Abstract
The concept of orphan drugs for treatment of orphan genetic diseases is perceived enthusiastically at present, and this is leading to research investment on the part of governments, disease-specific foundations and industry. This review attempts to survey the potential to use traditional pharmaceuticals as opposed to biopharmaceuticals to treat single-gene disorders. The available strategies include the use of antisense oligonucleotides (ASOs) to alter splicing or knock-down expression of a transcript, siRNAs to knock-down gene expression and drugs for nonsense mutation read-through. There is an approved drug for biallelic knock-down of the APOB gene as treatment for familial hypercholesterolemia. Both ASOs and siRNAs are being explored to knock-down the transthyretin gene to prevent the related form of amyloidosis. The use of ASOs to alter gene-splicing to treat spinal muscular atrophy is in phase 3 clinical trials. Work is progressing on the use of ASOs to activate the normally silent paternal copy of the imprinted UBE3A gene in neurons as a treatment for Angelman syndrome. A gene-activation or gene-specific ramp-up strategy would be generally helpful if such could be developed. There is exciting theoretical potential for converting biopharmaceutical strategies such gene correction and CRISPR-Cas9 editing to a synthetic pharmaceutical approach.
Introduction
In this review, the term pharmaceutical will be used to refer to traditional small-molecule drugs, usually derived from chemical synthesis as distinct from biopharmaceuticals, which include protein administration as in enzyme replacement therapy, monoclonal antibodies, most gene therapy vectors and cell therapy. The intent is to survey efforts to develop gene-specific treatments for Mendelian disorders using chemically synthesized compounds (primarily oligonucleotides). Of special interest are strategies that use the sequence specificity of nucleic acid hybridization such that a method might be easily adapted for many different genes simply by changing the sequence of the oligonucleotide. The most transportable example might be knock-down of expression using an oligonucleotide to achieve specificity toward a gene or transcript. At this time, it is reasonable to say that it is generally feasible to develop an oligonucleotide as a knock-down therapeutic for any gene in the genome. Correcting function using small molecules directed against proteins such as inhibitors, activators and chaperons will not be discussed, although these may also be allele-specific.
Oligonucleotide Therapies for Gene- or Transcript-specific Knock-down
This is the most broadly applicable strategy that is immediately accessible. It includes antisense knock-down as well as RNAi/siRNA knock-down. In principle, this approach could be used to treat any gene duplication pathology that involves dosage-sensitivity to over-expression. In some cases, a single gene is documented to be the dosage-sensitive product as best exemplified by over-expression of PMP22 in CMT1A associated with a 1.5 Mb duplication encompassing numerous genes. In other cases, the dosage-sensitive gene is thought to be known but the presence of multiple genes in the duplicated region leaves some ambiguity as to whether one gene entirely explains the increased dosage phenotype as in the case of duplications of the MECP2 region in males and duplications of the RAI1 region in Potocki–Lupski syndrome. In the case of the duplication that is the reciprocal of the Williams syndrome deletion, the gene that is dosage-sensitive to over-expression is even less clear. Some genes might require intrathecal delivery as for MECP2 while in other instances systemic delivery might suffice as for CMT1A. A very recent publication describes the reversal of phenotypes in Mecp2 duplication mice using ASOs (1). Most genes are not dosage-sensitive for over-expression, so if a duplication includes 10–20 genes, only 1 or 2 may be mediating the over-expression phenotype. One can also speculate about whether gene-specific knock-down might be applied in various forms of aneuploidy with trisomy 21 being the most obvious candidate. If it was feasible to knock-down 3–10 genes on chromosome 21, what might be the potential benefits to infants with Down syndrome?
An important variable for the overall feasibility for any knock-down treatment is whether there is irreversible pathology established prior to birth. Again CMT1A is a very favorable example where the pathology arises in adult life. In cases of prenatal damage, in utero therapy would only be an option if examples of postnatal success were well-documented. In addition, expanded carrier testing and non-invasive prenatal diagnosis leading to the options of pre-implantation genetic diagnosis or avoidance of affected births through termination of pregnancy would interface with the frequency of these therapies.
Antisense Oligonucleotides versus siRNA
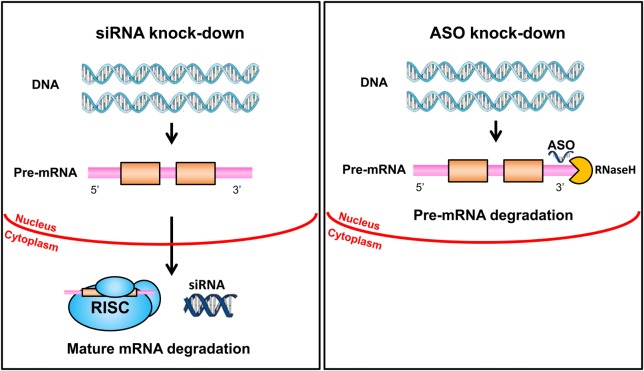
Two strategies are being pursued for knock-down of transcripts (Fig. 1). One utilizes antisense, single-stranded oligonucleotides that hybridize to RNA transcripts and lead to their degradation by ribonuclease H primarily acting in the nucleus. These ASOs are chemically modified to optimize their pharmacokinetic properties and functionality (Fig. 2). The second makes the use of RNA interference (RNAi) technology in which double-stranded siRNA preparations activate the RNA-induced silencing complex (RISC) pathway to degrade transcripts in the cytoplasm. Isis Pharmaceuticals has focused on antisense ASOs, and their approach is described in greater detail in many publications including a general review (3), a detailed discussion of pharmacodynamics (4), a review of approaches to pre-mRNA processing defects (5) and a current article in this journal on Huntington disease (HD) and spinocerebellar ataxia (6). Antisense oligonucleotides (ASOs) and siRNAs are competing strategies that could potentially be applied to almost any gene or transcript. The siRNAs are double-stranded RNA molecules of 20–25 base pairs in length designed to knock-down a gene using parts of the RNAi pathway. The potential to develop RNAi-related drugs was embraced enthusiastically in the 2000 decade, but fell out of favor to some extent thereafter as reviewed in 2011 and 2014 (7,8). More recently, two companies (Alnylam and Dicerna) (9) and some academic groups (10) have renewed interest especially in regard to treatment of transthyretin (TTR) amyloidosis. In addition, Alnylam is focusing on knock-down of PCSK9 as a strategy for treating various forms of hypercholesterolemia (11).
Figure 1.
Comparison of siRNA and ASO knock-down. On the left, double-stranded silencing RNA oligonucleotides reach the cytoplasm where they engage the RNA-RISC, which is part of the RNAi pathway. This leads to the degradation of mature mRNA. On the right, single-stranded ASOs reach the nucleus where they bind to pre-mRNA and engage RNase H, which degrades the transcript.
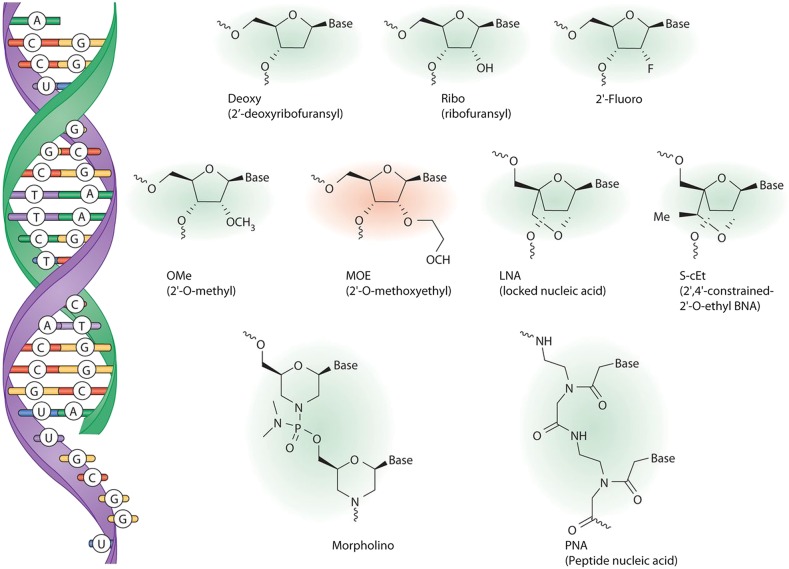
Figure 2.
Nucleotide analogues used in ASO drugs. ASOs (green) bind to the target RNA (purple) by Watson–Crick base pairing (left). Chemical structures of various nucleotides or nucleotide analogues commonly used in antisense drugs are shown. The ASO developed by Isis Pharmaceuticals to improve SMN2 splicing has the 2′-methoxyethyl modification (red). Reproduced from Rigo et al. (2) with permission.
Strategies for Allele-specific Knock-down
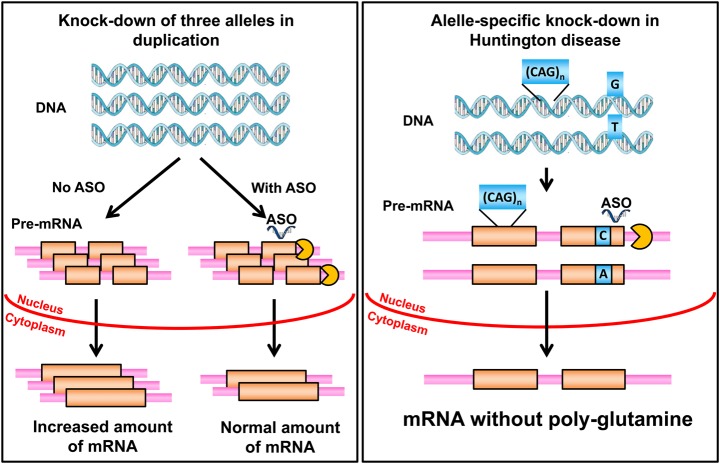
There are many human genetic disorders where allele-specific knock-down would be extremely desirable (Fig. 3). In general, this involves mutations that have some form of dominant gain-of-function or harmful effect where silencing the mutant alleles would be highly beneficial. Ideally, this would apply to genes where converting a dominant gain-of-function mutation to an inactive allele would result in a milder phenotype or even a cure. In some cases, there is certainty as to what the phenotype for heterozygous loss-of-function mutation might be. A good example is lethal osteogenesis imperfecta where dominant gain-of-function mutations cause a lethal phenotype, while the loss of function leads to a much milder form of osteogenesis. Whole classes of disorders are candidates for this form of therapy including especially the polyglutamine triplet repeat disorders (but not fragile X) and many rasopathy phenotypes caused by dominant gain-of-function point mutations. Allele-specific knock-down would most often involve silencing expression of a gene with a point mutation and could be relevant to achondroplasia, lethal osteogenesis imperfecta, rasopathies, cardiac arrhythmias, cardiomyopathies, some forms of epilepsy with ion channel mutations and many other disorders. For the polyglutamine triplet repeat mutations, the molecular challenge is different, requiring the knock-down of the allele with the expanded repeat at a specific locus while not significantly disturbing expression of the other allele with a normal length and also not disturbing expression of other genes with the triplet repeat structure.
Figure 3.
Comparison of all-allelic knock-down to allele-specific knock-down. On the left, a duplication such as occurs for PMP22 in CMT1A is depicted. An ASO is used to degrade a fraction of the pre-mRNA coming from all three alleles to decrease the amount of mature mRNA to the level that would ordinarily be produced by two copies of the gene. On the right, a triplet repeat expansion mutation encoding a polyglutamine tract in the protein is depicted. An ASO, at the site of a SNP in the transcript, is designed to hybridize with the allele that is in cis with the triplet repeat mutation. This leads to preferential degradation of the pre-mRNA from the mutant allele.
Hyperlipidemia
Mipomersen (Kynamro) is an antisense knock-down drug that targets the gene for apolipoprotein B (12). The design is for biallelic knock-down. It is approved for use in the USA for treatment of homozygous familial hypercholesterolemia and is administered by weekly subcutaneous injection (13). An ASO, designed to knock-down apolipoprotein C-III, has been shown to lower triglycerides patients with hypertriglyceridemia with and without the genetic deficiency of lipoprotein lipase (14,15).
Huntington Disease
A significant amount of research has been performed for HD, and a detailed discussion has just appeared in this journal (6) reviewing over 25 studies related to HD. Two quite different approaches, biallelic and allele-specific, are available to knock-down of the HTT gene, which encodes mutated huntingtin protein. Biallelic knock-down may be more straightforward and is the first approach to reach patients. In July 2015, Isis Pharmaceuticals, Inc. announced the initiation of a ‘Phase 1/2a clinical study of ISIS-HTTRx in patients with HD’. ISIS-HTTRx is designed to reduce the production of all forms of the huntingtin protein. Biallelic knock-down raises the question of what the effect of homozygous deficiency might be in an adult human. Homozygous null mice die between e8.5 and e10.5 of gestation (16), but this does not answer the question of how adult onset deficiency might affect a patient, although there is some evidence that normal huntingtin provides beneficial activity for the neuronal function and maintenance (17). There is also the possibility that the knock-down of both the normal and the mutant allele by 50% might delay the onset of symptoms indefinitely, and thus provide clinical benefit without causing new symptoms related to the huntingtin deficiency.
However, the allele-specific knock-down of the expanded allele would presumably be more attractive, and preclinical studies using this approach are encouraging (Fig. 3) (18). One strategy is to implement the knock-down in an allele-specific manner using naturally occurring SNPs in the gene. One group (18) concluded using this approach that ‘two allelic variants of an HD-SNP could provide a therapeutic option for all persons with HD; allele-specifically for roughly half, and non-specifically for the remainder’. The HD work provides a good precedent for how allele-specific knock-down might be achieved for other genes. In the case of point mutations that will vary widely from case-to-case, one could attempt to use the mutation itself to mediate the allele-specific knock-down, but not all mutations are favorable for this approach (19) in which cases heterozygous SNPs in the gene may still allow allele-specific effects by knocking down the SNP allele in cis with the deleterious mutation. For a highly polymorphic SNP, the mutation would often be in cis or trans on a random basis. ASOs targeted to intronic sequences can be active for knock-down, although activity is strongly influenced by splicing efficiency (20). This opens the potential for using polymorphisms in introns to achieve the allele-specific knock-down.
Angelman Syndrome
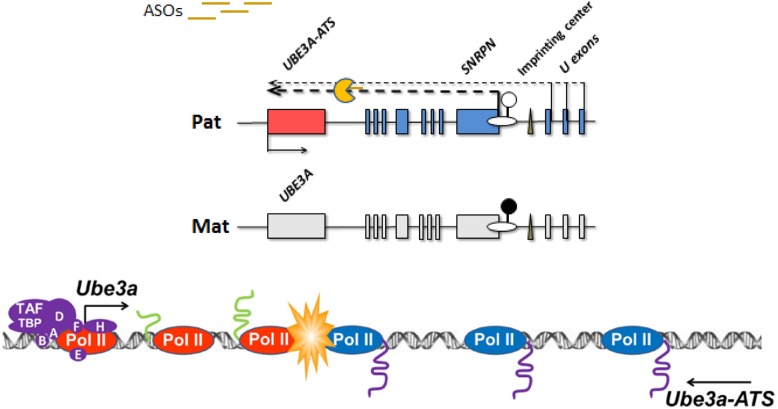
Angelman syndrome involves the UBE3A gene, which is subject to genomic imprinting with the silencing of the paternal allele in neurons. UBE3A encodes an ubiquitin ligase whose exact functions in the neuron are still relatively obscure. Accumulated evidence suggests the silencing of the paternal allele is mediated through an unusually long antisense paternal transcript that is transcribed in a convergent direction relative to UBE3A transcription (Fig. 4). There is evidence that terminating the antisense transcript allows the completion of the sense transcription of the paternal allele (21).
Figure 4.
ASO knock-down of natural occurring antisense in Angelman syndrome. The upper panel depicts a deleted or otherwise inactivated maternal allele for the SNRPN to UBE3A genomic interval. A very long transcript starting in the SNRPN region has a UBE3A antisense component at its distal position. ASOs to the upstream of the antisense region lead to the degradation of the antisense. Transcription is initiated, but not fully elongated at the paternal copy of the UBE3A promoter. In the lower panel, a collision model, which agrees with available data blocks the full extension of UBE3A transcription thus silencing the paternal allele of UBE3A. The degradation of the antisense allows for the complete transcription of the UBE3A gene. Pol II in blue represents antisense transcription while Pol II in red represents sense transcription.
Because Angelman syndrome is caused by loss-of-function mutations in the maternal allele of UBE3A, one potential therapeutic strategy would be to activate neuronal expression of the normally silent paternal allele. As noted above, the paternal allele is normally silenced by the expression of an antisense transcript. The first report of antisense knock-down activating expression of the paternal allele found that topoisomerase inhibitors could knock-down Ube3a antisense and lead to ‘unsilencing’ of the paternal allele in mouse cells (22). The topoisomerase inhibitors knock-down very long transcripts preferentially and are not specific to the Ube3a antisense (23). In addition, they have significant toxicity. In a search for a less toxic alternative, we have collaborated with Isis to explore a curious circumstance for up-regulating paternal expression of the Ube3a gene. Remarkably, it is feasible at least in mice to use an ASO to knock-down the naturally occurring antisense transcript (antisense to the natural antisense) thereby activating expression of the normally silenced Ube3a transcript from the paternal chromosome. This provides a targeted modification that appears not to disturb the expression of nearby genes including paternally expressed genes such as snoRNAs that are necessary for the prevention of Prader–Willi syndrome. This potential to knock-down only the very distal region of a very long transcript leaving its upstream functions intact is very fortuitous. Although a unique example, this represents a potential therapeutic strategy for oligonucleotide-based gene activation, which may have other applications as the knowledge of non-coding RNA-mediated gene regulation increases.
Splicing and Exon Skipping in Spinal Muscular Atrophy and Duchenne Muscular Dystrophy
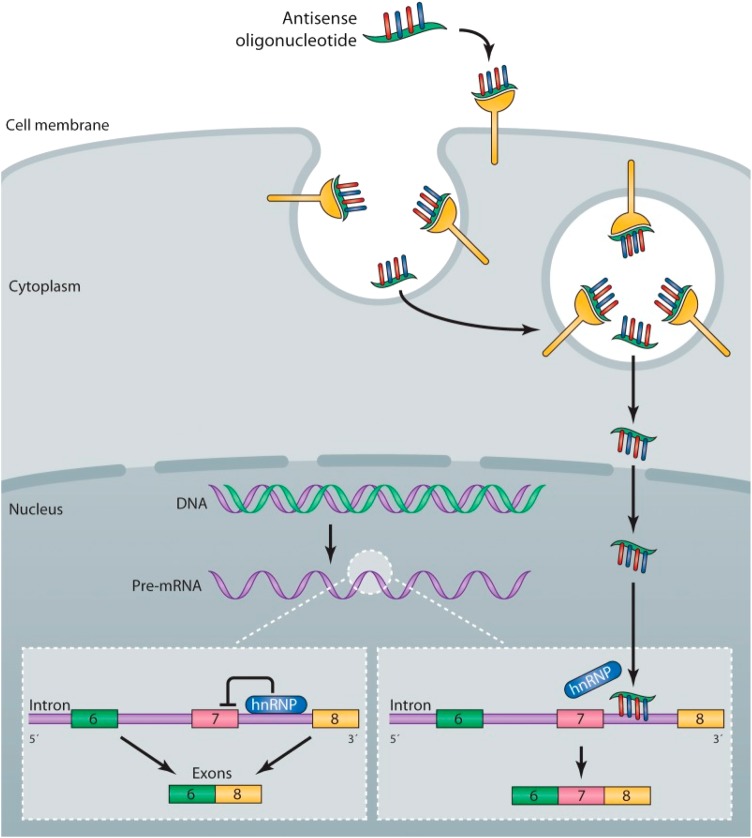
In addition to knock-down, there are examples where oligonucleotides can be used to modify splicing in a therapeutically favorable manner. One excellent example of this approach involves spinal muscular atrophy caused by biallelic deletions or loss-of-function mutations of the SMN1 gene (Fig. 5). For this gene, there is a highly homologous SMN2 gene nearby. However, the SMN2 gene is not very effective functionally because of natural skipping of exon 7. It was shown in preclinical studies that ASOs targeted to the intron can block the binding of a splicing-related protein and promote retention of exon 7 in SMN2, thus providing partial correction of the SMN1/SMN2 protein deficiency (Fig. 5) (24). In this case, ASO is fully modified with 2′-methoxyethyl to prevent cleavage by nucleases (Fig. 1). Spinal muscular atrophy (SMA) represents a disorder where clinical trials are further advanced than in many other cases. The clinicaltrials.gov website lists two Phase 2 and two Phase 3 trials recruiting subjects at this time for studies of an oligonucleotide designated as ISIS 396443 (ISIS SMNRx) (Table 1). Results, suggesting some clinical benefits, have been presented at international meetings and are described on the Isis corporate website, but are not published at this time.
Figure 5.
Mechanism of the action of an antisense drug that modulates SMN2 splicing. Single-stranded ASO are taken up into cells by an endocytic process via interaction with proteins expressed on the surface of cells. The ASOs escape the endosome and enter the nucleus, where they bind to the SMN2 pre-mRNA. The binding of the ASO to the RNA displaces an hnRNP protein that normally represses splicing of exon 7, resulting in the production of a mature mRNA that includes exon 7, which is translated into the full-length SMN protein. Reproduced from Rigo et al. (2) with permission.
Table 1.
Examples of gene-specific pharmaceutical approaches from clinicaltrials.gov
| Gene | Disorder | Intervention or study type | Sponsor | Clinical trial | Clinicaltrial.org | Status | Drug |
|---|---|---|---|---|---|---|---|
| HD | Huntington | ASO all-allelic knock-down | Isis | Phase 1/2 | NCT02519036 | ISIS HTTRx | |
| TTR | Transthyretin amyloidosis | siRNA | Alnylam | Phase 3 | NCT01960348 | Recruiting | ALN-TTR02 |
| TTR | Transthyretin amyloidosis | ASO all-allelic knock-down | Isis | Phase 3 | NCT01737398 | ||
| Enrolling | ISIS-TTR Rx | ||||||
| CFTR | CF | Nonsense read-through | PTC therapeutics | Phase 2 | NCT00458341 | Completed | PTC124 |
| SMN1/SMN2 | SMA | ASO splicing | Isis | Phase 3 | NCT02193074 | Recruiting | ISIS-SMN Rx |
| SMN1/SMN2 | SMA | ASO splicing | Isis | Phase 2 | NCT02386553 | Recruiting | ISIS-SMN Rx |
| SMN1/SMN2 | SMA | ASO splicing | Isis | Phase 2 | NCT02462759 | Recruiting | ISIS-SMN Rx |
| SMN1/SMN2 | SMA | ASO splicing | Isis | Phase 3 | NCT02594124 | Enrolling | ISIS-SMN Rx |
| APOB | Familial hypercholesterolemia | ASO all-allelic knock-down of APOB | Isis | Phase 3 | NCT00607373 | Completed | Mipomersen |
DMD see Bengtsson et al. (25) this series of reviews.
Another review in this series is focused on gene therapy for Duchenne muscular dystrophy (DMD) and other muscle disorders (25). That review includes a table of over 30 clinical trials for DMD including exon-skipping and nonsense mutation read-through, although they are not discussed. Efforts to utilize exon-skipping and nonsense mutation read-through qualify as pharmaceutical strategies. Two drugs designed to promote exon-skipping have entered clinical trials. Drisapersen is a 2′O-methyl-phosphorothioate oligonucleotide developed by Prosensa Holding N.V. Eteplirsen is a phosphorodiamidatemorpholino oligomer developed by Sarepta Therapeutics, Inc. Both drugs promote skipping of exon 51 that would benefit ∼14% of DMD patients. A clinical trial of eteplirsen reported in 2014 suggested limited clinical benefit (26). An update on results with these two drugs (27) suggests minimal benefit with drisapersen and perhaps some slowing of deterioration with eteplirsen.
Nonsense Read-through for Cystic Fibrosis, DMD and Other Disorders
The nonsense read-through strategy is not oligonucleotide based, and therefore, is not directly gene-specific but it can have preferential effects on genes harboring deleterious nonsense mutations. Nonsense mutations or premature termination codons cause disease by virtue of their occurrence in thousands of disease genes. They truncate protein synthesis and generally produce loss-of-function mutations. The potential for drug-mediated suppression or read-through of nonsense codons was first described in the 1960s with streptomycin, and in recent decades, numerous publications have delineated the ability of various aminoglycoside antibiotics to cause a reduction in translation fidelity at both sense and nonsense codons, thus allowing the insertion of an amino acid at the position of a nonsense codon (28). There are numerous drawbacks in using aminoglycosides as therapies for nonsense mutations, which led to screening to identify other compounds that would cause read-through at nonsense codons. One screening study identified PTC124 as a non-aminoglycoside compound that could promote read-through with minimal effect on naturally occurring termination codons (29). Numerous preclinical studies have been carried out examining the ability of PTC124, also known as ataluren. Ataluren can be administered orally, and it has been studied most extensively for patients with nonsense mutations causing cystic fibrosis (CF) or DMD. It is an approved drug in the European Union (30) for treatment of patients with nonsense mutations causing DMD. However, this was a conditional approval ‘despite limitations in the robustness of the efficacy data presented’. Thus, benefits were relatively marginal. A randomized, double-blind, placebo-controlled phase 3 trial of ataluren in cystic fibrosis reported in 2014 showed questionable efficacy (31). Overall, attempts to use nonsense-mediated read-through as a treatment approach have led to limited success.
The Critical Need for a Gene-activation/Ramp-up Method
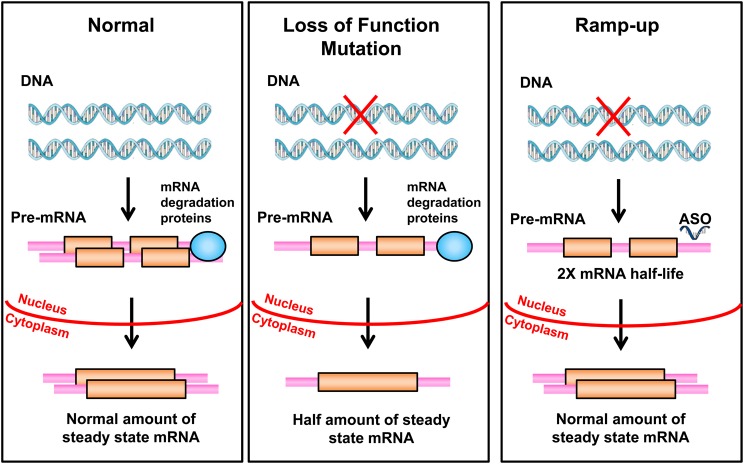
Many human genetic disorders are caused by heterozygous loss-of-function mutations, often referred to as dosage-sensitive genes where haploinsufficiency causes a disease phenotype. Prominent examples likely causing prenatal damage and therefore less amenable to this approach include mutations in ARID1B, EHMT1, SNYGAP1, SHANK3, EP300 and NSD1; ideal conditions with later onset would include loss-of-function mutations in LDLR and ANKRD11, deletions of CHRNA7, various forms of porphyria and contiguous gene deletions such as Smith–Magenis and Williams syndromes. The heritable cancer predispositions such as BRCA1/2 and Li–Fraumeni syndrome would probably not be well-treated by this approach because two independent normal copies of the gene are needed for optimal protection. This paradigm is limited to autosomal genes and excludes X-linked genes. For any of these mutations, it would potentially be curative to double the rate of transcription, double the half-life of the mRNA or double the half-life of the protein produced from the normal allele (Fig. 6). If this could be accomplished using an oligonucleotide in a gene-specific manner, it could be widely applied. Some of the loci involve both multi-gene deletions and point mutations, but up-regulating the normal allele should be beneficial in all cases regardless of the type of the loss-of-function mutation.
Figure 6.
A proposed ramp-up strategy. A heterozygous deletion or other inactivating mutation is depicted in the right panel. A hypothetical process whereby an ASO would bind to the 3′-UTR of mature mRNA and by some yet to be developed technology, double its half-life is proposed. This would lead to the production of a normal level of the protein.
Gene Correction
The concept of gene correction received a wave of attention in the 1990s (32,33). The strategy is to use various forms of oligonucleotide complexes to target the site of a specific mutation based on hybridization and induce some form of DNA repair event to correct the mutation to the normal sequence. Basic research especially in the use of triplex-forming peptide nucleic acids continues to the present (34), and there is a recent report of the correction of the F508del CFTR mutation in cultured human cells and in CF mutant mice (35). There are multiple more recent reports using CRISPR-Cas9 methods to correct mutations in iPSCs including for CF (36) and fragile X syndrome (37), but these cannot yet be considered pharmaceutical approaches since the Cas9 gene or protein must be delivered. Similar biopharmaceutical approaches using the CRISPR-Cas9 system in muscular dystrophy mice were reported (38).
Potential for the conversion of the CRISPR-Cas9 technology from a biopharmaceutical to a synthetic pharmaceutical strategy
The CRISPR-Cas9 technology has transformed the potential for genetic editing and manipulation in biology within a mere moment in time. A recent and most stimulating review is available from Sternberg and Doudna (39). Given the role of oligonucleotides in the treatment strategies described above and the essential involvement of RNA guides in the CRISPR-Cas9 system, it is tempting to ask whether the CRISPR technology can be harnessed as a small-molecule approach for gene-editing and gene regulation. Current approaches require introducing the protein or coding sequence for Cas9 or a related protein, but might it be possible to construct next-generation guide RNAs that would themselves have enzymatic activity or would recruit endogenous proteins to mediate a desired gene-editing? In one report (40), adult mice with a deficiency in the fumarylacetoacetate hydrolase (Fah) gene which causes tyrosinemia were gene corrected with the hydrodynamic injection of a donor DNA template and plasmid DNA encoding Cas9 and sgRNA. Although this injection involved three components, perhaps one could envision one moderate-sized single-stranded or double-stranded oligonucleotide being able to target a unique site in the genome and mediate the desired editing designed to treat single-gene disorders. The targeting potential could be used to implement up-regulation or down-regulation of a specific gene. The CRISPR-Cas9 system can be modified to inactivate its catalytic nuclease activity to create a dead Cas9 (dCas9), which can provide a docking mechanism that can bring along or otherwise engage regulatory elements. The dCas9 can be fused to transcriptional repressors and activators and potentially can up-regulate or down-regulate a gene of interest; see Sternberg and Doudna for references (39). This suggests the possibility of a ramp-up strategy as described above for a specific gene or allele. Sternberg and Doudna (39) also point out that ‘dead Cas9 can bind single-stranded RNA targets (41), suggesting that programmable manipulation of cellular RNA transcripts may become possible in the near future’.
Epigenetic Strategies
Initially there was a consideration to focus this review on epigenetic approaches to therapy. Depending ones definition of epigenetic, perhaps some of the knock-down strategies described above could be considered as epigenetic since the gene expression is altered, but the primary genomic sequence is unchanged. However, most current visions of epigenetic therapy involve relatively non-specific alterations of DNA methylation or histone modifications. If epigenetic approaches could be combined with oligonucleotides to achieve gene specificity, epigenetic approaches would become more attractive. Again the CRISPR-Cas9 system has been explored, and ‘dCas9 fusions to effector domains that install epigenetic markers may also enable the specific perturbation of epigenetic regulation (39)’.
Conflict of Interest statement. A.L.B. is Professor at Baylor College of Medicine (BCM), Chief Medical Officer, Baylor Miraca Genetic Laboratories (BMGL) and a consultant with Nurix, Inc. BMGL is a for-profit joint venture partially owned by BCM and offers commercial genetic laboratory testing. A.L.B. receives no personal compensation from Isis Pharmaceuticals but does receive grant support aimed at advancing oligonucleotide therapy for Angelman syndrome.
Funding
This work was supported by NIH grants R01 HD37283, an R01 project within the Baylor Intellectual Disabilities Research Center (1U54HD083092) and the Angelman Syndrome Foundation.
References
- 1.Sztainberg Y., Chen H., Swann J., Hao S., Tang B., Wu Z., Tang J., Wan Y.-W., Zhandong Liy Z., Rigo F., Zoghbi H. (2015) Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature, 528, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigo F., Hua Y., Krainer A.R., Bennett C.F. (2012) Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol., 199, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett C.F., Swayze E.E. (2010) RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol., 50, 259–293. [DOI] [PubMed] [Google Scholar]
- 4.Geary R.S., Norris D., Yu R., Bennett C.F. (2015) Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev., 87, 46–51. [DOI] [PubMed] [Google Scholar]
- 5.Rigo F., Seth P.P., Bennett C.F. (2014) Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol., 825, 303–352. [DOI] [PubMed] [Google Scholar]
- 6.Keiser M.S., Kordasiewicz H., McBride J. (2016) Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington's disease and spinocerebellar ataxia. Hum. Mol. Genet., 25, R53–R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson B.L., McCray P.B. Jr (2011) Current prospects for RNA interference-based therapies. Nat. Rev. Genet., 12, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Y., Wang C.C., Choy K.W., Du Q., Chen J., Wang Q., Li L., Chung T.K., Tang T. (2014) Therapeutic potentials of gene silencing by RNA interference: principles, challenges, and new strategies. Gene, 538, 217–227. [DOI] [PubMed] [Google Scholar]
- 9.(2015) Alnylam, Dicerna tussle over RNAi tech. Nat. Biotechnol., 33, 791. [DOI] [PubMed] [Google Scholar]
- 10.Niemietz C., Chandhok G., Schmidt H. (2015) Therapeutic oligonucleotides targeting liver disease: TTR amyloidosis. Molecules, 20, 17944–17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald K., Frank-Kamenetsky M., Shulga-Morskaya S., Liebow A., Bettencourt B.R., Sutherland J.E., Hutabarat R.M., Clausen V.A., Karsten V., Cehelsky J. et al. (2014) Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet, 383, 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merki E., Graham M.J., Mullick A.E., Miller E.R., Crooke R.M., Pitas R.E., Witztum J.L., Tsimikas S. (2008) Antisense oligonucleotide directed to human apolipoprotein B-100 reduces lipoprotein(a) levels and oxidized phospholipids on human apolipoprotein B-100 particles in lipoprotein(a) transgenic mice. Circulation, 118, 743–753. [DOI] [PubMed] [Google Scholar]
- 13.Thomas G.S., Cromwell W.C., Ali S., Chin W., Flaim J.D., Davidson M. (2013) Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: a randomized, double-blind, placebo-controlled trial. J. Am. Coll. Cardiol., 62, 2178–2184. [DOI] [PubMed] [Google Scholar]
- 14.Gaudet D., Brisson D., Tremblay K., Alexander V.J., Singleton W., Hughes S.G., Geary R.S., Baker B.F., Graham M.J., Crooke R.M., Witztum J.L. (2014) Targeting APOC3 in the familial chylomicronemia syndrome. N. Engl. J. Med., 371, 2200–2206. [DOI] [PubMed] [Google Scholar]
- 15.Gaudet D., Alexander V.J., Baker B.F., Brisson D., Tremblay K., Singleton W., Geary R.S., Hughes S.G., Viney N.J., Graham M.J. et al. (2015) Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N. Engl. J. Med., 373, 438–447. [DOI] [PubMed] [Google Scholar]
- 16.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. (1995) Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet., 11, 155–163. [DOI] [PubMed] [Google Scholar]
- 17.Zuccato C., Valenza M., Cattaneo E. (2010) Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev., 90, 905–981. [DOI] [PubMed] [Google Scholar]
- 18.Skotte N.H., Southwell A.L., Ostergaard M.E., Carroll J.B., Warby S.C., Doty C.N., Petoukhov E., Vaid K., Kordasiewicz H., Watt A.T. et al. (2014) Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One, 9, e107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F. et al. (2013) Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res., 41, 9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers T.A., Crooke S.T. (2014) Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS One, 9, e108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng L., Person R.E., Huang W., Zhu P.J., Costa-Mattioli M., Beaudet A.L. (2013) Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet., 9, e1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H.S., Allen J.A., Mabb A.M., King I.F., Miriyala J., Taylor-Blake B., Sciaky N., Dutton J.W. Jr, Lee H.M., Chen X. et al. (2012) Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature, 481, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T. et al. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature, 501, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staropoli J.F., Li H., Chun S.J., Allaire N., Cullen P., Thai A., Fleet C.M., Hua Y., Bennett C.F., Krainer A.R. et al. (2015) Rescue of gene-expression changes in an induced mouse model of spinal muscular atrophy by an antisense oligonucleotide that promotes inclusion of SMN2 exon 7. Genomics, 105, 220–228. [DOI] [PubMed] [Google Scholar]
- 25.Bengtsson N.E., Seto J.T., Hall J.K., Chamberlain J.S., Odom G.L. (2016) Progress and prospects of gene therapy clinical trials for the muscular dystrophies. Hum. Mol. Genet., 25, R9–R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J. et al. (2013) Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol., 74, 637–647. [DOI] [PubMed] [Google Scholar]
- 27.Kole R., Krieg A.M. (2015) Exon skipping therapy for Duchenne muscular dystrophy. Adv. Drug Deliv. Rev., 87, 104–107. [DOI] [PubMed] [Google Scholar]
- 28.Karijolich J., Yu Y.T. (2014) Therapeutic suppression of premature termination codons: mechanisms and clinical considerations (review). Int. J. Mol. Med., 34, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S. et al. (2007) PTC124 targets genetic disorders caused by nonsense mutations. Nature, 447, 87–91. [DOI] [PubMed] [Google Scholar]
- 30.Haas M., Vlcek V., Balabanov P., Salmonson T., Bakchine S., Markey G., Weise M., Schlosser-Weber G., Brohmann H., Yerro C.P. et al. (2015) European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscul. Disord., 25, 5–13. [DOI] [PubMed] [Google Scholar]
- 31.Kerem E., Konstan M.W., De B.K., Accurso F.J., Sermet-Gaudelus I., Wilschanski M., Elborn J.S., Melotti P., Bronsveld I., Fajac I. et al. (2014) Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir. Med., 2, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole-Strauss A., Yoon K., Xiang Y., Byrne B.C., Rice M.C., Gryn J., Holloman W.K., Kmiec E.B. (1996) Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science, 273, 1386–1389. [DOI] [PubMed] [Google Scholar]
- 33.Kren B.T., Parashar B., Bandyopadhyay P., Chowdhury N.R., Chowdhury J.R., Steer C.J. (1999) Correction of the UDP-glucuronosyltransferase gene defect in the Gunn rat model of Crigler-Najjar syndrome type I with a chimeric oligonucleotide. Proc. Natl Acad. Sci. USA, 96, 10349–10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNeer N.A., Schleifman E.B., Glazer P.M., Saltzman W.M. (2011) Polymer delivery systems for site-specific genome editing. J. Control Release, 155, 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeer N.A., Anandalingam K., Fields R.J., Caputo C., Kopic S., Gupta A., Quijano E., Polikoff L., Kong Y., Bahal R. et al. (2015) Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun., 6, 6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firth A.L., Menon T., Parker G.S., Qualls S.J., Lewis B.M., Ke E., Dargitz C.T., Wright R., Khanna A., Gage F.H., Verma I.M. (2015) Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep., 12, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park C.Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W. (2015) Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep., 13, 234–241. [DOI] [PubMed] [Google Scholar]
- 38.Xu L., Park K.H., Zhao L., Xu J., El R.M., Gao Y., Zhu H., Ma J., Han R. (2015) CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther., doi:10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternberg S.H., Doudna J.A. (2015) Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol. Cell, 58, 568–574. [DOI] [PubMed] [Google Scholar]
- 40.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol., 32, 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connell M.R., Oakes B.L., Sternberg S.H., East-Seletsky A., Kaplan M., Doudna J.A. (2014) Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 516, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]