Clec4a4-DTR mice will be hereafter referred as WT, Clec9A-DTR, and Clec4a4-DTR mice if not otherwise stated). Using these experimental conditions, only mild colitis was induced in the controls, whereas in Clec9A-DTR mice, severe symptoms were observed already at earlier stages of colitis. In fact, Clec9A-DTR mice showed large weight loss over the course of DSS treatment (Figure 4a). The observed body weight reduction in Clec9A-DTR mice correlated with a shortened colon length (Figure 4b), increased rectal bleeding (Figure 4c), and enhanced CD11b+Ly6G+ neutrophil and CD11b+Ly6C+ inflammatory monocyte infiltrations in colon (Figure 4d,e). On the other hand, under the same low-dose DSS treatment, Clec4a4-DTR mice displayed hardly any weight lost, no measurable colon shortening, and weak rectal bleeding. In fact, there was a clear trend that Clec4a4-DTR mice showed more resistance to colitis than WT mice (Figure 4a–c). This “protective” trend was observed not only at low DSS concentrations, but was also confirmed when we increased DSS to 5%. The body weight of WT mice was reduced significantly by 25%, whereas it hardly changed for Clec4a4-DTR mice. On the contrary, at concentration of 5% DSS, Clec9A-DTR mice succumbed to the treatment (Figure 4g).
Figure 4.
Ablation of CD103+CD11b− cells enhances susceptibility to dextran sodium sulfate (DSS)-induced colitis. Wild-type (WT), Clec9A–diphtheria toxin receptor (DTR), and Clec4a4-DTR mice were injected with 20 ng g−1 DT following the schedule described in Methods. Then, 2% DSS was supplied at day 0 ad libidum in the drinking water for 7 consecutive days followed by drinking water at day 8. (a) Body weight was monitored daily over a period of 15 days. Black circles: DT-treated WT control; black triangles: DT-treated Clec9A-DTR; black squares: DT-treated Clec4a4-DTR. Each group: n=8. Values represent the mean±s.d. Three independent experiments were performed with the same numbers of animals. (b) Measurement of colon length at day 8 (cm) of control WT mice (white bar) and DSS-treated DT-injected WT (black bar), Clec4a4-DTR (striped bar), and Clec9A-DTR (gray bar) mice. Each group: n=8. Values represent the mean±s.d. (c) Fecal samples of DT-injected WT controls (black circles), Clec4a4-DTR (black squares), and Clec9A-DTR (black triangles) mice were collected at day 8 upon DSS treatment and scored for blood content. Each group: n>9 mice. Student's t-test significance: ***P>0.001, NS, not significant. (d, e) Increased influx of inflammatory myeloid cells in the absence of Clec9A+CD103+CD11b− dendritic cell (DC) subset at early stages of inflammation (day 4 (d4)). Colon of DT-injected WT, Clec4a4-DTR, and Clec9A-DTR mice were analyzed at day 0 and 4 days upon start of DSS treatment. Representative dot plots of three mice indicating CD11bLy6G+ and CD11bLy6C+ cells (d) and bar chart of 3–4 mice±s.d. Student's t-test significance: **P>0.005. (f) Intestinal permeability as determined by quantifying the amount of fluorescein isothiocyanate (FITC)–dextran levels (μg ml−1) in the serum after its oral gavage. DT-injected WT, Clec9A-DTR, or Clec4a4-DTR mice were tested at days 4 (filled symbols) and 10 (empty symbols) from the beginning of DSS treatment. For each group, 7–9 mice were analyzed. Student t-test significance: *P>0.01; **P>0.005. (g) Clec9A DTR mice do not survive a 5%, high-dose DSS treatment. WT, Clec9A-DTR, and Clec4a4-DTR mice were treated with 5% DSS for 7 consecutive days followed by drinking water at day 8. Body weight was monitored daily over a period of 13 days. Mice were killed when body weight was <75% of their original weight. Black circles: DT-treated WT control; black triangles: DT-treated Clec9A-DTR; white circles: DT-treated Clec4a4-DTR. Each group: n=4. Values represent the mean±s.d.
Next we monitored the epithelial integrity in the presence or absence of different DC subsets. To do this DT-treated or untreated Clec9A- or Clec4a4-DTR transgenic mice were fed with 2% DSS for 7 days and epithelial permeability was scored at days 4 and 10 (3 days after the termination of DSS treatment) with fluorescein isothiocyanate (FITC)–dextran introduced by gavage. As predicted, at day 10, Clec9A-DTR-ablated mice showed greatly increased leakage of FITC–dextran in serum. Interestingly, epithelium of Clec4a4-DTR mice seemed to stay intact whereas that of WT mice showed signs of leakage (Figure 4f).
Taken together, CD103+CD11b−-ablated mice were highly susceptible to DSS-induced colitis, whereas no obvious inflammation was seen without DSS in this short DT treatment schedule in steady-state conditions (data not shown). On the other hand, ablation of CD103+CD11b+ and partial depletion of CX3CR1high macrophages in the Clec4a4-DTR mouse conferred resistance in the development of DSS-induced colon inflammation. The protection was not mediated by the absence of CX3CR1high cells because a CD169-DTR mouse21 in which this particular gut macrophage subpopulation can be ablated is susceptible to colitis with all typical signs: shortened colon, increased bleeding, and intestinal permeability (Figure 5a–e).
Figure 5.
Depletion of CX3CR1high macrophages leads to severe intestinal inflammation. (a) Flow cytometry analysis of different macrophage and dendritic cell (DC) subsets. CX3CR1-GFP-CD169-DTR and CX3CR1-GFP-WT mice were injected with DT (20 ng per g body weight) and analyzed the following day for the ablation profile of different CD11chighMHC IIhigh DCs and CD11cintMHC II high macrophages, respectively. For DC profiling, anti-CD103 and anti-CD11b were used, whereas for macrophage profiling, cells were stained with anti-CD64 and monitored for CX3CR1 GFP expression. (b–e) Ablation of CX3CR1high macrophages enhances susceptibility to dextran sodium sulfate (DSS)-induced colitis. Wild-type (WT) and CD169–diphtheria toxin receptor (DTR) mice were injected with 20 ng g−1 DT following the schedule described in Methods. (b) Body weight was monitored daily over a period of 15 days. Open circles: DT-treated WT control; filled circles: DT-treated CD169-DTR. Each group: n=5. Values represent the mean±s.d. Two independent experiments were performed with the same numbers of animals. (c) Fecal samples of DT-injected WT controls (open circles) and CD169-DTR (filled circles) mice were collected at day 8 upon DSS treatment and scored for blood content. Each group: n>5 mice. Student's t-test significance: ***P>0.001. (d) Measurement of colon length at day 8 (cm) of control WT mice (gray bar) and DSS-treated DT-injected WT (white bar) or CD169 DTR (black bar) mice. Each group: n=5. Values represent the mean±s.d. (e) Intestinal permeability as determined by quantifying the amount of fluorescein isothiocyanate (FITC)–dextran levels (μg ml−1) in the serum after its oral gavage. DT-injected WT (open circles) and CD169-DTR mice (filled circles) were tested at days 4 and 10 from the beginning of DSS treatment. For each group, 5–9 mice were analyzed.
Ablation of Clec9A+ DCs affects the expression of several IFN-γ-inducible genes in IECs
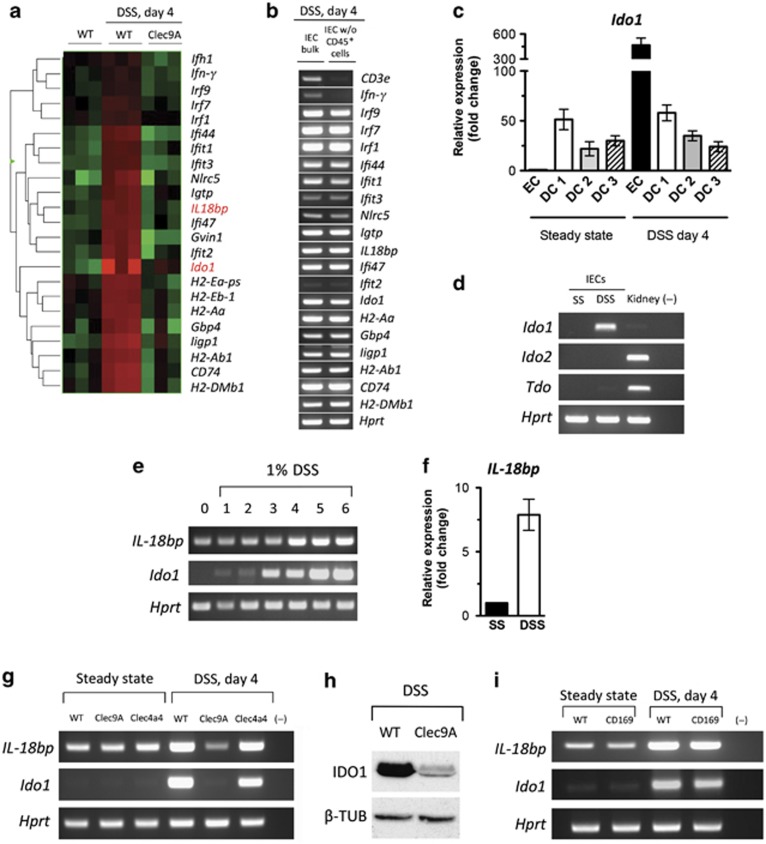
An elaborate interplay between gut microbiota, epithelial cell layer, and immune cells controls gut homeostasis and constrains overexuberant inflammatory responses. Beside the passive role as a physical barrier, the IECs express antimicrobial peptides and enzymes, essential for resistance against invasive bacteria as well as for maintenance of intestinal tolerance. To assess a possible IEC contribution to the severe DSS-induced inflammation observed in Clec9A-DTR mice, we next performed microarray-based comparisons of gene expression in IECs collected from untreated control WT, DSS-treated WT, and Clec9A-DTR mice. Interestingly, microarray analyses of the intestinal epithelial fraction from DSS-treated WT mice revealed a clear upregulation of IFN-γ and a series of IFN-γ-inducible genes, such as IFN-γ-induced GTPases (e.g., Gvin1, Gbp4, Igtp, ligp1), IFN-γ-induced proteins (e.g., Ifit1, Ifit2, Ifit3, Ifit44), IFN-γ-induced regulatory factors (e.g., Irf1, Irf7, and Irf9), NOD-like receptor family CARD domain containing 5 (Nlrc5), IFN-γ-induced major histocompatibility complex (MHC) class II-related proteins (e.g., H2-DMb1, H2-Ab1, H2-Aa, H2-Eb1, Cd74), as well as anti-inflammatory proteins (Ido1 and IL-18bp) (Figure 6a). Validation of the lymphocyte-depleted IEC fraction showed that all genes, except IFN-γ, were IEC specific (Figure 6b). By comparing the gene expression profiles between DSS-treated WT control and Clec9A-DTR mice, we observed that all IFN-γ-induced genes were downregulated in Clec9A-DTR mice (Figure 6a) that underlines the surprising role of gut CD103+CD11b−Clec9A+ DCs in regulating the intestinal IFN-γ response during DSS-induced colitis.
Figure 6.
Epithelial expressed interferon-γ (IFN-γ)-inducible genes are strongly affected by ablation of CD103+CD11b− dendritic cells (DCs). (a) Heat map showing differential expression of selected genes regulated by IFN-γ of colon intestinal epithelial cells (IECs) obtained from wild-type (WT) untreated mice and dextran sodium sulfate (DSS)-treated day 4 WT and Clec9A–diphtheria toxin receptor (DTR) mice (n=3). (b) Gene validation comparing bulk IECs and CD45+ lymphocyte-depleted IECs obtained from DSS-treated animals. IECs were isolated from the colon as described in Methods and loaded on a Percoll gradient to separate the lymphocytes from the epithelial fraction. RNA and subsequently complementary DNA (cDNA) was prepared and validated for Cd3, Ifn-γ, and a series of Ifn-γ-induced genes, including Ido1 and IL-18bp. One representative sample is shown. (c) Quantitative real-time PCR (qPCR) analysis of Ido1 expression in different intestinal DC subsets and IECs at steady state (SS) and 4 days after DSS treatment. N=3±s.e.m. (d) Indoleamine 2,3 dioxygenase (IDO1) is the major tryptophan-degrading enzyme in the colonocytes. IECs obtained from distal part of the colon of DSS-treated WT mice (day 4) were analyzed for Ido1, Ido2, and Tdo expression by semiquantitative real-time PCR (RT-PCR) analysis. Hprt was used as an endogenous mRNA control. Results are representative of 3–4 pooled colons. (e) Ido1 and IL-18bp expression profile during DSS treatment in IECs. WT mice were treated with 1% DSS over 6 days. Colonocytes were isolated from the distal part of three mice every day and monitored by RT-PCR for Ido1 and IL-18bp mRNA expression. (f) qPCR analysis of IL-18bp expression in IECs at steady state and 4 days after DSS treatment. N=3±s.e.m. (g) RT-PCR analysis of Ido1 and IL-18bp in IECs obtained from pooled colons of DT-injected untreated or DSS-treated (day 4) WT, Clec9A-DTR, and Clec4a4-DTR mice. PCR results are representative of three independent IEC isolations. (h) IDO1 protein expression in IECs pooled from three DSS-treated WT or Clec9A DTR mice (day 4). Representative immunoblots for epithelial IDO1 (45 kDa) and β-tubulin control (50 kDa) are shown. (i) Absence of CX3CR1high macrophages does not affect expression of IDO1 and interleukin-18-binding protein (IL-18bp) in IECs during colitis. RT-PCR analysis of Ido1and IL-18bp in IECs obtained from DT-injected untreated or DSS-treated (day 4) WT and CD169-DTR mice. PCR results are representative of three independent IEC isolations.
Absence of Clec9A+CD103+CD11b− DCs leads to diminished expression of IDO1 and IL-18bp in IECs during early stages of colitis
Our gene array results indicate a marked downregulation of two anti-inflammatory molecules, the enzyme Ido1 and the decoy protein IL-18bp, in DSS-treated Clec9A-DTR mice (Figure 6a).
It is well documented that the immune modulatory activity of IDO1 is critical in limiting DSS-induced inflammation.22, 23 As IDO1 is expressed in mononuclear cells, especially in DCs, and in other cells such as epithelial cells, we first compared the levels of Ido1 expression between different LP DC subsets and colon IECs. At steady-state conditions, CD103+CD11b− DCs are the major Ido1-expressing cells in the colon, but after DSS exposure, Ido1 mRNA expression in IECs exceeded by almost 10-fold the level of DC expression (Figure 6c). IDO1 was also confirmed as the major enzyme involved in the tryptophan catabolism in the gut, as the expression of two other enzymes involved, Ido2 and tryptophan 2,3 dioxygenase (Tdo), were not detectable in IECs at steady state as well as during DSS treatment (Figure 6d). Notably, tissue damage caused by DSS-induced Ido1 expression in IECs within 24 h and its expression was subsequently maintained over the 6 days tested (Figure 6e). Because of this pronounced DSS-induced upregulation of Ido1 mRNA in colon IECs and the massive downregulation in Clec9A-DTR mice, we validated the gene array results by semiquantitative PCR analysis as well as by western blot. PCR analysis revealed hardly detectable expression of Ido1 mRNA at steady state in all three mice groups, whereas a sharp increase could be observed at early stages of inflammation in WT control and in Clec4a4-DTR mice (Figure 6g). Interestingly and consistent with the inflammation-prone phenotype of Clec9A-DTR mice, we found that Ido1 was downregulated at both RNA and protein levels when Clec9A+CD103+CD11b− DCs were depleted in mice treated with DSS (Figure 6g,h).
The neutralization of the proinflammatory cytokine IL-18 through IL-18bp is also critical in limiting DSS-induced inflammation.24 Differently to Ido1 mRNA, basal levels of IL-18bp mRNA are detectable in IECs at steady state, but like Ido1, IL-18bp is upregulated over time when the epithelial injury is induced (Figures 6e,f) and its increase is IFN-γ dependent like that of Ido1. Because of this augmented expression during induced inflammation, we also tested whether the levels of IL-18bp are influenced by the absence of one of the DC subsets in steady state or during the colitic response. PCR analysis revealed low levels of IL-18bp at steady state in all three groups of mice, with a sharp increase at early stages of inflammation in WT control and in Clec4a4-DTR mice (Figure 6g). Consistently with our gene array data, IECs obtained from Clec9A-DTR mice display clearly diminished expression of IL-18bp mRNA (Figure 6g).
To determine whether epithelial expression of Ido1 and IL-18bp was controlled by the immunosuppressive CX3CR1high macrophage subpopulation, we also assessed inflammation-dependent epithelial Ido1 and IL-18bp mRNA expression levels in CD169-DTR mice. Surprisingly, ablation of CX3CR1high macrophages did not affect Ido1 or IL-18bp expression as comparable mRNA levels to WT were measured in IECs (Figure 6i).
Altogether, these results highlight the unique property of CD103+CD11b− DCs in regulating the expression levels of IFN-γ-inducible immune regulatory molecules synthesized by IECs important for gut homeostasis. Consistently with our observation, IFN-γ-deficient mice are highly susceptible to DSS-treatment that underlines an essential protective function of IFN-γ in controlling early phases of intestinal inflammation. In fact, IFN-γ mice had to be terminated at day 8 because of massive body weight loss (<70%) and severe rectal bleeding (Figure 7a,b).
Figure 7.
IFN-γ −/− mice show enhanced susceptibility to dextran sodium sulfate (DSS)-induced colitis. Wild-type (WT) and interferon-γ (IFN-γ)−/− mice were treated as described in Methods. (a) Body weight was monitored daily over a period of 11 days. IFN-γ−/− mice were killed at day 8 because of severe body weight loss (>30%). White circles: CB57/BL6 control; black circles: IFN-γ−/− mice. Each group: n=5. Values represent the mean±s.d. Two independent experiments were performed with the same numbers of animals. (b) Fecal samples of CB57/BL6 control and IFN-γ −/− mice were collected at day 7 upon DSS treatment and scored for blood content. Each group: n>7 mice. Student's t-test significance: ****P>0.0001.
CD103+CD11b− DCs modulate IFN-γ production by LP CD4+ and CD8+ T cells and intraepithelial CD8+ T cells
As Ido1 and IL-18bp expression is reported to be controlled by IFN-γ,25, 26 we first confirmed the expression of the IFN-γ receptor in IECs and CMT-93 colon epithelial cell line, as shown in Figure 8a, and then corroborated the IFN-γ-dependent Ido1 and IL-18bp upregulation in the CMT-93 cells (Figure 8b). Consistently, IECs obtained from DSS-treated IFN-γ −/− mice lacked the upregulation of Ido1 and IL-18bp typically observed after the chemical treatment in WT mice (Figure 8c).
Figure 8.
IDO1 and IL-18bp expression is modulated by interferon-γ (IFN-γ). (a) Colonocytes express IFN-γ receptor (IFN-γR). The ex vivo isolated colonocytes and CMT-93 colon epithelial cell line were analyzed by semiquantitative real-time PCR (RT-PCR) analysis for IFN-γ receptor expression. Hprt was used as an endogenous mRNA control. (b) IDO1 and IL-18bp expression is induced by IFN-γ. CMT-93 cells were stimulated overnight with 100 U ml−1 IFN-γ and analyzed for Ido1 and IL-18bp expression by semiquantitative RT-PCR analysis. (c) IFN-γ−/− mice do not upregulate Ido1 and IL-18bp epithelial expression upon dextran sodium sulfate (DSS) treatment. Intestinal epithelial cells (IECs) were collected from untreated or DSS-treated wild-type (WT) and IFN-γ−/− mice and evaluated by semiquantitative RT-PCR. One representative sample from each experimental group of three mice is shown. SS, steady state. (d) Clec9A–diphtheria toxin receptor (DTR) mice have a decreased proportion of IFN-γ-expressing lamina propria (LP) T cells and intraepithelial lymphocytes (IELs). Representative flow cytometry plots of LP and IELs harvested from wild-type (WT) and Clec9A-DTR mice 4 days after DSS treatment and stained for CD4, CD8, and γ/δ T cell receptor (TCR), respectively (representative fluorescence-activated cell sorting (FACS) dot plot, right panel) and stained for intracellular IFN-γ. Quantification of LP CD4+ T cells, LP CD8+ T cells, LP CD4−CD8− T-cell fraction, γ/δ+, and CD8+ IELs expressing IFN-γ. N=6–8 mice pooled from 2 independent experiments±s.e.m. Student's t-test significance: *P>0.01, ***P>0.001, NS, not significant. (e) Quantitative real-time PCR (qPCR) analysis of IL-15, IL-12p40, IL-12p35, and IL-13p19 expression in distinct colon dendritic cell (DC) subsets obtained from control WT mice: CD103+CD11b−, CD103+CD11b+, and CD103−CD11b+. Data are representative of 3 independent experiments with 10 mice pooled in each group.
The observation that CD103+CD11b− DCs control the levels of IFN-γ-inducible genes in IECs prompted us to characterize the cellular source of IFN-γ. As shown in Figure 8d, we analyzed different T-cell populations localized in the colon LP or epithelial layer for their capacity to produce IFN-γ during early stages of DSS treatment and tested whether its secretion is controlled by CD103+CD11b− DCs. At steady state, intestinal T cells do not secrete IFN-γ, but an intestinal T cell-mediated IFN-γ response was induced in response to DSS treatment as shown in Figures 6b and 8d. Notably, we found that in the absence of this particular DC subset LP, CD4+ T and CD8+ T cells as well as intraepithelial CD8+ T cells were significantly impaired in their capacity to produce IFN-γ (Figure 8d), a reduction that correlates with the reduced levels of IFN-γ-inducible genes in IECs during early stages of intestinal inflammation. No significant difference in IFN-γ production was observed in the non-CD4/CD8 T-cell LP fraction.
Finally, we sought to identify the cytokines that link CD103+CD11b− DCs to the production of IFN-γ by intestinal lymphocytes. Interestingly, quantitative real-time PCR analysis of isolated MHCII+CD11chigh myeloid cell subsets (CD103+CD11b−, CD103+CD11b+, CD103−CD11b−) in colon revealed a differential expression pattern of cytokines. Only CD103+CD11b− DCs expressed IL-12p35/IL-12p40 (IL-12) and IL-15, both cytokines involved in supporting IFN-γ production of intestinal lymphocytes,27, 28 whereas CD103+CD11b+ DCs expressed IL-23 p19/IL-12p40 (IL-23) (Figure 8e). DSS-mediated epithelial injury expanded the numbers of CD103+CD11b+ DCs by almost twofold, but surprisingly, no additional enhancement of IL12p35 and IL-15 mRNA levels was observed after 4 days of DSS challenge (Supplementary Figure S3), although we cannot exclude a transient cytokine increase during the first days of DSS treatment.
Collectively, these results suggest that under tissue injury conditions mediated via DSS, expansion of IL-12- and IL-15-producing CD103+CD11b− DCs modulates the secretion of IFN-γ by intestinal lymphocytes that then triggers the expression of IFN-γ-inducible epithelial genes, including the well-characterized anti-inflammatory molecules like IDO1 and IL-18bp that contribute in containing intestinal inflammation.
To test whether the reduced levels of IFN-γ-induced proteins such as IDO1 and IL-18bp contribute to colitis-prone phenotype observed in CD103+CD11b− DC-ablated mice, we treated WT and Clec9A-DTR mice with immunostimulatory oligonucleotides (ISS-ODNs) that have been shown to trigger IFN-γ-response and to limit disease severity in experimental colitis.29, 30 Two injections of ISS-ODNs increased the IFN-γ levels in both mouse strains, WT and Clec9A-DTR, supporting the effectiveness of the treatment (Figure 9a). ISS-ODN-mediated IFN-γ increase not only restored epithelial Ido1 and IL-18bp expression in Clec9A-DTR mice (Figure 9b), but also clearly reverted the severe inflammatory intestinal response caused by the absence of CD103+CD11b− DCs. In fact, Clec9A-DTR mice did not show the typical severe weight loss observed upon DSS (2%) treatment in non-ISS-ODN-injected animals and behaved similarly to WT control mice (Figure 9c).
Figure 9.
Immunostimulatory oligonucleotide (ISS-ODN) treatment limits the colitis severity in Clec9A–diphtheria toxin receptor (DTR) mice. DT-injected wild-type (WT) and Clec9A-DTR mice were injected intraperitoneally (i.p.) 10 μg of ISS-ODN at the start of the dextran sodium sulfate (DSS) treatment (2%) and 4 days later. (a) Interferon-γ (IFN-γ) response was measured in the serum collected at day 4. (b) Epithelial Ido1 and IL-18bp expression profile at steady-state or under DSS treatment. Representative samples of three WT and Clec9A-DTR mice are shown. (c) The body weight was monitored daily over a period of 10 days. Black circles: DT-treated WT control; white circles: DT-treated WT control+ISS-ODN; black squares: DT-treated Clec9A-DTR; white squares: DT-treated Clec9A-DTR+ISS-ODN; Each group: n=6 mice from two independent experiments. Values represent the mean±s.d. ND, not detectable.
DISCUSSION
Functional mucosal barrier is lost in IBDs because of an uncontrolled inflammatory cascade arising from a number of interdependent and possibly sequential events involving both nonimmune cells, such as IECs, and immune cells, including DCs, ILCs, innate γ/δ T cells, and regulatory T cells.31 Despite extensive literature on this field, we still have gaps in our understanding of which mediator(s) play essential roles in disease onset. As DCs have been implicated as one of the major contributors in DSS-mediated colonic inflammation,32 we dissected in this study the contribution of distinct intestinal DC subsets in controlling intestinal inflammation and showed that CD103+CD11b− DC subset has a unique role in protecting the mucosal barrier.
In the gut LP, a complex network of myeloid cells, including several distinct subsets of DCs and macrophages,33, 34 regulates the intestinal homeostasis.35 Intestinal myeloid cells clearly differ, not only by their phenotype and by distinct developmental requirements, but also in their in vivo function. Based on our gene array analysis, we are confident to cluster colonic CD103+CD11b− and CD103+CD11b+ cells as bona fide DCs and CD103−CD11b+ cells as macrophage lineage because of their specific DC- or macrophage-related expression profiles, confirming the observations of others.36, 37, 38 Recently, when analyzed more in detail, CD103−CD11b+ cells could be further separated clearly into a DC and a macrophage fraction based on phenotype, gene profile, and kinetics.39, 40 Both bona fide colon DCs substantially differ not only in their developmental programming, but possibly also in their biological functions as they express different patterns of, e.g., cytokines, chemokines, and pathogen recognition receptors.
Therefore, the comparison between two Clec9A- and Clec4a4-DTR mouse strains provided us valuable tools to further investigate the contribution of each DC subpopulation in gut homeostasis.
The differential expression of Clec9A and Clec4a4 lectins in DC subpopulations allowed us to visualize their anatomical location in the large intestine. Interestingly, at steady state, DC subsets are colocalized in isolated ILFs, whereas in the LP only abundant numbers of CD103+CD11b− DCs are present. On the other hand, the CD103+CD11b+ DC cells were barely detectable in the LP. Therefore, our data suggest that the recently described motile CD103+CX3CR1− DC subset in the small intestine that occupies the gut epithelium, samples bacteria from the intestinal lumen, and emigrates to the MLN via CCR7 upregulation41 is the Clec9A-expressing CD103+CD11b− DC subset. In addition, CD11cintMHCII+CX3CR1+ macrophages situated close to the intestinal epithelium represent a clear, phenotypically distinct subpopulation that outnumber the CD103+CD11b− DCs by 3–4-fold.14, 42, 43
To investigate the contribution of each DC subset in intestinal physiology and disease, we induced mild DSS colitis in WT, Clec9A-DTR, and Clec4a4-DTR mice. Surprisingly, although Clec4a4-DTR mice did not show any colitis symptoms, Clec9A-DTR mice showed severe clinical symptoms marked by diarrhea, bloody stools, and consistent weight loss upon mild DSS treatment. This underlines for the first time the crucial role of CD103+CD11b− DCs in controlling intestinal inflammation. However, this finding contradicts with the phenotype described for Batf3−/− mice, lacking CD103+CD11b− DCs,9 that did not show any exacerbated inflammation during DSS-mediated colitis. Possible reasons for this could be mouse genetic background (129SvEv vs. BALB/c), gene knockout vs. DT-induced cell lineage ablation, or even different commensal gut microbiota. Moreover, a potential cytokine-driven alternative pathway in CD8+/CD103+ DC development observed in Batf3−/− mice in response to infections cannot be excluded in response to DSS challenge.44 Interestingly, so far, only gut macrophages35, 45, 46 and IL-23-secreting CD103+CD11b+ DCs47 have been considered to be key players in the maintenance of gut defense and homeostasis. In particular, IL-10-conditioned intestinal CX3CR1+ macrophages were shown to contribute in maintenance of intestinal integrity,48, 49 as CX3CR1-deficient mice with reduced numbers of CX3CR1+ macrophages resulted in severe colitis, bacterial translocation, and colitogenic Th17 responses.15 Our results underline the fundamental contribution of CD103+CD11b− DCs as additional members in arsenal in regulating intestinal homeostasis and protecting the gut mucosae.
Intestinal homeostasis involves IECs that provide physical segregation of commensal bacteria as well as integration of the microbial signals.31 Expression of pathogen recognition receptors, including members of the Toll-like receptor and NOD-like receptor family, allows them to sense commensal and pathogen-derived signals promoting epithelial homeostasis and repair as well as immune regulatory mucosal responses. As a result, antimicrobial peptides and mucus secretion reinforce the biochemical barrier.50, 51 The physical and biochemical barrier provided by IECs is not only modulated by the commensal microbial community, but also by underlying mucosal immune cells. Myeloid cells, including macrophages and DCs, ILCs, and T effector cells, often via their secreted cytokines, regulate through complex cellular networks, intestinal epithelia host defenses, and barrier functions. For example, IL-23 produced by DCs regulates IL-22 secretion, a cytokine that mediates epithelial cell proliferation and wound healing,52 and controls epithelial antimicrobial peptide responses, important for the retention of both commensal and pathogens in the outer mucus layer.53 Other cytokines, such as IFN-γ , IL-17, and IL-10, can contribute to the pathogenesis of IBD by suppressing or aggravating intestinal inflammation and its associated clinical symptoms. As often the case for other pleiotropic cytokines, IFN-γ may also have multifaceted functions in controlling mucosal inflammation. On one hand, it can exert proinflammatory functions by exacerbating mucosal inflammation.54 On the other hand, in particular, at early stages of inflammation, IFN-γ may also have important homeostatic functions, for example, by stimulating Paneth cells to release antimicrobial peptides,55 by modulating anti-inflammatory molecules like IDO1,30, 56 decoy proteins such as IL-18bp,24, 26 as well as by controlling goblet cell function.57 Furthermore, IFN-γ-induced epithelial MHC class II expression has been shown to be protective against colitis.58 The protective role of IFN-γ during DSS-mediated intestinal inflammation was also confirmed in IFN-γ-deficient mice that, in our hands, were highly susceptible to the chemical treatment. In addition, as observed in Clec9A-DTR mice, IFN-γ-deficient mice do not upregulate epithelial IDO1 and IL-18bp in response to DSS. Strikingly, our results are in complete disagreement with the colitis-resistant phenotype observed by Ito et al.;54 however their results were obtained using a different IFN-γ-deficient mouse strain (Tagawa et al.59 vs. Dalton et al.60). In addition, they used another molecular weight of DSS (5 vs. 40 kDa) to induce chemical-induced epithelial injury.
Here, we have identified a novel mechanism of how a specific DC subset controls intestinal inflammation through the modulation of a series of IFN-γ-inducible genes in IECs, including IFN-γ-regulated factors (IRFs), MHC class II molecules, and related molecules such as invariant chain (CD74). Of particular relevance is the regulation of immunosuppressive molecules such as IDO1 and IL-18bp that are typically upregulated during intestinal inflammation. In fact, in the absence of CD103+CD11b− DCs, the level of IDO1, the rate-limiting enzyme of tryptophan catabolism, in IECs plummets that is often associated with IBDs (e.g., ulcerative colitis and Crohn's disease)22, 61 including intestinal malignancies.62, 63 Its expression levels have even been correlated with the severity of gastrointestinal diseases.30, 56, 64 Immunosuppressive effects of IDO1 are linked with decreased local concentrations of tryptophan and enhanced levels of its metabolites such as kynunerines that induce direct cell growth arrest and T-cell unresponsiveness.65, 66 At steady-state conditions, we and others have shown that gut DCs, in particular the CD103+CD11b− subset, are the major IDO1-expressing cells in the colon, maintaining intestinal homeostasis via the balance between FoxP3+ regulatory T cells and T helper type T helper 1/17 effector cells.64, 67 However, during intestinal inflammation, IECs, but not DCs, represent the major source of IDO1 activity possibly with the function to counterbalance tissue-damaging responses and to protect the epithelium from invading microbes. In effect, immunohistological analyses confirmed a specific IDO1 expression pattern in inflamed mucosa, in particular, where epithelia cells flank ulcers and border crypt abscesses.68
Furthermore, we could also observe that in the colitis-prone Clec9A-DTR mice another important decoy protein was reduced in IECs, namely the IL-18bp.69 Although the function of IL-18 in gut inflammation is controversial,70 it is clear that blocking its activity attenuates intestinal damage.24, 71 The imbalance of the IL-18/ IL-18bp ratio could lead to higher levels of active IL-18 resulting in exacerbation of intestinal inflammation that we observe in Clec9A-DTR mice. Interestingly, the dysregulation of microRNA-controlled insulin-like growth factor binding protein 5 (IGFBP5) in LP myofibroblasts lead to increased severity of DSS-induced colitis because of decreased availability of IGF that is needed for colonic epithelial repair.72 Therefore, cytokine-binding proteins seem to emerge as major regulators of epithelial integrity.
Through its capacity to secrete IL-12 and IL-15, the CD103+CD11b− subset clearly affects IFN-γ production of LP CD4+ and CD8+ T cells as well as of intraepithelial CD8+ T cells. Importantly, when higher IFN-γ levels are restored by immunostimulatory DNA, the colitis-prone phenotype of Clec9A-DTR mice can be reverted.
In summary, here we propose CD103+CD11b− DCs as the major regulators of intestinal homeostasis by controlling IFN-γ-induced anti-inflammatory proteins in IECs such as IDO1 and IL-18bp. Our results highlight CD103+CD11b− DCs in addition to the anti-inflammatory CX3CR1hi macrophages as the key myeloid cells safeguarding the normal gut homeostasis.
METHODS
Mouse strains. Clec9A-DTR mice were recently generated in our laboratory as described in Piva et al.73 using a BAC (bacterial artificial chromosome) recombineering approach. The Clec4a4-DTR mouse strain was obtained by gene targeting. In short, the IRES-DTR cassette followed by removable selection marker (PGK-NeoR) was inserted after the stop codon in the 3′ untranslated region of Clec4a4 gene. After electroporation of the targeting construct, several BALB/c ES colonies carrying desired DTR insertion within 3′ untranslated region of Clec4a4 were established. Selected ES clones were subsequently used for blastocyst microinjection, leading to generation of chimeric animals and ultimately germline transmission of the modified allele. As controls, aged-matched transgenic negative littermates were used, as indicated. All original transgenic mouse strains are of BALB/c background (Supplementary Figure S4).
CX3CR1-GFP transgenic mice were originally generated by Dr D Littman (New York, NY) and were kindly provided by Florent Ginhoux (SIgN, A*Star, Singapore, Singapore). IFN-γ −/− mice (strain B6.129S7-Ifngtm1Ts/J) were obtained from The Jackson Laboratory (Bar Harbor, ME).
All transgenic mice were bred and housed under specific pathogen-free conditions in the Nanyang Technological University animal facility. This study was carried out in strict accordance with the recommendations of the NACLAR (National Advisory Committee for Laboratory Animal Research) guidelines under the Animal & Birds (Care and Use of Animals for Scientific Purposes) Rules of Singapore. The protocol ARF SBS/NIE 0158AZ was approved by the institutional animal care and use committee of the Nanyang Technological University of Singapore.
Antibodies and flow cytometry. Fluorochrome-labeled anti-CD45, anti-CD3, anti-CD4, anti-CD8, anti-γ/δ T cell receptor, anti-CD11c, anti-CD103, anti-MHC class II, anti-Ly6C, anti-Ly6G, anti-Clec4a4 (also called 33D1 antigen or DCIR2,12 and anti-Clec9A (also known as DNGR-1)11, 74 were purchased from BioLegend (San Diego, CA). Stained cells were analyzed on a LSRII or Fortessa FACS (BD Biosciences, San Jose, CA) and data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Depletion and isolation of intestinal DC subsets and T cells in colon and MLNs. WT, Clec9A-DTR, and Clec4a4-DTR mice were injected intraperitoneally with 20 ng g−1 DT at day −1. At day 0, colon and MLNs were collected. The colon was flushed with cold phosphate-buffered saline (PBS), and cut longitudinally in small pieces that were subsequently washed in Hanks' balanced salt solution containing 1 mM DTT at 37 °C under shaking for 20 min. To remove the epithelium, colon pieces were incubated in Hank's balanced salt solution containing 1.3 mM EDTA under shaking conditions at 37 °C for 1 h. To enrich intraepithelial lymphocytes (IELs), a 67/44% Percoll (GE Healthcare Life Sciences, Singapore) gradient was performed. To isolate LP DCs, the remaining colon pieces were digested in serum-free Iscove's modified Dulbecco's medium and 0.1 mg ml−1 Collagenase D (Roche Applied Science, Basel, Switzerland) at 37 °C for another additional 1 h and 30 min. Digested pieces were gently passed five times through a cell strainer. The leukocyte population containing T cells and DCs was enriched by a 70/40% Percoll gradient. Low-density cells at the interface were harvested and further processed for stainings. MLNs were processed for DC isolation as described in Ruedl et al.75 via collagenase digestion.
Experimental acute colitis model: DSS treatment. Female WT (negative littermates), Clec9A-DTR, and Clec4a4-DTR mice were injected at day −1 with 20 ng g−1 DT and during the DSS treatment every 3 days. 2% DSS (50,000 Da, MP Biomedical, Santa Ana, CA) was supplied at day 0 ad libidum in the drinking water for 7 consecutive days with fresh DSS supplied every 3 days. At day 8, the DSS was replaced with drinking water. Body weight was monitored daily and fecal samples were collected between days 5 and 8. There were 6 mice per group in two independent experiments, for a total of 12 total mice per group.
Because of the direct toxic effect of DSS on epithelial cells that leads to a complete loss of surface epithelium, we induced a rather moderate colitis by administration of a low concentration of 2% DSS. At this dosage we did not observe a loss of surface epithelium in control mice at day 4 of DSS treatment, thus allowing the assessment of the barrier function of an intact epithelial layer.
Measurement of fecal blood. Fecal blood content was measured in fecal pellets collected at day 8 using the Hemoccult SENSA (Beckman Coulter, Brea, CA) following the manufacturer's instructions.
Analysis of inflammatory cell infiltrations during acute colitis. Colon LP cells of WT, Clec9A-DTR, and Clec4a4-CX3CR1GFP DTR mice were isolated as described before, stained with PerCP-Cy5.5-labeled anti-CD11b, PE-labeled anti-Ly6C, and APC-labeled Ly6G antibodies, and analyzed by flow cytometry.
Intracellular IFN-γ cytokine staining. Isolated colon LP cells and IELs were stimulated with phorbol 12-myristate 13-acetate/ionomycin (2 h) and with Brefeldin A for additional 2 h and stained with anti-CD45, -CD3, -CD4, -CD8, and -γ/δ T cell receptor-specific antibodies. Subsequently, cells were permeabilized and fixed using the FOXP3 Staining Buffer Set according to the manufacturer's instructions (eBioscience, San Diego, CA) and intracellularly stained with PE-labeled anti-IFN-γ. Stained cells were analyzed by flow cytometry gating of CD45- and CD3-expressing cells. Representative dot plots showing CD4/CD8 staining profile for LP cells and CD8/γ/δ staining profile for IELs are shown.
FITC–dextran intestinal permeability assay. Intestinal permeability was assessed by oral administration of FITC–dextran (4 kDa, Sigma-Aldrich, St Louis, MO), a macromolecule that is used as a permeability probe. All mice were gavaged at days 0, 4, and 10 after the start of DSS treatment with 60 mg per 100 g body weight FITC–dextran 4 h before blood collection. Whole blood was obtained by cardiac puncture at the time of killing, and FITC–dextran translocation was measured in sera by fluorometry. Dilutions of FITC–dextran in serum/PBS were used as a standard curve, and absorption was measured in a fluorometer at 488/535 nm emission.
Preparation of colonic DCs and microarray analysis. Colon MHC+CD11chigh cells were sorted on the basis of CD103 and CD11b expression as follows: CD103+CD11b−, CD103+CD11b+, and CD103−CD11b+. Three independent colon isolations for each group (eight mice pooled/group) were performed. Total RNA was extracted using the Arcturus Picopure RNA Isolation Kit (Applied Biosystems, Life Technologies, Singapore). RNA integrity was assessed by Agilent Bioanalyzer and the RNA Integrity Number was calculated. All RNA samples had the RNA Integrity Number of ≥9.4. Target single-stranded DNA was prepared starting from 5 ng of total RNA using the WT Ovation Pico RNA Amplification and the Encore Biotin Module (NuGEN, San Carlos, CA). SPIA (Single Primer Isothermal Amplification) complementary DNA (cDNA) was prepared, fragmented, and labeled according to the manufacturer's instructions. Fragmented target single-stranded DNA at 5 μg was hybridized to the Affymetrix GeneChip Mouse Gene 1.0 ST Arrays for 17 h at 45 °C. The arrays were then washed and stained using the Affymetrix Fluidics Station 450 and scanned using GeneChip Scanner 3000. Array images were analyzed using Expression Console and comparison analyses were carried out according to the instructions provided by Affymetrix. Arrays were normalized with RMA/quantile normalization, and differential gene analysis determined with limma. These analyses (and the principal component analysis plot) were performed in R 2.15.2/bioconductor, through Pipeline Pilot. Heat maps were generated in TM4 (Boston, MA).
Isolation of colonic IECs, RNA preparation, and microarray analysis. IECs were isolated from the distal part of the colon by shaking colon pieces in PBS containing 1.3 mM EDTA at 37 °C for 1 h. Cell pellets were then lysed in Trizol (Life Technologies) and total RNA were extracted following the manufacturer's instructions. Isolated RNA was then reconstituted in RNase-free water, digested with DNase I to remove contaminating genomic DNA, and subsequently purified using chloroform–isopropanol precipitation. Precipitated RNA was washed with 75% ethanol and reconstituted in RNase-free water.
For microarray analysis, total RNA was labeled with Low Input Quick Amp Labeling Kit, One-Color (Agilent p/n 5190-2305) following the manufacturer's instructions (One-Color Microarray-Based Gene Expression, Analysis Low Input Quick Amp Labeling, version 6.5). Briefly, 100 ng of total RNA was converted into double-stranded cDNA by priming with an oligo-dT primer containing the recognition site for T7 RNA polymerase. In vitro transcription with T7 RNA polymerase was used to produce cyanine 3-CTP-labeled cRNA. Then, 600 ng of labeled cRNA was hybridized onto Agilent SurePrint G3 Human GE 8 × 60K Microarray for 17 h at 65 °C, 10 r.p.m. in Agilent hybridization oven. After hybridization, the microarray slide was washed in gene expression wash buffer 1 for 1 min at room temperature and another 1 min in gene expression wash buffer 2 at 37 °C before scanning on Agilent High Resolution Microarray Scanner (C-model). Raw signal data were extracted from the TIFF image with Agilent Feature Extraction Software (V10.7.1.1).
Semiquantitative and quantitative real-time PCR. cDNA was generated using Superscript reverse transcription (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Real-time PCR was performed using Tprofessional thermocycler TRIO (Biometra, Goettingen, Germany). Primer sequences were as follows: Ido1; forward (Fwd): 5′-CTGTATGAGGGGGTCTGGGA-3′ and reverse (Rev): 5′-GACTGGGGGAGCTGACTCTA-3′ (207 bp). IL-18bp; Fwd: 5′-GAGCACAGGAACACAAGCAC-3′ and Rev: 5′-GGGCCTGCTGATCTGGATAC-3′ (221 bp). IFN-γ Receptor; Fwd: 5′-GTGGTTGCTCCTCTTACCGTC-3′ and Rev: 5′-GCCTCCAGAGCCTTTGTTTCT-3′ (302 bp). Hprt; Fwd: 5′-GCTGGTGAAAAGGACCTCT-3′ and Rev: 5′-CACAGGACTAGAACACCTGC-3′ (249 bp).
Quantitative real-time PCR was performed on cDNA using SYBR green chemistry (KabaBiosystem, Woburn, WA). Reactions were run on a real-time quantitative PCR system (Illumina, San Diego, CA). Samples were normalized to β-actin, represent the median of triplicate analyses, and are displayed as a fold change from naive WT controls unless otherwise stated. Primer sequences were as follows: Ido1; Fwd: 5′-GTGGAAATCGCAGCTTCTCC-3′ and Rev: 5′-ACTGGTAGCTATGTCGTGCA-3′. IL-18bp; Fwd: 5′-CTGGAAGAACTGAGCCCCAC-3′ and Rev: 5′-CGGAGCTGTCTTCAACCCAT-3′. β-Actin; Fwd: 5′-AAGGCCAACCGTGAAAAGAT-3′ and Rev: 5′-CCTGTGGTACGACCAGAGGCATACA-3′.
Colon epithelial cell line. Murine colon epithelial cell line CMT-93 was obtained from ATCC (Manassas, VA) and grown as monolayer at 37 °C in complete Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Trypsin-treated cells were plated in 12-well plates and treated with 100 U ml−1 recombinant IFN-γ for 24 h. Total RNA was extracted from CMT-93 cells and transcribed to cDNA as described above.
Immunofluorescence microscopy. The distal part of the colon was opened longitudinally, washed in cold PBS, embedded, snapped frozen in Tissue-Tek OCT medium (Sakura, Finetek, Alphen aan den Rijn, The Netherlands, and processed for 8 μm thick cryosections. After fixation in acetone, sections were rehydrated in PBS containing 2% fetal calf serum and stained overnight at 4 °C using different monoclonal antibodies as indicated in the correspondent legends. Stained sections were then washed in PBS containing 2% fetal calf serum and mounted in DAPI (4′,6-diamidino-2-phenylindole)-containing antifade mountant (VECTASHIELD, Vector Laboratories, Burlingame, CA). In the case of colons obtained from CX3CR1 transgenic mice, mice were perfused with 2% paraformaldehyde with 30% sucrose in PBS before colon dissection. All images were obtained with identical settings at × 20 objective magnification as indicated in the legend using confocal microscope (Zeiss LSM510 META inverted, Goettingen, Germany).
Western blotting. IECs isolated from 3 DSS-treated WT or Clec9A-DTR mice were pooled and lysed on ice in a lysis buffer consisting of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitors. After centrifugation at 10,000 g for 15 min at 4 °C, supernatants were collected. Protein concentrations were determined using NanoDrop Spectrophotometer (Wilmington, DE). Normalized samples were run on 10% Tris-glycine SDS-polyacrylamide gels using the Mini-Sub Cell GT system (Bio-Rad, Hercules, CA) and transferred onto nitrocellulose membranes (Bio-Rad). The membranes were subsequently blocked in PBS supplemented with 0.05% (v/v) Tween-20 (Sigma-Aldrich Pte. Ltd., Singapore) and 3% (w/v) nonfat milk (Bio-Rad) overnight at 4 °C and then incubated for 1 h with the primary antibody rat anti-mouse IDO1 (BioLegend) or polyclonal β-tubulin (Santa Cruz Biotechnology, Dallas, TX) antibody, respectively. The membranes were rinsed with PBS/Tween-20 and incubated with the corresponding HPRT-labeled secondary antibodies. The presence of Ido1 (45 kDa) and tubulin (50 kDa) was confirmed by the enhanced chemiluminescence detection system (SignalFire, ECL reagent, Cell Signaling Technology, Danvers, MA).
Treatment with immunostimulatory DNA (ISS-ODN). Animals were treated with ISS-ODN (5′-TGACTGTGAACGTTCGAGATGA-3′) as described in Ciorba et al.30 Briefly, WT and Clec9A-DTR mice were injected with DT at day −1 and day 4 and treated with 2% DSS at day 0. ISS-ODN (10 μg) was injected intraperitoneally at day 0 and day 4. To confirm the efficacy of the ISS-ODN treatment, IFN-γ levels were measured in sera of treated animals through conventional enzyme-linked immunosorbent assay at day 4.
Statistical analysis. Statistical analysis was performed using GraphPad Prism software (La Jolla, CA). All values are expressed as the average±s.d. or s.e.m. as indicated in the legend. All experiments were repeated as at least two to three independent experiments. Samples were analyzed using Student's t-test (two tailed). A P-value of <0.05 was considered to be significant.
The microarray data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE58446.
Acknowledgments
We thank Monika Tetlak for the excellent mouse management and Shi Hui Foo Ivy for microarray sample preparation. This work is devoted to Erich Ruedl. This work was supported by National Medical Research Council grants NMMR/1253/2010, NMRC/CBRG/0023/2012, and MOE2014-T2-1-011 to C.R.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
Author contributions
A.R.B.M.M. and P.T. performed the experiments and interpreted the data; J.S., S.C.L., and Y.A.S. contributed to specific experiments; M.P. performed bioinformatics analysis; F.Z. analyzed and discussed the microarray data; K.K. and C.R. designed the experiments, interpreted the data, and wrote the manuscript.
Supplementary Material
References
- Brown, E.M., Sadarangani, M. & Finlay, B.B. The role of the immune system in governing host-microbe interactions in the intestine. Nat. Immunol. 14, 660–667 (2013). [DOI] [PubMed] [Google Scholar]
- Macdonald, T.T. & Monteleone, G. Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925 (2005). [DOI] [PubMed] [Google Scholar]
- Ponda, P.P. & Mayer, L. Mucosal epithelium in health and disease. Curr. Mol. Med. 5, 549–556 (2005). [DOI] [PubMed] [Google Scholar]
- Schmitz, H. et al. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 116, 301–309 (1999). [DOI] [PubMed] [Google Scholar]
- Peeters, M. et al. Clustering of increased small intestinal permeability in families with Crohn's disease. Gastroenterology 113, 802–807 (1997). [DOI] [PubMed] [Google Scholar]
- Hashimoto, D., Miller, J. & Merad, M. Dendritic cell and macrophage heterogeneity in vivo. Immunity 35, 323–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic, M., Mortha, A., Muller, P.A. & Merad, M. Mononuclear phagocyte diversity in the intestine. Immunol. Res. 54, 37–49 (2012). [DOI] [PubMed] [Google Scholar]
- Bogunovic, M. et al. Origin of the lamina propria dendritic cell network. Immunity 31, 513–525 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson, B.T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F. et al. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho, D. et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 458, 899–903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak, D. et al. Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007). [DOI] [PubMed] [Google Scholar]
- Persson, E.K. et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Contreras, O. et al. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J. Clin. Invest. 121, 4787–4795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, B., Saurer, L., Schenk, M., Dickgreber, N. & Mueller, C. CX3CR1 defines functionally distinct intestinal mononuclear phagocyte subsets which maintain their respective functions during homeostatic and inflammatory conditions. Eur. J. Immunol. 41, 773–779 (2011). [DOI] [PubMed] [Google Scholar]
- Cerovic, V., Bain, C.C., Mowat, A.M. & Milling, S.W. Intestinal macrophages and dendritic cells: what's the difference? Trends Immunol. 35, 270–277 (2014). [DOI] [PubMed] [Google Scholar]
- Perse, M. & Cerar, A. Dextran sodium sulphate colitis mouse model: traps and tricks. J. Biomed. Biotechnol. 2012, 718617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml, B.U. et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell 154, 843–858 (2013). [DOI] [PubMed] [Google Scholar]
- Zigmond, E. et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090 (2012). [DOI] [PubMed] [Google Scholar]
- Purnama, C. et al. Transient ablation of alveolar macrophages leads to massive pathology of influenza infection without affecting cellular adaptive immunity. Eur. J. Immunol. 44, 2003–2012 (2014). [DOI] [PubMed] [Google Scholar]
- Gurtner, G.J., Newberry, R.D., Schloemann, S.R., McDonald, K.G. & Stenson, W.F. Inhibition of indoleamine 2,3-dioxygenase augments trinitrobenzene sulfonic acid colitis in mice. Gastroenterology 125, 1762–1773 (2003). [DOI] [PubMed] [Google Scholar]
- Schuhmann, D. et al. Interfering with interferon-gamma signalling in intestinal epithelial cells: selective inhibition of apoptosis-maintained secretion of anti-inflammatory interleukin-18 binding protein. Clin. Exp. Immunol. 163, 65–76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar, P.V. et al. Interleukin 18 is a primary mediator of the inflammation associated with dextran sulphate sodium induced colitis: blocking interleukin 18 attenuates intestinal damage. Gut 50, 812–820 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S.V. & Schultze, J.L. New insights into IDO biology in bacterial and viral infections. Front. Immunol. 5, 384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgin, V., Novick, D. & Rubinstein, M. The promoter of IL-18 binding protein: activation by an IFN-gamma -induced complex of IFN regulatory factor 1 and CCAAT/enhancer binding protein beta. Proc. Natl. Acad. Sci. USA 99, 16957–16962 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, E.C. Interleukin 15 is a potent stimulant of intraepithelial lymphocytes. Gastroenterology 115, 1439–1445 (1998). [DOI] [PubMed] [Google Scholar]
- Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 38, 769–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz, D. et al. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 122, 1428–1441 (2002). [DOI] [PubMed] [Google Scholar]
- Ciorba, M.A. et al. Induction of IDO-1 by immunostimulatory DNA limits severity of experimental colitis. J. Immunol. 184, 3907–3916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, L.W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- Berndt, B.E., Zhang, M., Chen, G.H., Huffnagle, G.B. & Kao, J.Y. The role of dendritic cells in the development of acute dextran sulfate sodium colitis. J. Immunol. 179, 6255–6262 (2007). [DOI] [PubMed] [Google Scholar]
- Persson, E.K., Scott, C.L., Mowat, A.M. & Agace, W.W. Dendritic cell subsets in the intestinal lamina propria: ontogeny and function. Eur. J. Immunol. 43, 3098–3107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner, A. & Jung, S. Development and function of dendritic cell subsets. Immunity 40, 642–656 (2014). [DOI] [PubMed] [Google Scholar]
- Farache, J., Zigmond, E., Shakhar, G. & Jung, S. Contributions of dendritic cells and macrophages to intestinal homeostasis and immune defense. Immunol. Cell. Biol. 91, 232–239 (2013). [DOI] [PubMed] [Google Scholar]
- Miller, J.C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watchmaker, P.B. et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 15, 98–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, C.C. et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6, 498–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, C.L. et al. CCR2(+)CD103(-) intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 8, 327–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farache, J. et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess, J.H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005). [DOI] [PubMed] [Google Scholar]
- Chieppa, M., Rescigno, M., Huang, A.Y. & Germain, R.N. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med. 203, 2841–2852 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, E. & Jung, S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 34, 162–168 (2013). [DOI] [PubMed] [Google Scholar]
- Mantovani, A. & Marchesi, F. IL-10 and macrophages orchestrate gut homeostasis. Immunity 40, 637–639 (2014). [DOI] [PubMed] [Google Scholar]
- Kinnebrew, M.A. et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval, D.S. et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014). [DOI] [PubMed] [Google Scholar]
- McGuckin, M.A., Linden, S.K., Sutton, P. & Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278 (2011). [DOI] [PubMed] [Google Scholar]
- Bevins, C.L. & Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011). [DOI] [PubMed] [Google Scholar]
- Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava, S. et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, R. et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 146, 330–338 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin, H.F. et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-gamma. J. Exp. Med. 211, 1393–1405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu, M. et al. IDO1 plays an immunosuppressive role in 2,4,6-trinitrobenzene sulfate-induced colitis in mice. J. Immunol. 191, 3057–3064 (2013). [DOI] [PubMed] [Google Scholar]
- Songhet, P. et al. Stromal IFN-gammaR-signaling modulates goblet cell function during Salmonella Typhimurium infection. PLoS One 6, e22459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelemann, C. et al. Interferon-gamma induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One 9, e86844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa, Y., Sekikawa, K. & Iwakura, Y. Suppression of concanavalin A-induced hepatitis in IFN-gamma(-/-) mice, but not in TNF-alpha(-/-) mice: role for IFN-gamma in activating apoptosis of hepatocytes. J. Immunol. 159, 1418–1428 (1997). [PubMed] [Google Scholar]
- Dalton, D.K., Pitts-Meek, S., Keshav, S., Figari, I.S., Bradley, A. & Stewart, T.A. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259, 1739–1742 (1993). [DOI] [PubMed] [Google Scholar]
- Wolf, A.M. et al. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin. Immunol. 113, 47–55 (2004). [DOI] [PubMed] [Google Scholar]
- Cherayil, B.J. Indoleamine 2,3-dioxygenase in intestinal immunity and inflammation. Inflamm. Bowel Dis. 15, 1391–1396 (2009). [DOI] [PubMed] [Google Scholar]
- Ciorba, M.A. Indoleamine 2,3 dioxygenase in intestinal disease. Curr. Opin. Gastroenterol. 29, 146–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoli, G. et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 59, 595–604 (2010). [DOI] [PubMed] [Google Scholar]
- Terness, P. et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J. Exp. Med. 196, 447–457 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumento, G., Rotondo, R., Tonetti, M., Damonte, G., Benatti, U. & Ferrara, G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 196, 459–468 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. & Zhu, B.T. Indoleamine 2,3-dioxygenase tissue distribution and cellular localization in mice: implications for its biological functions. J. Histochem. Cytochem. 58, 17–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinande, L. et al. Inflamed intestinal mucosa features a specific epithelial expression pattern of indoleamine 2,3-dioxygenase. Int. J. Immunopathol. Pharmacol. 21, 289–295 (2008). [DOI] [PubMed] [Google Scholar]
- Dinarello, C.A., Novick, D., Kim, S. & Kaplanski, G. Interleukin-18 and IL-18 binding protein. Front. Immunol. 4, 289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund, B. Interleukin-18 in intestinal inflammation: friend and foe? Immunity 32, 300–302 (2010). [DOI] [PubMed] [Google Scholar]
- Siegmund, B. et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-gamma and TNF-alpha production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1264–R1273 (2001). [DOI] [PubMed] [Google Scholar]
- Chivukula, R.R. et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 157, 1104–1116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva, L., Tetlak, P., Claser, C., Karjalainen, K., Renia, L. & Ruedl, C. Cutting edge: Clec9A+ dendritic cells mediate the development of experimental cerebral malaria. J. Immunol. 189, 1128–1132 (2012). [DOI] [PubMed] [Google Scholar]
- Caminschi, I. et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112, 3264–3273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedl, C., Kopf, M. & Bachmann, M.F. CD8(+) T cells mediate CD40-independent maturation of dendritic cells in vivo. J. Exp. Med. 189, 1875–1884 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.