Abstract
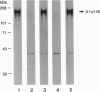
We have cloned a gene encoding a DNA-binding protein by Southwestern screening of a murine cDNA library with a double-stranded oligonucleotide containing the sequence from the bidirectional promoter of the alpha 1 and alpha 2 collagen IV genes. The middle portion of this 1131-amino acid protein has a region homologous to bacterial DNA ligases, and the more carboxyl portion contains several domains homologous to p40, p38, p37, and p36.5 subunits of activator 1 (A1, also called replication factor C), a human replication protein complex. Western blotting revealed that antiserum generated against part of the recombinant protein reacted specifically with the 145-kDa component of the purified human A1 complex, indicating that it is the murine counterpart of the A1 p145. Characterization of the DNA-binding activity of the recombinant fusion protein by gel mobility-shift assay revealed that it had a preference for a run of pyrimidines on one strand. Deletion analysis using recombinant proteins revealed that the DNA ligase-like domain was required for DNA-binding activity. The finding that the region required for the binding of murine A1 p145 to DNA has similarity to a domain found in DNA ligases suggests that this region may be utilized by both proteins in recognizing DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barany F., Gelfand D. H. Cloning, overexpression and nucleotide sequence of a thermostable DNA ligase-encoding gene. Gene. 1991 Dec 20;109(1):1–11. doi: 10.1016/0378-1119(91)90582-v. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. Molecular characterisation of the DNA ligase gene, CDC17, from the fission yeast Schizosaccharomyces pombe. Eur J Biochem. 1987 Feb 2;162(3):659–667. doi: 10.1111/j.1432-1033.1987.tb10688.x. [DOI] [PubMed] [Google Scholar]
- Biswas E. E., Ewing C. M., Biswas S. B. Characterization of the DNA-dependent ATPase and a DNA unwinding activity associated with the yeast DNA polymerase alpha complex. Biochemistry. 1993 Mar 30;32(12):3020–3026. doi: 10.1021/bi00063a013. [DOI] [PubMed] [Google Scholar]
- Bruggeman L. A., Burbelo P. D., Yamada Y., Klotman P. E. A novel sequence in the type IV collagen promoter binds nuclear proteins from Engelbreth-Holm-Swarm tumor. Oncogene. 1992 Aug;7(8):1497–1502. [PubMed] [Google Scholar]
- Burbelo P. D., Martin G. R., Yamada Y. Alpha 1(IV) and alpha 2(IV) collagen genes are regulated by a bidirectional promoter and a shared enhancer. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9679–9682. doi: 10.1073/pnas.85.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2516–2520. doi: 10.1073/pnas.89.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Studies of the cloned 37-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5211–5215. doi: 10.1073/pnas.89.12.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherney B. W., McBride O. W., Chen D. F., Alkhatib H., Bhatia K., Hensley P., Smulson M. E. cDNA sequence, protein structure, and chromosomal location of the human gene for poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8370–8374. doi: 10.1073/pnas.84.23.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Tsunasawa S., Sakiyama F., Nakata A. Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):1–7. doi: 10.1007/BF00330179. [DOI] [PubMed] [Google Scholar]
- Kameshita I., Matsuda Z., Taniguchi T., Shizuta Y. Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J Biol Chem. 1984 Apr 25;259(8):4770–4776. [PubMed] [Google Scholar]
- Kaytes P., Wood L., Theriault N., Kurkinen M., Vogeli G. Head-to-head arrangement of murine type IV collagen genes. J Biol Chem. 1988 Dec 25;263(36):19274–19277. [PubMed] [Google Scholar]
- Kim C., Snyder R. O., Wold M. S. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992 Jul;12(7):3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867–19870. [PubMed] [Google Scholar]
- Kurosaki T., Ushiro H., Mitsuuchi Y., Suzuki S., Matsuda M., Matsuda Y., Katunuma N., Kangawa K., Matsuo H., Hirose T. Primary structure of human poly(ADP-ribose) synthetase as deduced from cDNA sequence. J Biol Chem. 1987 Nov 25;262(33):15990–15997. [PubMed] [Google Scholar]
- Lauer G., Rudd E. A., McKay D. L., Ally A., Ally D., Backman K. C. Cloning, nucleotide sequence, and engineered expression of Thermus thermophilus DNA ligase, a homolog of Escherichia coli DNA ligase. J Bacteriol. 1991 Aug;173(16):5047–5053. doi: 10.1128/jb.173.16.5047-5053.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kwong A. D., Pan Z. Q., Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase delta. J Biol Chem. 1991 Jan 5;266(1):594–602. [PubMed] [Google Scholar]
- Lee S. H., Pan Z. Q., Kwong A. D., Burgers P. M., Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem. 1991 Nov 25;266(33):22707–22717. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- Nossal N. G. Protein-protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J. 1992 Feb 1;6(3):871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. Accessory protein function in the DNA polymerase III holoenzyme from E. coli. Bioessays. 1992 Feb;14(2):105–111. doi: 10.1002/bies.950140206. [DOI] [PubMed] [Google Scholar]
- O'Donnell M., Onrust R., Dean F. B., Chen M., Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993 Jan 11;21(1):1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onrust R., Stukenberg P. T., O'Donnell M. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J Biol Chem. 1991 Nov 15;266(32):21681–21686. [PubMed] [Google Scholar]
- Pan Z. Q., Chen M., Hurwitz J. The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):6–10. doi: 10.1073/pnas.90.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shark K. B., Conway T. Cloning and molecular characterization of the DNA ligase gene (lig) from Zymomonas mobilis. FEMS Microbiol Lett. 1992 Sep 1;75(1):19–26. doi: 10.1016/0378-1097(92)90450-3. [DOI] [PubMed] [Google Scholar]
- Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Uchida K., Morita T., Sato T., Ogura T., Yamashita R., Noguchi S., Suzuki H., Nyunoya H., Miwa M., Sugimura T. Nucleotide sequence of a full-length cDNA for human fibroblast poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1987 Oct 29;148(2):617–622. doi: 10.1016/0006-291x(87)90921-1. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., LaMarco K. L., Johnson P. F., Landschulz W. H., McKnight S. L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988 Jul;2(7):801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- Wickner S. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3511–3515. doi: 10.1073/pnas.73.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. C., Reddy M. K., von Hippel P. H. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry. 1992 Sep 22;31(37):8675–8690. doi: 10.1021/bi00152a001. [DOI] [PubMed] [Google Scholar]
- Zahradka P., Ebisuzaki K. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur J Biochem. 1982 Oct;127(3):579–585. [PubMed] [Google Scholar]