Abstract
Phosphoinositides (PIs) form a minor class of phospholipids with crucial functions in cell physiology, ranging from cell signalling and motility to a role as signposts of compartmental membrane identity. Phosphatidylinositol 3‐phosphates are present at the plasma membrane and within the endolysosomal system, where they serve as key regulators of both cell signalling and of intracellular membrane traffic. Here, we provide an overview of the metabolic pathways that regulate cellular synthesis of PI 3‐phosphates at distinct intracellular sites and discuss the mechanisms by which these lipids regulate cell signalling and membrane traffic. Finally, we provide a framework for how PI 3‐phosphate metabolism is integrated into the cellular network.
Keywords: autophagy, endolysosomal system, membrane traffic, phosphatidylinositol 3‐phosphate, signalling
Subject Categories: Membrane & Intracellular Transport, Metabolism, Signal Transduction
Introduction
Phosphoinositides (PIs) are a minor class of comparably short‐lived membrane phospholipids that mediate crucial cellular and organismal functions including signalling, gating of ion channels, cytoskeleton regulation and motility, development, as well as the regulation of intracellular membrane traffic (Di Paolo & De Camilli, 2006; Balla, 2013). PI 4‐phosphates such as phosphatidylinositol 4‐phosphate [PI(4)P] and phosphatidylinositol 4,5‐bisphosphate [PI(4,5)P2] are concentrated along both the exocytic pathway and the plasma membrane. PI 3‐phosphates can be produced at the cell surface during signalling, but they are mainly found as prominent components and hallmarks of the endosomal system (Balla, 2013; Raiborg et al, 2013).
Phosphatidylinositol 3‐phosphates are generated via the phosphorylation of the inositol ring of phosphatidylinositol and its derivatives at the 3‐position, giving rise to phosphatidylinositol 3‐phosphate [PI(3)P], phosphatidylinositol 3,4‐bisphosphate [PI(3,4)P2], phosphatidylinositol 3,5‐bisphosphate [PI(3,5)P2], and phosphatidylinositol 3,4,5‐trisphosphate [PI(3,4,5)P3]. The distinctive subcellular localization displayed by the different phosphatidylinositol 3‐phosphate species, as well as the rapidly reversible nature of phosphorylation, positions them as key regulators of processes at intracellular membranes, from signalling cascades downstream of extracellular receptor activation to intracellular membrane trafficking (Balla, 2013; Raiborg et al, 2013) (Fig 1). At the plasma membrane, PI(3,4,5)P3 and PI(3,4)P2 are generated upon extracellular stimulation (Cantley, 2002; Vanhaesebroeck et al, 2010a), and PI(3,4)P2 synthesis was recently described as an essential mediator of the late stages of clathrin‐mediated endocytosis (CME) (Posor et al, 2013). Additionally, there is a pool of PI(3,4)P2 on endomembranes, the function of which has yet to be established (Watt et al, 2004). PI(3)P is the predominant species found on early endosomes and plays an important role in autophagy, while PI(3,5)P2 is present on late endocytic/multivesicular bodies, lysosomal and autophagic compartments (Mayinger, 2012). The generation of these distinctive pools of PI 3‐phosphates (Fig 1) occurs via three classes of PI 3‐kinases (PI3Ks) that exhibit different substrate specificities as well as subcellular localizations, and via a single PI 5‐kinase, PIKfyve, that catalyses PI(3,5)P2 synthesis from PI(3)P (Balla, 2013; Raiborg et al, 2013). Conversely, turnover of PI 3‐phosphates is accomplished by several types of PI phosphatases, most notably the myotubularin family of PI 3‐phosphatases (Hsu & Mao, 2015).
Figure 1. Localization of 3‐phosphoinositides in the cell, and their relevant kinases and phosphatases.
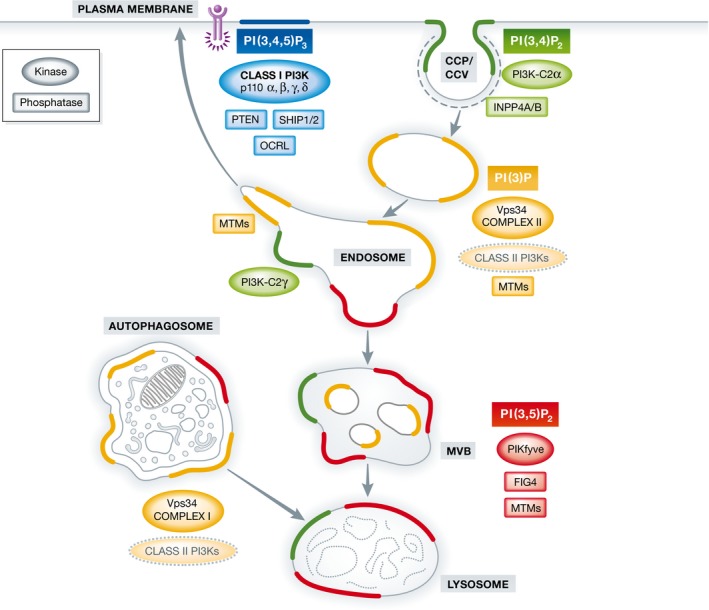
At the plasma membrane, receptor tyrosine kinase (purple) activation results in the production of PI(3,4,5)P3 via class I PI3Ks (with α, β, γ, and δ isoforms). PI(3,4,5)P3 is a substrate for the PI 3‐phosphatase PTEN and for the PI 5‐phosphatases SHIP1/2 and OCRL. Class II PI3Kα generates a plasma membrane pool of PI(3,4)P2 necessary for clathrin‐coated pit (CCP) maturation and formation of free clathrin‐coated vesicles (CCV) during endocytosis. PI(3)P is a defining and essential feature of early endosomes and is generated primarily by the class III PI3K Vps34 complex II with a possible contribution of class II PI3Ks (encircled by dashed line), either by direct PI(3)P synthesis or indirectly via PI(3,4)P2 hydrolysis by the PI 4‐phosphatases INPP4A/B. As endosomes mature into late endosomes/multivesicular bodies (MVBs), PI(3)P is converted into PI(3,5)P2 via the PI(3)P 5‐kinase PIKfyve. Endosomes and lysosomes also harbour a poorly understood pool of PI(3,4)P2. In the liver, this endosomal PI(3,4)P2 pool is synthesized by class II PI3Kγ. Autophagosomes contain PI(3)P produced by the autophagosome‐specific Vps34 complex I, and possibly by the class II PI3Ks (dashed line). Endosomal recycling to the cell surface requires PI(3)P hydrolysis by myotubularin phosphatases (MTMs) such as MTM1 and the concomitant generation of PI(4)P (not shown) to enable exocytosis. PI(3,5)P2 turnover at MVBs and/or lysosomes is mediated by MTMs together with the PI(3,5)P2 5‐phosphatase Fig4.
Getting started: Synthesis of PI 3‐phosphates
Phosphorylation of the phosphatidylinositol D3 OH group depends on distinct classes of PI3Ks either at the plasma membrane or at endosomes, thereby generating a membrane identity tag for the endosomal system. All PI3Ks contain a “signature motif” consisting of the catalytic kinase domain, a helical domain as well as a membrane‐binding C2 domain (Vanhaesebroeck et al, 2010a; Vadas et al, 2011; Balla, 2013). There are eight different PI3Ks, grouped into three classes based on the conserved domains found outside the signature motif, their association with regulatory subunits, as well as their preferred lipid substrates (Fig 2). Class I PI3Ks mainly produce PI(3,4,5)P3 in vivo, class II generates PI(3,4)P2 and PI(3)P, and class III exclusively forms PI(3)P. A single orthologue of each class can be found in C. elegans and D. melanogaster, while yeast contain only a class III PI3K (Vanhaesebroeck et al, 2010a; Vadas et al, 2011; Jean & Kiger, 2014).
Figure 2. Schematic representation of the kinases and phosphatases that act on PI 3‐phosphates.
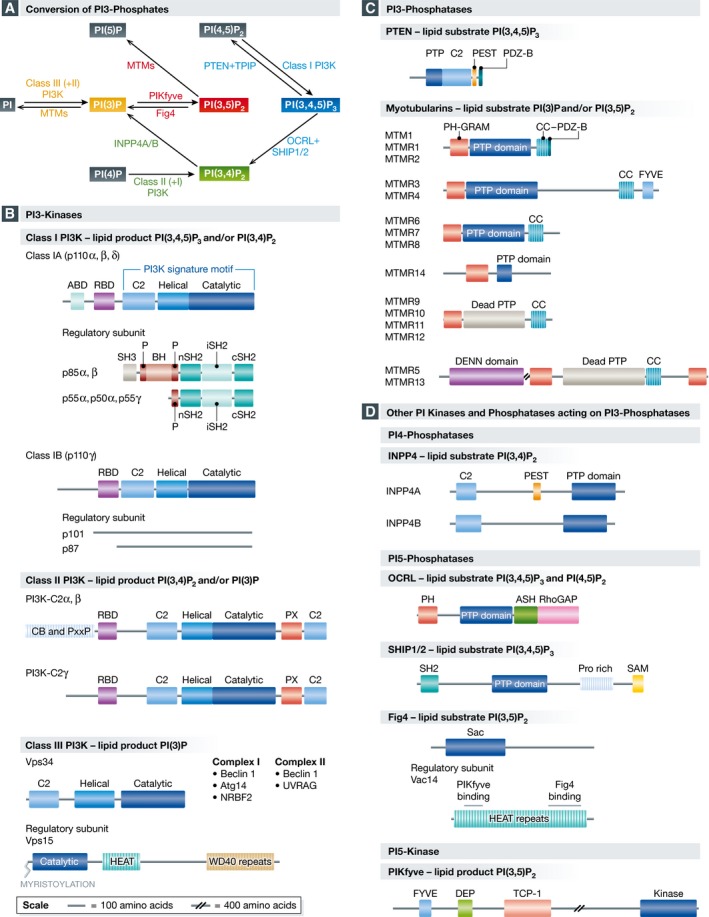
(A) Interconversion of PI 3‐phosphates via kinases (closed arrowheads) and phosphatases (open arrowheads). (B) PI 3‐kinases, (C) PI 3‐phosphatases and (D) other PI kinases and phosphatases acting on PI3‐phosphates. The various modules and domains are drawn roughly to scale. Adaptor binding domain (ABD); ASPM‐SPD2‐Hydin (ASH); Bar cluster region homology domain (BH); Clathrin‐binding region (CB); Coiled‐coil (CC) domain; Differentially expressed in normal and neoplastic cells (DENN) domain; Fab1, YOTB, Vac1 and EEA1 (FYVE); HEAT repeat domain (HEAT); Intervening coiled‐coil domain (iSH2); PDZ‐domain binding sequence (PDZ‐B); Phosphatase domain (PTP); Phox homology (PX); Pleckstrin homology (PH)‐glucosyltransferases, rab‐like GTPase activators and myotubularins (GRAM) domain; Proline, glutamine, serine, threonine (PEST) sequence; Proline‐rich region (P, Pro Rich); Ras binding domain (RBD); Rho GAP‐like domain (RhoGAP); Sac phosphatase domain (Sac); Sterile alpha motif (SAM); Src homology 2 (SH2) domain; Src homology 3 (SH3) domain; SH3‐domain binding proline‐rich segment (PxxP); T‐complex protein 1 (TCP‐1) homology domain.
Class I PI 3‐kinases
Class I PI3Ks are tightly regulated enzymes that predominantly produce PI(3,4,5)P3. They function as heterodimers, consisting of one of four possible catalytic subunits (p110α, β, δ or γ) (Hiles et al, 1992) that associate with one of two classes of regulatory subunits, the p85 class (p85α and its splice variants p55α or p50α, p85β and p55γ) and the p101/p87 class. The catalytic subunits are grouped based on the regulatory subunit they associate with: class IA subunits (p110α, β and δ) associate with the p85 class, while the class IB subunit (p110γ) associates with p101 or p87 (Vadas et al, 2011; Jean & Kiger, 2014) (Fig 2). The assembled class I PI3Ks act downstream of activated receptor tyrosine kinases (RTKs) and heterotrimeric guanine nucleotide‐binding protein (G protein)‐coupled receptors (GPCRs) to control signalling cascades involved in multiple processes such as cell growth, metabolism, proliferation, survival and repression of autophagy (Vanhaesebroeck et al, 2010a; Jean & Kiger, 2014). The class IA catalytic subunits (p110α and δ in vivo) are activated following RTK stimulation, resulting in the production of PI(3,4,5)P3 at the plasma membrane and the recruitment of effectors that bind this PI, notably the protein kinase Akt (also referred to as protein kinase B, PKB). Active Akt controls many downstream pathways such as the mechanistic target of rapamycin complex‐1 (mTORC1) pathway, the pro‐apoptotic factor BAD and the FOXO transcription factors (Vanhaesebroeck et al, 2010a; Dibble & Cantley, 2015). Contrary to this, the p110γ and p110β catalytic subunits are activated by the Gβγ heterodimers downstream of GPCR activation (Stephens et al, 1994; Stoyanov et al, 1995; Vanhaesebroeck et al, 2010a). Other inputs that can activate class I PI3Ks include small GTPases, for example active Ras and its family members, which bind the Ras binding domain (RBD) of the p110α, δ and γ catalytic subunits, or Rab5 that associates with the RBD of p110β. The four catalytic subunits have distinct yet overlapping functions and exhibit some degree of tissue specificity: p110α and β are expressed ubiquitously, while p110γ and δ are mainly found in immune cells (Vanhaesebroeck et al, 1997; reviewed in Vanhaesebroeck et al, 2010a). Because of their crucial role in controlling growth and proliferation, activating mutations targeting the class I PI3K signalling cascades are associated with cancer, and PI3Ks therefore represent important drug targets (Vanhaesebroeck et al, 2010b).
Class II PI 3‐kinases
Class II PI3Ks are monomeric enzymes and represent the least studied class of PI3Ks with functions in both signalling and membrane traffic. A single orthologue is found in Drosophila and C. elegans, while three isoforms are present in mammals. The α and β isoforms are expressed ubiquitously, while γ is restricted mainly to the liver. The main difference between the three mammalian isoforms resides in divergent N‐terminal extensions that are likely autoinhibitory and mediate protein–protein interactions (Fig 2). Unlike the class I and class III PI3Ks, class II enzymes have no known regulatory subunits but are instead regulated by interacting proteins such as clathrin, which binds to the inhibitory N‐terminal region of PI3KC2α and β, or small GTPases that associate with the RBD (e.g. Rab5 in the case of PI3KC2γ). Class II PI3Ks are also sensitive to signalling inputs, such as phosphorylation, ubiquitination, and possibly calcium (Kok et al, 2009; Sasaki et al, 2009; Vanhaesebroeck et al, 2010a; Jean & Kiger, 2014; Braccini et al, 2015).
Accumulating evidence implicates class II PI3K isoforms in coupling cell signalling to membrane traffic. This involves a role for PI3K intracellular localization or the presence of additional regulatory factors in triggering the formation of PI(3)P (Yoshioka et al, 2012; Franco et al, 2014) and/or PI(3,4)P2 (Leibiger et al, 2010; Posor et al, 2013; Braccini et al, 2015). The α isoform has been demonstrated to preferentially synthesize PI(3,4)P2 both in vitro and at plasma membrane endocytic pits in vivo, a site where PI(3,4)P2 is required for membrane constriction prior to endocytic vesicle fission (Posor et al, 2013). Defective CME and/or endosomal trafficking may also underlie impaired angiogenesis and vascular barrier function in endothelial‐specific PI3KC2α knock‐out mice (Yoshioka et al, 2012). In agreement with a role for class II PI3Ks in PI(3,4)P2 synthesis, it was recently shown that the liver‐specific γ isoform produces an endosomal pool of PI(3,4)P2 required for sustained Akt2 activation following insulin stimulation (Braccini et al, 2015). The class II α (Brown et al, 1999; Leibiger et al, 2010) and β isoforms (Arcaro et al, 2002) have also been linked to growth factor and nutrient signalling, though their precise function as well as the lipid product involved in these pathways remains to be determined. In addition to their function in PI(3,4)P2 synthesis at the cell surface and at endosomes, class II PI3Ks either directly or indirectly contribute to the maintenance of intracellular PI(3)P pools that regulate endocytic trafficking and autophagy (Jean et al, 2012; Devereaux et al, 2013; Franco et al, 2014). Further understanding of the diverse roles of the class II PI3Ks will be aided by the identification of new effectors and roles for their poorly understood lipid product, PI(3,4)P2.
Class III PI 3‐kinase
Vps34 is the only PI 3‐kinase in yeast (Schu et al, 1993) and was initially identified in a yeast screen for genes regulating sorting from the endosome to the lysosome (Herman & Emr, 1990). Membrane targeting of the catalytic subunit Vps34 is largely mediated via a constitutive interaction with the N‐terminally myristoylated adaptor protein Vps15/p150 (Fig 2). Vps34 is present in two complexes in both yeast and mammalian cells, each complex having separate locations and functions. Complex I is involved in autophagy and contains Vps15/p150, Beclin‐1 (equivalent to yeast Vps30), ATG14L (Atg14 in yeast) and NRBF2 (Atg38 in yeast) (Cao et al, 2014; Lu et al, 2014), while the endosomal complex II contains Vps15/p150, Beclin‐1/Vps30 and UVRAG (termed Vps38 in yeast) (Kihara et al, 2001; Funderburk et al, 2010; Rostislavleva et al, 2015).
The formation of these distinct complexes underlies the multitude of functions exerted by class III PI3K in intracellular trafficking, for example in endosomal fusion and sorting, trafficking from endosomes to lysosomes, phagocytosis, autophagy, as well as cytokinesis. For example, recruitment of complex II to early endosomes involves the association of Vps15/p150 via its WD40 repeat domain with Rab5‐GTP, resulting in the production of a pool of PI(3)P that drives endosomal association of effectors such as the endosomal sorting complex required for transport (ESCRT). In contrast, complex I—which contains the autophagy protein ATG14L—is responsible for the role of Vps34 in PI(3)P production on autophagosome precursor membranes (Ktistakis et al, 2012; Raiborg et al, 2013; Jean & Kiger, 2014). Contrary to its ability to positively regulate autophagy at the level of the autophagosome membrane, Vps34 also has a putative function in the negative regulation of autophagy via its involvement in amino acid sensing (Nobukuni et al, 2005; Gulati et al, 2008) and mTORC1 activation through the PI(3)P‐dependent recruitment of phospholipase D1 (PLD1) (Yoon et al, 2011; Bridges et al, 2012). Providing a further layer of complexity, mTORC1 activity specifically inhibits the autophagy promoting Atg14L‐containing complex I (Yuan et al, 2013), while it activates complex II via phosphorylation of UVRAG during the process of autophagosome–lysosome reformation following recovery from starvation (Munson et al, 2015). Consequently, the different Vps34 complexes not only have opposing effects in the cell but can potentially negatively regulate one another.
Fab1/PIKfyve
In addition to PI(3)P, endosomes, late endosomes/multivesicular bodies (MVBs) and likely lysosomes contain a functionally important pool of PI(3,5)P2 on their limiting membranes synthesized by the PI 5‐kinase PIKfyve (termed Fab1 in yeast) (Sbrissa et al, 1999; Ikonomov et al, 2002; McCartney et al, 2014). Fab1/PIKfyve, an evolutionary conserved protein, is recruited to endosomes by its FYVE (Fab1, YOTB, Vac1 and EEA1) domain‐mediated interaction with PI(3)P and forms part of a multiprotein complex that tightly controls lipid kinase activity (Fig 2). Mutations in PIKfyve and thus PI(3,5)P2 production have been linked to neurological disorders such as amyotrophic lateral sclerosis (ALS) and Charcot–Marie–Tooth disease (McCartney et al, 2014).
Turning over: Hydrolysis of PI 3‐phosphates by PI phosphatases
PI 3‐phosphate turnover is generally accomplished by PI 3‐phosphatases (Fig 2) that comprise the myotubularin family, PTEN, and TPIP, all of which contain a catalytic CX5R motif in their active site (Hsu & Mao, 2015). Interestingly, all of these PI 3‐phosphatases have been linked to human diseases as outlined below.
Myotubularin 3‐phosphatases
Myotubularin (MTM1) was first identified as a gene mutated in patients with X‐linked myotubular myopathy (also called X‐linked centronuclear myopathy, XLCNM) and, despite sequence similarity with protein tyrosine phosphatases, was subsequently demonstrated to act as a PI 3‐phosphatase that hydrolyses PI(3)P and/or PI(3,5)P2 (Blondeau et al, 2000; Taylor et al, 2000). In addition to myotubular myopathy, mutations in MTMR2 and MTMR13 are found in two forms of Charcot–Marie–Tooth neuropathy (CMT4B1 and 4B2) (Nicot & Laporte, 2008). The myotubularins are restricted to eukaryotes with 15 family members in mammals (MTM1 and MTMR1–14) (Fig 2), six of which are inactive pseudophosphatases lacking the catalytic cysteine within the CX5R motif. Heterodimers composed of active and inactive isoforms have commonly been observed, with the phosphatase‐inactive form being able to modulate the activity and localization of the active form (Laporte et al, 2003; Hnia et al, 2012; Hsu & Mao, 2015). In addition, the phosphatase‐inactive MTMR5/13 proteins harbour a DENN domain Rab guanine‐nucleotide exchange factor (GEF) that functionally activates Rab21 on PI(3)P endosomes, while their association with an active MTM allows for the hydrolysis of endosomal PI(3)P (Jean et al, 2012). Myotubularins function in a broad range of endolysosomal trafficking events. MTM1 and MTMR2 are found on early and late endosomes, respectively, where they can act on partially distinct pools of PI(3)P (Cao et al, 2007, 2008; Ketel et al, 2016). Another key site of action of the myotubularins is the autophagosomal system with the myotubularins MTMR3, the MTMR8/9 complex and MTMR14/JUMPY regulating the pools of PI(3)P and PI(3,5)P2 that are necessary for the initiation and progression of autophagy (Vergne et al, 2009; Taguchi‐Atarashi et al, 2010; Zou et al, 2012; Wu et al, 2014).
Myotubularins have also been implicated in regulating signalling events. As an example, MTM1‐deficient muscles from knock‐out mice display elevated mTORC1 activity that persists after starvation resulting in a defect in autophagy (Fetalvero et al, 2013). Contrary to this, MTM1 knock‐down in HeLa cells and primary human skeletal myotubes results in defective growth factor stimulus‐dependent Akt activation and reduced mTORC1 activity, leading to activation of caspase‐dependent pro‐apoptotic signalling (Razidlo et al, 2011). The reason for these conflicting results is not clear; however, it does appear that MTM1 acts both upstream and downstream of the Akt pathway. Laporte and colleagues found an increase in both phospho‐ and total Akt in muscles of MTM1‐null mice and an increase in phospho‐mTOR. The fact that the increase in mTOR phosphorylation only occurred at a later stage in the disease pathology than alterations in Akt suggests that these branches of the pathway are uncoupled. The authors also found dysregulation of autophagy with an increase in autophagosome formation at both early and late disease stages, while autophagy‐mediated degradation was only perturbed at late disease stages, suggesting this may be caused by the increased mTOR activity also only seen at late stages (Al‐Qusairi et al, 2013).
PTEN and TPIP 3‐phosphatases
PTEN (phosphatase and tensin homologue located on chromosome 10) is a tumour suppressor that dephosphorylates the D3 position of PI(3,4,5)P3 (Fig 2), thus antagonizing class I PI3K signalling by preventing Akt activation. PTEN suppression of Akt leads to reduced mTORC1 activity and, hence, crucially regulates energy metabolism, cell survival and proliferation. PTEN is also one of the most commonly mutated tumour suppressors with the loss of the lipid phosphatase activity driving tumorigenicity (Di Cristofano et al, 1998; Song et al, 2012). Recent data indicate that in addition to the plasma membrane, PTEN can operate at endocytic vesicles to inactivate Akt via a mechanism that involves PI(3)P binding to the PTEN C2 domain (Naguib et al, 2015). These data thus illustrate that the production of one species of PI 3‐phosphate (i.e. PI(3)P) can mediate the removal of another (i.e. PI(3,4,5)P3).
The function of PTEN must be tightly controlled as even small variations in activity can have dramatic effects on cell growth and therefore on tumorigenicity (Salmena et al, 2008; Song et al, 2012). The function of PTEN is regulated by a variety of post‐transcriptional and post‐translational mechanisms, for example by multiple C‐terminal phosphorylation sites, ubiquitination, acetylation, SUMOylation and regulation by microRNAs (Fragoso & Barata, 2015). In addition to PTEN, some cells and tissues express the poorly characterized PTEN homologue TPIP (TPTE and PTEN homologous inositol lipid phosphatase) (Sasaki et al, 2009). Despite their conserved phosphatase activity, unlike PTEN, TPIP does not affect Akt function (Walker et al, 2001). As TPIP displays sequence homology to voltage‐sensing phosphatases (VSPs), its phosphatase activity could potentially be regulated by changes in membrane potential (Kurokawa et al, 2012).
Other phosphatases acting on PI 3‐phosphates
Multiple phosphorylated PI 3‐phosphates such as PI(3,4)P2, PI(3,5)P2 and PI(3,4,5)P3 are also substrates for PI phosphatases other than the PI 3‐phosphatases (Hsu & Mao, 2015). For example, PI(3,4)P2 is converted to PI(3)P by the closely related CX5R motif‐containing PI(3,4)P2‐specific 4‐phosphatases INPP4A and INPP4B (Figs 2 and 3B). INPP4A/B negatively regulates Akt signalling and INPP4B acts as a tumour suppressor in human cancers (Woolley et al, 2015), while loss of INPP4A leads to neurodegeneration in mutant mice (Sasaki et al, 2010). Furthermore, PI(3,4,5)P3 is subject to hydrolysis by the Lowe syndrome‐associated PI 5‐phosphatases OCRL, which also acts on PI(4,5)P2, and SHIP1/2, resulting in the production of PI(3,4)P2 on endosomes or the plasma membrane, respectively (Pirruccello & De Camilli, 2012). Lastly, the HEAT repeat protein Vac14 acts as a scaffold for the assembly of an endosomal complex (Zhang et al, 2012) comprising PIKfyve and the PI(3,5)P2‐specific phosphatase Fig4 (Fig 2), whose activity appears to be the main source of cellular PI(5)P. Interestingly, mutations of Fig4 are responsible for Yunis‐Varón syndrome, a familial epilepsy with polymicrogyria, and Charcot–Marie–Tooth type 4J neuropathy (CMT4J) (Menezes et al, 2014; Vaccari et al, 2015) due to altered PI(3,5)P2 metabolism (see below).
Figure 3. Interplay of PI 3‐phosphates in regulating signalling and membrane traffic.
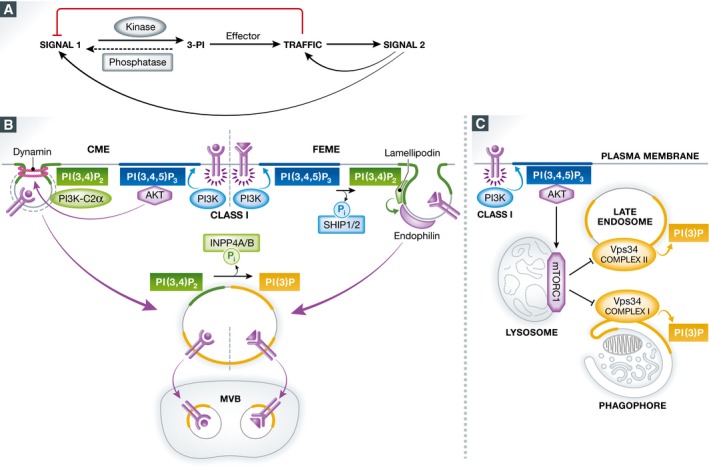
(A) Schematic representation of the role of PI 3‐phosphates (3‐PI) in the reciprocal regulation of cell signalling and membrane traffic. Signals (i.e. extracellular ligand that binds to its receptor) lead to activation of PI 3‐kinases and the 3‐PIs generated recruit effector proteins that often regulate membrane traffic, leading to the generation of a secondary signal. Such a downstream secondary signal may then regulate traffic and/or the response to the primary signal (e.g. by altering receptor levels or receptor‐associated signalling components). (B) Endocytosis of activated signalling receptors (purple) from the cell surface via clathrin‐mediated endocytosis (CME, left) or fast‐endophilin‐mediated endocytosis (FEME, right) involves PI(3,4)P2 and PI(3,4,5)P3 synthesis by class II PI3Kα (PI3KC2α) and class I PI3Ks. In FEME, SHIP1/2 converts PI(3,4,5)P3 to PI(3,4)P2, which serves to recruit lamellipodin and thereby endophilin. PI(3,4)P2 is hydrolysed to PI(3)P on endosomes by the PI(3,4)P2‐specific PI 4‐phosphatase INPP4A. Activation of Akt signalling by PI(3,4,5)P3 and PI(3,4)P2 stimulates CME via dynamin 1, while dysregulated CME causes Akt overactivation. Endosomal sorting of internalized signalling receptors to internal vesicles of multivesicular bodies (MVBs) terminates signalling. (C) mTORC1 activation on the lysosome is downstream of PI(3,4,5)P3 generated via class I PI3Ks and activated Akt. mTORC1 signalling represses PI(3)P generation via distinct Vps34‐containing class III PI3K complexes, thereby suppressing autophagosome formation and late endosome maturation.
Collectively, the cellular PI 3‐kinase and phosphatase activities allow for a rapid conversion of PI 3‐phosphate identities between the plasma membrane and the endolysosomal system.
Manipulating PI 3‐phosphate synthesis and turnover
Recent advances have allowed for the discovery of new pools of PI 3‐phosphates, as well as new methods to modulate subcellular PI 3‐phosphate levels in a spatiotemporally controlled manner. We will discuss some of the most recent advances regarding the acute manipulation of PI 3‐phosphate levels in Boxes Box 1: PI 3‐phosphate analogues, Box 2: Inducible acute manipulations by chemical dimerization and optogenetics, Box 3: Chemical inhibitors of PI 3‐phosphate signalling. For a more detailed overview, the reader is referred to excellent recent reviews (Idevall‐Hagren & De Camilli, 2015; Maekawa & Fairn, 2015).
Box 1: PI 3‐phosphate analogues.
Recent advances in chemical synthesis has resulted in the development of membrane‐permeable PI 3‐phosphate analogues whereby the charged phosphate groups are masked with a hydrolase‐sensitive acetoxymethylester group that is cleaved inside living cells within minutes (Laketa et al, 2009). These PI analogues have been successfully used in vitro and in vivo: for example, to delineate a requirement of PI(3,4,5)P3 for receptor tyrosine kinase recycling, the discovery that PI(3,4)P2 is an essential mediator of CME, and the finding that the PI(4,5)P2/PI(3,4,5)P3 ratio determines actomyosin contractility and plasma membrane expansion during tissue morphogenesis in Drosophila larvae (Posor et al, 2013; Laketa et al, 2014; Reversi et al, 2014). Caveats include the fact that modified PI 3‐phosphates insert non‐specifically into membranes and therefore may not reflect the heterogeneous spatial effects of endogenous PI 3‐phosphates, as well the fact that their activation inside the cell by ester hydrolysis is comparably slow, requiring several minutes. The recent development of photoactivatable PIs that can be deprotected by UV light in 30 s now allows for the possibility of both temporal and spatial control of PI 3‐phosphate levels. For example, it has been demonstrated that photo‐uncaging of PI(3)P is a sufficient signal to induce endosomal fusion (Subramanian et al, 2010; Mentel et al, 2011).
Box 2: Inducible acute manipulations by chemical dimerization and optogenetics.
The acute activation of enzymes, for example PI 3‐kinases or phosphatases, can be accomplished via the utilization of dimerization systems. Inducible acute manipulations provide a powerful tool in dissecting the spatial function of PI 3‐phosphates, notably how one species of PI 3‐phosphate can exert different functions on different membranes by specifically modulating its level at a given subcellular site, which is often difficult, sometimes impossible, to achieve via genetic approaches. For example, PI(3)P is produced both on early endosomes and autophagosomes via distinct, yet partially overlapping Vps34‐containing class III PI3K complexes (Kihara et al, 2001; Funderburk et al, 2010). PI3KC2α can act on both the plasma membrane to synthesize PI(3,4)P2 (Posor et al, 2013) and on endosomes, where it may synthesize PI(3)P (Falasca & Maffucci, 2012; Franco et al, 2014) and/or PI(3,4)P2.
An established method to acutely manipulate PI 3‐phosphate levels at defined subcellular sites is via the dimerization between FKBP12 with the FRB domain of mTOR, whereby a membrane‐targeting sequence can be fused to one domain and the enzyme to another, allowing the translocation to a specific membrane upon addition of rapamycin or analogues. Application of this technology established a crucial role for early endosomal PI(3)P in controlling endosome size and receptor sorting (Fili et al, 2006). Drawbacks, however, include the effects of rapamycin on mTOR signalling and the irreversibility of the dimerization. Recently, a reversible chemical dimerization system using a competitive ligand has been developed, allowing for the activation and subsequent arrest of PI 3‐kinase activity to determine the turnover of PI(3,4,5)P3 and its downstream effectors (Feng et al, 2014).
An independent rapidly reversible dimerization approach is based on optogenetics whereby protein heterodimerization is induced by light of specific wavelengths, and, hence, is reversible as protein heterodimers rapidly dissociate following cessation of the light pulse. This technique has been used to study the recruitment of class I PI 3‐kinases to the plasma membrane and subsequent recovery from their activation (Toettcher et al, 2011; Idevall‐Hagren et al, 2012). A great advantage of this approach is the ability to apply focal light pulses to rapidly induce PI 3‐phosphate synthesis or turnover locally within cells. On the other hand, the application of optogenetics limits the use of fluorescent proteins to red and far‐red variants, thereby putting restrictions on live imaging approaches.
Box 3: Chemical inhibitors of PI 3‐phosphate signalling.
The development of specific and selective inhibitors of the different classes and isoforms of PI3Ks is of great interest as a biological tool to better understand their diverse function, as well as for their potential clinical applications, most notably in anti‐cancer therapy and the treatment of inflammatory diseases.
Potent pan‐PI3K inhibitors wortmannin and LY294002 inhibit the p110 catalytic subunit of the class I PI3K; however, they inhibit other kinases (e.g. Vps34‐class III PI3K, PI3KC2β) at higher concentrations.
Idelalisib is a p110‐δ‐specific inhibitor used in the treatment of chronic lymphocytic leukemia (Zhu et al, 2015). Several other isoform‐selective class I PI3K inhibitors exist.
Specific Vps34 inhibitors, PIK‐III, SAR405, VPS34‐IN1, are also promising for therapeutic applications as potential anti‐cancer drugs. These inhibitors have also provided additional evidence for Vps34‐mediated PI(3)P production during vesicle trafficking, autophagy and signalling (Bago et al, 2014; Dowdle et al, 2014; Ronan et al, 2014).
Lead structures for compounds that selectively inhibit class II PI3Kγ is a promising first step in the development of specific inhibitors of the class II PI3Ks, the understanding of which has been hampered by a lack of structural information on these kinases, their substrate interactions and their regulation by allosteric activators or inhibitors (Freitag et al, 2015).
Inhibitors of the PI(3,4,5)P3 3‐phosphatase PTEN (e.g. SF1670, bpV(phen), bpV(pic), or VO‐OHpic) have been reported and shown to cause elevated PI(3,4,5)P3 levels and overactivation of the Akt pathway with corresponding effects on cell polarization, migration, akin to genetic PTEN loss‐of‐function models (Di Cristofano et al, 1998; Song et al, 2012; Mak & Woscholski, 2015), however that may inhibit other enzymes such as the PI 4‐phosphatases INPP4A/B (Spinelli et al, 2015).
Coupling PI 3‐phosphate synthesis and turnover by co‐regulation of phosphatase and kinase activities
While the presence of antagonistic PI kinases and phosphatases in the same complex may seem counterintuitive, they often act in concert to regulate the conversion of different PIs during membrane trafficking. Such a regulated PI kinase/phosphatase complex, in which the active partner changes in a spatiotemporally controlled manner, would allow for the switch in PI identity needed to terminate one event and commence another. This would allow a switch between the trafficking of a vesicle requiring one PI species and its fusion with its target membrane that may require another (Botelho, 2009). Several examples of such complexes have indeed been discovered. The perhaps best studied case is the conserved protein complex formed between the PI(3)P 5‐kinase PIKfyve and the PI(3,5)P2‐phosphatase Fig4, held together by the scaffolding protein Vac14 that is responsible for the biosynthesis and turnover of PI(3,5)P2 (Jin et al, 2008; Ikonomov et al, 2009). Loss of PIKfyve or Vac14 causes a loss or decrease in PI(3,5)P2 levels, respectively (Duex et al, 2006b; Zhang et al, 2007; Ikonomov et al, 2011). As the phosphatase Fig4 is required for PIKfyve activation, its deletion also results in PI(3,5)P2 depletion (Duex et al, 2006a; Chow et al, 2007). These data argue that the presence of both a kinase and a phosphatase in the same complex allows for tight regulation of endosomal PI(3,5)P2 levels. Another example is the association of the myotubularin PI 3‐phosphatases MTM1 and MTMR2 with the WD40 domain of the Vps15 subunit of the class III PI3K complex II on endosomes, which inhibits their phosphatase activity. The association of the Vps15‐containing class III PI3K complex with MTM1 or with endosomal GTP‐bound Rabs, required for PI3K activation, is mutually exclusive. These data suggest that the Rab GTPases together with MTMs serve as molecular switches for controlling the sequential synthesis and degradation of endosomal PI(3)P (Cao et al, 2007, 2008). Interestingly, the related MTMR13, a pseudophosphatase that interacts with and regulates active MTMs, has been found to associate with the class II PI3K (Pi3K68D) in Drosophila and with the mammalian class II PI3Kα, thereby coordinating the turnover of a PI(3)P pool that regulates cargo exit from endosomes (Jean et al, 2012). Similar modules comprising coupled PI kinase and phosphatase activities likely underlie regulated PI conversion within the endosomal system, for example to enable cargo exit from PI(3)P‐containing endosomes to the plasma membrane enriched in PI(4)P and PI(4,5)P2. Indeed, recent data demonstrate that surface delivery of endosomal cargo requires hydrolysis of PI(3)P by the phosphatidylinositol 3‐phosphatase MTM1. Removal of endosomal PI(3)P by MTM1 was shown to be accompanied by phosphatidylinositol 4‐kinase 2α‐dependent generation of PI(4)P and recruitment of the exocyst tethering complex to enable membrane fusion. These data suggest a close coupling between PI(3)P turnover and PI(4)P synthesis that is driven by both coincident detection and reciprocal interactions between PI 3‐phosphatase and PI 4‐kinase activities to allow for local PI conversion at endosomes. Defects in this PI conversion module likely underlie the muscle and non‐muscle defects observed in patients suffering from X‐linked centronuclear myopathy (myotubular myopathy) due to mutations in the MTM1 gene (Ketel et al, 2016) (Fig 1).
While these examples focus on PI(3)P conversion in endosomal trafficking, given the importance of PI conversion in all aspects of membrane trafficking, the identification of new PI kinase and phosphatase complexes will provide valuable insight into how these conversion cascades are regulated. There is also evidence for the formation of regulatory complexes between PI3Ks and phosphatases involved in signal transduction, as seen for the PI(3,4,5)P3 3‐phosphatase PTEN and the p85 regulatory subunit of class I PI3Ks. This interaction positively regulates the phosphatase activity of PTEN, providing a mechanism to couple Akt activation via p85‐PI3K‐mediated synthesis of PI(3,4,5)P3 with Akt inactivation via p85‐PTEN‐triggered PI(3,4,5)P3 hydrolysis (Barber et al, 2006; Rabinovsky et al, 2009; Chagpar et al, 2010).
And action! PI 3‐phosphate‐binding modules and effectors
PIPs mediate their effects at membranes via the recruitment of effector proteins containing PIP‐binding domains. The incorporation of common binding domains into proteins with otherwise diverse structures and activities allows PIPs to have a broad range of functions inside the cell. Effector proteins can have direct effects on the nanoscale organization and shape of membranes, for example via tubulation, fusion or fission of membranes. Moreover, they can be involved in intracellular transport, can coordinate or regulate the activation or inhibition of other proteins such as small GTPases, kinases or phosphatases and can serve as platforms for intracellular signalling cascades (Lemmon, 2003, 2008; Kutateladze, 2010). Among the major PI 3‐phosphate recognizing domains are the PH, the FYVE and the PX domains, though many other domains exist that can associate with PI 3‐phosphates, often in conjunction with other proteins or membrane lipids (Table 1).
Table 1.
Phosphatidylinositol 3‐phosphate binding domains
| Domain | Typical size (amino acids) | Structure | Lipid‐binding specificity | Examples |
|---|---|---|---|---|
| C2 | 120–130 | β‐sandwich, Ca2+ dependent (Guerrero‐Valero et al, 2009) | Anionic and neutral lipids (PtdSer, PtdChol), and PIPs | PTEN, Sch9, Class II PI3Ks, INPP4A and B |
| FYVE | 60–70 | Cysteine‐rich Zn2+ finger (Dumas et al, 2001) | PI(3)P | EEA1, Hrs, SARA, Alfy, FYVE‐CENT, MTMR3, MTMR4, PIKfyve, Bem1, FYCO1, Protrudin |
| PX | 130 | α and β structure (Bravo et al, 2001) | PI(3)P, occasionally other PIPs | Sorting nexins (SNXs), Class II PI3Ks, kinesin KIF16B, SNARE Vam7p, SGK3 |
| PH | 100–120 | β‐sandwich with C‐terminal α amphipathic helix (Thomas et al, 2002) | PIPs, some highly specific, others promiscuous | Akt, TAPP1, ARNO, PDK1, Lamellipodin |
| GRAM | 70 | β‐sandwich with C‐terminal α‐helix, similar to PH domain (Begley et al, 2006) | PI(3,5)P2 and PI(5)P | Myotubularins, glucosyltransferases, TBC domain‐containing proteins |
| GLUE | 130 | Split PH domain with two β‐sheets, one α‐helix (Teo et al, 2006) | PI(3)P (only example of a PH‐like domain that can bind PI(3)P) | Vps36 subunit of the ESCRT complex |
| PROPPIN/WD40 | 500/300 | β‐propeller (Baskaran et al, 2012) | PI(3,5)P2, in some cases PI(3)P | Atg18/WIPI proteins, Raptor |
Pleckstrin homology (PH) domains with high affinity binding to PI(3,4,5)P3 and PI(3,4)P2 play a key role in PI3K signalling from the plasma membrane following agonist stimulation of cell surface receptors, with Akt as the best studied example of such a PI3K downstream effector. Growth factor activation of receptors stimulates class I PI3Ks, resulting in the synthesis of PI(3,4,5)P3, that may be further converted to PI(3,4)P2 by SHIP1/2 during the sustained phase of signalling. Both PI(3,4,5)P3 and PI(3,4)P2 bind with high affinity to the PH domain of Akt, thereby triggering its plasma membrane recruitment and activation as well as the phosphorylation of downstream targets, that is components and regulators of mTORC1 signalling (Pearce et al, 2010; Hemmings & Restuccia, 2012). Apart from Akt, other known PH domain‐containing effectors of PI(3,4,5)P3 (and possibly PI(3,4)P2) comprise members of the guanine‐nucleotide exchange factors (GEFs) for Rho and ADP ribosylation factor (Arf) family GTPases that are activated downstream of class I PI3K stimulating receptors. Rho‐GEFs often contain a PH domain found in tandem with a Dbl homology (DH) domain that harbours GEF activity. Membrane recruitment by PIP binding of the PH module stimulates the GEF activity of the DH domain, for example to promote cortical actin assembly among other effector pathways (Snyder et al, 2001; Rossman et al, 2002; Baumeister et al, 2003, 2006). One example is the Rac‐GEF P‐Rex1, a large scaffold protein synergistically activated by PI(3,4,5)P3 and Gβγ (Welch et al, 2002). PH domains can also regulate GEFs by controlling their localization, as seen with Arf6‐GEF ARNO, which is recruited to the plasma membrane by binding of its PH domain to Arf6 and to PI(3,4,5)P3 (Venkateswarlu et al, 1998; Cohen et al, 2007). Furthermore, PH domains are found in some GTPase‐activating proteins (GAPs), suggesting a role in the inactivation of GTPases (Vitale et al, 2000; Venkateswarlu et al, 2004).
FYVE domains specifically associate with PI(3)P and have been found in a large variety of endosomal and autophagosomal proteins (Stenmark et al, 1996; Burd & Emr, 1998). FYVE domains mediate or contribute to endosomal or autophagosomal protein localization, frequently as part of coincidence detection. For example, membrane targeting of the early endosomal scaffold protein EEA1 (early endosomal antigen 1) requires coincidence detection of both PI(3)P and Rab5‐GTP to facilitate tethering and fusion of early endosomes (Simonsen et al, 1998; Christoforidis et al, 1999). Similar mechanisms mediate recruitment of other PI(3)P‐binding proteins. These include the degradative sorting adaptor Hrs (hepatocyte growth factor‐regulated tyrosine kinase substrate) (Raiborg et al, 2002), the FYVE domain‐containing adaptor SARA (Smad anchor for receptor activation), a mediator of transforming growth factor (TGF) β‐Smad signalling at endosomes (Miura et al, 2000; Itoh et al, 2002), the cytokinesis factor FYVE‐CENT (Sagona et al, 2010), the PI(3)P phosphatase MTMR4 (Lorenzo et al, 2006; Naughtin et al, 2010), ALFY (autophagy‐linked FYVE protein), a protein that targets ubiquitin‐containing protein aggregates for autophagic degradation (Simonsen et al, 2004) and the omegasome protein DFCP1 (double FYVE containing protein 1) (Polson et al, 2010).
Phox homology (PX) domains often preferentially associate with PI(3)Ps on endosomes; however, some family members exhibit a broader lipid specificity and localization (Seet & Hong, 2006; Teasdale & Collins, 2012). The largest group of PX domain‐containing proteins are the members of the sorting nexin (SNX) family of trafficking and signalling adaptors found at the plasma membrane and throughout the endosomal system (Cullen, 2008). A subset of SNXs in addition to their PI 3‐phosphate‐binding PX domain also contain a Bin‐Amphiphysin‐Rvs (BAR) domain, a dimeric helical protein module that can sense or induce membrane curvature, in particular when containing additional amphipathic helices (Peter et al, 2004). Other proteins containing PX domains include the class II PI3Ks and the endosomal kinesin KIF16B. The latter has been shown to link Vps34‐mediated PI(3)P production to the peripheral dispersion of endosomes (Hoepfner et al, 2005).
Other domains that can recognize and bind to PI 3‐phosphates are summarized in Table 1. These include the PH‐related GRAM (glucosyltransferases, Rab‐like GTPase activators and myotubularins) domain found in endosomal myotubularins (Doerks et al, 2000; Begley et al, 2003; Berger et al, 2003; Tsujita et al, 2004; Lorenzo et al, 2005) and the PI(3)P‐binding GLUE (GRAM‐like, ubiquitin binding in EAP45) domain present in the Vps36 subunit of the ESCRT‐II complex (Slagsvold et al, 2005; Teo et al, 2006). β‐propeller PROPPIN (β‐propellers that bind phosphoinositides) and WD40 domains found in autophagy proteins such as Atg18/WIPI2 (WD‐repeat PtdIns(3)P effector protein) can bind to PI 3‐phosphates as well (Strømhaug et al, 2004; Polson et al, 2010; Baskaran et al, 2012; Dooley et al, 2014). The Raptor subunit of mTORC1 associates via its WD40 domain with PI(3,5)P2 on lysosomes (Michell et al, 2006; Bridges et al, 2012). PI 3‐phosphate binding has also been reported for subsets of C2 domains, often in response to elevated intracellular calcium (Corbalan‐Garcia & Gómez‐Fernández, 2014). PI3KC2α contains a C2 domain that binds to PI(3,4)P2 and PI(4,5)P2, while the C2 domain of INNP4B has been shown to associate with PI(3,4,5)P3. These examples further illustrate how the localized production or consumption of one PI species can be regulated by another (Ferron & Vacher, 2006; Liu et al, 2006).
In many cases, the exact mode by which PI 3‐phosphates are recognized and exert their physiological effect is unknown. For example, the fusion of late endosomes, lysosomes and autophagosomes requires both PI(3)P and PI(4,5)P2 to promote SNARE complex assembly, as well as membrane tethering by the heterotetrameric HOPS complex (homotypic fusion and vacuole protein sorting) (Mima & Wickner, 2009; Xu & Wickner, 2010; Balderhaar & Ungermann, 2013; Solinger & Spang, 2013). Furthermore, the Rab7‐GEF Mon1‐Ccz1 binds and is recruited to early endosomes via PI(3)P, and the GEF activity itself is stimulated by membranes and PI(3)P (Nordmann et al, 2010; Poteryaev et al, 2010; Cabrera et al, 2014), though the precise PI(3)P‐binding determinants within Mon1‐Ccz1 remain to be unravelled.
These examples illustrate that nature has evolved mechanisms of PI 3‐phosphate recognition and function that extend well beyond the known repertoire of PI 3‐phosphate‐binding domains. In support of this view, recent proteomic studies have revealed a much larger PI‐binding interactome than predicted from the simple analysis of known PI‐binding domains (Jungmichel et al, 2014), suggesting that many more PI 3‐phosphate‐associated domains and effectors remain to be uncovered.
Integrating PI 3‐phosphate function: From signalling to membrane traffic and return
While PI 3‐phosphates—as discussed above—act as crucial second messengers activating signal transduction in response to extracellular stimuli, they also exhibit key roles in exo‐ and endocytic membrane traffic to and from the plasma membrane and within the endolysosomal system. During recent years, it has, however, become clear that the roles of PI 3‐phosphates in signalling from both the cell surface and from endomembranes (Vanhaesebroeck et al, 2010a) and in membrane traffic (Jean & Kiger, 2012; Balla, 2013) are intertwined and, hence, often cannot be separated (Fig 3A).
Receptor endocytosis in the regulation of signalling and cellular metabolism
Upon binding their ligand, activated plasma membrane receptors such as RTKs and GPCRs undergo endocytosis, either via CME or via clathrin‐independent internalization routes including macropinocytosis (Schmees et al, 2012; Levin et al, 2015) and fast‐endophilin‐mediated endocytosis (FEME) as described recently (Boucrot et al, 2015). Endocytosis most often serves to terminate receptor signalling and to modulate the cellular responsiveness to a given ligand by adjusting receptor surface levels. Following their traffic to early endosomes, internalized receptors can be sorted to intraluminal vesicles of MVBs or late endosomes. This results in termination of signalling via receptor segregation from the cytoplasm and via their subsequent degradation in lysosomes, thereby downregulating the future responsiveness of the cell to signalling receptor ligands (Fig 3B). Alternatively, receptors can be recycled back to the plasma membrane enabling cellular re‐sensitization to receptor ligands (Grant & Donaldson, 2009; Parachoniak & Park, 2012). Receptor endocytosis can in some cases also serve as a means to transmit extracellular signals over long distances via signalling endosomes, for example in neurons, where growth factor signals need to travel retrogradely from the synapse back to the soma over long distances (Sorkin & von Zastrow, 2009; Platta & Stenmark, 2011). Commencement of a signalling event at the plasma membrane, internalization of the receptor, its delivery to early endosomes, and subsequent sorting into the degradative or recycling pathways rely on the spatiotemporally controlled synthesis and turnover of defined pools of PI 3‐phosphates by the different classes of PI3Ks and phosphatases.
A major effect of the activation of RTKs and GPCRs is the generation of plasma membrane PI(3,4,5)P3 via class I PI3Ks (see above). Following their activation and downstream signalling events, activated receptors are internalized and PI(3,4,5)P3 is hydrolysed to prevent receptor hyperactivation and hyperproliferation of cells as seen in many forms of cancer (Sheppard et al, 2012). Recent data suggest differential roles for PI 3‐phosphates in coordinating receptor signalling, endocytosis and downstream receptor trafficking. A prime example is the epidermal growth factor receptor (EGFR), which following activation with different concentrations of ligand—at least in some cell types—can be internalized via CME, FEME (Boucrot et al, 2015) and macropinocytosis (Sorkin & Goh, 2009; Maekawa et al, 2014). EGFR internalization via CME can be driven either by low concentrations of its ligand or by exogenous supply of PI(3,4,5)P3 in the absence of EGF. In the latter case, receptors are exclusively recycled to the cell surface rather than sorted to MVBs for degradation. Recycling requires both PI(3,4,5)P3 and members of the PAR polarity complex proteins such as PARD3 (Laketa et al, 2014), suggesting that PI(3,4,5)P3 regulates receptor tyrosine kinase trafficking and may thereby enhance the cellular responsiveness to growth factors. How exactly elevated levels of PI(3,4,5)P3 promote CME of activated EGFR, while constitutive CME of transferrin appears to be unaffected, is unknown. One possibility is that PI(3,4,5)P3 serves as a substrate for 5‐phosphatases such as SHIP2 to generate a local pool of PI(3,4)P2, a lipid required for clathrin‐coated pit (CCP) maturation, at the plasma membrane (Posor et al, 2013). A similar mechanism may operate in FEME of activated EGFR and other RTKs and GPCRs, where class I PI3K‐mediated synthesis of PI(3,4,5)P3 downstream of receptor activation followed by its subsequent dephosphorylation into PI(3,4)P2 by SHIP1/2 has been shown to mediate recruitment of the PI(3,4)P2‐binding PH domain protein lamellipodin. Lamellipodin then engages the membrane‐deforming BAR domain protein endophilin and the actin cytoskeleton to drive clathrin‐independent receptor internalization (Fig 3B). Endophilin‐depleted cells display elevated mitogen‐associated protein kinase (MAPK) activity, while the response to high doses of growth factor (e.g. EGF) is dampened, perhaps as a result of compensatory macropinocytic EGFR internalization (Boucrot et al, 2015), a process that requires sequential breakdown of PI(3,4)P2 and PI(3)P by INPP4B and the myotubularin‐related 3‐phosphatases MTMR6 and 9 (Maekawa et al, 2014). Knock‐down of clathrin also results in elevated MAPK signalling, because the recruitment of the MAPK kinase MEK2 to endosomes is perturbed in clathrin‐depleted cells (Galperin & Sorkin, 2008). The work of Galperin and Sorkin indicates that the spatial segregation of MEK2 from EGFR signalling complexes at the level of endosomes depends on CME. These data imply that distinct endocytosis pathways differentially regulated by PI 3‐phosphates affect RTK signalling. Conversely, recent data suggest that crosstalk between RTK‐mediated Akt/GSK3β signalling and isoforms of dynamin regulates CME (Reis et al, 2015).
How specific receptors are internalized and signal appears to be at least in part caused by their direct association with components of the machinery for PI 3‐phosphate synthesis and turnover. Apart from the well‐studied signalling adaptor‐mediated association of class I PI3Ks with both RTKs and GPCRs (Naga Prasad et al, 2002), other members of the PI3K family have also been linked to specific receptors. In endothelial cells, the class II PI3KC2α is specifically required for the internalization of the receptors for vascular endothelial growth factor (VEGF) and sphingosine 1‐phosphate (S1P). As a consequence, VEGF and S1P signalling as well as endothelial function are impaired in endothelial cell‐specific PI3KC2α knock‐out mice, due to defective formation of VEGFR2‐ and S1PR‐positive signalling endosomes (Yoshioka et al, 2012; Biswas et al, 2013). Similar mechanisms may explain the role for PI3KC2α in TGFβ (Aki et al, 2015) and insulin signalling. In pancreatic β cells, PI3KC2α has been shown to generate PI(3,4)P2 downstream of insulin receptor activation, allowing for the selective activation of Akt1 to promote glucose‐stimulated insulin release (Leibiger et al, 2010). Although the exact role of PI3KC2α‐mediated PI(3,4)P2 synthesis has not been studied in all of these cases, it is tempting to speculate that receptor signalling and PI3KC2α activation are intimately connected to receptor internalization and/or endosomal trafficking. Consistent with this hypothesis, recent work has shown that CME of various receptors requires the local generation of PI(3,4)P2 at endocytic coated pits by PI3KC2α (Posor et al, 2013) to promote membrane constriction prior to endocytic vesicle fission (Daumke et al, 2014).
Receptor sorting within the endosomal system
While the production of PI(3,4)P2 is necessary for the completion of CME, it is the generation of PI(3)P that is essential for the termination of signalling events at the plasma membrane mediated via PI(3,4,5)P3. PTEN is recruited to endosomal vesicles via binding of its C2 domain to PI(3)P (Naguib et al, 2015), providing an example of how a downstream 3‐PI (PI(3)P) can negatively regulate an upstream one (PI(3,4,5)P3) via the regulation of a PI 3‐phosphatase. PTEN‐mediated turnover of PI(3,4,5)P3 may be linked to the formation of APPL endosomes. This subpopulation of signalling early endosomes contains the Akt‐interacting Rab5 effector proteins APPL1/2 (Miaczynska et al, 2004; Schenck et al, 2008) but is devoid of endosomal PI(3)P effectors such as EEA1 (Zoncu et al, 2009; but see Kalaidzidis et al, 2015 for a different view).
PI(3)P generated by class III PI3K Vps34 and possibly class II PI3Ks (Devereaux et al, 2013; Franco et al, 2014) is a defining feature of endosomes and of key importance for their function. Within the endosomal system, cargo may either be recycled to the cell surface, trafficked retrogradely to the trans‐Golgi network (TGN), or sorted to MVBs/late endosomes for lysosomal degradation. Surface recycling and retrograde traffic to the TGN both involve the formation of recycling tubules and the sorting of cargo into these tubules by lipid‐binding proteins of the sorting nexin (SNX) family. Tubule formation by SNX proteins depends on the ability of their PX‐BAR domain module to bind to PI(3)P and to form oligomeric self‐assemblies that remodel the endosomal membrane (van Weering & Cullen, 2014). Some SNXs are actively involved in cargo selection: SNX17, 31 and 27 harbour a PX and a FERM (4.1/ezrin/radixin/moesin) domain, the latter associating with and sorting PDZ‐domain‐containing cargo, such as β1‐integrin and β2‐adrenergic receptors (Lauffer et al, 2010; Böttcher et al, 2012; Tseng et al, 2014). Endosomal recycling of signalling and adhesion receptors thus regulates receptor surface expression and thereby the cellular responsiveness to ligands. Moreover, PI(3)P‐binding PX domain proteins including SNXs play a role in directing endosomal trafficking via the recruitment of molecular motors. For example, endosomal progression from early to recycling endosomes involves recruitment of the dynein adaptor KIBRA by the PI(3)P‐binding PX domain protein SNX4 (Traer et al, 2007). Outward traffic of cargo from endosomes to the cell surface depends on the plus end‐directed endosomal motor kinesin KIF16B, which is tethered to endosomes via a PI(3)P‐binding PX domain (Hoepfner et al, 2005).
Transmembrane cargo (e.g. receptors) degradation occurs via sorting into intraluminal vesicles (ILVs) of maturing endosomes to generate MVBs/late endosomes that will ultimately fuse with lysosomes. Endosomal maturation is accompanied by Rab conversion from early endosomal Rab5 to late endosomal Rab7, a process that depends on the recruitment and stimulation of the Rab7 GEF Mon1‐Ccz1 by PI(3)P (Nordmann et al, 2010; Poteryaev et al, 2010; Cabrera et al, 2014) as well as active cargo sorting into ILVs by the ESCRT complex. The ESCRT machinery contains 4 protein complexes, ESCRT 0, I, II and III that are recruited and work in a sequential manner (Raiborg & Stenmark, 2009). ESCRT 0 is recruited to the membrane via the PI(3)P‐binding FYVE domain of its Hrs subunit, which recognizes ubiquitinated cargo such as activated growth factor receptors destined for degradation (Raiborg et al, 2001). The ubiquitinated receptors are then passed off to the ESCRT‐I and II complexes, with ESCRT‐II also binding to PI(3)P via the GLUE domain of its Vps36/Eap45 subunit. Finally, ESCRT‐II recruits the ESCRT‐III complex, which forms spirals that drive membrane deformation and fission of ILVs (Slagsvold et al, 2005; Chiaruttini et al, 2015). It is the balance between receptor degradation in lysosomes and recycling to the cell surface that modulates the sensitivity of cells to receptor ligands.
Receptor signalling from early endosomes
In addition to regulating membrane traffic within the endolysosomal system, PI(3)P also regulates signalling from early endosomes. TGFβ signalling relies on the recruitment of the PI(3)P‐binding FYVE domain‐containing proteins SARA (SMAD anchor for receptor activation) or endofin to early endosomes, allowing for the phosphorylation of the receptor target SMAD2 and the translocation of the SMAD2–SMAD4 complex to the nucleus to regulate gene transcription (Tsukazaki et al, 1998; Chen et al, 2007). Similarly, mitogenic signalling has been shown to involve the APPL subpopulation of early endosomes as an intermediate relay station en route to the nucleus (Miaczynska et al, 2004; Schenck et al, 2008). Conversely, signalling events can control the production of PI(3)P by Vps34, in turn modulating signalling. For example, pheromone activation of the GPCR Ste2p in S. cerevisiae causes the α subunit of its trimeric G protein complex Gpa1 to bind to and activate the Vps15 subunit of class III Vps34‐PI3K. The resulting increase in endosomal PI(3)P levels serves to recruit the FYVE domain adaptor bud emergence protein 1, an essential component of MAPK signalling from endosomes (Slessareva et al, 2006).
Recent data indicate that early endosomes in addition to PI(3)P also capitalize on a local pool of PI(3,4)P2 synthesized by class II PI3Ks such as PI3KC2γ for signalling. PI3KC2γ associates with active Rab5 to control endosomal Akt2 activation downstream of insulin signalling in hepatocytes (Braccini et al, 2015). Whether similar mechanisms operate in all cell types remains an interesting question for future studies.
Nutrient signalling and the control of autophagy by PI 3‐phosphates
Perhaps the most prominent example of the integration of membrane traffic and signalling within the endolysosomal system is the close association of lysosomal function with the mechanistic target of rapamycin (mTOR) signalling pathway and the control of autophagy by mTOR signalling (Fig 3C). The lysosome is a master regulator directing the cell into either an anabolic or catabolic state by serving as an integrator of the cellular nutrient and energy status. Disruptions in lysosomal signalling are increasingly found to be causative in diseases such as diabetes, cancer and various neurological disorders (Zoncu et al, 2011b; Ferguson, 2015). The surface of the lysosome serves as the site of action for two evolutionarily ancient, key signalling complexes, the mTORC1 and the AMP‐activated protein kinase (AMPK). These complexes are antagonistically regulated to promote anabolic or catabolic processes, respectively (Hardie, 2014; Zhang et al, 2014). mTOR is a serine/threonine protein kinase that when active promotes growth by positively regulating protein translation through the activation of S6 kinase, as well as by inhibiting autophagy via the inhibition of ULK1 (Unc‐51‐like kinase) (Laplante & Sabatini, 2012). mTORC1 acts as a signal integrator at the lysosome, ensuring that all the components necessary for growth are present prior to its activation. The recruitment of mTORC1 to the lysosome, where it will be activated, is mediated via the cellular amino acid status (Sancak et al, 2008; Zoncu et al, 2011a; Bar‐Peled et al, 2013). Once at the lysosome, the activation of mTORC1 is under control of growth factor signalling. The generation of PI(3,4,5)P3 by class I PI3K with the subsequent activation of Akt (see above) results in the downstream activation of mTORC1 on the lysosome (Inoki et al, 2002; Vander Haar et al, 2007). In addition to the role of the class I PI3K in mTORC1 regulation, both class II and III PI3Ks have been implicated in mTORC1 regulation, though their exact roles merit further study. Amino acid stimulation of Vps34 results in the generation of PI(3)P, which in turn recruits phospholipase D1 (PLD1) via its PI(3)P‐binding PX domain. PLD1 then generates a pool of phosphatidic acid that binds to the FKBP12‐rapamycin‐binding domain of mTOR, triggering dissociation of the inhibitory DEPTOR subunit of mTORC1 (Byfield et al, 2005; Nobukuni et al, 2005; Gulati et al, 2008; Yoon et al, 2011, 2015; Bridges et al, 2012). Both the alpha isoform of the class II PI3K and PIKfyve have also been implicated in mTORC1 activation through their sequential activity in generating PI(3,5)P2 that binds the WD40 domain of the Raptor subunit of mTORC1 (Bridges et al, 2012). Furthermore, at least in yeast, PI(3,5)P2 has an additional role in the activation of TORC1 via the recruitment of the TORC1 substrate Sch9 (the homologue of mammalian S6K), which contains a PI(3,5)P2‐binding C2 domain (Jin et al, 2014). While much is known about the activation of mTORC1, it remains unclear how its inactivation is accomplished, although the process is likely to be actively controlled (Demetriades et al, 2014; Menon et al, 2014). Given the prominent role of PI 3‐phosphates in the endolysosomal system, it will be interesting to further explore whether and how these molecules may control the mTORC1 activation status and how such control may be linked to the late endosomal/lysosomal localization and function (Korolchuk et al, 2011).
A key process controlled by lysosomal signalling is macroautophagy (also referred to as autophagy), a major pathway for the degradation of cytoplasmic material. Autophagy is initiated by the engulfment of bulk cytosol (containing protein aggregates) and/or organelles by a cup‐shaped double‐membrane sheet known as the phagophore. The phagophore closes on itself to form the autophagosome, which delivers its contents to the lysosome for degradation by lysosomal hydrolases (Ravikumar et al, 2010). Autophagy is reciprocally regulated by class I and class III PI3Ks (O'Farrell et al, 2013). Receptor‐mediated class I PI3K signalling activates mTORC1 during times of nutrient abundance, leading to an inhibitory phosphorylation on ULK1 and, thus suppression of autophagy. The serine/threonine kinase ULK1 (termed Atg1 in yeast) together with ATG13, FIP200 and ATG101 form part of a multiprotein complex, which plays a pivotal role in the earliest steps of the autophagy process (Stanley et al, 2014). ULK1 also regulates early trafficking steps in autophagy, such as the activation of Rab12 (Xu et al, 2015), and stimulates Vps34 complex I to generate a local pool of PI(3)P on autophagic membranes. The latter is required for formation of the phagophore and for membrane expansion via phosphorylation of the Beclin‐1 subunit (Russell et al, 2013). mTORC1 also inhibits later stages of autophagy via an inhibitory phosphorylation on UVRAG, a component of Vps34 complex II, thereby suppressing autophagosome and endosome maturation (Kim et al, 2015). However, another study found that mTORC1 phosphorylation of UVRAG at different sites activates Vps34, thus generating a pool of PI(3)P on lysosomes necessary for autophagosome–lysosome reformation following starvation (Munson et al, 2015), indicating that the regulation of Vps34 activity by mTORC1 activity is complex. Together, these findings illustrate how class III PI3K Vps34 is regulated downstream of class I PI3K signalling to integrate endolysosomal membrane traffic and autophagic protein turnover with mTORC1 signalling from the lysosomal surface.
A number of PI(3)P effectors on autophagosomes have been identified. During early stages of autophagy, the FYVE domain‐containing PI(3)P‐binding protein DFCP1/TAFF1 associates with the nascent limiting membrane (termed “omegasome”), although the exact role of DCP1/TAFF1 there is unknown (Axe et al, 2008). A major class of PI 3‐P effectors in autophagy are the Atg18/WIPI/PROPPIN proteins, which are defined by a 7‐bladed β‐propeller that binds PI(3)P and PI(3,5)P2 (Nair et al, 2010; Baskaran et al, 2012). There are 4 human WIPI proteins, which have essential but incompletely understood roles in autophagy that may at least be partially distinct (Proikas‐Cezanne et al, 2015). WIPI2 was recently identified as the missing link between PI(3)P production and autophagosome formation as it recruits the ATG12–ATG5–ATG16L complex necessary for the conjugation of the essential autophagy protein LC3 to phosphatidylethanolamine at the omegasome (Dooley et al, 2015). Another FYVE domain‐containing PI(3)P effector is Alfy, which has a role in selective autophagy by linking ubiquitinated protein aggregates destined for degradation to the autophagic machinery (Simonsen et al, 2004; Clausen et al, 2010; Filimonenko et al, 2010). Furthermore, autophagosome biogenesis is promoted by the PI(3)P‐binding PX‐BAR domain protein SNX18, which has been shown to mediate delivery of Atg16L1‐ and LC3‐positive membranes to autophagosome precursors (Knævelsrud et al, 2013). While a crucial role for Vps34 in autophagy is undisputed, there is also evidence for Vps34‐independent autophagy, whereby PI(3)P could be produced either directly or indirectly by other lipid kinases and phosphatases, for example by the class II PI3Ks (Devereaux et al, 2013). Furthermore, a recent study suggests that PI(5)P synthesized by PIKfyve can promote autophagy via known PI(3)P effectors in glucose starvation‐induced autophagy (Vicinanza et al, 2015).
Given the importance of dysregulated autophagy in diseases such as cancer and neurodegenerative disorders, a better understanding of the roles of PI kinases, phosphatases, as well as their products and effectors, provides an exciting avenue for future research.
Future directions
We are only beginning to understand that the seemingly diverse functions of PI 3‐phosphates in the regulation of membrane traffic and cell signalling pathways are owed at least in part to their spatiotemporally controlled formation and turnover at defined subcellular sites. Hence, the same PI 3‐phosphate can exhibit different effects when present at distinct intracellular locations, perhaps even within distinct nanodomains present in the same organelle. Classical genetic loss‐of‐function approaches may therefore not be suited to capture this level of complexity as PI3Ks and phosphatases may also be recruited to several locations and/or activated locally. With the advent of new chemical (e.g. caged PI 3‐phosphates, Box 1) and optogenetic approaches (Box 2), paired with genome engineering to enable expression of tagged proteins at near endogenous levels, the tools are emerging to tackle this complexity and to pave the way for a new era of research on these fascinating lipids. A continuing challenge that remains is the analysis and visualization of such local pools of PI 3‐phosphates. It is likely that the development of ever more sophisticated fluorescent reporters as well as the direct visualization of tagged PI 3‐phosphate effector proteins will allow us to unravel more of nature's PI 3‐phosphate secrets. Finally, we predict that the development of specific inhibitors (Box 3) and sophisticated compounds for local inhibition of defined PI 3‐phosphate‐metabolizing enzymes will allow to untangle the complex interplay between the various PI 3‐phosphates and their roles in integrating cell signalling and membrane traffic between the plasma membrane and the endolysosomal system. Hopefully, these tools will also enable the treatment of some of the most devastating disorders linked to altered PI 3‐phosphate metabolism and signalling ranging from cancer to hereditary muscle disease and neurodegeneration.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Dr. Michael Krauß for a critical reading of the manuscript. The authors' own work is supported by Alexander von Humboldt Foundation and Banting Postdoctoral Fellowships (to A.L.M.) and a grant from the German Research Foundation to V.H. (SFB740/C08).
The EMBO Journal (2016) 35: 561–579
References
- Aki S, Yoshioka K, Okamoto Y, Takuwa N, Takuwa Y (2015) Phosphatidylinositol 3‐kinase class II α‐isoform PI3K‐C2α is required for transforming growth factor β‐induced Smad signaling in endothelial cells. J Biol Chem 290: 6086–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Qusairi L, Prokic I, Amoasii L, Kretz C, Messaddeq N, Mandel JL, Laporte J (2013) Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin‐proteasome pathways. FASEB J 27: 3384–3394 [DOI] [PubMed] [Google Scholar]
- Arcaro A, Khanzada UK, Vanhaesebroeck B, Tetley TD, Waterfield MD, Seckl MJ (2002) Two distinct phosphoinositide 3‐kinases mediate polypeptide growth factor‐stimulated PKB activation. EMBO J 21: 5097–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR (2014) Characterization of VPS34‐IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3‐phosphate‐binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3‐kinase. Biochem J 463: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C (2013) CORVET and HOPS tethering complexes ‐ coordinators of endosome and lysosome fusion. J Cell Sci 126: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93: 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DF, Alvarado‐Kristensson M, González‐García A, Pulido R, Carrera AC (2006) PTEN regulation, a novel function for the p85 subunit of phosphoinositide 3‐kinase. Sci STKE 2006: pe49 [DOI] [PubMed] [Google Scholar]
- Bar‐Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM (2013) A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH (2012) Two‐site recognition of phosphatidylinositol 3‐phosphate by PROPPINs in autophagy. Mol Cell 47: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister MA, Martinu L, Rossman KL, Sondek J, Lemmon MA, Chou MM (2003) Loss of phosphatidylinositol 3‐phosphate binding by the C‐terminal Tiam‐1 pleckstrin homology domain prevents in vivo Rac1 activation without affecting membrane targeting. J Biol Chem 278: 11457–11464 [DOI] [PubMed] [Google Scholar]
- Baumeister MA, Rossman KL, Sondek J, Lemmon MA (2006) The Dbs PH domain contributes independently to membrane targeting and regulation of guanine nucleotide‐exchange activity. Biochem J 400: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley MJ, Taylor GS, Kim SA, Veine DM, Dixon JE, Stuckey JA (2003) Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot‐Marie‐Tooth syndrome. Mol Cell 12: 1391–1402 [DOI] [PubMed] [Google Scholar]
- Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE (2006) Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc Natl Acad Sci USA 103: 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P, Schaffitzel C, Berger I, Ban N, Suter U (2003) Membrane association of myotubularin‐related protein 2 is mediated by a pleckstrin homology‐GRAM domain and a coiled‐coil dimerization module. Proc Natl Acad Sci USA 100: 12177–12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Yoshioka K, Asanuma K, Okamoto Y, Takuwa N, Sasaki T, Takuwa Y (2013) Essential role of class II phosphatidylinositol‐3‐kinase‐C2α in sphingosine 1‐phosphate receptor‐1‐mediated signaling and migration in endothelial cells. J Biol Chem 288: 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau F, Laporte J, Bodin S, Superti‐Furga G, Payrastre B, Mandel JL (2000) Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3‐kinase and phosphatidylinositol 3‐phosphate pathway. Hum Mol Genet 9: 2223–2229 [DOI] [PubMed] [Google Scholar]
- Botelho RJ (2009) Changing phosphoinositides “on the fly”: how trafficking vesicles avoid an identity crisis. BioEssays 31: 1127–1136 [DOI] [PubMed] [Google Scholar]
- Böttcher RT, Stremmel C, Meves A, Meyer H, Widmaier M, Tseng HY, Fässler R (2012) Sorting nexin 17 prevents lysosomal degradation of β1 integrins by binding to the β1‐integrin tail. Nat Cell Biol 14: 584–592 [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira AP, Almeida‐Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT (2015) Endophilin marks and controls a clathrin‐independent endocytic pathway. Nature 517: 460–465 [DOI] [PubMed] [Google Scholar]
- Braccini L, Ciraolo E, Campa CC, Perino A, Longo DL, Tibolla G, Pregnolato M, Cao Y, Tassone B, Damilano F, Laffargue M, Calautti E, Falasca M, Norata GD, Backer JM, Hirsch E (2015) PI3K‐C2γ is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun 6: 7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL (2001) The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3‐phosphate. Mol Cell 8: 829–839 [DOI] [PubMed] [Google Scholar]
- Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR (2012) Phosphatidylinositol 3,5‐bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell 23: 2955–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RA, Domin J, Arcaro A, Waterfield MD, Shepherd PR (1999) Insulin activates the alpha isoform of class II phosphoinositide 3‐kinase. J Biol Chem 274: 14529–14532 [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD (1998) Phosphatidylinositol(3)‐phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell 2: 157–162 [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM (2005) hVps34 is a nutrient‐regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 280: 33076–33082 [DOI] [PubMed] [Google Scholar]
- Cabrera M, Nordmann M, Perz A, Schmedt D, Gerondopoulos A, Barr F, Piehler J, Engelbrecht‐Vandré S, Ungermann C (2014) The Mon1‐Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin‐fold‐Rab interface and association with PI3P‐positive membranes. J Cell Sci 127: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC (2002) The phosphoinositide 3‐kinase pathway. Science 296: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Cao C, Laporte J, Backer JM, Wandinger‐Ness A, Stein MP (2007) Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic 8: 1052–1067 [DOI] [PubMed] [Google Scholar]
- Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger‐Ness A (2008) Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Bio Cell 19: 3334–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM (2014) NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J 15: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagpar RB, Links PH, Pastor MC, Furber LA, Hawrysh AD, Chamberlain MD, Anderson DH (2010) Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3‐kinase. Proc Natl Acad Sci USA 107: 5471–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Wang Z, Ma J, Zhang L, Lu Z (2007) Endofin, a FYVE domain protein, interacts with Smad4 and facilitates transforming growth factor‐beta signaling. J Biol Chem 282: 9688–9695 [DOI] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo‐Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A (2015) Relaxation of loaded ESCRT‐III spiral springs drives membrane deformation. Cell 163: 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH (2007) Mutation of Fig4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397: 621–625 [DOI] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, Johansen T (2010) p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6: 330–344 [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG (2007) Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell 18: 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan‐Garcia S, Gómez‐Fernández JC (2014) Signaling through C2 domains: more than one lipid target. Biochim Biophys Acta 1838: 1536–1547 [DOI] [PubMed] [Google Scholar]
- Cullen PJ (2008) Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol 9: 574–582 [DOI] [PubMed] [Google Scholar]
- Daumke O, Roux A, Haucke V (2014) BAR domain scaffolds in dynamin‐mediated membrane fission. Cell 156: 882–892 [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA (2014) Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux K, Dall'Armi C, Alcazar‐Roman A, Ogasawara Y, Zhou X, Wang F, Yamamoto A, De Camilli P, Di Paolo G (2013) Regulation of mammalian autophagy by class II and III PI 3‐kinases through PI3P synthesis. PLoS ONE 8: e76405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon‐Cardo C, Pandolfi PP (1998) Pten is essential for embryonic development and tumour suppression. Nat Genet 19: 348–355 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC (2015) Regulation of mTORC1 by PI3K signaling. Trends Cell Biol 25: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Strauss M, Brendel M, Bork P (2000) GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane‐associated proteins. Trends Biochem Sci 25: 483–485 [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA (2014) WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12‐5‐16L1. Mol Cell 17: 238–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley HC, Wilson MI, Tooze SA (2015) WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy 11: 190–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella‐Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF et al (2014) Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16: 1069–1070 [DOI] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS (2006a) Phosphoinositide 5‐phosphatase Fig4p is required for both acute rise and subsequent fall in stress‐induced phosphatidylinositol 3,5‐bisphosphate levels. Eukaryot Cell 5: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS (2006b) The Vac14p‐Fig4p complex acts independently of Vac7p and couples PI3, 5P2 synthesis and turnover. J Biol Chem 172: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG (2001) Multivalent endosome targeting by homodimeric EEA1. Mol Cell 8: 947–958 [DOI] [PubMed] [Google Scholar]
- Falasca M, Maffucci T (2012) Regulation and cellular functions of class II phosphoinositide 3‐kinases. Biochem J 443: 587–601 [DOI] [PubMed] [Google Scholar]
- Feng S, Laketa V, Stein F, Rutkowska A, MacNamara A, Depner S, Klingmüller U, Saez‐Rodriguez J, Schultz C (2014) A rapidly reversible chemical dimerizer system to study lipid signaling in living cells. Angew Chem Int Ed Engl 53: 6720–6723 [DOI] [PubMed] [Google Scholar]
- Ferguson SM (2015) Beyond indigestion: emerging roles for lysosome‐based signaling in human disease. Curr Opin Cell Biol 35: 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, Vacher J (2006) Characterization of the murine Inpp4b gene and identification of a novel isoform. Genes 376: 152–161 [DOI] [PubMed] [Google Scholar]
- Fetalvero KM, Yu Y, Goetschkes M, Liang G, Valdez RA, Gould T, Triantafellow E, Bergling S, Loureiro J, Eash J, Lin V, Porter JA, Finan PM, Walsh K, Yang Y, Mao X, Murphy LO (2013) Defective autophagy and mTORC1 signaling in myotubularin null mice. Mol Cell Biol 33: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fili N, Calleja V, Woscholski R, Parker PJ, Larijani B (2006) Compartmental signal modulation: endosomal phosphatidylinositol 3‐phosphate controls endosome morphology and selective cargo sorting. Proc Natl Acad Sci USA 103: 15473–15478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P‐binding protein Alfy. Mol Cell 38: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso R, Barata JT (2015) Kinases, tails and more: regulation of PTEN function by phosphorylation. Methods 77–78: 75–81 [DOI] [PubMed] [Google Scholar]
- Franco I, Gulluni F, Campa CC, Costa C, Margaria JP, Ciraolo E, Martini M, Monteyne D, De Luca E, Germena G, Posor Y, Maffucci T, Marengo S, Haucke V, Falasca M, Perez‐Morga D, Boletta A, Merlo GR, Hirsch E (2014) PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell 28: 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag A, Prajwal P, Shymanets A, Harteneck C, Nürnberg B, Schächtele C, Kubbutat M, Totzke F, Laufer SA (2015) Development of first lead structures for phosphoinositide 3‐kinase‐C2γ inhibitors. J Med Chem 58: 212–221 [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z (2010) The Beclin 1‐VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 20: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin E, Sorkin A (2008) Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin‐dependent endocytosis. Traffic 9: 1776–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG (2009) Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol 10: 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero‐Valero M, Ferrer‐Orta C, Querol‐Audí J, Marin‐Vicente C, Fita I, Gómez‐Fernández JC, Verdaguer N, Corbalán‐García S (2009) Structural and mechanistic insights into the association of PKCalpha‐C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci USA 106: 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G (2008) Amino acids activate mTOR complex 1 via Ca2 + /CaM signaling to hVps34. Cell Metab 7: 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG (2014) AMPK–sensing energy while talking to other signaling pathways. Cell Metab 20: 939–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings BA, Restuccia DF (2012) PI3K‐PKB/Akt pathway. Cold Spring Harb Perspect Biol 4: a011189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK, Emr SD (1990) Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol 10: 6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiles ID, Otsu M, Volinia S, Fry MJ, Gout I, Dhand R, Panayotou G, Ruiz‐Larrea F, Thompson A, Totty NF, Hsuan JJ, Courtneidge SA, Parker PJ, Waterfield MD (1992) Phosphatidylinositol 3‐kinase: structure and expression of the 110 kd catalytic subunit. Cell 70: 419–429 [DOI] [PubMed] [Google Scholar]
- Hnia K, Vaccari I, Bolino A, Laporte J (2012) Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol Med 18: 317–327 [DOI] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M (2005) Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell 121: 437–450 [DOI] [PubMed] [Google Scholar]
- Hsu F, Mao Y (2015) The structure of phosphoinositide phosphatases: insights into substrate specificity and catalysis. Biochim Biophys Acta 1851: 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall‐Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P (2012) Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA 109: E2316–E2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall‐Hagren O, De Camilli P (2015) Detection and manipulation of phosphoinositides. Biochim Biophys Acta 1851: 736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Mlak K, Kanzaki M, Pessin J, Shisheva A (2002) Functional dissection of lipid and protein kinase signals of PIKfyve reveals the role of PtdIns 3,5‐P2 production for endomembrane integrity. J Biol Chem 277: 9206–9211 [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Fenner H, Shisheva A (2009) PIKfyve‐ArPIKfyve‐Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem 284: 35794–35806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A (2011) The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve‐/‐ embryos but normality of PIKfyve+/‐ mice. J Biol Chem 286: 13404–13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657 [DOI] [PubMed] [Google Scholar]
- Itoh F, Divecha N, Brocks L, Oomen L, Janssen H, Calafat J, Itoh S, Pt DP (2002) The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF‐beta/Smad signalling. Genes Cells 7: 321–331 [DOI] [PubMed] [Google Scholar]
- Jean S, Cox S, Schmidt EJ, Robinson FL, Kiger A (2012) Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol Biol Cell 23: 2723–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S, Kiger AA (2012) Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol 13: 463–470 [DOI] [PubMed] [Google Scholar]
- Jean S, Kiger AA (2014) Classes of phosphoinositide 3‐kinases at a glance. J Cell Sci 127: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J 27: 3221–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, Weisman LS (2014) Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell 25: 1171–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Sylvestersen KB, Choudhary C, Nguyen S, Mann M, Nielsen ML (2014) Specificity and commonality of the phosphoinositide‐binding proteome analysed by quantitative mass spectrometry. Cell Rep 6: 578–591 [DOI] [PubMed] [Google Scholar]
- Kalaidzidis I, Miaczynska M, Brewińska‐Olchowik M, Hupalowska A, Ferguson C, Parton RG, Kalaidzidis Y, Zerial M (2015) APPL endosomes are not obligatory endocytic intermediates but act as stable cargo‐sorting compartments. J Cell Biol 211: 123–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Müller R, Subramanian D, Schultz C, Laporte J, Haucke V (2016) A phosphoinositide conversion mechanisms for exit from endosomes. Nature 529: 408–412 [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001) Two distinct Vps34 phosphatidylinositol 3‐kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH (2015) mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 57: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knævelsrud H, Søreng K, Raiborg C, Håberg K, Rasmuson F, Brech A, Liestøl K, Rusten TE, Stenmark H, Neufeld TP, Carlsson SR, Simonsen A (2013) Membrane remodeling by the PX‐BAR protein SNX18 promotes autophagosome formation. J Cell Biol 202: 331–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K, Geering B, Vanhaesebroeck B (2009) Regulation of phosphoinositide 3‐kinase expression in health and disease. Trends Biochem Sci 34: 115–127 [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O'Kane CJ, Deretic V, Rubinsztein DC (2011) Lysosomal positioning coordinates cellular nutrient response. Nat Cell Biol 13: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis NT, Manifava M, Schoenfelder P, Rotondo S (2012) How phosphoinositide 3‐phosphate controls growth downstream of amino acids and autophagy downstream of amino acid withdrawal. Biochem Soc Trans 40: 37–43 [DOI] [PubMed] [Google Scholar]
- Kurokawa T, Takasuga S, Sakata S, Yamaguchi S, Horie S, Homma KJ, Sasaki T, Okamura Y (2012) 3′ Phosphatase activity toward phosphatidylinositol 3,4‐bisphosphate [PI(3,4)P2] by voltage‐sensing phosphatase (VSP). Proc Natl Acad Sci USA 109: 10089–10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG (2010) Translation of the phosphoinositide code by PI effectors. Nat Chem Biol 6: 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laketa V, Zarbakhsh S, Morbier E, Subramanian D, Dinkel C, Brumbaugh J, Zimmermann P, Pepperkok R, Schultz C (2009) Membrane‐permeant phosphoinositide derivatives as modulators of growth factor signaling and neurite outgrowth. Chem Biol 16: 1190–1196 [DOI] [PubMed] [Google Scholar]
- Laketa V, Zarbakhsh S, Traynor‐Kaplan A, Macnamara A, Subramanian D, Putyrski M, Mueller R, Nadler A, Mentel M, Saez‐Rodriguez J, Pepperkok R, Schultz C (2014) PIP3 induces the recycling of receptor tyrosine kinases. Sci Signal 7: ra5 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2012) mTOR Signaling. Cold Spring Harb Perspect Biol 4: a011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Bedez F, Bolino A, Mandel JL (2003) Myotubularins, a large disease‐associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 15: R285–R292 [DOI] [PubMed] [Google Scholar]
- Lauffer BE, Melero C, Temkin P, Lei C, Hong W, Kortemme T, von Zastrow M (2010) SNX27 mediates PDZ‐directed sorting from endosomes to the plasma membrane. J Cell Biol 190: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibiger B, Moede T, Uhles S, Barker CJ, Creveaux M, Domin J, Berggren PO, Leibiger IB (2010) Insulin‐feedback via PI3K‐C2alpha activated PKBalpha/Akt1 is required for glucose‐stimulated insulin secretion. FASEB J 24: 1824–1837 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2003) Phosphoinositide recognition domains. Traffic 4: 201–213 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid‐binding domains. Nat Rev Mol Cell Biol 9: 99–111 [DOI] [PubMed] [Google Scholar]
- Levin R, Grinstein S, Schlam D (2015) Phosphoinositides in phagocytosis and macropinocytosis. Biochim Biophys Acta 1851: 805–823 [DOI] [PubMed] [Google Scholar]
- Liu L, Song X, He D, Komma C, Kita A, Virbasius JV, Huang G, Bellamy HD, Miki K, Czech MP, Zhou GW (2006) Crystal structure of the C2 domain of class II phosphatidylinositide 3‐kinase C2alpha. J Biol Chem 281: 4254–4260 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Urbé S, Clague MJ (2005) Analysis of phosphoinositide binding domain properties within the myotubularin‐related protein MTMR3. J Cell Sci 118: 2005–2012 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Urbé S, Clague MJ (2006) Systematic analysis of myotubularins: heteromeric interactions, subcellular localisation and endosome related functions. J Cell Sci 119: 2953–2959 [DOI] [PubMed] [Google Scholar]
- Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, Li M, Yue Z (2014) NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L‐linked phosphatidylinositol‐3 kinase III activity. Nat Commun 22: 3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Terasaka S, Mochizuki Y, Kawai K, Ikeda Y, Araki N, Skolnik EY, Taguchi T, Arai H (2014) Sequential breakdown of 3‐phosphorylated phosphoinositides is essential for the completion of macropinocytosis. Proc Natl Acad Sci USA 111: E978–E987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Fairn GD (2015) Molecular probes to visualize the location, organization and dynamics of lipids. J Cell Sci 127: 4801–4812 [DOI] [PubMed] [Google Scholar]
- Mak LH, Woscholski R (2015) Targeting PTEN using small molecule inhibitors. Methods 77–78: 63–68 [DOI] [PubMed] [Google Scholar]
- Mayinger P (2012) Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta 1821: 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS (2014) Phosphatidylinositol 3,5‐bisphosphate: low abundance, high significance. BioEssays 36: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes MP, Waddell L, Lenk GM, Kaur S, MacArthur DG, Meisler MH, Clarke NF (2014) Whole exome sequencing identifies three recessive Fig4 mutations in an apparently dominant pedigree with Charcot‐Marie‐Tooth disease. Neuromuscul Disord 24: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD (2014) Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel M, Laketa V, Subramanian D, Gillandt H, Schultz C (2011) Photoactivatable and cell‐membrane‐permeable phosphatidylinositol 3,4,5‐trisphosphate. Angew Chem Int Ed Engl 50: 3811–3814 [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler‐Joseph S, Habermann B, Wilm M, Parton RG, Zerial M (2004) APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116: 445–456 [DOI] [PubMed] [Google Scholar]
- Michell RH, Heath VL, Lemmon MA, Dove SK (2006) Phosphatidylinositol 3,5‐bisphosphate: metabolism and cellular functions. Trends Biochem Sci 31: 52–63 [DOI] [PubMed] [Google Scholar]
- Mima J, Wickner W (2009) Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc Natl Acad Sci USA 106: 16191–16196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K (2000) Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol Cell Biol 20: 9346–9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG (2015) mTOR activates the VPS34‐UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 34: 2272–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naga Prasad SV, Laporte SA, Chamberlain D, Caron MG, Barak L, Rockman HA (2002) Phosphoinositide 3‐kinase regulates beta2‐adrenergic receptor endocytosis by AP‐2 recruitment to the receptor/beta‐arrestin complex. J Cell Biol 158: 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A, Bencze G, Cho H, Zheng W, Tocilj A, Elkayam E, Faehnle CR, Jaber N, Pratt CP, Chen M, Zong WX, Marks MS, Joshua‐Tor L, Pappin DJ, Trotman LC (2015) PTEN functions by recruitment to cytoplasmic vesicles. Mol Cell 58: 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Cao Y, Xie Z, Klionsky DJ (2010) Roles of the lipid‐binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem 285: 11476–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughtin MJ, Sheffield DA, Rahman P, Hughes WE, Gurung R, Stow JL, Nandurkar HH, Dyson JM, Mitchell CA (2010) The myotubularin phosphatase MTMR4 regulates sorting from early endosomes. J Cell Sci 123: 3071–3083 [DOI] [PubMed] [Google Scholar]
- Nicot AS, Laporte J (2008) Endosomal phosphoinositides and human diseases. Traffic 9: 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G (2005) Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH‐kinase. Proc Natl Acad Sci USA 102: 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht‐Vandré S, Ungermann C (2010) The Mon1‐Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 20: 1654–1659 [DOI] [PubMed] [Google Scholar]
- O'Farrell F, Rusten TE, Stenmark H (2013) Phosphoinositide 3‐kinases as accelerators and brakes of autophagy. FEBS J 280: 6322–6337 [DOI] [PubMed] [Google Scholar]
- Parachoniak CA, Park M (2012) Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol 22: 231–240 [DOI] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR (2010) The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11: 9–22 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Pirruccello M, De Camilli P (2012) Inositol 5‐phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem Sci 37: 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Stenmark H (2011) Endocytosis and signaling. Curr Opin Cell Biol 23: 393–403 [DOI] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA (2010) Mammalian Atg18 (WIPI2) localizes to omegasome‐anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522 [DOI] [PubMed] [Google Scholar]
- Posor Y, Eichhorn‐Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noé F, Haucke V (2013) Spatiotemporal control of endocytosis by phosphatidylinositol‐3,4‐bisphosphate. Nature 499: 233–237 [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A (2010) Identification of the switch in early‐to‐late endosome transition. Cell 141: 497–508 [DOI] [PubMed] [Google Scholar]
- Proikas‐Cezanne T, Takacs Z, Dönnes P, Kohlbacher O (2015) WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci 128: 207–217 [DOI] [PubMed] [Google Scholar]
- Rabinovsky R, Pochanard P, McNear C, Brachmann SM, Duke‐Cohan JS, Garraway LA, Sellers WR (2009) p85 Associates with unphosphorylated PTEN and the PTEN‐associated complex. Mol Cell Biol 29: 5377–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H (2001) FYVE and coiled‐coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 114: 2255–2263 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin‐coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Schink KO, Stenmark H (2013) Class III phosphatidylinositol 3‐kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J 280: 2730–2742 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia‐Arencibia M, Green‐Thompson ZW, Jimenez‐Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90: 1383–1435 [DOI] [PubMed] [Google Scholar]
- Razidlo GL, Katafiasz D, Taylor GS (2011) Myotubularin regulates Akt‐dependent survival signaling via phosphatidylinositol 3‐phosphate. J Biol Chem 286: 20005–20019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CR, Chen PH, Srinivasan S, Aguet F, Mettlen M, Schmid SL (2015) Crosstalk between Akt/GSK3β signaling and dynamin‐1 regulates clathrin‐mediated endocytosis. EMBO J 34: 2132–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversi A, Loeser E, Subramanian D, Schultz C, De Renzis S (2014) Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J Cell Biol 205: 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, Marquette JP, El‐Ahmad Y, Filoche‐Romme B, Schio L, Garcia‐Echeverria C, Goulaouic H, Pasquier B (2014) A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 10: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J (2002) A crystallographic view of interactions between Dbs and Cdc42: PH domain‐assisted guanine nucleotide exchange. EMBO J 21: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350: aac7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL (2013) ULK1 induces autophagy by phosphorylating Beclin‐1 and activating VPS34 lipid kinase. Nat Cell Biol 15: 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H (2010) PtdIns(3)P controls cytokinesis through KIF13A‐mediated recruitment of FYVE‐CENT to the midbody. Nat Cell Biol 12: 362–371 [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP (2008) Tenets of PTEN tumor suppression. Cell 133: 403–414 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar‐Peled L, Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signalling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A (2009) Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res 48: 307–343 [DOI] [PubMed] [Google Scholar]
- Sasaki J, Kofuji S, Itoh R, Momiyama T, Takayama K, Murakami H, Chida S, Tsuya Y, Takasuga S, Eguchi S, Asanuma K, Horie Y, Miura K, Davies EM, Mitchell C, Yamazaki M, Hirai H, Takenawa T, Suzuki A, Sasaki T (2010) The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 465: 497–501 [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Shisheva A (1999) PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5‐phosphoinositides. Effect of insulin. J Biol Chem 274: 21589–21597 [DOI] [PubMed] [Google Scholar]
- Schenck A, Goto‐Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M (2008) The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell 133: 486–497 [DOI] [PubMed] [Google Scholar]
- Schmees C, Villaseñor R, Zheng W, Ma H, Zerial M, Heldin CH, Hellberg C (2012) Macropinocytosis of the PDGF β‐receptor promotes fibroblast transformation by H‐RasG12V. Mol Biol Cell 23: 2571–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD (1993) Phosphatidylinositol 3‐kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260: 88–91 [DOI] [PubMed] [Google Scholar]
- Seet LF, Hong W (2006) The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta 1761: 878–896 [DOI] [PubMed] [Google Scholar]
- Sheppard K, Kinross KM, Solomon B, Pearson RB, Phillips WA (2012) Targeting PI3 kinase/AKT/mTOR signaling in cancer. Crit Rev Oncog 17: 69–95 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippé R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394: 494–498 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H (2004) Alfy, a novel FYVE‐domain‐containing protein associated with protein granules and autophagic membranes. J Cell Sci 117: 4239–4251 [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H (2005) Eap45 in mammalian ESCRT‐II binds ubiquitin via a phosphoinositide‐interacting GLUE domain. J Biol Chem 280: 19600–19606 [DOI] [PubMed] [Google Scholar]
- Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG (2006) Activation of the phosphatidylinositol 3‐kinase Vps34 by a G protein alpha subunit at the endosome. Cell 126: 191–203 [DOI] [PubMed] [Google Scholar]
- Snyder JT, Rossman KL, Baumeister MA, Pruitt WM, Siderovski DP, Der CJ, Lemmon MA, Sondek J (2001) Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J Biol Chem 276: 45868–45875 [DOI] [PubMed] [Google Scholar]
- Solinger JA, Spang A (2013) Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J 280: 2743–2757 [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP (2012) The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 13: 283–296 [DOI] [PubMed] [Google Scholar]
- Sorkin A, Goh LK (2009) Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res 315: 683–696 [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M (2009) Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 10: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli L, Lindsay YE, Leslie NR (2015) PTEN inhibitors: an evaluation of current compounds. Adv Biol Regul 57: 102–111 [DOI] [PubMed] [Google Scholar]
- Stanley RE, Ragusa MJ, Hurley JH (2014) The beginning of the end: how scaffolds nucleate autophagosome biogenesis. Trends Cell Biol 24: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D'Arrigo A (1996) Endosomal localization of the autoantigen EEA1 is mediated by a zinc‐binding FYVE finger. J Biol Chem 271: 24048–24054 [DOI] [PubMed] [Google Scholar]
- Stephens L, Smrcka A, Cooke FT, Jackson TR, Sternweis PC, Hawkins PT (1994) A novel phosphoinositide 3 kinase activity in myeloid‐derived cells is activated by G protein βγ subunits. Cell 77: 83–93 [DOI] [PubMed] [Google Scholar]
- Stoyanov B, Volinia S, Hanck T, Rubio I, Loubtchenkov M, Malek D, Stoyanova S, Vanhaesebroeck B, Dhand R, Nirnberg B, Gierschik P, Seedorf K, Hsuan JJ, Waterfield MD, Wetzker R (1995) Cloning and characterization of a G protein‐activated human phosphoinositide‐3 kinase. Science 269: 690–693 [DOI] [PubMed] [Google Scholar]
- Strømhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 15: 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian D, Laketa V, Müller R, Tischer C, Zarbakhsh S, Pepperkok R, Schultz C (2010) Activation of membrane‐permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat Chem Biol 6: 324–326 [DOI] [PubMed] [Google Scholar]
- Taguchi‐Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T (2010) Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 11: 468–478 [DOI] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE (2000) Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3‐phosphate. Proc Natl Acad Sci USA 97: 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM (2012) Insights into the PX (phox‐homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J 441: 39–59 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (2006) ESCRT‐I core and ESCRT‐II GLUE domain structures reveal role for GLUE in linking to ESCRT‐I and membranes. Cell 125: 99–111 [DOI] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DM (2002) High‐resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)‐trisphosphate. Curr Biol 23: 1256–1262 [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD (2011) Light‐based feedback for controlling intracellular signaling dynamics. Nat Methods 8: 837–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ (2007) SNX4 coordinates endosomal sorting of TfnR with dynein‐mediated transport into the endocytic recycling compartment. Nat Cell Biol 9: 1370–1380 [DOI] [PubMed] [Google Scholar]
- Tseng HY, Thorausch N, Ziegler T, Meves A, Fässler R, Böttcher RT (2014) Sorting nexin 31 binds multiple β integrin cytoplasmic domains and regulates β1 integrin surface levels and stability. J Mol Biol 426: 3180–3194 [DOI] [PubMed] [Google Scholar]
- Tsujita K, Itoh T, Ijuin T, Yamamoto A, Shisheva A, Laporte J, Takenawa T (2004) Myotubularin regulates the function of the late endosome through the gram domain‐phosphatidylinositol 3,5‐bisphosphate interaction. J Biol Chem 279: 13817–13824 [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell 95: 779–791 [DOI] [PubMed] [Google Scholar]
- Vaccari I, Carbone A, Previtali SC, Mironova YA, Alberizzi V, Noseda R, Rivellini C, Bianchi F, Del Carro U, D'Antonio M, Lenk GM, Wrabetz L, Giger RJ, Meisler MH, Bolino A (2015) Loss of Fig4 in both Schwann cells and motor neurons contributes to CMT4J neuropathy. Hum Mol Genet 24: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadas O, Burke JE, Zhang X, Berndt A, Williams RL (2011) Structural basis for activation and inhibition of class I phosphoinositide 3‐kinases. Sci Signal 4: re2 [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH (2007) Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 9: 316–323 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, Zvelebil MJ, Higashi K, Volinia S, Downward J, Waterfield MD (1997) P110delta, a novel phosphoinositide 3‐kinase in leukocytes. Proc Natl Acad Sci USA 94: 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavaré JM, Cullen PJ (1998) Insulin‐dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3‐kinase. Curr Biol 8: 463–466 [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Brandom KG, Lawrence JL (2004) Centaurin‐alpha1 is an in vivo phosphatidylinositol 3,4,5‐trisphosphate‐dependent GTPase‐activating protein for ARF6 that is involved in actin cytoskeleton organization. J Biol Chem 279: 6205–6208 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet‐Guibert J, Graupera M, Bilanges B (2010a) The emerging mechanisms of isoform‐specific PI3K signalling. Nat Rev Mol Cell Biol 11: 329–341 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Vogt PK, Rommel C (2010b) PI3K: from the bench to the clinic and back. Curr Top Microbiol Immunol 347: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas‐Cezanne T, Laporte J, Deretic V (2009) Control of autophagy initiation by phosphoinositide 3‐phosphatase Jumpy. EMBO J 28: 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC (2015) PI(5)P regulates autophagosome biogenesis. Mol Cell 57: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N, Patton WA, Moss J, Vaughan M, Lefkowitz RJ, Premont RT (2000) GIT proteins, A novel family of phosphatidylinositol 3,4, 5‐trisphosphate‐stimulated GTPase‐activating proteins for ARF6. J Biol Chem 275: 13901–13906 [DOI] [PubMed] [Google Scholar]
- Walker SM, Downes CP, Leslie NR (2001) TPIP: a novel phosphoinositide 3‐phosphatase. Biochem J 360: 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Kimber WA, Fleming IN, Leslie NR, Downes CP, Lucocq JM (2004) Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4‐bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem J 377: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JR, Cullen PJ (2014) Membrane‐associated cargo recycling by tubule‐based endosomal sorting. Semin Cell Dev Biol 31: 40–47 [DOI] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument‐Bromage H, Tempst P, Hawkins PT, Stephens LR (2002) P‐Rex1, a PtdIns(3,4,5)P3‐ and Gbetagamma‐regulated guanine‐nucleotide exchange factor for Rac. Cell 108: 809–821 [DOI] [PubMed] [Google Scholar]
- Woolley JF, Dzneladze I, Salmena L (2015) Phosphoinositide signaling in cancer: INPP4B Akt(s) out. Trends Mol Med 21: 530–532 [DOI] [PubMed] [Google Scholar]
- Wu Y, Cheng S, Zhao H, Zou W, Yoshina S, Mitani S, Zhang H, Wang X (2014) PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep 15: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wickner W (2010) Phosphoinositides function asymmetrically for membrane fusion, promoting tethering and 3Q‐SNARE subcomplex assembly. J Biol Chem 285: 39359–39365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Fotouhi M, McPherson PS (2015) Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep 16: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Du G, Backer JM, Frohman MA, Chen J (2011) Class III PI‐3‐kinase activates phospholipase D in an amino acid‐sensing mTORC1 pathway. J Cell Biol 195: 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Rosenberger CL, Wu C, Truong N, Sweedler JV, Chen J (2015) Rapid mitogenic regulation of the mTORC1 inhibitor, DEPTOR, by phosphatidic acid. Mol Cell 58: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Yoshida K, Cui H, Wakayama T, Takuwa N, Okamoto Y, Du W, Qi X, Asanuma K, Sugihara K, Aki S, Miyazawa H, Biswas K, Nagakura C, Ueno M, Iseki S, Schwartz RJ, Okamoto H, Sasaki T, Matsui O et al (2012) Endothelial PI3K‐C2α, a class II PI3K, has an essential role in angiogenesis and vascular barrier function. Nat Med 18: 1560–1569 [DOI] [PubMed] [Google Scholar]
- Yuan HX, Russell RC, Guan KL (2013) Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress‐induced autophagy. Autophagy 9: 1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS (2007) Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5‐bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA 104: 17518–17523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCartney AJ, Zolov SN, Ferguson CJ, Meisler MH, Sutton MA, Weisman LS (2012) Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P2 and PI(5)P. EMBO J 31: 3442–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu JW, Yin H, Lin SY, Lin SC (2014) The lysosomal v‐ATPase‐Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20: 526–540 [DOI] [PubMed] [Google Scholar]
- Zhu J, Hou T, Mao X (2015) Discovery of selective phosphatidylinositol 3‐kinase inhibitors to treat hematological malignancies. Drug Discov Today 20: 988–994 [DOI] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Balkin DM, Pirruccello M, Toomre D, De Camilli P (2009) A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell 136: 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar‐Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM (2011a) mTORC1 senses lysosomal amino acids through an inside‐out mechanism that requires the vacuolar H(+)‐ATPase. Science 334: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM (2011b) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang C, Marjanovic J, Kisseleva MV, Majerus PW, Wilson MP (2012) Myotubularin‐related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8. Proc Natl Acad Sci USA 109: 9539–9544 [DOI] [PMC free article] [PubMed] [Google Scholar]


