Abstract
Among major reactive oxygen species (ROS), hydrogen peroxide (H2O2) exhibits dual roles in plant metabolism. Low levels of H2O2 modulate many biological/physiological processes in plants; whereas, its high level can cause damage to cell structures, having severe consequences. Thus, steady-state level of cellular H2O2 must be tightly regulated. Glutathione peroxidases (GPX) and ascorbate peroxidase (APX) are two major ROS-scavenging enzymes which catalyze the reduction of H2O2 in order to prevent potential H2O2-derived cellular damage. Employing bioinformatics approaches, this study presents a comparative evaluation of both GPX and APX in 18 different plant species, and provides valuable insights into the nature and complex regulation of these enzymes. Herein, (a) potential GPX and APX genes/proteins from 18 different plant species were identified, (b) their exon/intron organization were analyzed, (c) detailed information about their physicochemical properties were provided, (d) conserved motif signatures of GPX and APX were identified, (e) their phylogenetic trees and 3D models were constructed, (f) protein-protein interaction networks were generated, and finally (g) GPX and APX gene expression profiles were analyzed. Study outcomes enlightened GPX and APX as major H2O2-scavenging enzymes at their structural and functional levels, which could be used in future studies in the current direction.
Keywords: ROS, signal transduction, antioxidant, peroxisome, chloroplast, mitochondria
Introduction
Reactive oxygen species (ROS), once perceived as toxic by-products, were known to cause oxidative damage in cells (Mittler et al., 2004; Suzuki and Mittler, 2006). Later, novel regulatory roles of these species were revealed in a wide range of biological processes such as cell signaling, growth, development, programmed cell death, and plant responses to various biotic/abiotic stress factors (Mullineaux and Karpinski, 2002; Uzilday et al., 2014). H2O2 is an endogenous ROS species known to play a dual role in plants, where it is beneficial at low concentrations but lethal at higher levels (Petrov and Van Breusegem, 2012). Nevertheless, at steady state levels, H2O2 acts as signaling molecule inducing the signal transduction mechanism to produce various cellular responses. Interestingly, pre-treatment of plants with H2O2 makes them more tolerant to biotic/abiotic stresses (Hossain et al., 2015). H2O2 was also noted for its regulatory functions in photosynthesis, cell cycle, development, senescence, and apoptosis (Mittler et al., 2004; Petrov and Van Breusegem, 2012). H2O2 has been accepted as a central component of signal transduction pathways in plant-adaptation to altered environmental conditions as it is both the only ROS with high permeability across membranes (that enables the transport of signals to distant sites) and its high stability when compared to other ROS with ~1 ms half-life (Bienert et al., 2007; Dynowski et al., 2008; Petrov and Van Breusegem, 2012). On the other hand, when the delicate balance between production and scavenging of H2O2 is disturbed, its overproduction results in significant damage to cell structures (Anjum et al., 2015; Sofo et al., 2015). To overcome H2O2-related cellular damage, aerobic organisms have developed various antioxidant machineries with enzymatic and non-enzymatic components. Ascorbate peroxidase (APX), glutathione peroxidase (GPX), and catalase (CAT) are the main enzymes responsible for suppressing toxic levels of H2O2 (Apel and Hirt, 2004). However, APX may have pivotal roles in ROS-scavenging because even very low concentrations are sufficient for H2O2 decomposition (Anjum et al., 2014; Sofo et al., 2015).
APX (EC, 1.11.1.11) belongs to the plant-type heme peroxidase superfamily in plants (Lazzarotto et al., 2011). Genome-wide studies demonstrated that APX in higher plants is encoded by multigenic families. Arabidopsis was reported to contain nine APX genes; whereas, rice has eight and tomato seven (Chew et al., 2003; Teixeira et al., 2004; Najami et al., 2008). Different isoforms are classified into sub-families according to their subcellular localization. Transmembrane domains in N- and C- terminal regions, as well as organelle-specific target molecules are the primary determinants in target localization of APXs (Ishikawa et al., 1998; Negi, 2011). Among nine APX genes identified in Arabidopsis, three were found to be encoded in cytosol whereas the other six were distributed in stroma, thylakoid, and peroxisome (Chew et al., 2003; Mittler et al., 2004). In rice, chloroplastic isoforms were expressed by three genes, cytosolic and peroxisomal forms were both encoded by two genes, and one gene was for the mitochondrial APX (Teixeira et al., 2006; Anjum et al., 2014). APX activity was also reported to increase under various stress conditions. For example, APX is differentially upregulated in response to heavy metal, drought, water, and heat stress (Sharma and Dubey, 2005; Koussevitzky et al., 2008; Yang et al., 2008; Anjum et al., 2014). In a previous study, Arg-38, Glu-65, Asn-71, and Asp208 residues were reported to be conserved among the entire APX family and known to be important in ligand (heme)-binding (Welinder, 1992). In addition to enzymatic properties, structural investigations on catalytic domains of the enzymes have been also performed. Three-dimensional structures of cAPX, sAPX, and their substrates showed the relationship between loop structure and stability in the absence of ascorbate (AsA; Yabuta et al., 2000; Anjum et al., 2014). The mitochondrial and chloroplastic APXs (< 30 s) have shorter half inactivation times (>1 h) compared to cytosolic and peroxisomal isoforms, which makes them more sensitive in either low concentrations or the absence of AsA (Caverzan et al., 2012; Anjum et al., 2014). Another important enzyme in H2O2-scavenging is the GPX from the non-heme containing peroxidase family (Bela et al., 2015). In Arabidopsis, eight GPX genes were reported (Milla et al., 2003; Koua et al., 2009). Based on in silico analysis, GPXs were predicted in chloroplast, mitochondria, cytosol, and ER localizations (Rouhier and Jacquot, 2005), and demonstrated high level of sequence similarity with strictly conserved cysteines and motifs (Dietz, 2011). Plant GPXs have cysteine residue in their active site (Koua et al., 2009), which is functional in both glutathione (GSH) and thiol peroxidase classes of the non-heme family. GPXs were also reported to be involved in stress responses. Many studies have demonstrated the significant increase in mRNA levels of GPXs under various abiotic/abiotic stress conditions such as oxidative stress, pathogen attack, metal, cold, drought, and salt (Navrot et al., 2006; Diao et al., 2014; Fu, 2014; Gao et al., 2014). For example, GPX genes were found to be upregulated under excess H2O2 and cold stresses in rice (Passaia et al., 2013). Transcriptome analysis indicated high level of GPX transcripts in dehydrated Glycine max samples (Criqui et al., 1992; Ferreira Neto et al., 2013). Several transgenic studies also supported the proposed function of GPXs. For example, the overexpression of GPX in its transgenic tomato resulted in higher tolerance against abiotic stress (Herbette et al., 2011). In addition to stress response, GPXs are also thought to regulate cellular redox homeostasis by modulating the thiol-disulfide balance (Bela et al., 2015). GPX expression was found to be highly upregulated to maintain redox homeostasis under oxidative stress which helped Brassica rapa to adapt long-term spaceflight (Sugimoto et al., 2014).
A scan of contemporary literature reveals a paucity of information on the identification and comparative analysis of GPX and APX in model and economically important food crops. Given the above, employing bioinformatics approaches, efforts were made in this study (a) to identify potential GPX and APX genes/proteins from 18 different plant species, (b) to analyze their exon/intron organization, (c) to provide detailed information about their physico-chemical properties, (d) to identify conserved motif signatures of GPX and APX, (e) to construct their phylogenetic trees and 3D models, (f) to generate protein-protein interaction networks, and finally (g) to analyze GPX and APX gene expression profiles.
Materials and methods
Retrieval of GPX and APX genes/proteins
Eight Arabidopsis GPX reference protein sequences such as GPX1 (P52032.2), GPX2 (O04922.1), GPX3 (O22850.1), GPX4 (Q8L910.1), GPX5 (Q9LYB4.1), GPX6 (O48646.2), GPX7 (Q9SZ54.2), and GPX8 (Q8LBU2.1), as well as and eight Arabidopsis APX reference sequences such as APX1 (Q05431.2), APX2 (Q1PER6.3), APX3 (Q42564.1), APX4 (P82281.2), APX5 (Q7XZP5.2), APX6 (Q8GY91.1), APXT (Q42593.2), and APXS (Q42592.2) were obtained from UniProtKB/Swiss-Prot database of NCBI (Romiti, 2006). These reference sequences were queried in proteome datasets of selected 18 plant species: Arabidopsis thaliana (L.) Heynh., Brachypodium distachyon (L.) P. Beauv., Brasica rapa L., Chlamydomonas reinhardtii P. A. Dang., Cucumis sativus L., Eucalyptus grandis W. Hill ex Maiden, Glycine max (L.) Merr., Gossypium raimondii Ulbr., Medicago truncatula Gaertn., Oryza sativa L., Phaseolus vulgaris L., Physcomitrella patens (Hedw.) Bruch & Schimp., Populus trichocarpa Torr. & A.Gray ex. Hook., Prunus persica (L.) Batsch, Solanum lycopersicum L., Sorghum bicolor (L.) Moench, Vitis vinifera L., and Zea mays L., all found in the Phytozome v.10.3 database (Goodstein et al., 2012). After sequences were obtained, the Hidden Markov Model (HMM) search of protein sequences were performed by Pfam (http://pfam.sanger.ac.uk) to confirm the protein domain families (Finn et al., 2016). Species were arbitrarily selected to represent the main plant groups such as monocots, dicots, and lower plants.
Analysis of GPX and APX genes/proteins
Physicochemical properties of GPX and APX proteins were determined by using ProtParam tool (Gasteiger et al., 2005). Sub-cellular localization was predicted by CELLO (Yu et al., 2006) and WoLF PSORT (Horton et al., 2007) servers. Exon-intron organization of GPX/APX genes was analyzed by using a GSDS server (Hu et al., 2014). The Conserved motif structure of GPX/APX sequences was analyzed using the MEME tool with the following parameter settings: maximum number of motifs to find, 5; minimum width of motif, 6 and maximum width of motif, 50 (Bailey et al., 2009). Protein sequences were aligned by ClustalW (Thompson et al., 1994) and phylogenies were constructed by MEGA 6 (Tamura et al., 2013) with the maximum likelihood (ML) method for 1.000 bootstraps. The gene duplication events were detected using the following criteria: (a) length of alignable sequence covers >75% of the longer gene, and (b) similarity of aligned regions is >75% (Gu et al., 2002). The expression data of APX and GPX genes at anatomical and developmental levels were retrieved from the Genevestigator database (Hruz et al., 2008). 3D models of APX/GPXs were predicted by using the Phyre2 server (Kelley and Sternberg, 2009). Model validation was performed by Rampage Ramachandran plot analysis (Lovell et al., 2003). 3D structure comparisons were done by calculating RMSD values of models using the CLICK server employing α-carbon superposition (Nguyen et al., 2011). Putative interaction partners of APX/GPXs were predicted with the STRING server (Franceschini et al., 2013) and an interactome network was generated using cytoscape (Smoot et al., 2011).
Results and discussion
H2O2 plays double roles in plants and modulates various crucial metabolic processes (Petrov and Van Breusegem, 2012). However, its increased levels can cause severe damage to cell structures; hence, steady-state level of cellular H2O2 is required to be tightly regulated (Anjum et al., 2014, 2015; Sofo et al., 2015). GPX and APX are two major ROS-scavenging enzymes which catalyze the reduction of H2O2 to prevent H2O2-derived cellular damage. In order to understand the structural, functional as well as evolutionary aspects of GPX and APX, employing bioinformatics approaches, this study attempted to present comparative analyses of putative GPX and APX homologs identified from18 plant species.
Analysis of GPXs
Retrieval of GPX genes/proteins
Eight potential Arabidopsis GPX protein sequences, namely GPX1-8, obtained from the UniProtKB/Swiss-Prot database of NCBI were used as queries in Phytozome database to retrieve the very close homologs of GPX sequences in 18 plant species. In the selection of GPX homologs from blastp hits, very strict criteria (only the highest hit sequence) was applied to avoid the redundant sequences and alternative splices of the same gene. A total of 87 GPX sequences were identified from the protein datasets of 18 plant species. These include; 8 genes for A. thaliana, 4 genes for B. distachyon, 8 genes for B. rapa, 1 gene for C. reinhardtii, 6 genes for C. sativus, 5 genes for E. grandis, 5 genes for G. max, 6 genes for G. raimondii, 5 genes for M. truncatula, 5 genes for O. sativa, 5 genes for P. vulgaris, 2 genes for P. patens, 5 genes for P. trichocarpa, 5 genes for P. persica, 5 genes for S. lycopersicum, 4 genes for S. bicolor, 5 genes for V. vinifera, and 3 genes for Z. mays (Table 1). Then, genomic, transcript, CDS, and protein sequences of identified 87 GPX sequences were retrieved for further analyses.
Table 1.
List of H2O2-scavenging enzyme glutathione peroxidase (GPX) homologs from 18 plant species and their primary gene/protein features.
| Species name | Phytozome gene ID | Gene/protein features of GPX sequences | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein domain familya | Domain family description | Exon no. | Protein length | MW (KDa) | Theor. pI | Localization CELLOb | Localization WoLF PSORTb | ||
| Arabidopsis thaliana (L.) Heynh. | AT1G63460 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 19.0 | 5.11 | Cyto | Cyto |
| AT2G25080 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 236 | 26.0 | 9.42 | Chlo/Mito | Chlo | |
| AT2G31570 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 169 | 18.9 | 5.60 | Cyto | Cyto | |
| AT2G43350 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 206 | 23.2 | 9.24 | Mito/Plas | Chlo/Mito | |
| AT2G48150 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.3 | 8.87 | Cyto | Mito | |
| AT3G63080 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 173 | 19.3 | 9.28 | Extr/Chlo/Nucl | Chlo | |
| AT4G11600 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 232 | 25.5 | 9.38 | Mito/Chlo | Mito | |
| AT4G31870 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 233 | 25.7 | 9.53 | Chlo | Chlo | |
| Brachypodium distachyon (L.) P.Beauv. | Bradi1g47140 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 226 | 24.4 | 9.57 | Chlo | Chlo |
| Bradi1g61930 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 198 | 22.4 | 7.56 | Cyto | Cyto | |
| Bradi3g51010 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 240 | 25.9 | 9.05 | Chlo | Chlo | |
| Bradi5g18000 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.4 | 6.31 | Cyto/Chlo | Nucl/Chlo | |
| Brasica rapa L. | Brara.B02692 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 229 | 25.2 | 9.21 | Mito | Chlo/Mito |
| Brara.C02198 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 197 | 21.9 | 8.55 | Extr/Plas | Extr | |
| Brara.E00003 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.2 | 9.05 | Extr/Cyto | Chlo | |
| Brara.E01208 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 169 | 18.9 | 6.34 | Cyto | Cyto | |
| Brara.G01994 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 176 | 19.5 | 9.15 | Extr/Cyto/Nucl | Chlo | |
| Brara.I01234 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.9 | 5.00 | Cyto | Cyto | |
| Brara.I04448 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 233 | 25.8 | 9.29 | Mito/Chlo/Extr | Chlo | |
| Brara.K00392 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 232 | 25.9 | 9.60 | Mito/Extr | Chlo | |
| Chlamydomonas reinhardtii P.A.Dang. | Cre03.g197750 | GSHPx (PF00255) | Glutathione peroxidase | 7 | 200 | 21.9 | 9.39 | Mito | Chlo |
| Cucumis sativus L. | Cucsa.084960 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 176 | 19.7 | 8.86 | Cyto | Chlo |
| Cucsa.094950 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 204 | 23.4 | 8.55 | Plas/Extr | Chlo | |
| Cucsa.184280 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.0 | 8.66 | Cyto/Extr | Cyto | |
| Cucsa.271420 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 241 | 26.4 | 9.5 | Chlo | Chlo | |
| Cucsa.303050 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 241 | 26.8 | 9.28 | Mito | Chlo/Mito | |
| Cucsa.303070 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.2 | 5.21 | Cyto | Cyto | |
| Eucalyptus grandis W. Hill ex Maiden | Eucgr.A00257 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 202 | 22.8 | 7.62 | Extr/Chlo | Chlo/Vacu |
| Eucgr.C02602 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 247 | 26.9 | 9.53 | Chlo | Chlo | |
| Eucgr.D01856 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.4 | 5.16 | Cyto | Cyto | |
| Eucgr.E00579 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 250 | 27.3 | 9.16 | Chlo | Chlo | |
| Eucgr.K03389 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 18.9 | 9.02 | Cyto | Chlo/Nucl | |
| Glycine max (L.) Merr. | Glyma.03G151500 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.0 | 9.45 | Mito/Cyto | Chlo |
| Glyma.05G240100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 199 | 22.7 | 7.54 | Extr | Extr | |
| Glyma.08G013900 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.9 | 5.09 | Cyto | Chlo | |
| Glyma.11G024100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.5 | 5.88 | Cyto | Cyto | |
| Glyma.17G223900 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 234 | 26.3 | 9.40 | Mito/Chlo | Chlo | |
| Gossypium raimondii Ulbr. | Gorai.001G038600 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 242 | 26.6 | 9.30 | Chlo | Chlo |
| Gorai.004G083200 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 171 | 19.1 | 9.24 | Nucl/Cyto/Extr | Nucl | |
| Gorai.004G087300 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 208 | 23.6 | 5.51 | Extr | Extr | |
| Gorai.004G211400 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 166 | 18.4 | 6.73 | Cyto | Chlo | |
| Gorai.006G186100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.7 | 6.73 | Cyto | Cyto | |
| Gorai.008G246600 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 19.1 | 4.59 | Cyto | Chlo | |
| Zea mays L. | GRMZM2G012479 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 230 | 24.9 | 9.55 | Mito | Chlo |
| GRMZM2G144153 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.4 | 6.58 | Cyto | Chlo/Nucl | |
| GRMZM2G329144 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.2 | 7.58 | Cyto | Chlo | |
| Vitis vinifera L. | GSVIVG01010737001 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.6 | 5.53 | Cyto | Cyto |
| GSVIVG01011101001 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.0 | 9.22 | Cyto | Mito | |
| GSVIVG01019765001 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.2 | 5.01 | Cyto | Cyto | |
| GSVIVG01019766001 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.6 | 6.73 | Cyto | Chlo/Extr | |
| GSVIVG01035981001 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 207 | 22.9 | 9.16 | Chlo/Mito | Chlo | |
| Oryza sativa L. | LOC_Os02g44500 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 238 | 25.8 | 9.42 | Chlo | Chlo |
| LOC_Os03g24380 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 169 | 19.2 | 8.80 | Cyto | Cyto | |
| LOC_Os04g46960 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.4 | 8.33 | Cyto | Chlo | |
| LOC_Os06g08670 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 234 | 25 | 9.51 | Mito/Chlo | Chlo | |
| LOC_Os11g18170 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 212 | 22.9 | 7.62 | Chlo/Extr | Chlo | |
| Medicago truncatula Gaertn. | Medtr1g014210 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 236 | 26.4 | 9.32 | Mito/Chlo | Chlo |
| Medtr7g094600 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.2 | 9.18 | Cyto/Mito | Nucl | |
| Medtr8g098400 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 172 | 19.3 | 4.82 | Cyto | Chlo | |
| Medtr8g098410 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 233 | 25.8 | 9.27 | Mito | Chlo | |
| Medtr8g105630 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.9 | 8.32 | Plas | Chlo | |
| Physcomitrella patens (Hedw.) Bruch & Schimp. | Phpat.004G103100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 247 | 26.7 | 9.24 | Chlo/Extr | Chlo |
| Phpat.017G045400 | GSHPx (PF00255) | Glutathione peroxidase | 1 | 170 | 19.1 | 8.30 | Cyto | Cyto | |
| Phaseolus vulgaris L. | Phvul.001G041100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 262 | 29.7 | 9.68 | Mito | Chlo |
| Phvul.001G149000 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.8 | 9.31 | Cyto/Nucl | Nucl | |
| Phvul.002G157200 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.0 | 4.97 | Cyto | Chlo/Nucl | |
| Phvul.002G288700 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 230 | 25.6 | 8.76 | Chlo/Mito | Chlo | |
| Phvul.002G322400 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 198 | 22.5 | 5.94 | Extr | Extr | |
| Populus trichocarpa Torr. & A.Gray ex. Hook. | Potri.001G105100 | GSHPx (PF00255) | Glutathione peroxidase | 5 | 170 | 19.3 | 4.78 | Cyto | Cyto |
| Potri.003G126100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 238 | 26.2 | 9.29 | Mito/Chlo | Chlo | |
| Potri.006G265400 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 232 | 25.3 | 9.48 | Chlo/Mito | Chlo | |
| Potri.007G126600 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 203 | 22.8 | 6.83 | Extr | Extr/Vacu | |
| Potri.014G138800 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 18.9 | 9.15 | Cyto/Extr | Chlo/Cyto | |
| Prunus persica (L.) Batsch | ppa010584m.g | GSHPx (PF00255) | Glutathione peroxidase | 6 | 244 | 26.7 | 9.33 | Chlo | Chlo |
| ppa010771m.g | GSHPx (PF00255) | Glutathione peroxidase | 6 | 237 | 25.9 | 9.20 | Mito | Chlo | |
| ppa011682m.g | GSHPx (PF00255) | Glutathione peroxidase | 6 | 200 | 22.7 | 8.27 | Extr/Cyto | Extr | |
| ppa012378m.g | GSHPx (PF00255) | Glutathione peroxidase | 6 | 172 | 19.4 | 8.97 | Cyto | Nucl/Cyto | |
| ppa012416m.g | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.4 | 4.86 | Cyto | Chlo | |
| Sorghum bicolor (L.) Moench | Sobic.001G365800 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 171 | 19.3 | 8.79 | Cyto | Chlo |
| Sobic.006G173900 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 168 | 18.4 | 6.58 | Cyto | Chlo/Nucl | |
| Sobic.010G067100 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 232 | 24.9 | 9.50 | Mito/Chlo | Chlo | |
| Sobic.K022000 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 205 | 22.6 | 5.68 | Cyto/Extr | Mito/Chlo | |
| Solanum lycopersicum L. | Solyc06g073460.2 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 167 | 18.9 | 6.37 | Cyto | Chlo |
| Solyc08g006720.2 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 238 | 26.2 | 9.18 | Chlo | Chlo | |
| Solyc08g080940.2 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 239 | 26.7 | 9.16 | Mito | Chlo | |
| Solyc09g064850.2 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.0 | 9.33 | Mito/Extr | Chlo | |
| Solyc12g056240.1 | GSHPx (PF00255) | Glutathione peroxidase | 6 | 170 | 19.3 | 4.97 | Cyto | Cyto | |
Protein domain families were searched in Pfam database.
Cyto, Cytosolic; Chlo, Chloroplastic; Mito, Mitochondrial; Vacu, Vacuolar; Nucl, Nuclear; Extr, Extracellular; Plas, Plasma membrane.
More than one localization specified in a single column also shows the significance of other entries in order.
Sequence analysis of GPX genes/proteins
A total of 87 GPX homologs were identified in the protein datasets of 18 plant species using Arabidopsis GPX1-8 for homology search. Identified GPX homologs belonged to the GSHPx (PF00255) protein family. They encoded a polypeptide of 166–262 amino acids residues (average length 197.5) and 18.4–29.7 kDa molecular weight with 4.59–9.60 pI value. The sequence variations in analyzed GPXs primarily derived from the “transit peptide” residues between organelle and non-organelle related GPXs (Table 1). Studies of molecular cloning and sequencing in A. thaliana have reported that chloroplastic GPX1 and GPX7 consisted of 236 and 233 amino acids, respectively; the first 1–64 residues in GPX1 and 1–69 residues in GPX7 from N-terminal site contained the transit peptides (Mullineaux et al., 1998; Lin et al., 1999; Mayer et al., 1999). Arabidopsis GPX2 and GPX4 were reported to be 169 and 170 residues, respectively with cytosolic localization: thereby, they did not contain any transit peptide (Lin et al., 1999). Arabidopsis GPX3 and GPX6 were 206 and 232 residues, respectively, with mitochondrial localizations; the first 1–12 amino acids in GPX3 and 1–54 residues in GPX6 covered the transit peptide (Lin et al., 1999; Mayer et al., 1999). Arabidopsis GPX5 was 173 residues with probable ER or Plasma membrane localization, without transit peptide (Erfle et al., 2000). Arabidopsis GPX8 comprised of 167 amino acids with cytosolic or nuclear localization, without transit peptide (Theologis et al., 2000). In the present study, alignment analysis revealed that in chloroplastic/mitochondrial-related GPXs, the transit peptide sequences formed the first 50–90 amino acid residues from the N-terminal site while in extra cellular/plasma membrane-related GPXs, residues of the first 20–50 amino acid from N-terminal region contained the putative transit peptide. However, cytosolic sequences lacked of any putative transit residues (Supplementary Figure S1). Thus, analyzed GPX sequences were roughly categorized in three main groups based on their sequence length; the chloroplastic/mitochondrial related GPXs comprised the longer sequences (i), extra cellular/plasma membrane related GPXs formed the medium-sized sequences (ii), and cytosolic related GPXs included the shorter sequences (iii). In addition, the regions corresponding to the transit peptide sites in analyzed sequences did not demonstrate any particular patterns. The less-conserved transit peptide residues could be related with the functional diversities of GPXs at various targets. However, despite the variations in sequence length and transit peptide residues, transcripts of GPX homologs mainly contained the six exons. Therefore, it is reasonable to claim that analyzed GPX sequences could have highly-conserved protein sequences, preserved during the formation of various GPXs. The alignment analysis of 87 GPX protein homologs also confirmed this claim, demonstrating the presence of more conserved residues in the main sites of the active enzyme (Supplementary Figure S2). Moreover, to discern the conserved motif patterns in GPX sequences, the most conserved five motif sequences were searched in sets of 87 GPX homologs using MEME tool (Table 2). Motif 1 and 3 were the 50 amino acid residues, while the motif 2 was 41, motif 4 was 15, and motif 5 was 6 residues in length. Motif 1 and 3 were related with the GSHPx (PF00255) protein family and present in almost all GPX homologs. The presence of long conserved residues and their relation with the GSHPx family could indicate the highly conserved structures of GPX sequences at these sites between/among species.
Table 2.
Most conserved five motifs of glutathione peroxidase (GPX) homologs in 18 plant species.
| Motif | Width | Identified site no. | Sequence | Protein domain familya |
|---|---|---|---|---|
| 1 | 50 | 87 of 87 | KYKDQGFEILAFPCNQFGGQEPGTNEEIQQFACTRFKAEYPIFDKVDVNG | GSHPx (PF00255) |
| 2 | 41 | 87 of 87 | FGDRIKWNFTKFLVDKEGHVVDRYAPTTSPLQIEKDIQKLL | Not found |
| 3 | 50 | 86 of 87 | KSIHDFTVKDIRGNDVDLSIYKGKVLLIVNVASQCGMTNSNYTELNHLYE | GSHPx (PF00255) |
| 4 | 15 | 87 of 87 | NAAPLYKFLKSSKWG | Not found |
| 5 | 6 | 63 of 87 | MAASHS | Not found |
Protein domain families have been searched in Pfam database.
Furthermore, alignment analysis also demonstrated that Asn (N), Gly (G), Arg (R), Pro (P), Thr (T), Tyr (Y), Lys (K), Ala-Ser (AS), Cys-Gly (CG), Phe-Pro (FP), Glu-Pro (EP), Leu-Lys (LK), Lys-Phe (KF), Asn-Gly (NG), Asn-Gln-Phe (NQF), and Trp-Asn-Phe (WNF) residues were strictly conserved in all aligned sequences, indicating their potential functions in enzyme activity and/or stability (Supplementary Figure S3). To infer a functional relationship between these conserved residues and GPX sequences, we searched for the known catalytic residues of model organism Arabidopsis GPXs in the UniProtKB/Swiss-Prot database of NCBI (www.ncbi.nlm.nih.gov/protein). The following residues were designated in the database as potential catalytic residues for Arabidopsis GPX1-8: GPX1 (Cys-111, Gln-146, Trp-200), GPX2 (Cys-41, Gln-76, Trp-130), GPX3 (Cys-80, Gln-115, Trp-169), GPX4 (Cys-44, Gln-79, Trp-133), GPX5 (Cys-46, Gln-81, Trp-135), GPX6 (Cys-105, Gln-140, Trp-194), GPX7 (Cys-108, Gln-143, Trp-197), and GPX8 (Cys-41, Glu-76, Trp-130). Interestingly, residues that correspond to these catalytic sites in other analyzed sequences were found to be strictly conserved (Supplementary Figure S3). This shows that active sites of the enzyme are considerably conserved between species.
Phylogenetic analysis of GPXs
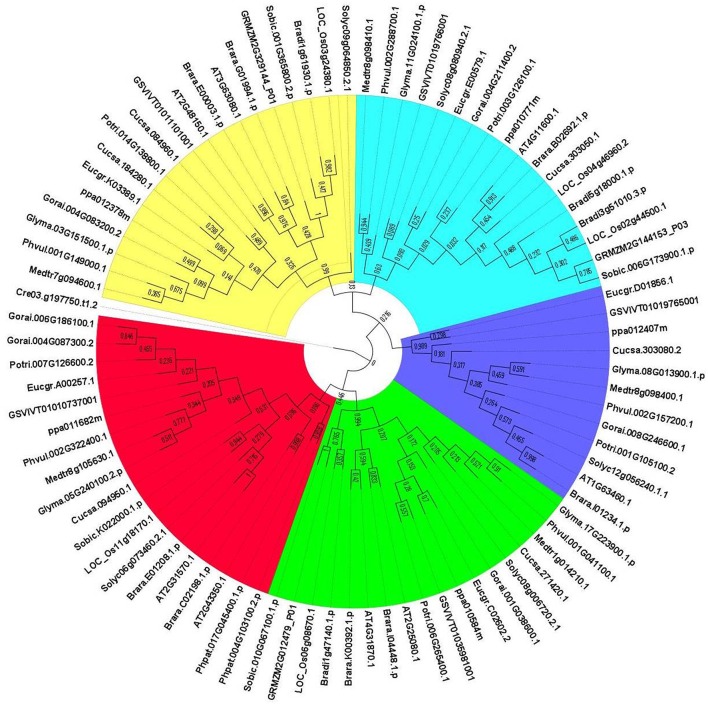
The evolutionary relationships between identified GPX sequences were analyzed by MEGA 6 using the Maximum Likelihood (ML) method with 1000 bootstraps. The constructed phylogeny included all 87 GPX homologs to discover the phylogenetic distribution of sequences (Figure 1). The tree was divided into six major groups based on the tree topology, and each group was indicated with a different color segment. The red segment included cytosolic, extra cellular, and plasma membrane related GPXs, the green segment contained mitochondrial and chloroplast related GPXs, the blue segment only had cytosolic GPXs, the cyan segment included cytosolic and chloroplast/mitochondrial related GPXs, the yellow segment contained cytosolic/nuclear related GPXs, and the non-colored segment had lower plant Chlamydomonas GPX with a chloroplastic/mitochondrial relation. Annotation of each segment based on the consensus of two subcellular localization servers, CELLO and WoLF PSORT, as well as tree topology for ambiguous sequences. Mainly cytosolic, nuclear, extra cellular and plasma membrane related GPXs were clustered together, while chloroplast/mitochondrial related GPXs also cluster together. Therefore, the presence or absence of transit peptide residues was the main contributing entity in the phylogenetic distribution of GPX sequences. In addition, the presence of sequences with different subcellular localizations in the same group inferred the possibility of gene duplication events in the formation of various GPX sequences. Duplications in plant genomes could be either as small-scale such as tandem and segmental duplications, or as large-scale such as whole-genome duplications (Ramsey and Schemske, 1998). The segmental duplications are observed in different chromosomes whereas tandem duplications occur in the same chromosome (Liu et al., 2011). Several segmental duplications were identified between GPX pairs (Table 3). The presence of segmental duplications, particularly between sequences with various subcellular localizations may partly explain the possibility of gene duplication events in GPX formations.
Figure 1.
Phylogenetic tree of glutathione peroxidase (GPX) homologs from 18 plant species. Tree was constructed by MEGA 6 using Maximum likelihood (ML) method with 1000 bootstraps. Segment classification based on the consensus of two subcellular localization servers, CELLO and WoLF PSORT as well as tree topology for ambiguous sequences. Red segment includes cytosolic, extra cellular, and plasma membrane related GPXs, green segment contains mitochondrial and chloroplast related GPXs, blue segment only have cytosolic GPXs, cyan segment includes cytosolic, and chloroplast/mitochondrial related GPXs, yellow segment contains cytosolic/nuclear related GPXs, and non-colored segment has lower plant Chlamydomonas GPX with chloroplastic/mitochondrial relation.
Table 3.
The segmental duplication events in some glutathione peroxidase (GPX) pairs.
| Species name | Segmental duplication pairs |
|---|---|
| Arabidopsis thaliana (L.) Heynh. | AT2G25080-AT4G31870 |
| AT2G48150-AT3G63080 | |
| Brachypodium distachyon (L.) P.Beauv. | Bradi5g18000-Bradi3g51010 |
| Brasica rapa L. | Brara.E00003-Brara.G01994 |
| Brara.I04448-Brara.K00392 | |
| Gossypium raimondii Ulbr. | Gorai.004G087300-Gorai.006G186100 |
| Gorai.004G211400-Gorai.008G246600 | |
| Vitis vinifera L. | GSVIVG01019765001-GSVIVG01019766001 |
| Oryza sativa L. | LOC_Os04g46960-LOC_Os02g44500 |
| Physcomitrella patens (Hedw.) Bruch & Schimp. | Phpat.017G045400-Phpat.004G103100 |
| Prunus persica (L.) Batsch | ppa012416m.g-ppa010771m.g |
Expression profile analysis of GPXs
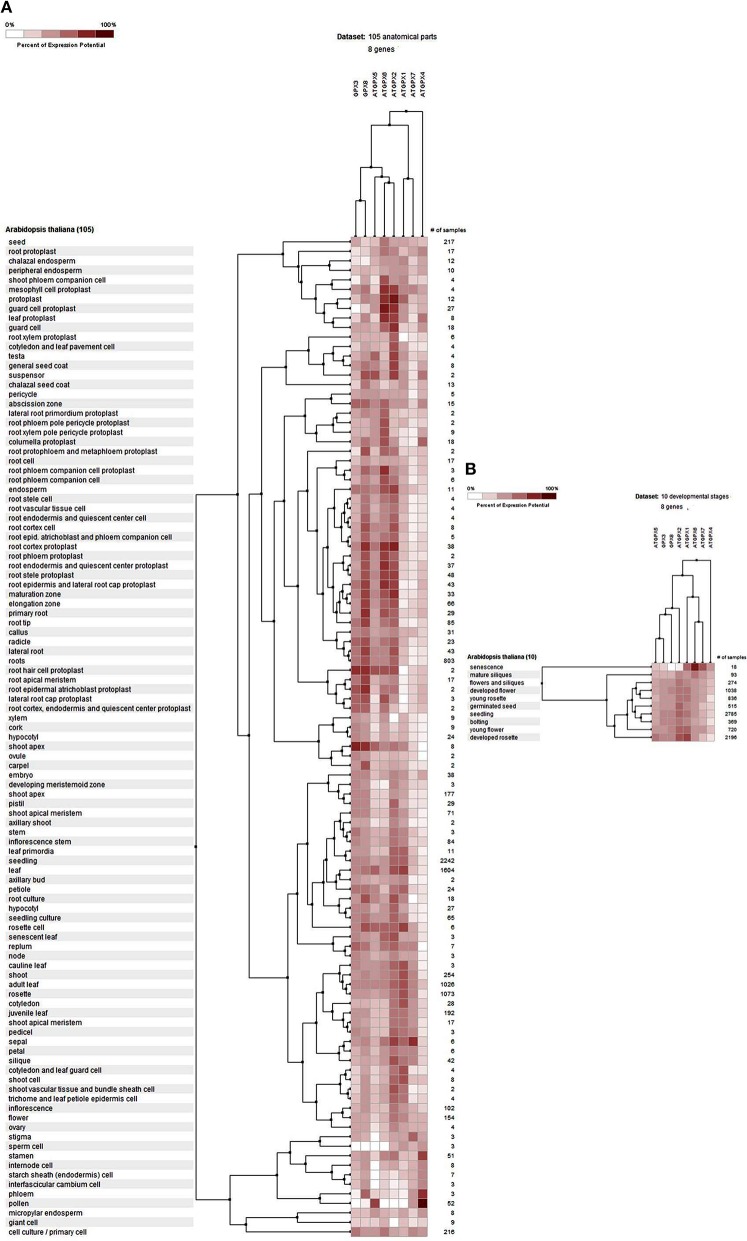
The potential expression profile of GPX genes was analyzed at 105 anatomical parts and 10 developmental stage levels using model organism A. thaliana GPXs from Genevestigator platform (Figure 2). Eight Arabidopsis genes, namely GPX1 (AT2G25080), GPX2 (AT2G31570), GPX3 (AT2G43350), GPX4 (AT2G48150), GPX5 (AT3G63080), GPX6 (AT4G11600), GPX7 (AT4G31870), and GPX8 (AT1G63460) were retrieved from the “Affymetrix Arabidopsis ATH1 Genome Array” platform using the Genevestigator interface, and conditions and genes with similar profiles were comparatively analyzed using the Hierarchical clustering tool with the Euclidean distance method. At an anatomical part level (Figure 2A), analyzed GPX1-8 genes were expressed in almost all 105 anatomical tissues in Arabidopsis plants. However, various root and leaf protoplast cells, seed-related tissues, and active growth zones demonstrated significantly higher GPX activity. This indicates that stress factors and/or active metabolism could lead to the up-regulation of various GPX genes in different tissues. Many studies have also showed that balance between production and scavenging of H2O2 could be disturbed by various biotic/abiotic stress factors or perturbations such as drought, salinity, pathogen attack, oxidative state of the cells (Apel and Hirt, 2004; Anjum et al., 2014, 2015; Sofo et al., 2015). Besides, a number of studies were also available demonstrating the functional roles of GPXs in plant stress resistance/tolerance. For example, a GPX gene in Pennisetum glaucum enhanced the drought and salinity stress tolerance (Islam et al., 2015). Citrus GPX3 was reported to be essential in detoxification of ROS-induced cellular stresses as well as in Alternaria alternata pathogenesis (Yang et al., 2016). Silencing of mitochondrial GPX1 in O. sativa demonstrated the impaired photosynthesis in response to salinity (Lima-Melo et al., 2016). Glutathione transferases and peroxidases were reported as key components in Arabidopsis salt stress-acclimation (Horváth et al., 2015). GPX1 and GPX3 in legume root nodules played a protective function against salt stress, oxidative stress, and membrane damage (Matamoros et al., 2015). Therefore, GPXs, which are the antioxidant enzymatic components of the cells, are consequently induced to suppress/eliminate the toxic levels of H2O2. The increased GPX activities in analyzed Arabidopsis tissues could thereby be derived from the increased H2O2 or H2O2-related products. In addition, clustering analysis of GPX genes showed that GPX2, 3, 5, 6, and 8 demonstrate relatively similar expression profiles compared to those of GPX1, 4, and 7. At the developmental level (Figure 2B), the expression profiles of Arabidopsis GPX1-8 genes were analyzed at 10 developmental stages, including senescence, mature siliques, flowers and siliques, developed flower, young rosette, germinated seed, seedling, bolting, young flower, and developed rosette. GPX1-8 were relatively expressed in all developmental stages. However, the expression of GPXs in the senescence stage demonstrated slightly different patterns, particularly the mitochondrial GPX6 gene had the highest expression profile compared to other developmental stages. This may have been caused by senescence-related cellular deteriorations, leading to the substantial metabolic or physiological changes that significantly affect the overall metabolic energy consumption. Therefore, it seems that the expression profiles of GPXs are highly associated with the metabolic state of the cells.
Figure 2.
Expression profile of Arabidopsis glutathione peroxidase GPX1-8 genes at 105 anatomical parts (A) and 10 developmental stage levels (B). Genes and conditions with similar profiles were comparatively analyzed using hierarchical clustering tool with Euclidean distance method at Genevestigator platform.
3D structure analysis of GPXs
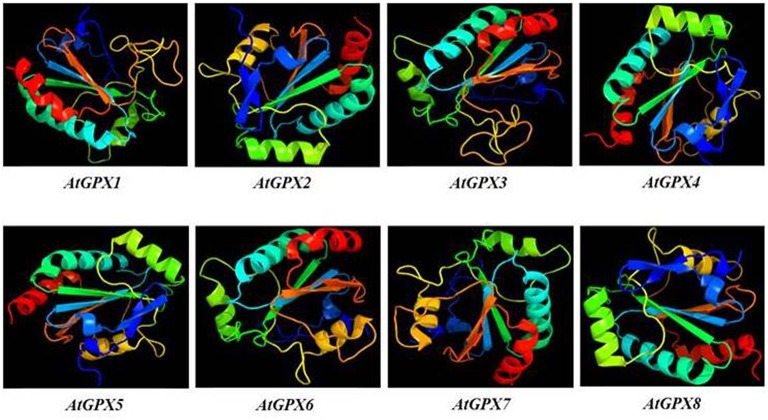
3D models of putative GPXs were constructed by using Phyre2 server for eight identified Arabidopsis GPX1-8 gene sequences (Figure 3). These sequences were: AT2G25080.1 (GPX1), AT2G31570.1 (GPX2), AT2G43350.1 (GPX3), AT2G48150.1 (GPX4), AT3G63080.1 (GPX5), AT4G11600.1 (GPX6), AT4G31870.1 (GPX7), and AT1G63460.1 (GPX8). In modeling, three templates such as 2F8A:A (GPX1, GPX3, GPX6, and GPX7), 2V1M:A (GPX2 and GPX5), and 2P5Q:A (GPX4 and GPX8) were used to maximize the alignment coverage, percentage identity and confidence for submitted sequences. Predicted models demonstrated the ≥98% of residues in allowed region in Ramachandran plot analysis, indicating that constructed models were fairly in good quality. To find out the structural divergence/similarity in generated models, the structures were superposed to calculate the percentage of structural overlap and RMSD values (Table 4). GPX1-GPX3, GPX4-GPX8, and GPX6-GPX7 pairs demonstrated the highly conserved structural overlap (100%) with 0.14, 0.00, and 0.03 RMSD values, respectively. The each designated pair also belonged to either chloroplastic/mitochondrial or cytosolic form, indicating their functional similarities with minor specifications. In addition, GPX1-GPX6 and 7, GPX2-GPX5, and GPX3-GPX6 and 7 pairs showed very high structural similarity with ≥94 structural overlaps. Despite the highly conserved structures of Arabidopsis GPX members, some minor variations were also present. It seems that these divergences in GPXs may not change the protein-3D structure, however, they could attribute the new functional roles to catalytic activities.
Figure 3.
3D models of predicted Arabidopsis glutathione peroxidase GPX1-8 sequences. Models were constructed by using Phyre2 server for AT2G25080.1 (GPX1), AT2G31570.1 (GPX2), AT2G43350.1 (GPX3), AT2G48150.1 (GPX4), AT3G63080.1 (GPX5), AT4G11600.1 (GPX6), AT4G31870.1 (GPX7), and AT1G63460.1 (GPX8) sequences, and colored by rainbow from N- to C-terminus.
Table 4.
Structural overlap (%)/RMSD values in superposed Arabidopsis glutathione peroxidases (GPXs).
| GPX1 | GPX2 | GPX3 | GPX4 | GPX5 | GPX6 | GPX7 | GPX8 | |
|---|---|---|---|---|---|---|---|---|
| GPX1 | – | 88.68/1.03 | 100.00/0.14 | 91.19/1.10 | 89.94/0.91 | 94.34/0.24 | 94.34/0.24 | 91.19/1.10 |
| GPX2 | 88.05/0.89 | – | 88.68/0.93 | 93.75/0.66 | 99.38/0.15 | 91.82/0.92 | 91.82/0.92 | 94.38/0.71 |
| GPX3 | 100.00/0.14 | 89.94/1.10 | – | 90.57/0.90 | 90.57/0.95 | 94.34/0.28 | 94.34/0.28 | 90.57/0.90 |
| GPX4 | 89.94/1.07 | 93.75/0.67 | 91.19/0.96 | – | 95.65/0.67 | 94.97/0.97 | 94.97/0.97 | 100.00/0.00 |
| GPX5 | 90.57/0.89 | 99.38/0.15 | 90.57/0.91 | 95.65/0.66 | – | 94.34/0.91 | 94.34/0.91 | 95.65/0.67 |
| GPX6 | 94.34/0.24 | 92.45/1.04 | 94.34/0.28 | 95.60/1.07 | 94.97/1.04 | – | 100.00/0.03 | 95.60/1.06 |
| GPX7 | 94.34/0.24 | 92.45/1.05 | 94.34/0.28 | 95.60/1.06 | 94.97/1.04 | 100.00/0.03 | – | 95.60/1.06 |
| GPX8 | 91.19/0.99 | 95.00/0.75 | 91.19/0.98 | 100.00/0.00 | 95.65/0.67 | 94.97/0.97 | 94.97/0.97 | – |
Red-color highlighted pairs show the highly conserved structural overlaps.
Interaction partner analysis of GPXs
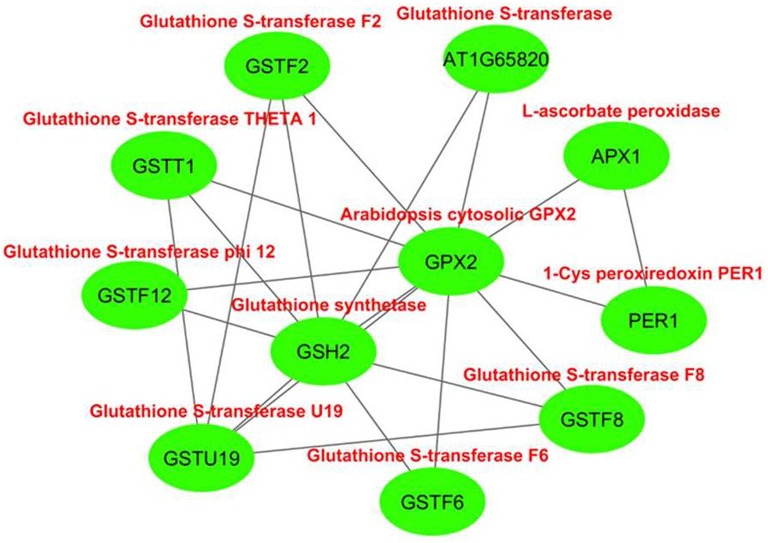
The interactome network was constructed for 10 putative interactors of Arabidopsis cytosolic GPX2, using Cytoscape with STRING data (Figure 4). APX1 (L-ascorbate peroxidase), GSH2 (glutathione synthetase), GSTF6 (glutathione S-transferase F6), GSTT1 (glutathione S-transferase THETA 1), PER1 (1-Cys peroxiredoxin PER1), AT1G65820 (glutathione S-transferase), GSTF12 (glutathione S-transferase phi 12), GSTF2 (glutathione S-transferase F2), GSTF8 (glutathione S-transferase F8), and GSTU19 (glutathione S-transferase U19) proteins were predicted as the main interaction partners of Arabidopsis cytosolic GPX2. APX1 is a type of H2O2-scavenging enzyme and a central component in the reactive oxygen gene network (Storozhenko et al., 1998; Fourcroy et al., 2004). GSH2 involves in the second step of the glutathione synthesis pathway from L-cysteine and L-glutamate (Wang and Oliver, 1996). GSTF6 functions in camalexin biosynthesis, is involved in the conjugation of reduced glutathione to various exogenous/endogenous hydrophobic electrophiles, and has a detoxification role for certain herbicides (Su et al., 2011). GSTT1, GSTF8, and GSTU19 are reported to have glutathione S-transferase or peroxidase activity. They further conjugate the reduced glutathione to various exogenous/endogenous hydrophobic electrophiles and play a detoxification role for certain herbicides (Wagner et al., 2002). PER1 is an antioxidant protein involved in the inhibition of germination under stress (Haslekås et al., 1998). AT1G65820 is a glutathione S-transferase. GSTF12 is involved in the transport of anthocyanins and proanthocyanidins into the vacuole (Kitamura et al., 2004). GSTF2 plays a role in binding and transport of small bioactive products and defense-related compounds under stress (Smith et al., 2003). The analysis indicated that cytosolic GPX2 enzyme is closely related with various pathways involving in antioxidant and secondary metabolite metabolisms, thereby supporting the functional role of GPXs in H2O2-scavenging and plant defense.
Figure 4.
Predicted 10 interaction partners of Arabidopsis cytosolic glutathione peroxidase GPX2. Interactome was generated using cytoscape with STRING data. APX1 (L-ascorbate peroxidase), GSH2 (glutathione synthetase), GSTF6 (glutathione S-transferase F6), GSTT1 (glutathione S-transferase THETA 1), PER1 (1-Cys peroxiredoxin PER1), AT1G65820 (glutathione S-transferase), GSTF12 (glutathione S-transferase phi 12), GSTF2 (glutathione S-transferase F2), GSTF8 (glutathione S-transferase F8), and GSTU19 (glutathione S-transferase U19) proteins were predicted as main interaction partners of Arabidopsis cytosolic GPX2.
Analysis of APXs
Retrieval of APX genes/proteins
Eight potential Arabidopsis APX protein sequences such as APX1-6, APXT, and APXS, obtained from the UniProtKB / Swiss-Prot database of NCBI, were used as queries in Phytozome database to retrieve the very close homologs of APX sequences in 18 plant species. In the selection of APX homologs from blastp hits, a very strict criterion (only the highest hit sequence) was applied to avoid redundant sequences and alternative splices of the same gene. A total of 120 APX sequences were identified from the protein datasets of 18 plant species. These were 8 genes for A. thaliana, 7 genes for B. distachyon, 8 genes for B. rapa, 4 genes for C. reinhardtii, 5 genes for C. sativus, 7 genes for E. grandis, 7 genes for G. max, 8 genes for G. raimondii, 7 genes for M. truncatula, 6 genes for O. sativa, 6 genes for P. vulgaris, 5 genes for P. patens, 7 genes for P. trichocarpa, 6 genes for P. persica, 7 genes for S. lycopersicum, 8 genes for S. bicolor, 6 genes for V. vinifera, and 8 genes for Z. mays (Table 5). Then, genomic, transcript, CDS, and protein sequences of 120 identified APX sequences were retrieved for further analyses.
Table 5.
List of H2O2-scavenging enzyme ascorbate peroxidase (APX) homologs from 18 plant species and their primary gene/protein features.
| Species name | Phytozome gene ID | Gene/protein features of GPX sequences | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein domain familya | Domain family description | Exon no. | Protein length | MW (KDa) | Theor. pI | Localization CELLOb | Localization WoLF PSORTb | ||
| Arabidopsis thaliana (L.) Heynh. | AT1G07890 | Peroxidase (PF00141) | Peroxidase | 8 | 250 | 27.5 | 5.72 | Cyto | Cyto |
| AT1G77490 | Peroxidase (PF00141) | Peroxidase | 12 | 426 | 46.0 | 6.81 | Chlo | Chlo | |
| AT3G09640 | Peroxidase (PF00141) | Peroxidase | 9 | 251 | 28.0 | 5.87 | Cyto | Cyto | |
| AT4G08390 | Peroxidase (PF00141) | Peroxidase | 10 | 372 | 40.4 | 8.31 | Chlo | Chlo | |
| AT4G09010 | Peroxidase (PF00141) | Peroxidase | 10 | 349 | 37.9 | 8.59 | Chlo/Mito | Chlo | |
| AT4G32320 | Peroxidase (PF00141) | Peroxidase | 10 | 329 | 36.2 | 8.99 | Chlo | Chlo | |
| AT4G35000 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.5 | 6.47 | Cyto | Cyto | |
| AT4G35970 | Peroxidase (PF00141) | Peroxidase | 9 | 279 | 30.8 | 8.80 | Cyto/Nucl | Cyto | |
| Brachypodium distachyon (L.) P.Beauv. | Bradi1g16510 | Peroxidase (PF00141) | Peroxidase | 9 | 256 | 27.7 | 5.28 | Cyto | Cyto |
| Bradi1g65820 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.4 | 5.71 | Cyto | Cyto | |
| Bradi3g40330 | Peroxidase (PF00141) | Peroxidase | 11 | 329 | 35.4 | 6.36 | Chlo | Chlo | |
| Bradi3g42340 | Peroxidase (PF00141) | Peroxidase | 9 | 289 | 31.5 | 7.70 | Cyto/Chlo | Cyto | |
| Bradi3g45700 | Peroxidase (PF00141) | Peroxidase | 12 | 439 | 47.3 | 5.61 | Chlo | Chlo | |
| Bradi5g10490 | Peroxidase (PF00141) | Peroxidase | 11 | 345 | 37.4 | 8.77 | Chlo/Mito | Chlo | |
| Bradi5g20670 | Peroxidase (PF00141) | Peroxidase | 10 | 333 | 36.1 | 8.71 | Mito | Chlo | |
| Brasica rapa L. | Brara.A00250 | Peroxidase (PF00141) | Peroxidase | 8 | 280 | 31.0 | 7.69 | Cyto | Cyto |
| Brara.A03521 | Peroxidase (PF00141) | Peroxidase | 9 | 251 | 28.1 | 6.41 | Cyto | Cyto | |
| Brara.C02583 | Peroxidase (PF00141) | Peroxidase | 9 | 348 | 37.9 | 8.59 | Chlo/Mito | Chlo | |
| Brara.G03518 | Peroxidase (PF00141) | Peroxidase | 10 | 439 | 47.5 | 7.70 | Chlo | Chlo | |
| Brara.I02406 | Peroxidase (PF00141) | Peroxidase | 10 | 354 | 38.8 | 7.12 | Chlo/Mito | Chlo | |
| Brara.I05334 | Peroxidase (PF00141) | Peroxidase | 7 | 250 | 27.5 | 5.61 | Cyto | Cyto | |
| Brara.K00318 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.7 | 6.67 | Cyto | Cyto | |
| Brara.K00699 | Peroxidase (PF00141) | Peroxidase | 10 | 327 | 36.1 | 8.72 | Chlo | Chlo | |
| Chlamydomonas reinhardtii P.A.Dang. | Cre02.g087700 | Peroxidase (PF00141) | Peroxidase | 10 | 327 | 35.6 | 8.67 | Mito/Chlo | Chlo/Mito |
| Cre05.g233900 | Peroxidase (PF00141) | Peroxidase | 8 | 347 | 36.4 | 9.23 | Chlo | Chlo/Mito | |
| Cre06.g285150 | Peroxidase (PF00141) | Peroxidase | 7 | 337 | 35.1 | 8.95 | Chlo/Mito | Chlo/Mito | |
| Cre09.g401886 | Peroxidase (PF00141) | Peroxidase | 10 | 372 | 39.4 | 8.63 | Chlo | Chlo | |
| Cucumis sativus L. | Cucsa.060660 | Peroxidase (PF00141) | Peroxidase | 11 | 413 | 44.8 | 7.09 | Chlo | Chlo |
| Cucsa.162470 | Peroxidase (PF00141) | Peroxidase | 8 | 249 | 27.3 | 7.74 | Chlo/Cyto | Nucl | |
| Cucsa.213340 | Peroxidase (PF00141) | Peroxidase | 9 | 249 | 27.3 | 5.43 | Cyto | Cyto | |
| Cucsa.311620 | Peroxidase (PF00141) | Peroxidase | 11 | 368 | 40.2 | 7.67 | Chlo | Chlo | |
| Cucsa.370590 | Peroxidase (PF00141) | Peroxidase | 9 | 286 | 31.4 | 6.41 | Cyto | Cyto | |
| Eucalyptus grandis W. Hill ex Maiden | Eucgr.A01180 | Peroxidase (PF00141) | Peroxidase | 9 | 249 | 27.4 | 6.07 | Cyto | Cyto |
| Eucgr.B02456 | Peroxidase (PF00141) | Peroxidase | 9 | 249 | 27.2 | 5.29 | Cyto | Cyto | |
| Eucgr.C01740 | Peroxidase (PF00141) | Peroxidase | 9 | 369 | 39.6 | 8.44 | Chlo | Chlo | |
| Eucgr.F00373 | Peroxidase (PF00141) | Peroxidase | 11 | 356 | 38.3 | 6.50 | Chlo | Chlo | |
| Eucgr.F04344 | Peroxidase (PF00141) | Peroxidase | 12 | 446 | 48.2 | 8.71 | Chlo | Chlo | |
| Eucgr.F04344 | Peroxidase (PF00141) | Peroxidase | 11 | 397 | 42.8 | 8.60 | Chlo | Chlo | |
| Eucgr.I01408 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.5 | 6.67 | Cyto/Chlo | Cyto | |
| Glycine max (L.) Merr. | Glyma.04G248300 | Peroxidase (PF00141) | Peroxidase | 11 | 386 | 41.9 | 7.06 | Chlo | Chlo |
| Glyma.06G068200 | Peroxidase (PF00141) | Peroxidase | 10 | 319 | 34.2 | 7.56 | Chlo | Chlo | |
| Glyma.06G114400 | Peroxidase (PF00141) | Peroxidase | 12 | 432 | 47.0 | 7.13 | Chlo | Chlo | |
| Glyma.11G078400 | Peroxidase (PF00141) | Peroxidase | 9 | 280 | 31.1 | 9.08 | Cyto/Mito | Cyto | |
| Glyma.12G032300 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.7 | 6.27 | Cyto | Cyto | |
| Glyma.12G073100 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.1 | 5.65 | Cyto | Cyto | |
| Glyma.14G177200 | Peroxidase (PF00141) | Peroxidase | 10 | 347 | 37.9 | 6.76 | Extr/Mito/Chlo | Chlo | |
| Gossypium raimondii Ulbr. | Gorai.002G196800 | Peroxidase (PF00141) | Peroxidase | 9 | 288 | 31.7 | 5.64 | Cyto | Cyto |
| Gorai.005G254100 | Peroxidase (PF00141) | Peroxidase | 9 | 288 | 31.9 | 6.67 | Cyto | Cyto | |
| Gorai.009G104500 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.5 | 5.73 | Cyto | Cyto | |
| Gorai.009G246900 | Peroxidase (PF00141) | Peroxidase | 11 | 385 | 41.7 | 8.89 | Chlo | Chlo | |
| Gorai.009G420500 | Peroxidase (PF00141) | Peroxidase | 9 | 251 | 27.8 | 6.01 | Cyto | Cyto | |
| Gorai.010G038200 | Peroxidase (PF00141) | Peroxidase | 11 | 355 | 38.8 | 7.53 | Chlo | Chlo | |
| Gorai.010G051400 | Peroxidase (PF00141) | Peroxidase | 12 | 422 | 46.0 | 6.77 | Chlo | Chlo | |
| Gorai.010G115200 | Peroxidase (PF00141) | Peroxidase | 10 | 334 | 36.2 | 8.17 | Chlo | Chlo | |
| Zea mays L. | GRMZM2G004211 | Peroxidase (PF00141) | Peroxidase | 9 | 290 | 32.0 | 7.72 | Cyto/Mito | Cyto |
| GRMZM2G006791 | Peroxidase (PF00141) | Peroxidase | 12 | 451 | 48.9 | 5.60 | Chlo | Chlo | |
| GRMZM2G047968 | Peroxidase (PF00141) | Peroxidase | 7 | 223 | 23.7 | 9.01 | Chlo/Cyto | Mito/Chlo | |
| GRMZM2G054300 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.3 | 5.56 | Cyto | Cyto | |
| GRMZM2G120517 | Peroxidase (PF00141) | Peroxidase | 11 | 339 | 37.0 | 8.86 | Mito | Chlo | |
| GRMZM2G137839 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.3 | 5.64 | Cyto | Cyto | |
| GRMZM2G156227 | Peroxidase (PF00141) | Peroxidase | 10 | 351 | 38.3 | 8.62 | Mito | Chlo | |
| GRMZM2G460406 | Peroxidase (PF00141) | Peroxidase | 8 | 289 | 31.6 | 7.73 | Cyto/Chlo | Cyto | |
| Vitis vinifera L. | GSVIVG01008846001 | Peroxidase (PF00141) | Peroxidase | 11 | 372 | 40 | 7.10 | Chlo | Chlo |
| GSVIVG01009079001 | Peroxidase (PF00141) | Peroxidase | 10 | 344 | 37.4 | 6.65 | Extr/Chlo | Chlo | |
| GSVIVG01024035001 | Peroxidase (PF00141) | Peroxidase | 9 | 289 | 31.7 | 7.72 | Chlo/Cyto | Cyto | |
| GSVIVG01025104001 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.5 | 5.71 | Cyto | Cyto | |
| GSVIVG01025551001 | Peroxidase (PF00141) | Peroxidase | 9 | 253 | 27.9 | 5.43 | Cyto | Cyto | |
| GSVIVG01035858001 | Peroxidase (PF00141) | Peroxidase | 10 | 330 | 35.9 | 6.47 | Chlo/Cyto | Chlo | |
| Oryza sativa L. | LOC_Os02g34810 | Peroxidase (PF00141) | Peroxidase | 12 | 478 | 51.1 | 5.36 | Chlo | Chlo |
| LOC_Os04g35520 | Peroxidase (PF00141) | Peroxidase | 11 | 359 | 38.3 | 8.76 | Chlo | Chlo | |
| LOC_Os04g51300 | Peroxidase (PF00141) | Peroxidase | 11 | 353 | 38.1 | 8.67 | Mito/Chlo | Chlo | |
| LOC_Os07g49400 | Peroxidase (PF00141) | Peroxidase | 9 | 251 | 27.1 | 5.18 | Cyto | Cyto | |
| LOC_Os08g41090 | Peroxidase (PF00141) | Peroxidase | 10 | 331 | 35.5 | 6.95 | Chlo | Chlo | |
| LOC_Os08g43560 | Peroxidase (PF00141) | Peroxidase | 9 | 291 | 31.7 | 7.74 | Chlo/Cyto/Mito | Cyto | |
| Medicago truncatula Gaertn. | Medtr3g088160 | Peroxidase (PF00141) | Peroxidase | 11 | 436 | 47.4 | 9.02 | Chlo | Chlo |
| Medtr3g088160 | Peroxidase (PF00141) | Peroxidase | 10 | 387 | 42.0 | 8.73 | Chlo | Chlo | |
| Medtr3g107060 | Peroxidase (PF00141) | Peroxidase | 10 | 320 | 34.7 | 8.08 | Chlo | Mito/Chlo | |
| Medtr4g061140 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.1 | 5.52 | Cyto | Cyto | |
| Medtr4g073410 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.7 | 6.26 | Cyto | Chlo/Cyto | |
| Medtr5g022510 | Peroxidase (PF00141) | Peroxidase | 9 | 281 | 31.4 | 8.74 | Cyto | Cyto | |
| Medtr5g064610 | Peroxidase (PF00141) | Peroxidase | 10 | 353 | 38.9 | 8.18 | Mito/Nucl | Chlo | |
| Physcomitrella patens (Hedw.) Bruch & Schimp. | Phpat.001G070500 | Peroxidase (PF00141) | Peroxidase | 11 | 358 | 38.4 | 7.56 | Chlo | Chlo |
| Phpat.001G104200 | Peroxidase (PF00141) | Peroxidase | 9 | 300 | 32.6 | 7.01 | Chlo | Cyto | |
| Phpat.001G162800 | Peroxidase (PF00141) | Peroxidase | 2 | 440 | 48.2 | 8.11 | Chlo | Chlo | |
| Phpat.017G025400 | Peroxidase (PF00141) | Peroxidase | 11 | 357 | 38.4 | 6.15 | Chlo | Chlo | |
| Phpat.020G011100 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.6 | 5.66 | Cyto | Cyto | |
| Phaseolus vulgaris L. | Phvul.008G176700 | Peroxidase (PF00141) | Peroxidase | 10 | 347 | 37.6 | 6.05 | Chlo/Extr | Chlo |
| Phvul.009G093000 | Peroxidase (PF00141) | Peroxidase | 10 | 317 | 34.2 | 8.38 | Chlo | Chlo | |
| Phvul.009G126500 | Peroxidase (PF00141) | Peroxidase | 12 | 436 | 47.8 | 8.67 | Chlo | Chlo | |
| Phvul.009G126500 | Peroxidase (PF00141) | Peroxidase | 11 | 387 | 42.4 | 8.51 | Chlo | Chlo | |
| Phvul.011G035000 | Peroxidase (PF00141) | Peroxidase | 9 | 287 | 31.6 | 7.10 | Cyto | Cyto | |
| Phvul.011G071300 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27 | 5.54 | Cyto | Cyto | |
| Populus trichocarpa Torr. & A.Gray ex. Hook. | Potri.004G174500 | Peroxidase (PF00141) | Peroxidase | 9 | 286 | 31.5 | 6.67 | Cyto | Cyto |
| Potri.005G161900 | Peroxidase (PF00141) | Peroxidase | 10 | 347 | 37.8 | 7.59 | Chlo/Mito | Chlo | |
| Potri.005G179200 | Peroxidase (PF00141) | Peroxidase | 10 | 345 | 37.8 | 5.98 | Cyto | Chlo/Mito | |
| Potri.006G132200 | Peroxidase (PF00141) | Peroxidase | 9 | 249 | 27.4 | 5.27 | Cyto | Cyto | |
| Potri.006G254500 | Peroxidase (PF00141) | Peroxidase | 10 | 337 | 36.7 | 8.44 | Chlo | Chlo | |
| Potri.009G015400 | Peroxidase (PF00141) | Peroxidase | 9 | 249 | 27.3 | 5.53 | Cyto | Cyto | |
| Potri.009G134100 | Peroxidase (PF00141) | Peroxidase | 9 | 286 | 31.4 | 7.06 | Cyto | Cyto | |
| Prunus persica (L.) Batsch | ppa006270m | Peroxidase (PF00141) | Peroxidase | 11 | 420 | 45.4 | 8.48 | Chlo | Chlo |
| ppa008008m | Peroxidase (PF00141) | Peroxidase | 10 | 349 | 38.4 | 6.09 | Mito/Chlo/Extr | Chlo | |
| ppa009582m | Peroxidase (PF00141) | Peroxidase | 9 | 286 | 31.4 | 6.21 | Cyto | Cyto | |
| ppa010413m | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.3 | 5.76 | Cyto | Cyto | |
| ppa010426m | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.6 | 5.37 | Cyto | Cyto | |
| ppa015878m | Peroxidase (PF00141) | Peroxidase | 10 | 319 | 34.3 | 6.24 | Chlo | Chlo | |
| Sorghum bicolor (L.) Moench | Sobic.001G410200 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.2 | 5.55 | Cyto | Cyto |
| Sobic.002G431100 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.1 | 5.18 | Cyto | Cyto | |
| Sobic.004G175500 | Peroxidase (PF00141) | Peroxidase | 13 | 473 | 51.1 | 5.03 | Chlo | Chlo | |
| Sobic.006G021100 | Peroxidase (PF00141) | Peroxidase | 9 | 476 | 52.1 | 8.97 | Nucl | Chlo | |
| Sobic.006G084400 | Peroxidase (PF00141) | Peroxidase | 11 | 344 | 37.2 | 8.60 | Mito/Chlo | Chlo | |
| Sobic.006G204000 | Peroxidase (PF00141) | Peroxidase | 11 | 395 | 42.9 | 8.74 | Mito/Chlo | Chlo | |
| Sobic.007G177000 | Peroxidase (PF00141) | Peroxidase | 8 | 289 | 31.5 | 7.73 | Cyto | Cyto | |
| Sobic.007G205600 | Peroxidase (PF00141) | Peroxidase | 10 | 333 | 36.2 | 7.58 | Chlo | Chlo | |
| Solanum lycopersicum L. | Solyc01g111510 | Peroxidase (PF00141) | Peroxidase | 8 | 287 | 31.6 | 7.10 | Cyto | Cyto |
| Solyc04g074640 | Peroxidase (PF00141) | Peroxidase | 10 | 345 | 37.6 | 7.60 | Chlo/Mito | Chlo | |
| Solyc06g005150 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.3 | 5.86 | Cyto | Cyto | |
| Solyc06g060260 | Peroxidase (PF00141) | Peroxidase | 10 | 345 | 37.8 | 8.48 | Chlo | Chlo | |
| Solyc08g059760 | Peroxidase (PF00141) | Peroxidase | 10 | 326 | 35.4 | 5.65 | Chlo | Chlo | |
| Solyc09g007270 | Peroxidase (PF00141) | Peroxidase | 9 | 250 | 27.6 | 5.63 | Cyto | Cyto | |
| Solyc11g018550 | Peroxidase (PF00141) | Peroxidase | 12 | 421 | 46.0 | 8.20 | Chlo | Chlo | |
Protein domain families were searched in Pfam database.
Cyto, Cytosolic; Chlo, Chloroplastic; Mito, Mitochondrial; Nucl, Nuclear; Extr, Extracellular.
More than one localization specified in a single column also shows the significance of other entries in order.
Sequence analysis of APX genes/proteins
A total of 120 APX homologs were identified in protein datasets of 18 plant species using Arabidopsis APX1-6, APXT, and APXS sequences by homology search. Identified APX sequences contained the peroxidase (PF00141) protein family domain. They encoded a protein of 197–478 amino acids residues (average length 323.9) and 23.7–52.1 kDa molecular weight with 5.03–9.23 pI value. The sequence variations in analyzed APXs demonstrated a correlation with their putative localizations, thereby indicated the presence of transit residues (Table 5). Molecular cloning studies from A. thaliana have demonstrated that APX1, APX2, and APX6 are polypeptides of 250, 251, and 329 amino acids, respectively, with cytosolic localizations but without transit peptide (Davletova et al., 2005; Jones et al., 2009; Aryal et al., 2011). APX3 and APX5 consisted of 287 and 279 amino acids, respectively, with peroxisomal localizations; however, sites of transit peptide residues are not precisely specified (Panchuk et al., 2002; Narendra et al., 2006; Bienvenut et al., 2012). APX4 is a 349 amino acids protein with chloroplastic localization, including 1–82 residues as transit peptide from the N-terminal site (Kieselbach et al., 2000; Panchuk et al., 2005; Aryal et al., 2011). APXT is a 426 amino acids chloroplastic protein, including 1–78 residues of transit peptide (Theologis et al., 2000; Panchuk et al., 2005). APXS consists of 372 amino acids with chloroplastic and/or mitochondrial localizations, but the exact site of the transit peptide is not specified (Jespersen et al., 1997; Mayer et al., 1999; Chew et al., 2003). In the present study, multiple-alignment of APX sequences revealed that chloroplastic/mitochondrial-related APXs contained the transit peptide residues in approximately 1–90 amino acids from the N-terminal site while cytosolic APXs did not have any putative transit peptide (Supplementary Figure S4). Thus, the analyzed APX sequences were gathered in two main groups based on subcellular localizations, such as chloroplastic/mitochondrial APXs (i) and cytosolic APXs (ii).
In addition, the regions corresponding to the transit peptide sites in analyzed sequences did not demonstrate any particular pattern. This could indicate that less conservancy in transit peptides may be associated with the functional diversities of APXs at various targets. Besides, APX transcripts mainly consisted of 8–12 exons, supporting the relatively less conserved structure of APXs compared to GPXs. However, alignment analysis also demonstrated the presence of a considerable degree of conserved residues in the main sites of enzyme (Supplementary Figure S5). Moreover, to analyze the availability of any conserved motif pattern/s in APX sequences, the most conserved five motif sequences of APX homologs were searched using MEME tool (Table 6). Motif 1 was 29 residues long, motif 2 and 4 were 21 residues, motif 3 was 32 residues, and motif 5 was 25 residues in length. However, only motifs 2 and 3 had a relation with the peroxidase (PF00141) protein family, and in this case were present in most of the sequences. This could indicate the highly conserved structures of APX sequences at those sites with peroxidase activity.
Table 6.
Most conserved five motifs of ascorbate peroxidase (APX) homologs in 18 plant species.
| Motif | Width | Identified site no. | Sequence | Protein domain familya |
|---|---|---|---|---|
| 1 | 29 | 120 of 120 | CHPIMLRLAWHDAGTYDKNTKTWGPNGSI | Not found |
| 2 | 21 | 101 of 120 | MGLNDQDIVALSGGHTLGRCH | Peroxidase (PF00141) |
| 3 | 32 | 119 of 120 | IITYADLYQLAGVVAVEVCGGPTIPMHCGRND | Peroxidase (PF00141) |
| 4 | 21 | 118 of 120 | DPEFRPWVEKYAEDQDAFFRD | Not found |
| 5 | 25 | 84 of 120 | ERSGFEQPWTVNWLKFDNSYFKEIL | Not found |
Protein domain families have been searched in Pfam database.
Furthermore, alignment analysis also demonstrated that Asp (D) and Gly-Gly (GG) residues are strictly conserved in all aligned sequences, indicating their potential functions in enzyme activity and/or stability (Supplementary Figure S6). To infer a functional relationship between these conserved residues and APX sequences, we searched for the known binding residues of model organism Arabidopsis APXs in the UniProtKB database (http://www.uniprot.org/uniprot/). The following residues were reported as potential active and metal binding residues for Arabidopsis GPX1-6, APXT, and APXS: APX1 (Arg-38, His-42, His-163, Thr-164, Thr-180, Asn-182, Ile-185, Asp-187), APX2 (Arg-39, His-43, His-163, Thr-164, Thr-180, Asn-182, Ile-185, Asp-187), APX3 (Arg-36, His-40, His-160, Thr-161, Thr-177, Asp-184), APX5 (Arg-35, His-39, His-158, Thr-159, Thr-175, Asp-182), APX6 (Arg-119, His-123, His-224), APXT (Arg-108, His-112, His-241, Thr-242, Thr-274, Asp-281), and APXS (Arg-129, His-133, His-262, Thr-263, Thr-295, Asp-302). These active and metal binding residues did not correspond to any of the strictly conserved residues in analyzed APX sequences but they were found to be conserved at considerable rates. However, when taken into consideration that some of the strictly conserved residues in analyzed GPX sequences correspond to the catalytic sites of the enzymes, we can make an inference that these strictly conserved residues in APX sequences may also be associated with the peroxidase activity of the enzyme.
Phylogenetic analysis of APXs
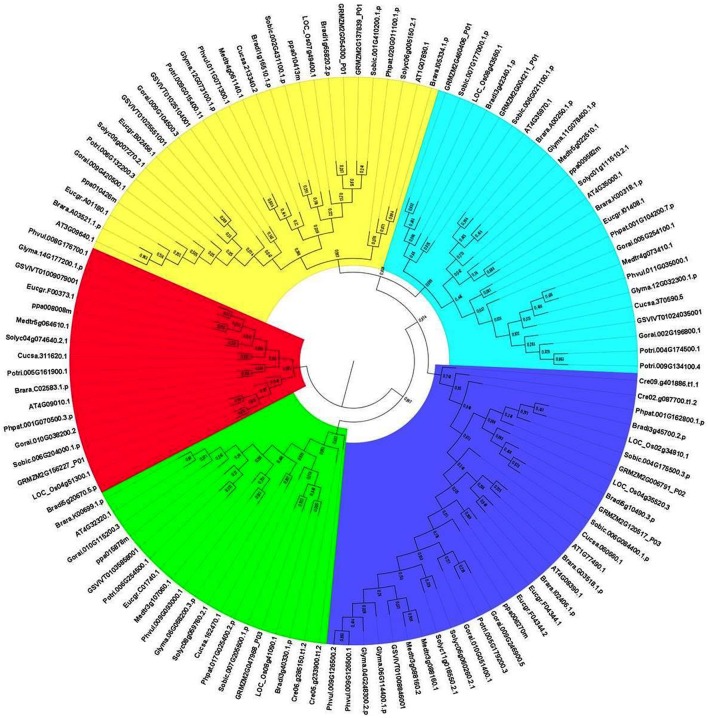
To analyze the evolutionary relationship between identified APX homologs, the phylogenetic tree was constructed by MEGA 6 using the Maximum Likelihood (ML) method with 1000 bootstraps (Figure 5). The constructed tree was divided into five major groups based on the tree topology, and each group was indicated with a different color segment. Blue, red, and green segments included the chloroplast/mitochondria-related APXs with relatively longer, medium and short sequences, respectively, whereas cyan and yellow segments mainly contained longer and shorter cytosolic APX sequences, respectively. Annotation of each segment was based on the consensus of two subcellular localization servers, CELLO and WoLF PSORT, as well as tree topology for ambiguous sequences. Overall, it was observed that cytosolic-related APXs clustered together, while alternatively chloroplast/mitochondrial-related APXs were together. In addition, in clustering of sequences at sub-branches was primarily based on the sequence length and monocot/dicot separation. However, there were considerable variations between sequences, even those belonging to the same subcellular localization. It is thought that these sequence variations could be attributed to the various functional diversities of APXs and/or be associated with different subcellular localizations. Moreover, some sequences were also available with different subcellular localizations in the same clade, indicating the possibility of gene duplication events in formation of some APX genes. The gene duplication events were searched based on the previously designated protocol (Gu et al., 2002). In doing so, several segmental and tandem duplications were identified between some APX pairs (Table 7). The identified segmental or tandem duplications in APX genes were observed between either chloroplastic and chloroplastic, or cytosolic and cytosolic forms. This could indicate the possibility of gene duplication events in the formation of close APX homologs.
Figure 5.
Phylogenetic tree of ascorbate peroxidase (APX) homologs from 18 plant species. Tree was constructed by MEGA 6 using Maximum likelihood (ML) method with 1000 bootstraps. Segment classification based on the consensus of two subcellular localization servers, CELLO and WoLF PSORT as well as tree topology for ambiguous sequences. Blue, red, and green segments include chloroplast/mitochondrial related APXs with mainly longer, medium and short sequences, respectively, while cyan and yellow segments contain longer and shorter cytosolic APX sequences, respectively.
Table 7.
The segmental and tandem duplications in some ascorbate peroxidase (APX) pairs.
| Duplication type | Species name | Duplicated pairs |
|---|---|---|
| Segmental duplication Pairs | Brachypodium distachyon (L.) P.Beauv. | Bradi5g10490-Bradi3g45700 |
| Eucalyptus grandis W. Hill ex Maiden | Eucgr.A01180-Eucgr.B02456 | |
| Glycine max (L.) Merr. | Glyma.06G114400-Glyma.04G248300 | |
| Glyma.11G078400-Glyma.12G032300 | ||
| Gossypium raimondii Ulbr. | Gorai.002G196800-Gorai.005G254100 | |
| Gorai.009G246900-Gorai.010G051400 | ||
| Vitis vinifera L. | GSVIVG01025104001- | |
| GSVIVG01025551001 | ||
| Oryza sativa L. | LOC_Os04g35520-LOC_Os02g34810 | |
| Populus trichocarpa Torr. & A.Gray ex. Hook. | Potri.004G174500-Potri.009G134100 | |
| Potri.006G132200-Potri.009G015400 | ||
| Prunus persica (L.) Batsch | ppa010431m.g-ppa010426m.g | |
| Sorghum bicolor (L.) Moench | Sobic.001G410200-Sobic.002G431100 | |
| Sobic.006G084400-Sobic.004G175500 | ||
| Sobic.007G177000-Sobic.006G021100, | ||
| Solanum lycopersicum L. | Solyc06g005150.2-Solyc09g007270.2 | |
| Solyc06g060260.2-Solyc11g018550.2 | ||
| Tandem duplication Pairs | Brachypodium distachyon (L.) P.Beauv. | Bradi1g16510-Bradi1g65820 |
| Gossypium raimondii Ulbr. | Gorai.009G104500-Gorai.009G420500 | |
| Zea mays L. | GRMZM2G006791-GRMZM2G120517 | |
| GRMZM2G054300-GRMZM2G137839 |
Expression profile analysis of APXs
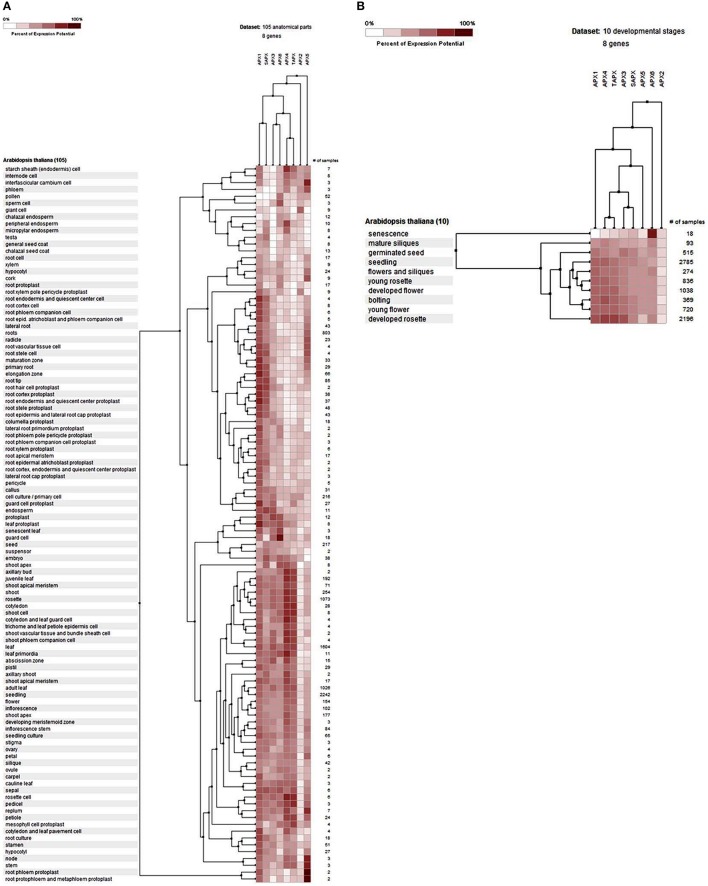
The gene expression profiles of APXs were analyzed at 105 anatomical parts and 10 developmental stage levels using model organism A. thaliana APXs from Genevestigator platform (Figure 6). Eight Arabidopsis genes, namely APX1 (AT1G07890), APX2 (AT3G09640), APX3 (AT4G35000), APX4 (AT4G09010), APX5 (AT4G35970), APX6 (AT4G32320), TAPX (AT1G77490), and SAPX (AT4G08390), were retrieved from the “Affymetrix Arabidopsis ATH1 Genome Array” platform using the Genevestigator interface. Thereafter, conditions and genes with similar profiles were comparatively analyzed using Hierarchical clustering tool with Euclidean distance method.
Figure 6.
Expression profile of Arabidopsis ascorbate peroxidase APX1-6, TAPX and SAPX genes at 105 anatomical parts (A) and 10 developmental stage levels (B). Genes and conditions with similar profiles were comparatively analyzed using hierarchical clustering tool with Euclidean distance method at Genevestigator platform.
At the anatomical level (Figure 6A), APX genes were expressed in almost all analyzed tissues of Arabidopsis with various folds. It was clear that the expression levels of genes were closely related with the expressed tissue type/s. For example, both cytosolic APX1 and chloroplastic/mitochondrial SAPX had significantly higher expression in actively growing zones, as well as many root and root protoplast-related structures. APX3, APX4, APX6, and TAPX were expressed in various shoot, bud, leaf, flower and seed related tissues at considerable rates. All these indicated that stress factors, actively growing tissues as well as normal physiological and metabolic changes could induce the expression of APX genes in tissue-dependent way. All these metabolic activities or their related consequences could exert the stresses to the cells. Many studies have further demonstrated that abiotic/abiotic stress factors such as heavy metal, drought, water, heat, cellular H2O2 level, oxidative state of the cell could increase the expression of APX genes to either suppress or eliminate the stressors (Ishikawa and Shigeoka, 2008; Koussevitzky et al., 2008; Yang et al., 2008; Petrov and Van Breusegem, 2012). For example, overexpression of Solanum melongena APX6 in transgenic O. sativa seedlings demonstrated high flood tolerance, reduced oxidative injury and fast plant growth rates (Chiang et al., 2015a). APX regulation by nitric oxide (NO) as a redox indicator in oxidative stress or as part of hormone induced signaling pathway in lateral root development were demonstrated (Correa-Aragunde et al., 2015). S-nitrosylation of Arabidopsis APX1 at Cys32 increased the H2O2 scavenging activity of enzyme, resulting in improved oxidative stress tolerance (Yang et al., 2015). Overexpression of APX and Cu/Zn SOD increased the drought resistance and recovery capacity from drought stress in Ipomoea batatas (Lu et al., 2015). Overexpressed Brassica campestris APX gene in transgenic Arabidopsis enhanced the heat tolerance via elimination of H2O2 (Chiang et al., 2015b). Therefore, increased APX activity in cells is an indicator of the presence of stress factors. At the developmental level (Figure 6B), the expression profile of Arabidopsis APXs was analyzed at 10 developmental stages: senescence, mature siliques, flowers and siliques, developed flower, young rosette, germinated seed, seedling, bolting, young flower, and developed rosette. In all developmental stages, APXs were relatively expressed. However, the expression pattern in senescence was slightly different from other developmental stages, notably cytosolic APX6 showed the highest expression. Interestingly, the Arabidopsis GPX6 gene also demonstrated the highest expression fold at senescence stage, inferring the possibility of functional similarities of these two enzymes. Overall, the expression profile and fold of APXs in various tissues and stages show that cells are constantly put under stress even with normal physiological and metabolic changes, requiring plants to eliminate these stressors.
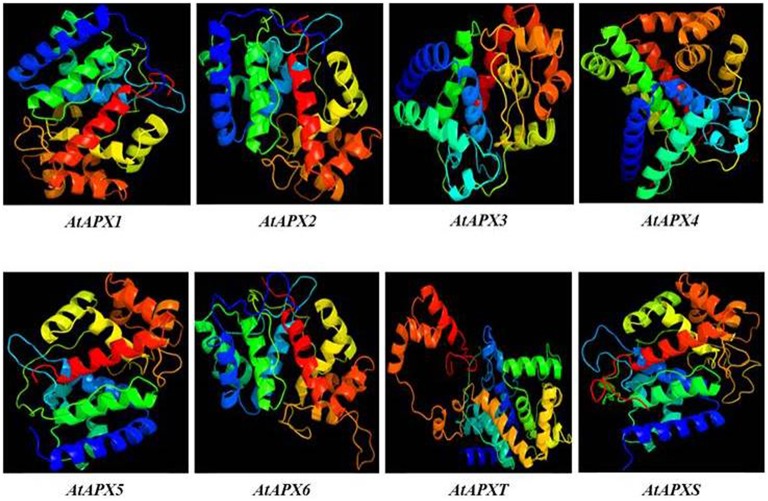
3D structure analysis of APXs
3D models of eight identified Arabidopsis APX sequences were constructed by using Phyre2 server (Figure 7). The modeled sequences were AT1G07890.1 (APX1), AT3G09640.1 (APX2), AT4G35000.1 (APX3), AT4G09010.1 (APX4), AT4G35970.1 (APX5), AT4G32320.1 (APX6), AT1G77490.1 (APXT), and AT4G08390.1 (APXS). In modeling, six templates such as 1APX:A (APX1), 1OAF:A (APX2, APX3 and APX5), 3RRW:B (APX4), 1BGP:A (APX6), 1ITK:B (APXT), and 1IYN:A (APXS) were used to maximize the alignment coverage, percentage identity, and confidence for the submitted sequences. Predicted models showed the ≥96% of residues were within the allowed region in Ramachandran plot, indicating that structures were acceptably high in quality. To analyze the divergence or similarity in generated models, the structures were superposed in order to calculate the percentage of structural overlap and RMSD values (Table 8). The superposition of APX sequences demonstrated that APX2-APX3, APX2-APX5, and APX3-APX5 pairs have highly conserved structural overlap (100%) with 0.00, 0.38, and 0.38 RMSD values, respectively. These conserved pairs primarily shared the cytosolic and/or peroxisomal localizations, inferring the possibility of a functional relationship between them. In addition, the APX1-APX2, 3, and 5 pairs had very high structural similarity with ≥99 structural overlaps. Therefore, it could be deduced that APX members topologically demonstrated highly conserved structures, despite their functional diversities in different cellular compartments.
Figure 7.
3D models of predicted Arabidopsis ascorbate peroxidase APX1-6, APXT, and APXS sequences. Models were constructed by using Phyre2 server for AT1G07890.1 (APX1), AT3G09640.1 (APX2), AT4G35000.1 (APX3), AT4G09010.1 (APX4), AT4G35970.1 (APX5), AT4G32320.1 (APX6), AT1G77490.1 (APXT), and AT4G08390.1 (APXS) sequences, and colored by rainbow from N- to C-terminus.
Table 8.
Structural overlap (%)/RMSD values in superposed Arabidopsis ascorbate peroxidases (APXs).
| APX1 | APX2 | APX3 | APX4 | APX5 | APX6 | APXS | APXT | |
|---|---|---|---|---|---|---|---|---|
| APX1 | – | 99.19/0.43 | 99.59/0.41 | 75.10/1.75 | 99.58/0.51 | 81.53/1.52 | 95.18/0.95 | 89.16/1.49 |
| APX2 | 99.19/0.41 | – | 100.00/0.00 | 75.0071.78 | 100.00/0.38 | 81.45/1.58 | 95.16/0.86 | 87.90/1.35 |
| APX3 | 99.59/0.41 | 100.00/0.00 | – | 75.31/1.85 | 100.00/0.38 | 82.30/1.56 | 97.12/0.86 | 88.48/1.38 |
| APX4 | 75.10/1.75 | 75.40/1.73 | 73.66/1.91 | – | 73.22/1.92 | 72.29/1.88 | 75.79/1.72 | 66.27/1.83 |
| APX5 | 99.58/0.48 | 100.00/0.38 | 100.00/0.38 | 74.48/1.85 | – | 81.17/1.67 | 97.49/0.90 | 89.12/1.31 |
| APX6 | 82.73/1.57 | 82.26/1.70 | 83.13/1.68 | 69.88/1.82 | 84.10/1.70 | – | 81.12/1.50 | 73.90/1.66 |
| APXS | 95.18/1.00 | 95.97/0.97 | 97.12/1.00 | 75.00/1.73 | 97.49/1.05 | 82.73/1.55 | – | 83.52/1.38 |
| APXT | 87.55/1.47 | 89.52/1.36 | 89.71/1.32 | 67.46/1.87 | 90.79/1.32 | 74.70/1.80 | 82.42/1.34 | – |
Red-color highlighted pairs show the highly conserved structural overlaps.
Interaction partner analysis of APXs
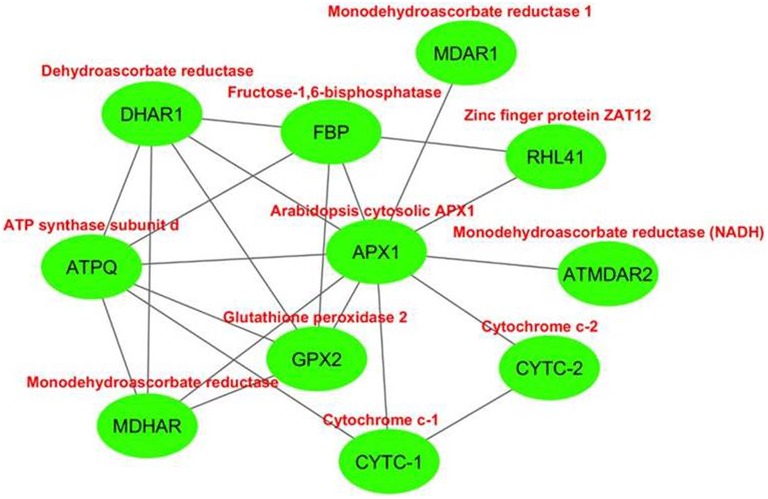
The interactome network was constructed for 10 putative interactors of Arabidopsis cytosolic APX1 using Cytoscape with STRING data (Figure 8). MDHAR (monodehydroascorbate reductase), GPX2 (glutathione peroxidase 2), DHAR1 (dehydroascorbate reductase), MDAR1 (monodehydroascorbate reductase 1), RHL41 (zinc finger protein ZAT12), ATPQ (ATP synthase subunit d), FBP (fructose-1,6-bisphosphatase), ATMDAR2 [monodehydroascorbate reductase (NADH)], CYTC-1 (cytochrome c-1), and CYTC-2 (cytochrome c-2) proteins were predicted as the main interaction partners of Arabidopsis cytosolic APX1. MDHAR, MDAR1 and ATMDAR2 catalyze the conversion of monodehydroascorbate to ascorbate (Chew et al., 2003). GPX2 is a type of H2O2-scavenging enzyme and a crucial component in reactive oxygen network (Tanaka et al., 2005). DHAR1 has dual functions: soluble protein, it demonstrates GSH-dependent thiol transferase and dehydroascorbate (DHA) reductase activities, and is involved in redox homeostasis. As a peripheral membrane protein, it functions as voltage-gated ion channel (Dixon et al., 2002; Sasaki-Sekimoto et al., 2005). RHL41 affects in modulation of light acclimation, and cold and oxidative stress responses (Rizhsky et al., 2004; Davletova et al., 2005). ATPQ functions in ATP production (Carraro et al., 2014). FBP is reported to be a key component in photosynthetic sucrose synthesis (Cho et al., 2012). CYTC-1 and CYTC-2 are electron carrier proteins related with mitochondrial electron transport chain (Welchen and Gonzalez, 2005). In light of putative interaction partner analysis, it was apparent that Arabidopsis cytosolic APX1 is either directly or indirectly associated with redox homeostasis, stress adaptation and photosynthesis/respiration-related pathways. This could also help in better understanding the functional role of APX1 in various plant defense mechanisms.
Figure 8.
Predicted 10 interaction partners of Arabidopsis cytosolic ascorbate peroxidase APX1. Interactome was generated using cytoscape with STRING data. MDHAR (monodehydroascorbate reductase), GPX2 (glutathione peroxidase 2), DHAR1 (dehydroascorbate reductase), MDAR1 (monodehydroascorbate reductase 1), RHL41 (zinc finger protein ZAT12), ATPQ (ATP synthase subunit d), FBP (fructose-1,6-bisphosphatase), ATMDAR2 (monodehydroascorbate reductase (NADH)), CYTC-1 (cytochrome c-1), and CYTC-2 (cytochrome c-2) proteins were predicted as main interaction partners of Arabidopsis cytosolic APX1.
Comparison of APX and GPX sequences
A strict homology search of Arabidopsis GPX1-8 sequences in proteome datasets of 18 specified plant species has given a total of 87 putative GPX sequences; however, homology search of Arabidopsis APX1-6, APXT, and APXS in proteome datasets of these species identified a total of 120 putative APXs (Tables 1, 5). Sequences of GPX homologs contained the GPX (PF00255) protein family domain while APX homologs included the peroxidase (PF00141) domain. GPX genes encoded a protein of 166–262 residues with 18.4–29.7 kDa molecular weight and 4.59–9.60 pI value, while APXs encoded a polypeptide of 197–478 residues with 23.7–52.1 kDa molecular weight and 5.03–9.23 pI value. GPX transcripts mainly contained six exons; whereas, APX usually had 8–12 exons, implicating the relatively less conserved structure of APXs compared to GPXs. Sequence variations in GPX and APX homologs primarily derived from the “transit peptide” residues between organelle and non-organelle related sequences. Besides, regions corresponding to transit peptide sites in APX/GPX sequences did not demonstrate any particular pattern, indicating the less conserved structure of transit peptides thereby the functional diversities of APXs/GPXs at various targets. In addition, multiple-alignment analyses demonstrated the presence of a considerable degree of conserved residues in main sites of both enzymes. In GPX phylogeny, cytosolic-, nuclear-, extra cellular,- and plasma membrane-related GPXs were relatedly clustered while chloroplast/mitochondrial-related GPXs grouped together. APX phylogeny also showed similar clustering pattern, in which cytosolic-related APXs were relatedly clustered while chloroplast/mitochondrial-related APXs were together. This indicates that presence/absence of “transit peptide” residues was the main determinant in phylogenetic distribution of APX/GPX sequences. Moreover, presence of sequences with different subcellular localizations in the same phylogenetic group inferred the possibility of gene duplication events in formation of some APX/GPX sequences. Several segmental duplications were identified in some GPX pairs, while several segmental and tandem duplications were available in some APX pairs. Expression profiles of GPX and APX genes in model organism Arabidopsis indicated that stress factors, actively growing tissues, even normal physiological, and metabolic changes could induce the expression of APX/GPX genes. Interactome analyses of Arabidopsis cytosolic APX1 and GPX2 also implicated that both enzymes are closely related with antioxidant and redox homeostasis, secondary metabolite metabolisms and stress adaptation thereby supporting the functional roles of APXs/GPXs in H2O2-scavenging and plant defense. Despite of some minor variations, APX and GPX members, they topologically demonstrated highly conserved structure.
Conclusions
The presence or absence of transit peptide residues are the main contributing factors in subcellular localization and phylogenetic distribution of APX/GPXs. The APX/GPX expression is highly associated with the metabolic state of the cells. In addition, there are grounds for belief that these two enzymes work together in various pathways such as antioxidant and secondary metabolite metabolisms, redox homeostasis, stress adaptation, and photosynthesis/respiration. This also supports the functional role of these enzymes in H2O2-scavenging, thereby implicating their importance in the plant defense. However, further molecular and physiological studies are required to elucidate the various functional roles of APX/GPX isoforms.
Author contributions
IK and EF contributed to the study conception and design. KK performed experiments and collected data. Data analysis and interpretation were performed by RV. IK, EF, KK, and RV prepared, and NA performed critical reading and revision of the manuscript. IO and EF supervised and MO coordinated this work. All the authors read and approved the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
NA gratefully acknowledges the partial financial supports received from FCT (Government of Portugal) through contract (SFRH/BPD/84671/2012), the Aveiro University Research Institute/CESAM (UID/AMB/50017/2013), “COMPETE” through Project n.° FCOMP-01-0124-FEDER-02800 (FCT PTDC/AGR-PRO/4091/2012), and to FCT/MEC through national funds, and the co-funding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00301
References
- Anjum N. A., Gill S. S., Gill R., Hasanuzzaman M., Duarte A. C., Pereira E., et al. (2014). Metal/metalloid stress tolerance in plants: role of ascorbate, its redox couple, and associated enzymes. Protoplasma 251, 1265–1283. 10.1007/s00709-014-0636-x [DOI] [PubMed] [Google Scholar]
- Anjum N. A., Sofo A., Scopa A., Roychoudhury A., Gill S. S., Iqbal M., et al. (2015). Lipids and proteins - major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. 22, 4099–4121. 10.1007/s11356-014-3917-1 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Aryal U. K., Krochko J. E., Ross A. R. (2011). Identification of phosphoproteins in Arabidopsis thaliana leaves using polyethylene glycol fractionation, immobilized metal-ion affinity chromatography, two-dimensional gel electrophoresis and mass spectrometry. J. Proteome Res. 11, 425–437. 10.1021/pr200917t [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bela K., Horváth E., Gallé Á., Szabados L., Tari I., Csiszár J. (2015). Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 176, 192–201. 10.1016/j.jplph.2014.12.014 [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Møller A. L., Kristiansen K. A., Schulz A., Møller I. M., Schjoerring J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. 10.1074/jbc.M603761200 [DOI] [PubMed] [Google Scholar]
- Bienvenut W. V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., et al. (2012). Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol. Cell Proteomics 11, M111.015131. 10.1074/mcp.m111.015131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro M., Giorgio V., Šileikytë J., Sartori G., Forte M., Lippe G., et al. (2014). Channel formation by yeast F-ATP synthase and the role of dimerization in the mitochondrial permeability transition. J. Biol. Chem. 289, 15980–15985. 10.1074/jbc.C114.559633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caverzan A., Passaia G., Rosa S. B., Ribeiro C. W., Lazzarotto F., Margis-Pinheiro M. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1019. 10.1590/S1415-47572012000600016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O., Whelan J., Millar A. H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. 10.1074/jbc.M307525200 [DOI] [PubMed] [Google Scholar]
- Chiang C. M., Chen L. F. O., Shih S. W., Lin K. H. (2015a). Expression of eggplant ascorbate peroxidase increases the tolerance of transgenic rice plants to flooding stress. J. Plant Biochem. Biotechnol. 24, 257–267. 10.1007/s13562-014-0265-7 [DOI] [Google Scholar]
- Chiang C. M., Chien H. L., Chen L. F. O., Hsiung T. C., Chiang M. C., Chen S. P., et al. (2015b). Overexpression of the genes coding ascorbate peroxidase from Brassica campestris enhances heat tolerance in transgenic Arabidopsis thaliana. Biol. Plant 59, 305–315. 10.1007/s10535-015-0489-y [DOI] [Google Scholar]
- Cho M. H., Jang A., Bhoo S. H., Jeon J. S., Hahn T. R. (2012). Manipulation of triose phosphate/phosphate translocator and cytosolic fructose-1, 6-bisphosphatase, the key components in photosynthetic sucrose synthesis, enhances the source capacity of transgenic Arabidopsis plants. Photosynth. Res. 111, 261–268. 10.1007/s11120-012-9720-2 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N., Foresi N., Lamattina L. (2015). Nitric oxide is an ubiquitous signal for maintaining redox balance in plant cells: regulation of ascorbate peroxidase as a case study. J. Exp. Bot. 66, 2913–2921. 10.1093/jxb/erv073 [DOI] [PubMed] [Google Scholar]
- Criqui M. C., Jamet E., Parmentier Y., Marbach J., Durr A., Fleck J. (1992). Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol. Biol. 18, 623–627. 10.1007/BF00040684 [DOI] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. 10.1105/tpc.104.026971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y., Xu H., Li G., Yu A., Yu X., Hu W., et al. (2014). Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 41, 4919–4927. 10.1007/s11033-014-3358-4 [DOI] [PubMed] [Google Scholar]
- Dietz K. J. (2011). Peroxiredoxins in plants and cyanobacteria. Antiox. Redox Signal. 15, 1129–1159. 10.1089/ars.2010.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. P., Davis B. G., Edwards R. (2002). Functional divergence in the glutathione transferase superfamily in plants identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 277, 30859–30869. 10.1074/jbc.M202919200 [DOI] [PubMed] [Google Scholar]
- Dynowski M., Schaaf G., Loque D., Moran O., Ludewig U. (2008). Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 414, 53–61. 10.1042/BJ20080287 [DOI] [PubMed] [Google Scholar]
- Erfle H., Ventzki R., Voss H., Rechmann S., Benes V., Stegemann J., et al. (2000). Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408, 820–822. 10.1038/35048706 [DOI] [PubMed] [Google Scholar]
- Ferreira Neto J. R. C., Pandolfi V., Guimaraes F. C. M., Benko-Iseppon A. M., Romero C., De Oliveira Silva R. L., et al. (2013). Early transcriptional response of soybean contrasting accessions to root dehydration. PLoS ONE 8:e83466. 10.1371/journal.pone.0083466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcroy P., Vansuyt G., Kushnir S., Inzé D., Briat J. F. (2004). Iron-regulated expression of a cytosolic ascorbate peroxidase encoded by the APX1 gene in Arabidopsis seedlings. Plant Physiol. 134, 605–613. 10.1104/pp.103.029876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., et al. (2013). STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, 808–815. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. Y. (2014). Cloning of a new glutathione peroxidase gene from tea plant (Camellia sinensis) and expression analysis under biotic and abiotic stresses. Bot. Stud. 55:7 10.1186/1999-3110-55-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Chen J., Ma T., Li H., Wang N., Li Z., et al. (2014). The glutathione peroxidase gene family in Thellungiella salsuginea: genome-wide identification, classification, and gene and protein expression analysis under stress conditions. Int. J. Mol. Sci. 15, 3319–3335. 10.3390/ijms15023319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook, ed Walker J. M. (Totowa, NJ: Humana; ), 571–607. [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Cavalcanti A., Chen F. C., Bouman P., Li W. H. (2002). Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262. 10.1093/oxfordjournals.molbev.a004079 [DOI] [PubMed] [Google Scholar]
- Haslekås C., Stacy R. A., Nygaard V., Culiáñez-Macià F. A., Aalen R. B. (1998). The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol. Biol. 36, 833–845. [DOI] [PubMed] [Google Scholar]
- Herbette S., de Labrouhe D. T., Drevet J. R., Roeckel-Drevet P. (2011). Transgenic tomatoes showing higher glutathione peroxydase antioxidant activity are more resistant to an abiotic stress but more susceptible to biotic stresses. Plant Sci. 180, 548–553. 10.1016/j.plantsci.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth E., Brunner S., Bela K., Papdi C., Szabados L., Tari I., et al. (2015). Exogenous salicylic acid-triggered changes in the glutathione transferases and peroxidases are key factors in the successful salt stress acclimation of Arabidopsis thaliana. Funct. Plant Biol. 42, 1129–1140. 10.1071/FP15119 [DOI] [PubMed] [Google Scholar]
- Hossain M. A., Bhattacharjee S., Armin S. M., Qian P., Xin W., Li H. Y., et al. (2015). Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant. Sci. 6:420. 10.3389/fpls.2015.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Shigeoka S. (2008). Recent advances in ascorbate biosynthesis and the physiological significance of ascorbate peroxidase in photosynthesizing organisms. Biosci. Biotechnol. Biochem. 72, 1143–1154. 10.1271/bbb.80062 [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Yoshimura K., Sakai K., Tamoi M., Takeda T., Shigeoka S., et al. (1998). Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol. 39, 23–34. 10.1093/oxfordjournals.pcp.a029285 [DOI] [PubMed] [Google Scholar]
- Islam T., Manna M., Reddy M. K. (2015). Glutathione Peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd 2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS ONE 10:e0143344. 10.1371/journal.pone.0143344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen H., Kjaersgård I. V., Ostergaard L., Welinder K. G. (1997). From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem. J. 326, 305–310. 10.1042/bj3260305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. M., MacLean D., Studholme D. J., Serna-Sanz A., Andreasson E., Rathjen J. P., et al. (2009). Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J. Proteomics. 72, 439–451. 10.1016/j.jprot.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kieselbach T., Bystedt M., Hynds P., Robinson C., Schröder W. P. (2000). A peroxidase homologue and novel plastocyanin located by proteomics to the Arabidopsis chloroplast thylakoid lumen. FEBS Lett. 480, 271–276. 10.1016/S0014-5793(00)01890-1 [DOI] [PubMed] [Google Scholar]
- Kitamura S., Shikazono N., Tanaka A. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37, 104–114. 10.1046/j.1365-313X.2003.01943.x [DOI] [PubMed] [Google Scholar]
- Koua D., Cerutti L., Falquet L., Sigrist C. J., Theiler G., Hulo N., et al. (2009). PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 37, D261–D266. 10.1093/nar/gkn680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Suzuki N., Huntington S., Armijo L., Sha W., Cortes D., et al. (2008). Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 283, 34197–34203. 10.1074/jbc.m806337200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarotto F., Teixeira F. K., Rosa S. B., Dunand C., Fernandes C. L., de Vasconcelos Fontenele A., et al. (2011). Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 191, 234–250. 10.1111/j.1469-8137.2011.03659.x [DOI] [PubMed] [Google Scholar]
- Lima-Melo Y., Carvalho F. E., Martins M. O., Passaia G., Sousa R. H., Neto M. C. L., et al. (2016). Mitochondrial GPX1 silencing triggers differential photosynthesis impairment in response to salinity in rice plants. J. Integr. Plant. Biol. [Epub ahead of print]. 10.1111/jipb.12464. [DOI] [PubMed] [Google Scholar]
- Lin X., Kaul S., Rounsley S., Shea T. P., Benito M. I., Town C. D., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. 10.1038/45471 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jiang H. Y., Chen W., Qian Y., Ma Q., Cheng B., et al. (2011). Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul. 63, 225–234. 10.1007/s10725-010-9519-0 [DOI] [Google Scholar]
- Lovell S. C., Davis I. W., Arendall W. B., III., de Bakker P. I., Word J. M., Prisant M. G., et al. (2003). Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450. 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Lu Y. Y., Deng X. P., Kwak S. S. (2015). Over expression of CuZn superoxide dismutase (CuZn SOD) and ascorbate peroxidase (APX) in transgenic sweet potato enhances tolerance and recovery from drought stress. Afr. J. Biotechnol. 9, 8378–8391. 10.5897/AJB10.926 [DOI] [Google Scholar]
- Matamoros M. A., Saiz A., Peñuelas M., Bustos-Sanmamed P., Mulet J. M., Barja M. V., et al. (2015). Function of glutathione peroxidases in legume root nodules. J. Exp. Bot. 66, 2979–2990. 10.1093/jxb/erv066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K., Schüller C., Wambutt R., Murphy G., Volckaert G., Pohl T., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777. 10.1038/47134 [DOI] [PubMed] [Google Scholar]
- Milla M. A. R., Maurer A., Rodriguez Huete A., Gustafson J. P. (2003). Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 36, 602–615. 10.1046/j.1365-313X.2003.01901.x [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trend Plant Sci. 9, 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Mullineaux P., Karpinski S. (2002). Signal transduction in response to excess light: getting out of the chloroplast. Curr. Opin. Plant Biol. 5, 43–48. 10.1016/S1369-5266(01)00226-6 [DOI] [PubMed] [Google Scholar]
- Mullineaux P. M., Karpinski S., Jiménez A., Cleary S. P., Robinson C., Creissen G. P. (1998). Identification of cDNAs encoding plastid−targeted glutathione peroxidase. Plant J. 13, 375–379. 10.1046/j.1365-313X.1998.00052.x [DOI] [PubMed] [Google Scholar]
- Najami N., Janda T., Barriah W., Kayam G., Tal M., Guy M., et al. (2008). Ascorbate peroxidase gene family in tomato: its identification and characterization. Mol. Genet. Genomics 279, 171–182. 10.1007/s00438-007-0305-2 [DOI] [PubMed] [Google Scholar]
- Narendra S., Venkataramani S., Shen G., Wang J., Pasapula V., Lin Y., et al. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57, 3033–3042. 10.1093/jxb/erl060 [DOI] [PubMed] [Google Scholar]
- Navrot N., Collin V., Gualberto J., Gelhaye E., Hirasawa M., Rey P., et al. (2006). Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 142, 1364–1379. 10.1104/pp.106.089458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi Y. K. (2011). Comparative in silico analysis of ascorbate peroxidase protein sequences from different plant species. J. Bioengg. Biomed. Sci. 1, 103 10.4172/2155-9538.1000103 [DOI] [Google Scholar]
- Nguyen M. N., Tan K. P., Madhusudhan M. S. (2011). CLICK - topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res. 39, W24–W28. 10.1093/nar/gkr393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk I. I., Volkov R. A., Schöffl F. (2002). Heat stress-and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129, 838–853. 10.1104/pp.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk I. I., Zentgraf U., Volkov R. A. (2005). Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222, 926–932. 10.1007/s00425-005-0028-8 [DOI] [PubMed] [Google Scholar]
- Passaia G., Fonini L. S., Caverzan A., Jardim-Messeder D., Christoff A. P., Gaeta M. L., et al. (2013). The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 208, 93–101. 10.1016/j.plantsci.2013.03.017 [DOI] [PubMed] [Google Scholar]
- Petrov V. D., Van Breusegem F. (2012). Hydrogen peroxide - a central hub for information flow in plant cells. AoB Plants 2012:pls014. 10.1093/aobpla/pls014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 29, 467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- Rizhsky L., Davletova S., Liang H., Mittler R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279, 11736–11743. 10.1074/jbc.M313350200 [DOI] [PubMed] [Google Scholar]
- Romiti M. (2006). Entrez Nucleotide and Entrez Protein FAQs. Entrez Sequences Help. Bethesda, MD: National Center for Biotechnology Information (US) Updated 2010 May 12. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK49541/ [Google Scholar]
- Rouhier N., Jacquot J.-P. (2005). The plant multigenic family of thiol peroxidases. Free Radical Biol. Med. 38, 1413–1421. 10.1016/j.freeradbiomed.2004.07.037 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y., Taki N., Obayashi T., Aono M., Matsumoto F., Sakurai N., et al. (2005). Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44, 653–668. 10.1111/j.1365-313X.2005.02560.x [DOI] [PubMed] [Google Scholar]
- Sharma P., Dubey R. S. (2005). Modulation of nitrate reductase activity in rice seedlings under aluminium toxicity and water stress: role of osmolytes as enzyme protectant. J. Plant Physiol. 162, 854–864. 10.1016/j.jplph.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Smith A. P., Nourizadeh S. D., Peer W. A., Xu J., Bandyopadhyay A., Murphy A. S., et al. (2003). Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S−transferase that interacts with flavonoids. Plant J. 36, 433–442. 10.1046/j.1365-313X.2003.01890.x [DOI] [PubMed] [Google Scholar]
- Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofo A., Scopa A., Nuzzaci M., Vitti A. (2015). Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Intl. J. Mol. Sci. 16, 13561–13578. 10.3390/ijms160613561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S., De Pauw P., Van Montagu M., Inzé D., Kushnir S. (1998). The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 118, 1005–1014. 10.1104/pp.118.3.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Xu J., Li Y., Lei L., Zhao L., Yang H., et al. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23, 364–380. 10.1105/tpc.110.079145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M., Oono Y., Gusev O., Matsumoto T., Yazawa T., Levinskikh M. A., et al. (2014). Genome-wide expression analysis of reactive oxygen species gene network in Mizuna plants grown in long-term spaceflight. BMC Plant Biol. 14:4. 10.1186/1471-2229-14-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Mittler R. (2006). Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol. Plant 126, 45–51. 10.1111/j.0031-9317.2005.00582.x [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Izawa S., Inoue Y. (2005). GPX2, encoding a phospholipid hydroperoxide glutathione peroxidase homologue, codes for an atypical 2-Cys peroxiredoxin in Saccharomyces cerevisiae. J. Biol. Chem. 280, 42078–42087. 10.1074/jbc.M508622200 [DOI] [PubMed] [Google Scholar]
- Teixeira F. K., Menezes-Benavente L., Galvão V. C., Margis R., Margis-Pinheiro M. (2006). Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 224, 300–314. 10.1007/s00425-005-0214-8 [DOI] [PubMed] [Google Scholar]
- Teixeira F. K., Menezes-Benavente L., Margis R., Margis-Pinheiro M. (2004). Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J. Mol. Evol. 59, 761–770. 10.1007/s00239-004-2666-z [DOI] [PubMed] [Google Scholar]
- Theologis A., Ecker J. R., Palm C. J., Federspiel N. A., Kaul S., White O., et al. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816–820. 10.1038/35048500 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilday B., Turkan I., Ozgur R., Sekmen A. H. (2014). Strategies of ROS regulation and antioxidant defense during transition from C3 to C4 photosynthesis in the genus Flaveria under PEG-induced osmotic stress. J. Plant Physiol. 171, 65–75. 10.1016/j.jplph.2013.06.016 [DOI] [PubMed] [Google Scholar]
- Wagner U., Edwards R., Dixon D. P., Mauch F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49, 515–532. 10.1023/A:1015557300450 [DOI] [PubMed] [Google Scholar]
- Wang C. L., Oliver D. J. (1996). Cloning of the cDNA and genomic clones for glutathione synthetase fromArabidopsis thaliana and complementation of agsh2 mutant in fission yeast. Plant Mol. Biol. 31, 1093–1104. 10.1007/BF00040827 [DOI] [PubMed] [Google Scholar]
- Welchen E., Gonzalez D. H. (2005). Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther-and meristem-specific expression of the Cytc-1 gene. Plant Physiol. 139, 88–100. 10.1104/pp.105.065920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder K. G. (1992). Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2, 388–393. 10.1016/0959-440X(92)90230-5 [DOI] [Google Scholar]
- Yabuta Y., Yoshimura K., Takeda T., Shigeoka S. (2000). Molecular characterization of tobacco mitochondrial L-galactono-γ-lactone dehydrogenase and its expression in Escherichia coli. Plant Cell Physiol. 41, 666–675. 10.1093/pcp/41.6.666 [DOI] [PubMed] [Google Scholar]
- Yang H., Mu J., Chen L., Feng J., Hu J., Li L., et al. (2015). S-Nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. 10.1104/pp.114.255216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. L., Yu P. L., Chung K. R. (2016). The glutathione peroxidase-mediated reactive oxygen species resistance, fungicide sensitivity and cell wall construction in the citrus fungal pathogen Alternaria alternata. Environ. Microbiol. [Epub ahead of print]. 10.1111/1462-2920.13125. [DOI] [PubMed] [Google Scholar]
- Yang Y., Han C., Liu Q., Lin B., Wang J. (2008). Effect of drought and low light on growth and enzymatic antioxidant system of Picea asperata seedlings. Acta Physiol. Plant. 30, 433–440. 10.1007/s11738-008-0140-z [DOI] [Google Scholar]
- Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. 10.1002/prot.21018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.