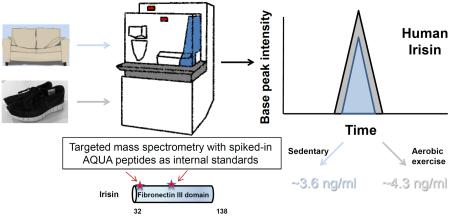
SUMMARY
Exercise provides many health benefits, including improved metabolism, cardiovascular health, and cognition. We have shown previously that FNDC5, a type I transmembrane protein, and its circulating form irisin, convey some of these benefits in mice. However, recent reports questioned the existence of circulating human irisin both because human FNDC5 has a non-canonical ATA translation start and claims that many human irisin antibodies used in commercial ELISA kits lack required specificity. In this paper we have identified and quantitated human irisin in blood using mass spectrometry with control peptides enriched with stable isotopes as internal standards. This precise state-of-the-art method shows that human irisin is mainly translated from its non-canonical start codon and circulates at ~3.6 ng/ml in sedentary individuals; this level is increased to ~4.3 ng/ml in individuals undergoing aerobic interval training. These data unequivocally demonstrate that human irisin exists, circulates and is regulated by exercise.
Keywords: FNDC5, irisin, mass spectrometry, exercise
INTRODUCTION
The health benefits of physical activity and exercise are well recognized (Hawley et al., 2014; Mann and Rosenzweig, 2012; Voss et al., 2013). Exercise is the first line of therapy for various metabolic diseases like diabetes and obesity, but exercise also improves outcomes in diseases involving other tissues, such as the heart and brain. We recently described a novel polypeptide that is secreted from skeletal muscle and is increased with exercise. Irisin is the shed extracellular domain of a transmembrane protein called FNDC5. FNDC5, when expressed from adenoviral vectors in mice, causes an elevation of irisin in the blood and improved metabolic health in recipient animals (Bostrom et al., 2012). It also stimulates the expression of a potential neuroprotective gene program in the brain, particularly in the hippocampus (Wrann et al., 2013). Several papers have studied the effects of exercise on circulating irisin in humans; positive associations between irisin plasma level and exercise have been observed in some but not all cohorts and modes of exercise (Daskalopoulou et al., 2014; Hofmann et al., 2014; Huh et al., 2014; Kraemer et al., 2014; Lee et al., 2014; Norheim et al., 2014). Data suggest that early sampling after exercise and high intensity training protocols are particularly effective at raising circulating irisin levels. Most of these studies have relied on commercial antibodies and ELISA assays.
Human FNDC5 has an atypical translation start codon ATA, in place of the more typical ATG. While it is now known that many eukaryotic mRNAs begin translation with non-ATG start codons (Ingolia et al., 2011; Ivanov et al., 2011; Peabody, 1989), two recent papers have claimed that this ATA codon in human FNDC5 represents a null mutation and therefore human irisin would not be produced (Albrecht et al., 2015; Raschke et al., 2013). These authors argue that if FNDC5 exists in humans, it is translated from a downstream ATG and hence, the irisin polypeptide is a “myth” and does not exist. In addition, these authors claim that the many papers measuring human irisin are all artifacts of poor antibody specificity (Albrecht et al., 2015; Erickson, 2013); this is despite the fact that Lee et al. had previously detected an irisin peptide in human plasma with mass spectrometry (Lee et al., 2014) In this paper we have investigated the presence of human irisin in blood using quantitative mass spectrometry. As internal standards, we synthesized irisin peptides and included a valine enriched in stable isotopes (six 13C atoms). The peptides were used to develop a quantitative platform for the measurement of human irisin; these data should facilitate future studies of this molecule in both mice and humans.
Results
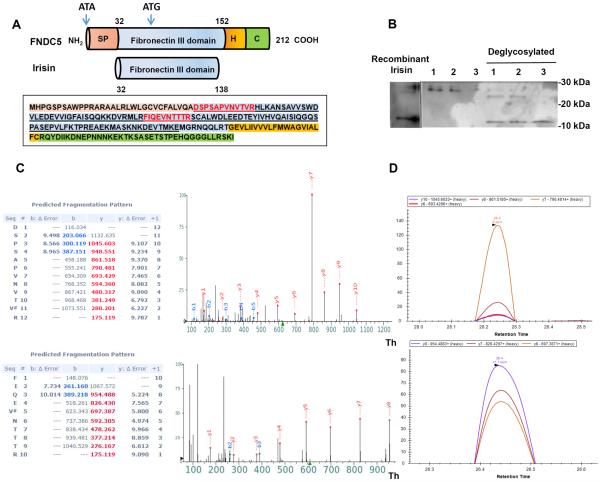
Two peptides were chosen as standards for this mass spectrometric analysis. These were both chosen because they are unique to the irisin sequence (FNDC5 ectodomain) and not encoded in any other proteins in the annotated human genome. As shown in Figure 1A, one peptide represents the most extreme N-terminal 12 amino acids (DSPSAPVNVTVR12) of the processed irisin molecule, coming immediately after the signal peptide (Fig. 1A). Importantly, this peptide is downstream of the non-canonical ATA codon but upstream of the first ATG codon in the FNDC5 mRNA. Therefore detection of this peptide would demonstrate use of the non-canonical start codon. A second tryptic peptide (48FIQEVNTTTR57) was chosen from the central portion of irisin, three amino acids downstream of the ATG. Plasma samples from human volunteers who had undergone aerobic interval training (see Methods) were used to develop this assay. These plasma samples were first treated with a commercial affinity resin to remove the very abundant albumin and immunoglobulins, so that these proteins would not hinder analysis of less abundant proteins (see Methods). Samples were then deglycosylated with the Protein Deglycosylation Mix from NEB, which contains PNGase F, O-Glycosidase, Neuraminidase, β1-4 Galactosidase, and β-N-acetylglucosaminidase, and results in complete deglycosylation. After electrophoresis, the anti-irisin antibody detected a band running at approximately 12 kDa, the predicted size of the irisin polypeptide (Fig. 1B). To characterize the synthetic heavy irisin peptides were subjected to LC-MS/MS analysis in both data dependent and parallel reaction monitoring (PRM) acquisition modes. As shown in Figure 1C, the intensity of the y ions series from the MS2 spectra for both peptides correspond to the rank order elution profile in the PRM acquisition mode, (Fig. 1D) validating that these ions can be used for identification and quantification of irisin.
Figure 1. Analysis of Irisin Peptides by Mass Spectrometry.
(A) Schematic representation of the FNDC5 protein structure (top) and irisin (bottom). SP = signal peptide, H = hydrophobic domain, C = c-terminal domain. Human FNDC5 sequence with corresponding domains colored. Human irisin sequence is underlined as well as synthetic AQUA peptides used in this study (red).
(B) Immunoblotting of irisin plasma samples from three subjects undergoing aerobic interval training with or without deglycosylation enzyme (Protein Deglycosylation Mix (NEB)) and deglycosylated recombinant irisin.
(C) MS2 spectra acquired using a Q Exactive mass spectrometer for the two synthetic AQUA peptides and their b-, y-ion series m/z values. Mass accuracy values are given in PPMs and “#” denotes the heavy valine residue.
(D) PRM elution profile for the y-ions for the AQUA peptides using Skyline software. Retention times for each peptide are labeled on the x-axis and y-axis represents the relative intensity for each y-ion peak. See also Figure S1.
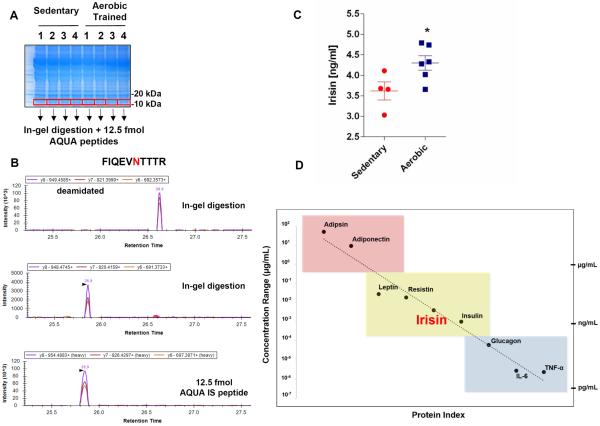
Next, for the quantification of irisin in human plasma by mass spectrometry, albumin and immunoglobulin depleted plasma from four sedentary and six aerobically interval trained subjects was deglycosylated and resolved by SDS-PAGE prior to in-gel trypsin digestion. After this, 12.5 femtomoles of each heavy peptide were spiked into the sample prior to absolute quantification (AQUA) of irisin (Fig. 2A) (Gerber et al., 2003). Of note, often with enzymatic deglycosylation of proteins there is a propensity for deamidation occurring on asparagine residues, increasing the mass of the residue by 0.984 Da and slightly delaying the reverse phase retention (Zielinska et al., 2010). Therefore, successful identification of human irisin peptides (as for other N-glycosylated plasma proteins) must take into account this mass shift. Deamidation modifications for both endogenous plasma irisin peptides are observed without dramatically changing the MS2 spectra (Fig. S1A) nor altering the PRM rank order elution profile (Fig 2B). Fragment ions for both peptides were quantified using Skyline version 3.1 (MacLean et al., 2010) and comparable levels of quantification for both peptides, downstream of the ATA start codon and the later ATG suggest irisin is mainly translated from its non-canonical start codon (Table 1, Fig. S1B, Fig. S2). We found that irisin levels are present at ~3.6 ng/ml in sedentary individuals and are significantly increased to ~ 4.3 ng/ml in individuals undergoing aerobic interval training (Fig. 2C, Table1).
Figure 2. Detection of Irisin in Human Plasma.
(A) SDS PAGE separation of 50 μg of plasma from each subject and visualized by coomassie staining. Molecular mass regions corresponding to completely deglycosylated irisin (10–15 kDa) were excised from six separate gels (300 μg from the original 100 μl plasma) for each subject and digested in-gel in the presence of 12.5 femtomoles of each internal standard AQUA peptide.
(B) PRM elution profile for internal tryptic irisin peptide (FIQEVNTTR) using Skyline software found in sedentary subject 1. Top panel is the deamidated asparagine form of the peptide found in the plasma, middle panel is the unmodified peptide found in the plasma and the bottom panel is 12.5 femtomoles of heavy internal standard (IS) AQUA peptide.
(C) Irisin levels in plasma from sedentary subjects (Sedentary) or subject undergoing aerobic interval training (Aerobic). Values are shown as mean±SEM. n=4 (Sedentary) and n=6 (Aerobic). * p= 0.0411 compared to sedentary subject group as determined by unpaired t-test, two-tailed.
(D) Depicted are several plasma proteins and their circulating concentrations ranging from the μg/ml (red), ng/mL (yellow) and pg/ml (blue) levels. We quantify circulating plasma irisin at a 3–5 ng/ml. See also Figure S2.
Table 1.
Quantification of Irisin in Plasma from Human Subjects Skyline software was used to quantify absolute amounts of irisin peptides from the plasma of sedentary and aerobically trained subjects. The 25 kDa glycosylated bioactive form of irisin was used to calculate its ng/ml concentrations in plasma.
| Sedentary | N-terminal peptide (femtomoles) | Internal (femtomoles) | combined average (femtomoles) | ng/mL |
|---|---|---|---|---|
| Subject 1 | 16.66 | 12.78 | 14.72 | 3.68 |
| Subject 2 | 11.98 | 12.3 | 12.14 | 3.03 |
| Subject 3 | 13.73 | 15.59 | 14.66 | 3.66 |
| Subject 4 | 16.21 | 16.7 | 16.455 | 4.11 |
| Average | 14.65 | 14.34 | 14.49 | 3.62 |
| Aerobic | N-terminal peptide (femtomoles) | Internal (femtomoles) | combined average (femtomoles) | ng/mL |
|---|---|---|---|---|
| Subject 1 | 13.98 | 14.78 | 14.38 | 3.66 |
| Subject 2 | 18.22 | 19.74 | 18.98 | 4.74 |
| Subject 3 | 16.11 | 22.22 | 19.165 | 4.79 |
| Subject 4 | 16.28 | 17.38 | 16.83 | 4.28 |
| Subject 5 | 16.68 | 19.2 | 17.94 | 4.48 |
| Subject 6 | 15.1 | 17.11 | 16.105 | 4.02 |
| Average | 16.06 | 18.41 | 17.23 | 4.33 |
Discussion
We have developed here a quantitative, precise, and unbiased assay for the detection of human irisin in plasma. This assay definitively shows that human irisin circulates and has a very similar or identical architecture to the mouse protein (Bostrom et al., 2012). Human irisin circulates at a level at or above the levels observed for many other important biological hormones, as shown in Figure 2C. It confirms earlier reports that have identified a unique peptide in human plasma by untargeted mass spectrometry, (Albrecht et al., 2015; Lee et al., 2014); but for the first time, it provides quantitation of the circulating levels of human irisin in an unbiased and antibody-independent manner. Irisin concentrations are present at ~3.6 ng/ml in sedentary individuals and are significantly increased to ~ 4.4 ng/ml in individuals undergoing aerobic interval training, We therefore also confirm our earlier report of irisin being regulated by endurance exercise in humans (Bostrom et al., 2012).
Several papers have called the start codon of the human FNDC5 gene, which is an ATA, rather than the more common ATG, a mutation. Indeed, these authors concluded that human FNDC5 is a non-coding “pseudogene” or that “the human species has an effective gene knockout of FNDC5” (Albrecht et al., 2015; Raschke et al., 2013). This claim was based on a transfection assay expressing human FNDC5 from a CMV-promoter driven plasmid, which yielded protein levels lower than human FNDC5 expressed with an ATG instead of an ATA from the same plasmid. However, several lines of reasoning stand against that claim. First, the high degree of conservation of the irisin amino acid sequence across most mammalian species (including humans) strongly argues against FNDC5 in humans being a pseudogene. Second, the simple fact that Raschke et al. detect human FNDC5 protein made from the ATA-FNDC5 sequence proves that human FNDC5 is not a pseudogene; these are generally defined as genes that have lost their protein-coding ability (Vanin, 1985). Third, their conclusion that low protein production from CMV-promoter driven plasmid expressed in HEK293 cells translates to inefficient FNDC5 translation in vivo is completely speculative since this experiment did not consider endogenous regulation of human FNDC5 in its native state. Indeed, non-canonical starts of translation are often indicative of complex regulation of translation (Chang and Wang, 2004; Starck et al., 2012). Fourth, as mentioned above, our detection here of equal amounts of peptide 1 and 2 in human plasma demonstrates that human irisin is, in fact, mainly translated from its non-canonical start codon and not the further downstream ATG.
The earlier report (Albrecht et al., 2015) had several serious methodological deficiencies. First, their failure to detect irisin in human serum at 12 kDa by Western blotting relied on deglycosylation by only one enzyme, namely PNGase F; however, this leads to only incomplete deglycosylation. PNGase F is an effective enzymatic method for removing almost all N-linked oligosaccharides but not other oligosaccharides. Hence, with PNGase F, no visible band will appear at 12kDa and the irisin signal will be diluted across the lane leading to apparent lower levels. In our previously published method, (Wrann et al., 2013) we used the Protein Deglycosylation Mix from NEB, which contains, in addition to PNGase F, O-Glycosidase, Neuraminidase, β1-4 Galactosidase, and β-N-acetylglucosaminidase; this leads to complete deglycosylation and the appearance of a 12 kDa bands in recombinant mammalian irisin and humans plasma by immunoblot (Figure 1).
Second, these authors (Albrecht et al., 2015) used a method of protein mass spectrometry called “shotgun proteomics”, which randomly samples peptides for detection from all the peptides contained in the sample. While the method has the potential to detect irisin, it would be suboptimal for detection because the peptides of interest can be missed in complex samples due to their low abundance. In these cases targeted proteomics is required. This allows the mass spectrometer to focus on the targeted peptides and ignore signal from co-eluting peptides. AQUA-based quantification concomitantly with PRM produces spectra that are highly specific because all potential product ions of a peptide and elution profile confirm the identity of the peptide.
Third, and perhaps most importantly, the authors report their own detection limits for irisin at about 100 ng/ml. However, since many reports of human irisin fall below this level (Kraemer et al., 2014; Kurdiova et al., 2014; Moraes et al., 2013; Wang et al., 2015; Zhang et al., 2014). Hence it is rather surprising that these authors concluded that human irisin did not exist or was a “myth”.
It is worth noting that limitations of own study include that the AQUA heavy peptides were added to the irisin preparations after the extraction of the proteins from the SDS-PAGE gel; we therefore cannot account for how much irisin protein was lost during the sample preparation (albumin/IgG removal, deglycosylation and retrieval from the gel band, etc.); the numbers reported here must therefore be considered a slight underestimation of the irisin levels. In our experience, typical losses during sample preparation range between 10–30%. In addition, this assay is relatively costly and relies on available mass spectrometry instrumentation and capabilities. However, while this assay is relatively low throughput, it should prove useful for benchmarking more high through-put assays as they are developed. Taken together, targeted mass spectrometry with the use of heavy irisin AQUA peptides settles the existence, the overall architecture of human irisin in the plasma, and its regulation by exercise.
EXPERIMENTAL PROCEDURES
Aerobic Interval Training
Plasma samples were collected from young healthy participants (n=6 males, 25±5 years, BMI= 24.3±2.5 kg/m2) following 12-weeks of high-intensity aerobic training. Training consisted of 3-days per week of intervals on a cycle ergometer (4 × 4 min >90% peak aerobic capacity + 3 min rest) separated by 2-days per week of walking on a treadmill (45 min at 70% peak aerobic capacity). All training was supervised at the Dan Abraham Healthy Living Center at Mayo Clinic, Rochester Minnesota. A separate sedentary group served as no-treatment control (n=4 males, 26±3 years, BMI=26.1±3.4 kg/m2). Written informed consent approved by the Mayo Clinic Institutional Review Board was given by all participants before entering the study.
Plasma collection and purification
Participants consumed a weight maintaining diet for 3-days then were admitted to the Clinical Research Unit at Mayo Clinic Hospital. The aerobic training group did not exercise during the 3-day meal period. Participants consumed a standardized evening meal then remained fasted overnight. Arterialized blood was collected at 0700h into sodium heparin tubes from a retrograde catheter in a dorsal hand vein (heated to 49° C). Plasma was separated by centrifugation and stored at −80°C unt il analysis. Human plasma specimens (100 μL) were depleted of albumin and IgG using the ProteoExtract-kit (Millipore) and subsequently concentrated using 3 kDa molecular weight cut-off spin-filter columns (Millipore). Deglycosylation of plasma was performed using Protein Deglycosylation Mix (New England Biolabs) as per the manufacturer's denaturing protocol. Deglycosylated plasma samples were reduced with 10 mM DTT and alkylated with 50 mM iodoacetamide prior to being resolved by SDS-PAGE using 4-12% NuPAGE Bis-Tris precast gels (Life Technologies).
Western blotting
For Western blotting analyses, 50 μg of protein was resolved for each plasma sample by SDS-PAGE. Gels were transferred to 0.2 μm pore size Protran (BA83) nitrocellulose membranes (Whatman) in 25 mM Tris, 200 mM glycine. Membranes were blocked with 10% nonfat dry milk in PBS containing 0.1% Tween (PBST) for 1 hour at room temperature. Membranes were then probed with anti-FNDC5 (Irisin) rabbit polyclonal antibody (1:1000) (IN102, Adipogen) overnight at 4 °C, washed with PB ST and incubated with horseradish peroxidase-conjugated secondary antibodies (1:3000) (Sigma) followed by chemiluminescent (Perkin Elmer Life Sciences) detection. Recombinant mammalian irisin was obtained from Ember Therapeutics.
In-gel digestion
For in-gel digestions, deglycosylated plasma samples (300 μg) were reduced with 10 mM DTT and alkylated with 50 mM iodoacetamide prior to being resolved by SDS-PAGE using 4-12% NuPAGE Bis-Tris precast gels (Life Technologies). Gels were coomassie stained and fragments were excised from the 10–15 kDa region. Gel pieces were destained and dehydrated with 100% acetonitrile, vacuumed dried, and resuspended with 50 mM ammonium bicarbonate (Sigma-Aldrich) with 500 ng sequencing grade trypsin (Promega) was added for an overnight incubation at 37°C (Shevchenko et al., 1996). Digests were quenched after 12 hr with 5% formic acid and de-salted using homemade stage tips as previously described (Rappsilber et al., 2007). Peptides were eluted with 70% acetonitrile and 1% formic acid, then dried using a speedvac, and resuspended in 10μl of 5% formic acid and 4% acetonitrile containing the heavy valine synthesized irisin peptides (Supplemental Table 1).
Liquid chromatography and tandem mass spectrometry
Mass spectrometry data was collected using a Q Exactive or LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) coupled with a Famos Autosampler (LC Packings) and an Accela 600 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated onto a 100 μm inner diameter microcapillary column packed with ~0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) followed by ~20 cm of Accucore C18 resin (1.6 μm, 150 Å, Thermo Fisher Scientific). For each analysis, we loaded ~2 μl onto the column. Peptides were separated using a 50-minute gradient of 8 to 30% acetonitrile in 0.125% formic acid with a flow rate of ~300 nL/min.
Parallel reaction monitoring acquisition
Parallel reaction monitoring (PRM) analyses were performed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific). A full MS scan from 400–700 Th at an orbitrap resolution of 70,000 (at m/z 200), AGC target 5 × 106 and a 500 ms maximum injection time. Full MS scans were followed by 25–50 PRM scans at 35,000 resolution (AGC target 5 × 106, 500 ms maximum injection time) as triggered by a scheduled inclusion list (Supplemental Table 2). The PRM method employed an isolation of target ions by a 2 Th isolation window, fragmented with normalized collision energy (NCE) of 25, MS/MS scans were acquired with a starting mass range of 100 m/z and acquired as a profile spectrum data type. Precursor and fragment ions were quantified using Skyline version 3.1 (MacLean et al., 2010).
Data dependent acquisition
For data dependent acquisitions using the Q Exactive, the scan sequence began with an Orbitrap MS1 spectrum with the following parameters: resolution 70,000, scan range 400–1400 Th, automatic gain control (AGC) target of 5 × 106, maximum injection time of 250 ms, and centroid spectrum data type. We selected the top twenty precursors for MS2 analysis which consisted of HCD high- energy collision dissociation with the following parameters: resolution 17,500, AGC 1 × 105, maximum injection time 60 ms, isolation window 2 Th, normalized collision energy (NCE) 25, and acquired as a centroid spectrum data type. The underfill ratio was set at 9%, which corresponds to a 1.5 × 105 intensity threshold. In addition, unassigned and singly charged species were excluded from MS2 analysis and dynamic exclusion was set to automatic.
For data dependent acquisitions using an LTQ Orbitrap Elite, the MS1 survey scan was performed in the orbitrap in the range of 300–1500 Th at a resolution of 3×104, followed by the selection of the ten most intense ions (TOP10) for CID-MS2 fragmentation using a precursor isolation width window of 2 Th. The AGC settings were 3 × 106 and 2.5 × 105 ions for survey and MS2 scans, respectively. Ions were selected for MS2 when their intensity reached a threshold of 500 counts and an isotopic envelope was assigned. Maximum ion accumulation times were set to 1000 ms for survey MS scans and to 250 ms for MS2 scans. Singly charged ion species and ions for which a charge state could not be determined were not subjected to MS2. Ions within a 10 ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 120 s.
Peptide and protein identification
Following mass spectrometry data acquisition, Thermo Fisher RAW files were converted into mzXML format and processed using a suite of software tools developed in-house for analysis of proteomics datasets. All precursors selected for MS/MS fragmentation were confirmed using algorithms to detect and correct errors in monoisotopic peak assignment and refine precursor ion mass measurements. All MS/MS spectra were then exported as individual DTA files and searched using the Sequest algorithm (Eng et al., 1994). These spectra were searched against a database containing sequences of all human proteins reported by Uniprot (Magrane and Consortium, 2011) in both forward and reversed orientations. Common contaminating protein sequences (e.g. human keratins, porcine trypsin) were included as well. The following parameters were selected to identify peptides from un-enriched peptide samples: 25 ppm precursor mass tolerance; 0.02 Da product ion mass tolerance; no enzyme digestion; up to two tryptic missed cleavages; variable modifications: oxidation of methionine (+15.994915) and deamidation of asparagine (0.984016); fixed modifications: carbamidomethylation of cysteine (+57.021464). AScore algorithm to quantify the confidence with which each deamidation modification could be assigned to a particular residue in each peptide (Beausoleil et al., 2006). Peptides with AScores above 13 were considered to be localized to a particular residue (p < 0.05).
Statistical analysis
Data analysis was performed using GraphPad Prism 6 software. As appropriate, two-tailed Student's t test. Significance was assigned to differences with a P value less than 0.05. Pooled data are presented as mean ± SEM.
Supplementary Material
ACKNOWLEGMENTS
C.D.W. was supported by a K99/R00-NIH Pathway to Independence (PI) Award (NS087096) and J.A.P. was supported by an NIH/NIDDK grant K01 (DK098285). This work is funded by the JPB Foundation and NIH grants (DK31405 and DK90861) to B.S. The human exercise project was supported by NIH grants R01AG09531 (KSN), T32DK007352 (MMR) and the Mayo Clinic CTSA grant UL1TR000135 from the National Center for Advancing Translational Sciences. We thank Jeffrey Knott from Cell Signaling Technology (Danvers, Ma) for synthesizing the heavy irisin AQUA peptides. Heavy Irisin AQUA peptides will be freely distributed upon request.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS M.P.J., C.D.W., S.P.G., and B.M.S. planned the majority of experiments and wrote the paper. M.P.J. developed the mass spec assay for the irisin identification with heavy peptides. C.D.W. developed the serum clean-up and de-glycosylation protocol and performed the statistical analysis. M.P.J. and S.P.G. performed the peptide fingerprinting identification of irisin. J.A.P., J.S., and K.K.G. contributed with technical assistance for the serum preparation, Western blotting, or mass spectrometry. M.M.R. and K.S.N. performed the human cohort study.
REFERENCES
- Albrecht E, Norheim F, Thiede B, Holen T, Ohashi T, Schering L, Lee S, Brenmoehl J, Thomas S, Drevon CA, et al. Irisin - a myth rather than an exercise-inducible myokine. Scientific reports. 2015;5:8889. doi: 10.1038/srep08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. Nature. 2012. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KJ, Wang CC. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. The Journal of biological chemistry. 2004;279:13778–13785. doi: 10.1074/jbc.M311269200. [DOI] [PubMed] [Google Scholar]
- Daskalopoulou SS, Cooke AB, Gomez YH, Mutter AF, Filippaios A, Mesfum ET, Mantzoros CS. Plasma irisin levels progressively increase in response to increasing exercise workloads in young, healthy, active subjects. European journal of endocrinology / European Federation of Endocrine Societies. 2014;171:343–352. doi: 10.1530/EJE-14-0204. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Irisin and FNDC5 in retrospect: An exercise hormone or a transmembrane receptor? Adipocyte. 2013;2:289–293. doi: 10.4161/adip.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ, Zierath JR. Integrative biology of exercise. Cell. 2014;159:738–749. doi: 10.1016/j.cell.2014.10.029. [DOI] [PubMed] [Google Scholar]
- Hofmann T, Elbelt U, Stengel A. Irisin as a muscle-derived hormone stimulating thermogenesis--a critical update. Peptides. 2014;54:89–100. doi: 10.1016/j.peptides.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Huh JY, Mougios V, Kabasakalis A, Fatouros I, Siopi A, Douroudos II, Filippaios A, Panagiotou G, Park KH, Mantzoros CS. Exercise-Induced Irisin Secretion Is Independent of Age or Fitness Level and Increased Irisin May Directly Modulate Muscle Metabolism Through AMPK Activation. The Journal of clinical endocrinology and metabolism. 2014;99:E2154–2161. doi: 10.1210/jc.2014-1437. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic acids research. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer RR, Shockett P, Webb ND, Shah U, Castracane VD. A transient elevated irisin blood concentration in response to prolonged, moderate aerobic exercise in young men and women. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2014;46:150–154. doi: 10.1055/s-0033-1355381. [DOI] [PubMed] [Google Scholar]
- Kurdiova T, Balaz M, Vician M, Maderova D, Vlcek M, Valkovic L, Srbecky M, Imrich R, Kyselovicova O, Belan V, et al. Effects of obesity, diabetes and exercise on Fndc5 gene expression and irisin release in human skeletal muscle and adipose tissue: in vivo and in vitro studies. The Journal of physiology. 2014;592:1091–1107. doi: 10.1113/jphysiol.2013.264655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Linderman JD, Smith S, Brychta RJ, Wang J, Idelson C, Perron RM, Werner CD, Phan GQ, Kammula US, et al. Irisin and FGF21 are cold-induced endocrine activators of brown fat function in humans. Cell metabolism. 2014;19:302–309. doi: 10.1016/j.cmet.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics (Oxford, England) 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database : the journal of biological databases and curation. 2011;2011 doi: 10.1093/database/bar009. bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N, Rosenzweig A. Can exercise teach us how to treat heart disease? Circulation. 2012;126:2625–2635. doi: 10.1161/CIRCULATIONAHA.111.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes C, Leal VO, Marinho SM, Barroso SG, Rocha GS, Boaventura GT, Mafra D. Resistance exercise training does not affect plasma irisin levels of hemodialysis patients. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2013;45:900–904. doi: 10.1055/s-0033-1354402. [DOI] [PubMed] [Google Scholar]
- Norheim F, Langleite TM, Hjorth M, Holen T, Kielland A, Stadheim HK, Gulseth HL, Birkeland KI, Jensen J, Drevon CA. The effects of acute and chronic exercise on PGC-1alpha, irisin and browning of subcutaneous adipose tissue in humans. The FEBS journal. 2014;281:739–749. doi: 10.1111/febs.12619. [DOI] [PubMed] [Google Scholar]
- Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. The Journal of biological chemistry. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Raschke S, Elsen M, Gassenhuber H, Sommerfeld M, Schwahn U, Brockmann B, Jung R, Wisloff U, Tjonna AE, Raastad T, et al. Evidence against a beneficial effect of irisin in humans. PloS one. 2013;8:e73680. doi: 10.1371/journal.pone.0073680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science (New York, NY) 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- Vanin EF. Processed pseudogenes: characteristics and evolution. Annual review of genetics. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Voss MW, Vivar C, Kramer AF, van Praag H. Bridging animal and human models of exercise-induced brain plasticity. Trends in cognitive sciences. 2013;17:525–544. doi: 10.1016/j.tics.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Zhang XW, Chen WK, Huang QX, Chen QQ. Relationship between serum irisin levels and urinary albumin excretion in patients with type 2 diabetes. Journal of diabetes and its complications. 2015;29:384–389. doi: 10.1016/j.jdiacomp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Wrann CD, White JP, Salogiannnis J, Laznik-Bogoslavski D, Wu J, Ma D, Lin JD, Greenberg ME, Spiegelman BM. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell metabolism. 2013;18:649–659. doi: 10.1016/j.cmet.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chen P, Chen S, Sun Q, Zeng QC, Chen JY, Liu YX, Cao XH, Ren M, Wang JK. The association of new inflammatory markers with type 2 diabetes mellitus and macrovascular complications: a preliminary study. European review for medical and pharmacological sciences. 2014;18:1567–1572. [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.